Abstract
The actions of thyroid hormone (TH) on lipid metabolism in the liver are associated with a number of genes involved in lipogenesis and lipid metabolism; however, the underlying mechanisms through which TH impacts on lipid metabolism remain to be elucidated. The present study aimed to investigate the effects of hyperthyroidism on the serum levels of the microRNA (miR) miR-206 and the role of miR-206 on TH-regulated lipid metabolism in liver cells. Serum was obtained from 12 patients diagnosed with hyperthyroidism and 10 healthy control subjects. Human hepatoblastoma (HepG2) cells were used to study the effects of triiodothyronine (T3) and miR-206 on lipid metabolism. Expression of miR-206 in serum and cells was determined by reverse transcription-quantitative polymerase chain reaction analysis. Lipid accumulation in HepG2 cells was assessed with Oil Red O staining. Suppression or overexpression of miR-206 was performed via transfection with a miR-206 mimic or miR-206 inhibitor. Serum miR-206 was significantly decreased in patients with hyperthyroidism compared with euthyroid controls. Treatment of HepG2 cells with T3 led to reduced total cholesterol (TC) and triglyceride (TG) content, accompanied by reduced miR-206 expression. Inhibition of endogenous miR-206 expression decreased intracellular TG and TC content in HepG2 cells. By contrast, overexpression of miR-206 in HepG2 partially prevented the reduction in TG content induced by treatment with T3. In conclusion, serum miR-206 expression is reduced in patients with hyperthyroidism. In addition, miR-206 is involved in T3-mediated regulation of lipid metabolism in HepG2 cells, indicating a role for miR-206 in thyroid hormone-induced disorders of lipid metabolism in the liver.
Keywords: microRNA-206, lipid metabolism, thyroid hormone, hyperthyroidism
Introduction
Hyperthyroidism is a pathological syndrome, characterized by high levels of circulating thyroid hormone (TH). Thyrotoxicosis is diagnosed by high serum concentrations of thyroxine (T4) and triiodothyronine (T3), and low serum concentrations of thyrotropin, additionally termed thyroid-stimulating hormone. Symptoms of overt hyperthyroidism include heat intolerance, palpitations, anxiety, fatigue, weight loss and muscle weakness, which are associated with lipid metabolism disorders (1). Accumulating evidence has demonstrated the important roles of TH in regulating lipid metabolism, with a number of genes involved in lipid metabolism in the liver, an important TH target tissue, being directly regulated by TH (2). The effects of TH on lipid metabolism in the liver are associated with a number of genes involved in lipogenesis and fatty acid oxidation (3). However, the underlying mechanisms associating TH with altered lipid metabolism remain to be elucidated.
MicroRNAs (miRNAs/miRs) are a class of small, noncoding, single-stranded RNA molecules containing ~22–25 nucleotides, that are able to bind to the 3′untranslated region (3′UTR) of their target mRNA, leading to translational inhibition or mRNA degradation (4). miRNAs are important regulators of biological processes, including metabolism, cell growth, apoptosis and carcinogenesis (5,6). Global miRNA and mRNA expression profiling has revealed a potential regulatory role for miRNAs in response to TH in the developing mouse liver (7). miR-1, miR-206, miR-133a and miR-133b expression was observed to be significantly increased in the livers of hypothyroid mice, while their expression was decreased in the livers of hyperthyroid mice (8). In addition, a number of miRNAs have been reported to be involved in cholesterol efflux and hepatic lipid metabolism (9). Therefore, miRNAs may serve a role in lipometabolism regulated by TH in hyperthyroidism.
The present study aimed to investigate the effects of increased TH on serum miR-206 expression in patients with hyperthyroidism. The role of the miRNA miR-206 in lipid metabolism was assessed in cultured human hepatoblastoma HepG2 cells in response to T3, with either overexpression or inhibition of endogenous miR-206 expression. It was hypothesized that miR-206 expression may be altered by TH and has an important role in T3-regulated lipid metabolism in liver cells.
