Summary
Membrane-less organelles are intracellular compartments specialized to carry out specific cellular functions. There is growing evidence supporting the possibility that such organelles form as a new phase, separating from cytoplasm or nucleoplasm. However, a main challenge to such phase separation models is that the initial assembly, or nucleation, of the new phase is typically a highly stochastic process, and does not allow for the spatiotemporal precision observed in biological systems. Here we investigate the initial assembly of the nucleolus, a membrane-less organelle involved in different cellular functions including ribosomal biogenesis. We demonstrate that the nucleolus formation is precisely timed in D. melanogaster embryos and follows the transcription of rRNA. We provide evidence that transcription of rRNA is necessary for overcoming the highly stochastic nucleation step in the formation of the nucleolus, through a seeding mechanism. In the absence of rDNA, the nucleolar proteins studied are able to form high concentration assemblies. However, unlike the nucleolus, these assemblies are highly variable in number, location and time at which they form. In addition, quantitative study of the changes in the nucleoplasmic concentration and distribution of these nucleolar proteins in the wild-type embryos is consistent with the role of rRNA in seeding the nucleolus formation.
Graphical abstract
Introduction
Cells are composed of different membrane-less organelles that provide specialized environments for cellular functions. However, not much is known about the mechanism by which their constituent macromolecules assemble to form them. The nucleolus is a quintessential membrane-less organelle that forms around the ribosomal DNA (rDNA) repeats. Different steps of ribosomal biogenesis occur in the nucleolus including transcription of rRNA by RNA polymerase I, processing of pre-rRNAs and the assembly of mature rRNAs with ribosomal proteins. Other functions of the nucleolus include response to cellular stresses and the regulation of the cell cycle [1–3]. Despite such pivotal roles of the nucleolus, mature eggs lack this organelle. Instead, at a precise time during embryogenesis the numerous components of the nucleolus including more than 700 different proteins [4–6] and RNAs assemble at a specific region of the rDNA repeats called the nucleolus organizer region (NOR). The assembly of the nucleolus is then repeated during the interphase of every subsequent cell cycle following its disassembly during mitosis.
In all metazoans studied thus far, the re-assembly of nucleolar proteins following cell division initiates with the formation of transient high concentration foci that are dispersed throughout the nucleoplasm [7–13]. These high concentration assemblies behave like liquids that are immiscible in the nucleoplasm [14]. Therefore, a model has been proposed in which nucleolus formation can be regarded as a liquid-liquid phase separation [15]. Based on this model, proteins with certain biophysical properties can separate out from the nucleoplasm to make a distinct environment specialized for particular cellular functions. Such thermodynamically-driven phase separations provide a means to concentrate certain proteins and RNAs that is energetically inexpensive compared to active assembly. In addition, unlike other multivalent assemblies, the liquid nature of the nucleolus allows for fast diffusion of components including substrates and products in and out of the organelle. Despite such advantages, the kinetics of the phase separation process is typically limited by the initial nucleation event, a highly stochastic process that does not allow for the spatiotemporal precision observed in biological systems.
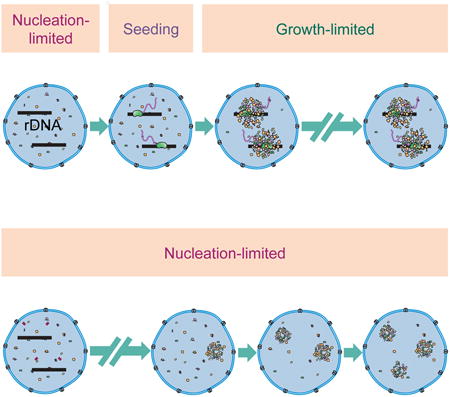
In any first order phase transition process, the initial nucleation step is a stochastic event in which thermal fluctuations in the system result in the formation of small assemblies of the new high-concentration phase (e.g. the nucleolus). In early stages, when these assemblies are smaller than a critical size, growth of the new phase results in an increase in the free energy, making this new phase unstable (Figure 1). This instability typically imposes a rate-limiting step on early nucleation in phase transitions, with classical examples being ice formation [16, 17] and in vitro assembly of microtubules [18]. However, the instability of the nucleation step can be circumvented if a seed is provided. In the presence of a seed, the formation of the new phase becomes growth-limited and the diffusion of constituent molecules governs the kinetics of self-assembly. In the case of membrane-less organelles, the kinetics of the initial nucleation step is widely unexplored. Previous studies have shown that exogenous transcription units can introduce new sites for the formation of different nuclear organelles [19–22]. However the necessity of such transcription sites in surmounting the stochastic nucleation step in the formation of nucleolar assemblies is not clear.
Figure 1. Nucleation and growth in phase separation processes.