Materials and methods
Patients
From October 2013 to March 2014 a total of 22 subjects recruited from October 2013 to March 2014 took part in the present study. A total of 12 patients diagnosed with hyperthyroidism were recruited from the Department of Endocrinology and 10 healthy subjects were recruited from the Physical Examination Center at The 82nd Hospital of Chinese People's Liberation Army (CPLA). Patients who had diabetes mellitus or other endocrine diseases, or with a body mass index >30 kg/m2, were excluded from taking part in the study. Patient details are shown in Table I. Patients taking any drugs known to influence TH metabolism were additionally excluded. The ethics committee of The 82nd Hospital of CPLA approved the present study. Written informed consent was obtained from all subjects.
Table I.
Clinical characteristics of study subjects.
| Patients with hyperthyroidism | Healthy subjects | P-value | P-value summary | |
|---|---|---|---|---|
| No. subjects (female) | 12 (8) | 10 (8) | – | – |
| Age, years | 49.0±7.5 | 42.7±12.1 | 0.1512 | ns |
| FT3, pg/ml | 9.9±0.5 | 3.5±0.1 | <0.0001 | – |
| FT4, ng/dl | 5.3±0.5 | 1.3±0.0 | <0.0001 | – |
| ALT, U/l | 23.8±1.9 | 18.6±2.1 | 0.0827 | ns |
| AST, U/l | 25.3±1.7 | 23.4±2.5 | 0.5143 | ns |
| TG, mmol/l | 1.22±0.31 | 1.71±0.50 | 0.3768 | ns |
| TC, mmol/l | 3.31±0.25 | 3.49±0.43 | 0.7080 | ns |
Data (mean ± standard error of the mean) were compared using an unpaired t-test. FT3, serum free triiodothyronine; FT4, serum free thyroxine; ALT, alanine aminotransferase; AST, aspartate aminotransferase; TG, triglyceride content; TC, total cholesterol; ns, not significant.
Clinical characteristics analysis
The clinical characteristics of the study population include in serum free triiodothyronine (FT3), serum free thyroxine (FT4), alanine aminotransferase (ALT), aspartate aminotransferase (AST), triglyceride content (TG) and total cholesterol (TC). These were measured by Olympus AU640 automatic biochemical analyzer (Olympus Corporation, Tokyo, Japan).
Cell culture and treatment
Human hepatoblastoma (HepG2) cell line is known to be misidentified, originally thought to be a hepatocellular carcinoma cell line but shown to be from an hepatoblastoma (10). HepG2 is a perpetual cell line, which was derived from the liver tissue of a 15-year-old Caucasian American male with a well-differentiated hepatocellular carcinoma. HepG2 cells are a suitable in vitro model system for the study of polarized human hepatocytes. HepG2 cells and their derivatives are also used as a model system for studies of liver metabolism and toxicity of xenobiotics. HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin, and 100 µg/ml streptomycin. For treatment with T3 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), cells were seeded in 12-well culture plates at a density of 3×105 cells/well, and maintained for 2 days incubated at 37°C in a humidified chamber containing 5% CO2. Following overnight incubation in serum-free DMEM, cells were treated with 100 nM T3 for 12, 24 or 36 h. For the investigation of triglyceride synthesis, the medium was supplied with a 0.25 mM free fatty acid (FFA) cocktail composed of lauric acid, myristic acid, linoleic acid, oleic acid and arachidonic acids (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Transient transfections
HepG2 cells, grown to 70–80% confluence, were transiently transfected with miR-206 mimic (sense, 5′-UGGAAUGUAAGGAAGUGUGUGG-3′ and antisense, 5′-ACACACUUCCUUACAUUCCAUU-3′) or miR-206 inhibitor (5′-CCACACACUUCCUUACAUUCCA-3′), or a corresponding mimic negative control inhibitor (5′-UUCUCCGAACGUGUCACGUTT-3′; Shanghai GenePharma Co., Ltd., Shanghai, China), using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. All the mimic and inhibitor containing FAM reporter fluorophores were transfected at 10 nM. Transfection efficiency was observed using an inverted fluorescent microscope (original magnification, ×400; Leica Microsystems GmbH, Wetzlar, Germany), and evaluated using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The mixtures were added to the culture, and the culture medium was changed after 8 h, and cultured in DMEM supplemented with 10% fetal bovine serum for 36 h further for subsequent experiments.