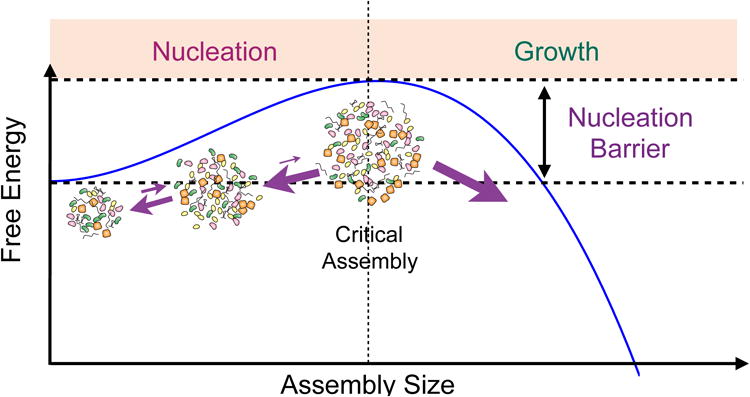
Birth of a new phase is called nucleation. Smaller size assemblies are unstable due to their large surface to volume ratio, which increases the negative effect of surface tension. Therefore, smaller assemblies shrink rather than grow. However, if the assemblies reach a critical size, they will readily grow. If the nucleation barrier is large, then the formation of new assembly becomes nucleation-limited. Otherwise, this process becomes growth limited.
In the present study, we investigate the initial nucleation step in the nucleolus assembly by performing a quantitative study in D. melanogaster embryos. We demonstrate that nucleolus formation is indeed precisely timed, and temporally follows the activation of rDNA transcription. To evaluate the contribution of rRNA in bypassing the stochastic nucleation step, we quantified the distribution of the nucleolar proteins in mutant embryos lacking rDNA repeats. Interestingly, the nucleolar proteins studied can still form high concentration assemblies in the absence of rDNA. However, higher variability is observed in the number, position, and time at which such assemblies form in the mutant embryos. Moreover, the observed changes in the concentration of the nucleolar proteins and the spatiotemporal pattern of the emergence of high-concentration foci in the wild-type embryos are also consistent with a model in which transcription of rRNA alters the assembly of the nucleolar components from a stochastic nucleation-limited process to a high-precision growth-limited event.
Results
Earliest Localization of Fibrillarin and RNA pol I to NORs at Nuclear Cycle 13
In order to visualize the de novo formation of the nucleolus in Drosophila embryos, we generated transgenic lines expressing two nucleolar markers tagged with fluorescent proteins: a subunit of RNA pol I, RpI135, tagged with eGFP, and fibrillarin tagged with TagRFP (hereafter RpI135-GFP and RFP-Fib). The transgenic flies expressing these two proteins were used to characterize the pattern of nuclear localization of RNA pol I and fibrillarin during early embryogenesis.
Figure 2A shows representative images of embryos expressing RpI135-GFP and RFP-Fib during nuclear cycles (n.c.) 10-14. Prior to n.c. 13, the fluorescent signal of RpI135-GFP is diffuse and uniform in the nucleus. The first localization of RpI135-GFP to bright sub-nuclear foci occurs at 6 ± 0.5 minutes into interphase of n.c. 13. A diffuse nucleoplasmic RpI135-GFP signal remains after the appearance of these bright foci. A similar behavior is observed for fibrillarin, which is involved in the processing of pre-rRNA (Movie 1) [23]. In all cycle 13 embryos, an initially uniform nuclear RFP-Fib fluorescent signal concentrates to sub-nuclear bright foci 5 ± 0.5 minutes into interphase and is seen in all subsequent cycles. In a small fraction of embryos (8 of 31 examined), a smaller, less easily detectable focus can be transiently observed in late cycle 12. In double labeling experiments, the initial bright foci of RpI135-GFP and RFP-Fib colocalize from the earliest points when localized signal is detected. The number of foci is consistent with the presence of one NOR per X and Y chromosome in Drosophila [24]. Our localization data with RpI135-GFP and RFP-Fib are similar to that of GFP-Nopp140 [25] and indicate that the large-scale recruitment of the nucleolar proteins to NORs happens at n.c. 13.
Figure 2. De novo nucleolus formation at n.c. 13.
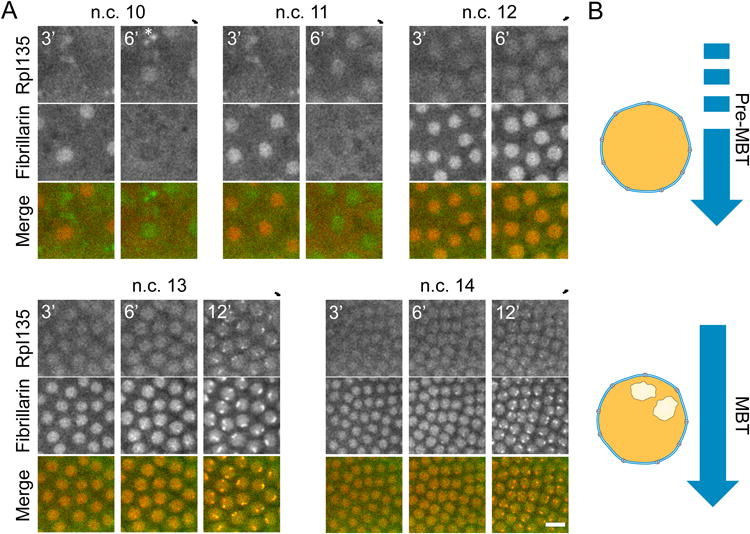
A Representative images of embryos expressing RpI135-GFP and RFP-Fib at n.c. 10-14. The nuclear signal for these two proteins is diffuse throughout the nucleus during n.c. 10-12, but both proteins colocalize to the nucleolus (bright foci) from n.c. 13. Asterisks show auto-fluorescence. Scale bar 10 μm. Numbers show the time in minutes from the end of mitosis. B. Schematic representation of the distribution of the nucleolar proteins during early embryogenesis. The nucleolus forms for the first time during mid-blastula transition (MBT), which coincides with the large-scale zygotic genome activation. Prior to MBT, no nucleolus can be detected in the Drosophila embryos. See also Movie 1.