RT-qPCR quantification of miR-206 expression
Cell/serum total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) or QIAzol reagent (Applied Biosystems; Thermo Fisher Scientific, Inc.), followed by isolation and purification using a miRNeasy Mini kit/miRNeasy serum kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, first-strand cDNA was synthesized by RT using a TaqMan miRNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression of miR-206 was analyzed by qPCR using TaqMan Universal Master Mix II in the ABI 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). All primers were provided in the TaqMan® MicroRNA Assays (cat. no. 4427975; Thermo Fisher Scientific, Inc.). PCR conditions were as follows: 50°C for 2 min, 95°C for 2 min, and then 40 cycles of 94°C for 15 sec and 60°C for 1 min. U6 small nucleolar RNA was used as an internal standard for normalization. miR-206 expression was calculated using the comparative quantification cycle (Cq) method (2−ΔCq; ΔCq = CqmiR-206 - CqU6), the Cq values were converted into RQ via the 2-ΔΔCq method (11), incorporating the calculated amplification efficiency for each primer pair.
Oil Red o staining
Oil Red O stock solution was prepared in isopropanol (0.25 g/100 ml) and heated to 100°C for 10 min. HepG2 cells grown to 70–80% confluence were washed with PBS and fixed using 4% paraformaldehyde for 30 min at room temperature. Cells were soaked in filtered 0.6% (w/v) Oil Red O solution (60% Oil Red O stock solution, 40% water) for 20 min at room temperature. Cells were washed with water to remove unbound dye and visualized using light microscopy (original magnification, ×400).
Statistical analysis
Each sample was analyzed in triplicate. All data are presented as the mean ± standard error of the mean. Student's t-test was used to analyze the differences between groups. The correlation between T4 parameters and miR-206 expression levels was analyzed using the Pearson product-moment correlation coefficient method. Comparisons among multiple groups were statistically performed using one-way analysis of variance followed by Dunnett's test. Two-tailed P<0.05 was considered to indicate a statistically significant difference. The data were analyzed using SPSS software version 17.0 (SPSS, Inc., Chicago, IL, USA).
Results
Serum miR-206 levels in patients with hyperthyroidism
The clinical characteristics of the study population are presented in Table I. Patients with hyperthyroidism exhibited increased serum free T3 (FT3) and free T4 (FT4) concentrations compared with the control subjects.
The RT-qPCR analysis demonstrated that the serum miR-206 levels in patients with hyperthyroidism were significantly decreased compared with euthyroid control subjects (Fig. 1). The present study further analyzed the correlation between TH and miR-206 expression in patients with hyperthyroidism. No apparent correlation was observed between miR-206 expression and serum FT3 or FT4 levels (data not shown).
Figure 1.

Serum miR-206 levels in patients with hyperthyroidism and euthyroid control subjects. Serum miR-206 levels were determined using reverse transcription-quantitative polymerase chain reaction analysis. The results are presented as the mean ± standard error of the man. *P<0.05 vs. euthyroid control. miR, microRNA.
T3 downregulates miR-206 expression in HepG2 cells
In order to clarify the effect of TH on miR-206 expression, HepG2 cells were treated with 100 nM T3 for 12, 24 or 36 h, with miR-206 expression examined using RT-qPCR analysis. The expression of miR-206 was significantly decreased 24 h subsequent to the treatment with T3, compared with HepG2 cells without T3 (Fig. 2). The expression of miR-206 remained reduced at 36 h following the addition of T3 (Fig. 2).
Figure 2.

T3 downregulates miR-206 expression in HepG2 cells. HepG2 cells were treated with 100 nM T3 for 12, 24, and 36 h (n=4 per group). The relative expression of miR-206 was analyzed using reverse transcription-quantitative polymerase chain reaction analysis. The data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. respective control. T3, triiodothyronine; miR, microRNA.
Effects of T3 on TG and TC synthesis in HepG2 cells
To validate the effects of T3 on lipid synthesis, HepG2 cells were cultured in DMEM supplemented with a 0.25 mM FFA cocktail and treated with 100 nM T3 for 12, 24 or 36 h. Oil Red O staining demonstrated that T3 suppressed lipid accumulation in HepG2 cells (Fig. 3A). The TG was significantly reduced 12 h subsequent to treatment with T3 (Fig. 3B), while the TC content was significantly reduced 24 h subsequent to treatment with T3 (Fig. 3C). The TG and TC content of the HepG2 cells remained reduced 36 h following treatment with T3.
Figure 3.
Effects of T3 on TG synthesis in HepG2 cells. HepG2 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 0.25 mM free fatty acid cocktail and treated with 100 nM T3 for 12, 24 and 36 h (n=4 per group). (A) TG composition was visualized using Oil Red O staining (original magnification, ×400). (B) The TG of the cells was determined enzymatically using glycerol-3-phosphate oxidase-PAP. (C) The TC content of the cells was determined enzymatically using cholesterol oxidase-PAP. Data are presented as the mean ± standard deviation. *P<0.05. TG, triglyceride content; TC, total cholesterol; PAP, phenol + aminophenazone; T3, triiodothyronine.
Effects of miR-206 on TG and TC composition in HepG2 cells
The transfection of HepG2 cells with miR-206 inhibitor confirmed that the inhibitor effectively knocked down miR-206 expression (Fig. 4A). The addition of the miR-206 inhibitor reduced the TG at 24 h, and TG and TC content at 36 h in HepG2 cells (Fig. 4B).
Figure 4.
Effects of miR-206 on TG and TC composition in HepG2 cells. (A) HepG2 cells were transfected with miR-206 inhibitor to induce knockdown of miR-206 expression. (B) TG and TC concentration was determined at 12, 24 and 36 h post-transfection (n=4 per group). *P<0.05. miR, microRNA; NC, negative control; TG, triglyceride content; TC, total cholesterol.
miR-206 partially rescues the TG reduction induced by treatment with T3 in HepG2 cells
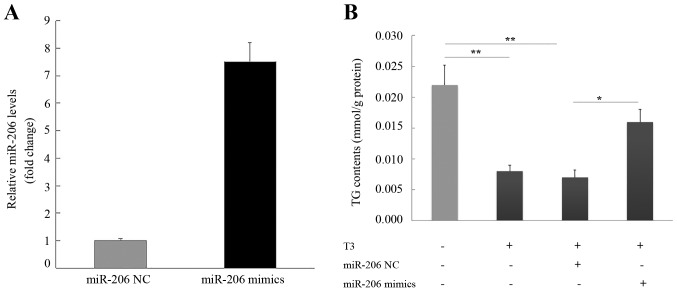
The transfection of HepG2 cells with the miR-206 mimic or mimic negative control confirmed that the miR-206 mimic increased miR-206 expression (Fig. 5A). HepG2 cells treated with T3 exhibited a reduced TG content that was restored with miR-206 overexpression (Fig. 5B). The addition of mimic negative control did not alter TG in T3-treated cells.
Figure 5.
miR-206 partially restores T3-induced reductions in TG. (A) HepG2 cells transfected with miR-206 mimic or mimic NC were treated with T3. (B) TG was determined enzymatically using glycerol-3-phosphate oxidase/phenol+aminophenazone. *P<0.05; **P<0.005. TG, triglyceride content; miR, microRNA; NC, negative control. T3, triiodothyronine.
Discussion
Appropriate TH levels are important for normal growth, development and metabolism in adults (12). It is well established that TH status is associated with body weight and energy expenditure (13). Hyperthyroidism, excess TH, promotes a hypermetabolic state characterized by increased resting energy expenditure and weight loss. Serum TG and TC are typically decreased in patients with hyperthyroidism. TH exerts direct and indirect actions on the regulation of TG and TC production, disposal and efflux (12,14,15), primarily through specific nuclear receptor-mediated T3 actions that modulate gene transcription. However, accumulating evidence has indicated that non-genomic mechanisms may be involved in the TH regulation of metabolism (16–18). Reports have indicated that miRNAs have a potential role in mediating the action of TH (7,19). The results of the present study demonstrated that miR-206 expression was significantly decreased in patients with hyperthyroidism compared with euthyroid subjects. Therefore, understanding the association between TH and miR-206 may further the understanding of the mechanisms underlying metabolic dysfunction in patients with hyperthyroidism.