Transcription of rDNA Initiates prior to Nucleolus Formation in Drosophila Embryos
To investigate whether the formation of a visible nucleolus is necessary for transcription of rDNA, we examined the expression of pre-rRNA during early embryogenesis. The maternal versus zygotic origin of the pre-rRNA was assessed by comparing the unfertilized eggs and 0-1 hr old embryos, where RNAs are mostly maternally provided, with embryos 3-4 hr post-fertilization in which zygotic genes are active [26, 27]. To measure pre-rRNA expression, cDNA from each group of embryos was analyzed by PCR for the presence of a segment of pre-rRNA, the internal transcribed segment 1 (ITS1), which is absent from the mature rRNA (Figure 3A). As depicted in Figure 3B, the ITS1 band is absent in the unfertilized eggs or in the 0-1 hr collection, and is only present in the 3-4 hr embryos. As controls for embryo staging, we measured expression of maternally-supplied hsp83 and zygotically-expressed fruhstart (z600) transcripts [27, 28], in comparison with ubiquitously expressed tubulin as a loading control. Our results indicate that in Drosophila embryos pre-rRNA is predominantly of zygotic origin.
Figure 3. The transcription of rDNA starts prior to the nucleolus formation in the Drosophila embryos.
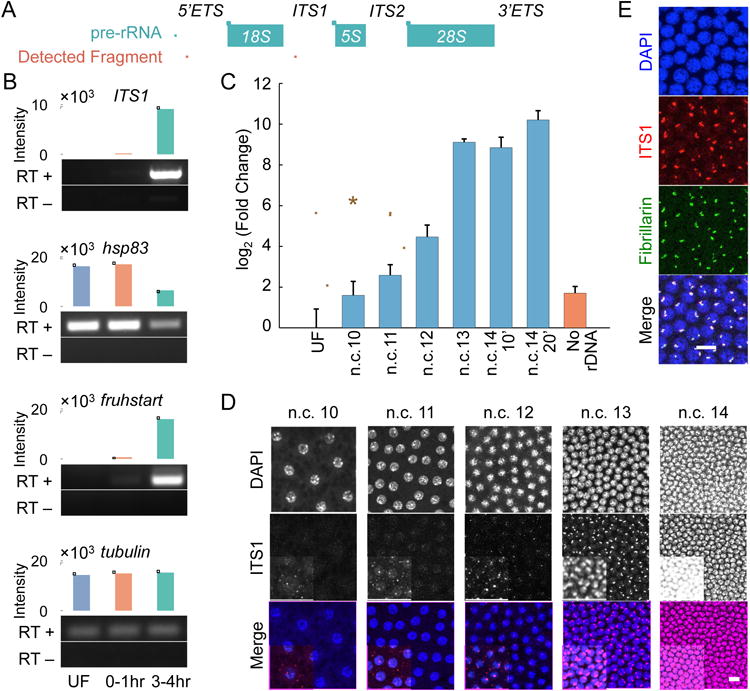
A. A schematic view of pre-rRNA, and the fragments used for detecting pre-rRNA. B. The pools of RNA present in wild-type embryos from three different collections were tested for the presence of pre-rRNA (ITS1), and three controls. The unfertilized embryos (UF) have only maternal RNA, the 0-1 hr collection has largely maternal or early zygotic RNA, and the 3-4 hr collection has mostly zygotically transcribed RNA. RT+ represents the samples that were reverse transcribed, while RT- is the negative control. hsp83 and fruhstart are maternal and zygotic, respectively. Tubulin is the loading control. C. The real-time PCR quantification of cDNAs from single embryo RNA extracts at different time points tested for the presence of 5′ETS shows a significant increase compared to the unfertilized embryos starting from n.c. 11. Data are presented as mean ± SEM of triplicate samples. Embryos were collected at seven minutes after the telophases of n.c. 10-12, 10 minutes of n.c. 13, and 10 and 20 minutes of n.c. 14. The mutants lacking rDNA were collected at n.c. 14. D. Likewise, FISH of ITS1 at n.c. 10-14 shows the emergence of bright foci starting from n.c. 11. For the purpose of presentation, the images in D and E have adjusted contrasts and changed saturation values, with the insets having lower saturation values. E. Immunolabeling of fibrillarin followed by FISH of ITS1 in embryos at n.c. 13. Scale bars 10 μm.
To determine more precisely when the transcription of rDNA is initiated we used fluorescent in situ hybridization (FISH) of ITS1 to detect pre-rRNA expression in n.c. 10-14 embryos. As shown in Figure 3D, pre-rRNA expression begins in one or two foci per nucleus starting at n.c. 11. We quantified the levels of nascent pre-rRNA in visually staged, precisely timed single embryos by quantitative real-time PCR (qRT-PCR) using primers complimentary to a region of 5′ external transcribed spacer (5′ETS, Figure 3A). There is a significant increase in the amount of pre-rRNA transcripts starting from n.c. 11 compared to unfertilized embryos (Figure 3C, P < 0.05 by Student's t-test). These results show that the transcription of rDNA starts at or prior to n.c. 11. The formation of a visible nucleolus at n.c. 13 coincides with an approximately 30-fold increase in the amount of pre-rRNA transcripts. Immunolabeling of fibrillarin followed by FISH of ITS1 in embryos at this stage indicates that the ITS1 puncta colocalize with fibrillarin bright foci (Figure 3E) confirming that the earliest visible accumulation of fibrillarin occurs at sites of pre-rRNA transcription.
Spatiotemporal Regulation of Nucleolus Formation by rDNA
To investigate the necessity of rDNA in overcoming the stochastic nucleation step in the formation of nucleolar assemblies, we followed the distribution of RpI135-GFP and RFP-Fib in embryos with compound chromosome X (C(1)DX/0, see Supplemental Experimental Procedures) that lack rDNA repeats. The amount of pre-rRNA in mutant embryos at n.c. 14 is < 0.4% of the amount observed in wild-type embryos (Figure 3C), and the expression level of the nucleolar proteins is unchanged (Figure S2B). Interestingly, even in the absence of rDNA, both RNA pol I and fibrillarin can form high-concentration assemblies that are similar to nucleoli, although smaller in size and initially less frequent (Figure 4A, Movie 2).
Figure 4. Fibrillarin and RNA pol I form high concentration assemblies in the absence of rDNA.
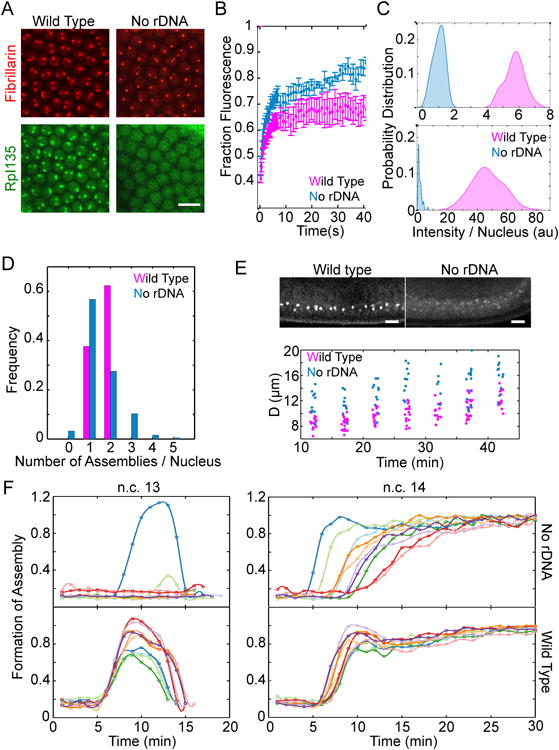
A. Representative images of the nuclei for wild type versus mutant embryos lacking rDNA at n.c. 14 show that both RFP-Fib and RpI135-GFP can form high concentration assemblies in the absence of rDNA. B. FRAP of Fibrillarin assemblies for the nucleoli and HANPs show that both are dynamic structures (rate constant for unbinding is 0.34 ± 0.03 s-1 for nucleoli and 0.27 ± 0.04 s-1 for HANPs). C. Histogram of the integrated intensity of nucleolus and HANPs normalized to the mean intensity of HANPs is shown for RFP-Fib (top) and RpI135-GFP (bottom). The probabilities for the case of RpI135-GFP in nucleolus is multiplied by 10 for better visualization. D. The fraction of nuclei showing different numbers of nucleoli in the wild-type or HANPs in the absence of rDNA is depicted for embryos after 15 minutes into n.c. 14. E. Top: Lateral view of the wild type and mutant embryos lacking rDNA at n.c. 14, expressing RFP-Fib. Bottom: quantification of the distance from the chorion of the nucleoli in a wild-type and HANPs in the mutant embryos lacking rDNA. F. Formation of fibrillarin high concentration assemblies for ten wild type (bottom) and ten mutant embryos lacking rDNA (top) over time at n.c. 13-14. Each color depicts an individual embryo. Time zero marks the end of mitosis. Scale bars 10 μm. See also Movie 2 and Figure S1.