The liver is an important target of TH in the regulation of energy metabolism and physiology (20,21). Hyperthyroidism frequently results in liver dysfunction, and hypothyroidism is associated with non-alcoholic fatty liver disease. Previous studies have suggested that T3 has the ability to reduce levels of lipids, including TC and TG (22,23). The results of the present study demonstrated that prolonged treatment with T3 led to reduced TG and TC content in HepG2 cells. In addition, it was observed that that treatment with T3 resulted in a downregulation of miR-206 expression. Increasing evidence has suggested that miRNAs are involved in the post-transcriptional regulation of genes in lipid metabolism (24,25). Previous studies have indicated that the regulatory role of miR-206 in TC handling is tissue-specific (26,27). It was demonstrated in the present study that the inhibition of endogenous miR-206 expression in HepG2 cells resulted in decreased intracellular TG and TC content. These results indicated that miR-206 may be involved in the TH-mediated regulation of lipid metabolism in HepG2 cells.
In order to confirm that miR-206 has a role in the TH-mediated regulation of lipid metabolism, HepG2 cells were transfected with an miR-206 mimic prior to being treated with T3. Overexpression of miR-206 in HepG2 cells increased lipid accumulation and attenuated the T3-induced downregulation of TG. A previous study demonstrated that expression of miR-206 increased in the liver of hypothyroid mice compared with controls (7). Given that TH deficiency is associated with an increased incidence of hepatic steatosis, these results suggested that miR-206 may serve an important role in the lipid dysregulation in the liver associated with hyperthyroidism and hypothyroidism.
The present study has limitations, in that the miR-206 levels were only determined in the serum of hyperthyroid patients and not in the liver. In addition, the use of HepG2 cells provided an in vitro model of lipid metabolism in liver cells and conclusions may not be directly applied to in vivo liver physiology. However, the results of the present study suggested that miRNAs may be a promising target for future investigations into disorders of lipid metabolism in the liver in cases of hyper- or hypothyroidism. Future studies, including the use of mouse models of hyperthyroidism, are required for in vivo investigation into the effects of miR-206 on lipid metabolism in the liver in a physiological context. Further larger studies may be used to study the effects of hyper- or hypothyroidism on miR-206 expression in human subjects.
In conclusion, the results of the present study demonstrated that patients with hyperthyroidism exhibited decreased serum miR-206 expression levels and that miR-206 was involved in the T3-mediated regulation of lipid metabolism in HepG2 cells. The results of the present study provided novel insights into the molecular mechanisms underlying TH regulation of lipid metabolism. Whether miR-206 is affected in the liver in patients with hyperthyroidism remains to be elucidated. Further experiments are required to clarify whether miR-206 has the potential to be either a biomarker or therapeutic target in hyperthyroidism.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Medical and Scientific Innovations Project of Chinese People's Liberation Army (grant no. 14SM030).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
YJZ, HW and YG collected data. YJZ, CZ and NZ conducted the experiments. YJZ, NZ, WK, RL, and XX analysed the data. CZ and YPZ wrote the paper. YPZ and XX designed the study.
Ethics approval and consent to participate
The Ethics Committee of the 82nd Hospital of CPLA approved the present study. Written informed consent was obtained from all subjects.
Consent for publication
Written informed consent was obtained from all subjects.
Competing interests
All authors declared that they have no conflict of interest with regard to this work.