Nucleoli are very dynamic structures, with high rates of exchange of proteins with the nucleoplasm [29]. To test whether the high-concentration assemblies of nucleolar proteins (HANPs) observed in the absence of rDNA have dynamic properties associated with nucleoli, or are kinetically arrested aggregates, we performed fluorescence recovery after photobleaching (FRAP). As depicted in Figure 4B, both HANPs and nucleoli recover within seconds, showing their dynamic nature.
Despite shared similarities, HANPs and bona fide nucleoli differ in some aspects (Movies 1-2). Firstly, the size of HANPs is smaller than that of the nucleoli, suggesting a role for rDNA in stabilizing the assembly of the nucleolar proteins through increasing favorable interactions between the nucleolar components (Figure 4C). Secondly, the nucleoli have an irregular structure, and appear to be punctate, whereas HANPs are round, as expected for a liquid phase. The irregularity in shape of the nucleolus may be due to the fact that only a small fraction of the rDNA genes at NORs are active at n.c. 14 in Drosophila embryos [30]. The small yet visible distance between the active rDNA genes would result in a punctate appearance, if each active site were to nucleate assembly of the nucleolar components.
Another difference is that the number of HANPs varies from zero to five per nucleus (Figure 4D). In wild-type embryos, only one or two nucleoli are observed per nucleus consistent with the number of NORs present. Moreover, while all nucleoli in the wild type embryos form at a distinct position, at the apical side of the nucleus, in the absence of rDNA, HANPs are more broadly dispersed throughout the nucleoplasm (Figure 4E). The normal position of the nucleolus in the wild type embryos is consistent with spatial organization of chromosomes in the blastoderm nuclei and the apical localization of the centromeric sequences that contain the NORs [31]. The seemingly random positioning of the HANPs in embryos that lack NORs suggests that these foci are not associated with specific sequences of DNA.
Even more remarkable is the difference in the time within which the assemblies form in the absence or presence of rDNA. Developmental events are precisely timed [32], including the formation of the nucleolus (Figure 4F). The variation in time at which the nucleoli form in the wild-type embryos is within the measurement errors (± 0.5 min, n = 10). However, in the absence of rDNA, this precision is lost by 10 fold at n.c. 14, and the time at which HANPs form varies between different embryos (variance = 6.66 ± 0.5 min, n = 10), and even between the nuclei of individual embryos (Figure 4F). This variability reduces the likelihood that HANPs will form during the short interphase of n.c. 13. Therefore in the absence of rDNA, the accumulation of the nucleolar proteins to sub-nuclear bodies becomes a highly stochastic event resulting in variable positioning and variable timing of the formation of HANPs.
Super-Saturation Concentrations of Nucleolar Proteins prior to Nucleolus Formation
In first order phase separation processes, the formation of the new high concentration phase (the nucleolus) is concentration dependent (Figure 5A). This means that below a critical concentration (Ccr), only one low concentration phase should exist (the nucleoplasmic phase), whereas above Ccr an additional high concentration phase should form. However, if nucleation of the new phase is limiting, nucleoplasmic concentrations higher than Ccr (super-saturation levels) can be reached before formation of the nucleolus, and upon a successful nucleation or seeding event, the nucleolus will grow until the nucleoplasmic concentration reaches Ccr.
Figure 5. Nucleolus formation is concentration dependent.
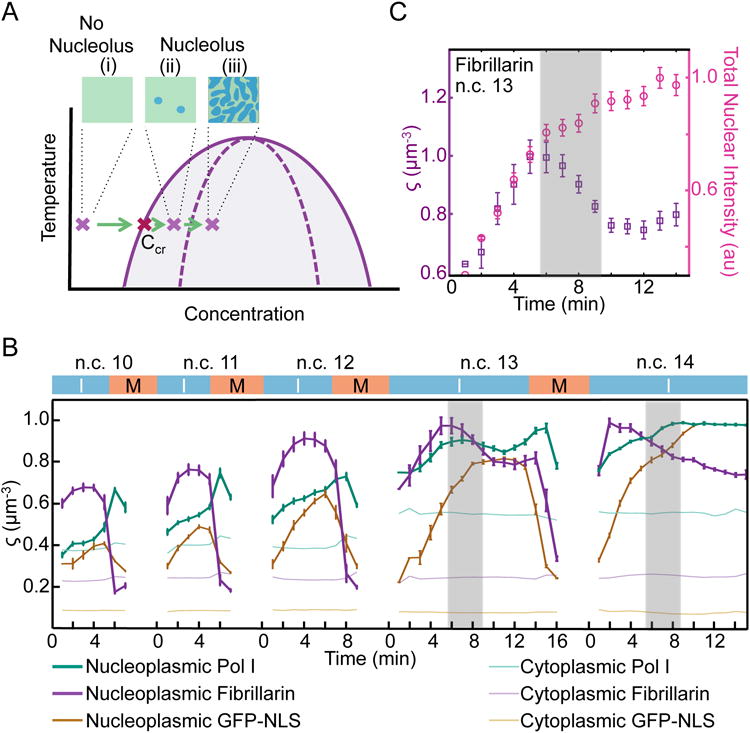
A. Phase separation processes are concentration dependent. The prediction of a first order phase separation is that at lower concentrations, only one low concentration phase, the nucleoplasm, should exist. After reaching a critical concentration (Ccr) a new high concentration phase, the nucleolus, should appear. The formation of this new phase is either nucleation-limited (ii) or growth-limited (iii). B. The normalized nucleoplasmic intensities (arbitrary units) per unit volume, excluding the nucleoli, for RFP-Fib, RpI135-GFP and GFP-NLS are shown as ς. The shaded areas in B and C depict the times that the nucleolus forms and coincides with a decrease in the nucleoplasmic concentration of RNA pol I and fibrillarin. C. Comparison of the ς for fibrillarin at n.c. 13 shown in B with its total intensity per nucleus. Although total nuclear amounts of fibrillarin increases until 14 min into n.c. 13, the nucleoplasmic concentration (ς) decreases upon nucleolus formation to reach a steady state. Both nucleoplasmic concentrations and total nuclear intensities have been normalized to their maximum values for presentation purposes. Zeros mark the end of mitosis (M) and the beginning of the interphase (I). Data are represented as mean ± SEM (n=10 for RpI135 and fibrillarin and n=3 for GFP-NLS). See also Figure S2.
We have characterized the concentration dependence of nucleolus formation by measuring nucleoplasmic concentrations of the two nucleolar proteins fibrillarin and RpI135 during early embryogenesis. We obtained these measurements at different time-points during n.c. 10-14 in terms of the nucleoplasmic intensities excluding the nucleolus, per unit volume for RpI135-GFP or RFP-Fib, denoted by ζ (arbitrary units / μm3). Time-lapse images of transgenic lines coexpressing H2Av were used to mark the progression of nuclear cycles. As shown in Figure 5B, there is a gradual increase in the nucleoplasmic concentrations of fibrillarin and RNA pol I during the late syncytial cycles. This increase continues until n.c. 13 when the nucleolus forms. These observations are consistent with a phase separation model in which the nucleolus is only observable at higher nucleoplasmic concentrations.
Interestingly, despite the constant increase in the total nuclear accumulation of fibrillarin and RNA pol I during the interphase of n.c. 13, the nucleoplasmic concentrations of these proteins decrease concurrently with the formation of the nucleolus (Figure 5B). No decrease in the nucleoplasmic concentration is observed for a control protein, GFP-NLS, which continues to rise and remains at high levels as the nucleolus forms until the onset of mitosis (Figure 5B). These results indicate that the nucleoplasmic concentrations of RNA pol I and fibrillarin are super-saturated prior to nucleolus formation, which is a characteristic of a nucleation-limited process. Following a triggering event –e.g. seeding by rDNA–the super-saturated proteins coalesce to form the nucleolus.
Local Inhomogeneities Emerge prior to the Nucleolus Formation at Cycle 14 but Not Cycle 13
rDNA could seed nucleolus formation either by a direct mechanism or through an intermediate. In the direct model, rDNA is a structural element of the seed, and therefore high-concentration assemblies of nucleolar proteins would be observed exclusively at NORs. Alternatively, if intermediate molecules such as rRNA transcripts were involved in decreasing the nucleation barrier, then it would be likely to observe high-concentration assemblies at places other than NORs. Resolving these two models requires detection of the nucleoli as well as quantification of local inhomogeneities in the distribution of the nucleolar proteins in the nucleoplasm prior to and during nucleolus formation. To facilitate detection of nucleoli in very early stages, we have developed an automated image analysis algorithm that detects the nucleoli at later time points when they can be detected unambiguously, and tracks them in preceding frames of the movie. We combined this algorithm to a method for calculating local inhomogeneities in the distribution of nucleoplasmic fluorescent intensities, based on gray-level co-occurrence matrix (GLCM) (see Supplemental Experimental Procedures) [33]. As depicted in Figure 6, at n.c. 14 the distribution of the nucleolar proteins becomes inhomogeneous prior to nucleolus formation, which means that at this cycle, high concentration assemblies emerge ubiquitously in the nucleoplasm. Upon progression of interphase, some of these small inhomogeneities disassemble (Figure S3A) while others coalesce to give rise to the nucleolus (Figure S3B). However, at n.c. 13 the decrease in the homogeneity coincides with nucleolus formation, and there is no detectable decrease in the homogeneity of the nucleoplasm, excluding the nucleolus. Therefore, although the initial assembly of the nucleolar proteins at n.c. 13 is strictly detectable at NORs, at n.c. 14 high concentration assemblies are observable throughout the nucleoplasm. This suggests that a product of rDNA rather than the template itself may be involved in seeding the formation of the nucleolus.
Figure 6. Local Inhomogeneities prior to the Nucleolus Formation at Cycle 14, but Not Cycle 13.