References
- 1.Cooper D. Hyperthyroidism. Lancet. 2003;362:459–468. doi: 10.1016/S0140-6736(03)14073-1. [DOI] [PubMed] [Google Scholar]
- 2.Boelaert K, Franklyn JA. Thyroid hormone in health and disease. J Endocrinol. 2005;187:1–15. doi: 10.1677/joe.1.06131. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheimer JH, Schwartz HL, Lane JT, Thompson MP. Functional relationship of thyroid hormone-induced lipogenesis, lipolysis and thermogenesis in rat. J Clin Invest. 1991;87:125–132. doi: 10.1172/JCI114961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-rna-mediated translation repression is determined by the promoter of the target gene; Proc Nail Acad Sci USA; 2008; pp. 8866–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen XM. MicroRNA signatures in liver diseases. World J Gastroentero. 2009;15:1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Dong H, Paquette M, Williams A, Zoeller RT, Wade M, Yauk C. Thyroid hormone may regulate mRNA abundance in liver by acting on micrornas. PLoS One. 2010;5:e12136. doi: 10.1371/journal.pone.0012136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visser WE, Heemstra KA, Swagemakers SM, Ozgür Z, Corssmit EP, Burggraaf J, VanIjcken WF, vanderSpek PJ, Smit JW, Visser TJ. Physiological thyroid hormone levels regulate numerous skeletal muscle transcripts. J Clin Endocr Metab. 2009;94:3487–3496. doi: 10.1210/jc.2009-0782. [DOI] [PubMed] [Google Scholar]
- 9.Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annu Rev Nutr. 2011;31:49–63. doi: 10.1146/annurev-nutr-081810-160756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep g2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Brent GA. Mechanisms of thyroid hormone action. J Clin Invest. 2012;122:3035–3043. doi: 10.1172/JCI60047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwen KA, Schröder E, Brabant G. Thyroid hormones and the metabolic syndrome. Eur Thyroid J. 2013;2:83–92. doi: 10.1159/000351249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb P. Thyroid hormone receptor and lipid regulation. Curr Opin Invest Drugs. 2010;11:1135–1142. [PubMed] [Google Scholar]
- 15.Shoemaker TJ, Kono T, Mariash CN, Evansmolina C. Thyroid hormone analogues for the treatment of metabolic disorders: New potential for unmet clinical needs? Endocr Pract. 2012;18:954–964. doi: 10.4158/EP12086.RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordeiro A, deSouza LL, Oliveira LS, Faustino LC, Santiago LA, Bloise FF, Ortiga-Carvalho TM, Almeida NA, Pazos-Moura CC. Thyroid hormone regulation of Sirtuin 1 expression and implications to integrated responses in fasted mice. J Endocrinol. 2013;216:181–193. doi: 10.1530/JOE-12-0420. [DOI] [PubMed] [Google Scholar]
- 18.Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, et al. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest. 2012;122:2428–2438. doi: 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap CS, Sinha RA, Ota S, Katsuki M, Yen PM. Thyroid hormone negatively regulates CDX2 and SOAT2 mRNA expression via induction of miRNA-181d in hepatic cells. Biochen Bioph Res Commun. 2013;440:635–639. doi: 10.1016/j.bbrc.2013.09.116. [DOI] [PubMed] [Google Scholar]
- 20.Ståhlberg N, Merino R, Hernández LH, Fernández-Pérez L, Sandelin A, Engström P, Tollet-Egnell P, Lenhard B, Flores-Morales A. Exploring hepatic hormone actions using a compilation of gene expression profiles. BMC Physiol. 2005;5:8. doi: 10.1186/1472-6793-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H, Yauk CL, Williams A, Lee A, Douglas GR, Wade MG. Hepatic gene expression changes in hypothyroid juvenile mice: Characterization of a novel negative thyroid-responsive element. Endocrinology. 2007;148:3932–3940. doi: 10.1210/en.2007-0452. [DOI] [PubMed] [Google Scholar]
- 22.Shibata A, Kawakami Y, Kimura T, Miyazawa T, Nakagawa K. α-tocopherol attenuates the triglyceride- and cholesterol-lowering effects of rice bran tocotrienol in rats fed a western diet. J Agr Food Chem. 2016;64:5361–5366. doi: 10.1021/acs.jafc.6b02228. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg IJ, Huang LS, Huggins LA, Yu S, Nagareddy PR, Scanlan TS, Ehrenkranz JR. Thyroid hormone reduces cholesterol via a non-LDL receptor-mediated pathway. Endocrinology. 2012;153:5143–5149. doi: 10.1210/en.2012-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickers KC, Sethupathy P, Baran-Gale J, Remaley AT. Complexity of microRNA function and the role of isomiRs in lipid homeostasis. J Lipid Res. 2013;54:1182–1191. doi: 10.1194/jlr.R034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F. MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in hepatocytes. Cell Signal. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Vinod M, Chennamsetty I, Colin S, Belloy L, De Paoli F, Schaider H, Graier WF, Frank S, Kratky D, Staels B, et al. MIR-206 controls LXRα expression and promotes LXR-mediated cholesterol efflux in macrophages. Biochim Biophys Acta. 2014;1841:827–835. doi: 10.1016/j.bbalip.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.