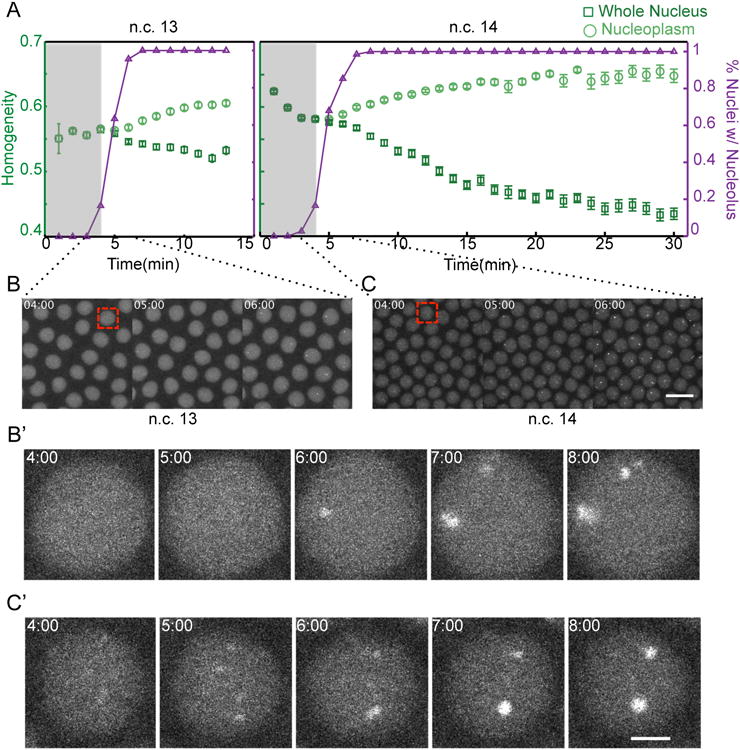
A. The emergence of local inhomogeneities in the whole nucleus and the nucleoplasm excluding the nucleolus has been calculated for fibrillarin as described in Supplemental Experimental Procedures. A decrease in the homogeneity shows the appearance of local inhomogeneities. As a comparison, the time at which the nucleoli are detectable are shown in terms of percent of nuclei with nucleolus (purple). Shaded areas highlight the time prior to nucleolus formation. The detectable decrease in the homogeneity at n.c. 13 is only due to the nucleolus formation, while at n.c. 14, inhomogeieties arise prior to nucleolus formation. B-C. Representative images of nuclei at n.c. 14 during the initial steps of nucleolus formation. While the only detectable bright foci at n.c. 13 are at NORs (C-C′), at n.c. 14 high-concentration assemblies appear ubiquitously in the nucleoplasm (D-D′). The nuclei in B′-C′ show the higher magnification of the regions depicted by red square in B-C, respectively. For the purpose of presentation, images have adjusted contrasts and saturation values. Scale bar for B-C, 10 μm and for B′-C′, 2 μm. See also Figure S3.
Previous studies have shown that several types of RNAs can nucleate de novo assembly of histone locus bodies, nuclear speckles, paraspeckles, and nuclear stress bodies [20]. To examine whether the loss of precision observed in the absence of rDNA emerges from the loss of rRNA transcripts or from the loss of the rDNA itself, we studied the effects of blocking rRNA synthesis in embryos with normal rDNA. Wild type fly embryos were injected with dsRNA against a subunit of RNA pol I, RpI12, resulting in an RNAi-mediated knockdown of RpI12 mRNA (Figure 7A). Reducing Rpl12 expression results in a two-fold decrease in the amount of nascent pre-rRNA transcripts in injected embryos (n = 25, P < 0.005 by Student's t-test, Figure 7B). In RpI12 RNAi embryos, the nucleoli are small or absent during n.c.13 (Figure 7C,E), and formation of the nucleolus is delayed by one cell cycle, to n.c.14 (Figure 7F). Therefore, the reduction in the rRNA transcription is concurrent with a delay in the nucleolus formation in embryos injected with anti-RpI12 RNAi (Figure 7F). In contrast, embryos injected with RNAi against RpI135 do not show reduction in the amount of pre-rRNA transcripts, possibly due to high levels of the maternal protein, and nucleolus formation remains unchanged (Figure 7A-D). The observed changes in nucleolus formation for embryos injected with RNAi against RpI12 is not likely to be due to altered cell cycle since the length of n.c. 13 is not affected [34]. We conclude that the observed delay in the formation of the nucleolus is due to the reduction in pre-rRNA transcripts that are key players in seeding the nucleolus formation.
Figure 7. Reduction in rRNA transcription delays nucleolus formation.
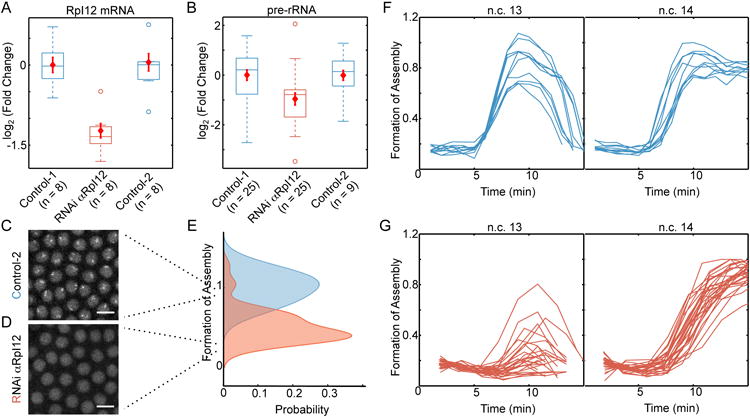
A. Injection of wild-type embryos with RNAi against RpI12 reduces the amount of this transcript to approximately 45% of that of un-injected embryos (Control-1) or embryos injected with RNAi against RpI135 (Control-2). B. The amount of pre-rRNA is determined by RT-qPCR on 25 individual control-1 embryos, 25 embryos injected with RNAi αRpI12, and nine of Control-2 embryos at 10 min into n.c. 13. The amount of pre-rRNA is reduced by nearly 2 fold in the embryos injected with RNAi for RpI12 compared to the two controls (P < 0.005 by Student's t-test). Red diamonds in A-B show mean ± SEM for each group. Representative images of n.c. 13 for a Control-2 embryo with normal nucleolus formation (C) and RpI12 RNAi embryo with delayed nucleolus formation (D). Scale bars, 10 μm. E. Formation of assemblies was measured for 39 individual RpI12 RNAi embryos (orange) and 39 un-injected embryos (blue) at 10 min into n.c. 13. Images shown in C and D score 0.9 and 0.3 for Formation of Assembly. F. Time evolution of formation of fibrillarin assemblies in the wild-type embryos (n=10, reproduced from Figure 4F) and G. RpI12 RNAi embryos (n=28) during n.c. 13-14. Each line depicts an individual embryo. Time zero marks the end of mitosis.
Discussion
Formation of membrane-less organelles requires coordinated recruitment of tens to hundreds of different macromolecules. Several features of the formation of such membrane-less organelles including P granules [35], centrosomes [36] and nucleolus [37, 38] are consistent with a phase transition model. However, phase separation processes are often limited by the initial nucleation event that is highly stochastic. We provide evidence that nucleolus formation in the wild-type embryos is not a highly variable nucleation-limited process. Rather, the transcription of rDNA renders the assembly process a well-controlled, growth-limited process. This accounts for the spatiotemporal precision of nucleolus formation observed in Drosophila embryos. In the absence of rDNA, the emergence of high concentration assemblies of the nucleolar components reduces to a nucleation-limited process with high spatiotemporal variability. Also, apparent supersaturated levels of fibrillarin and RNA pol I prior to nucleolus formation at n.c. 13 are consistent with the presence of a nucleation-limited process.
Several observations strongly suggest that pre-rRNA is the key player in seeding nucleolus formation. In all cases where nucleolus formation has been studied in metazoa, recruitment of nucleolar proteins occurs in the presence of unprocessed pre-rRNA. Our results show that the transcription of rDNA precedes the large-scale recruitment of the nucleolar proteins in Drosophila embryos. A similar chronological order has been observed for mouse embryos [39]. X. laevis embryos drive nucleolus formation prior to any detectable transcription by RNA polymerase I by loading eggs with maternally provided pre-rRNA [11, 40]. Likewise when nucleoli re-assemble after mitosis in mammalian tissue culture cells, pre-rRNA transcribed in the previous interphase is present [41]. Interestingly, the assembly of a synthetic nucleolus on an exogenous rDNA gene also requires transcription [19], and reducing transcription by RNA pol I can abolish nucleolus assembly in C. elegans [38], or delay the formation of the nucleolus in Drosophila embryos (Figure 7F-G here). Therefore pre-rRNA seems to play a pivotal role in seeding the assembly of the nucleolar proteins.
The presence of local inhomogeneities throughout the nucleoplasm during early n.c. 14 and their absence at n.c. 13 is consistent with the role of pre-rRNA in nucleating the assemblies (Figure 6). The amount of pre-rRNA at n.c. 13 is ≈30 times higher than n.c. 12 (Figure 3C), and a fraction of these transcripts may be carried over in the nucleus until the next cycle [41]. Therefore, far more rRNA transcripts are present at the beginning of n.c. 14 compared to n.c. 13. These “left-over” transcripts located throughout the nucleoplasm at n.c. 14 can seed the formation of assemblies ubiquitously in the nucleoplasm. Upon progression of interphase though, these local inhomogeneities are replaced by only two prominent assemblies located at NORs (Figure S3A-B). A subset of such small inhomogeneities disassemble during this time which could be due to the turnover of the seeding rRNA transcripts into ribosomes. Alternatively, in a process known as Ostwald ripening, reactivation of transcription can result in accumulation of further rRNA transcripts at NORs, which can in turn recruit more nucleolar components and destabilize smaller assemblies.
Our data on fibrillarin and RNA pol I identify intriguing differences in their localization patterns during early cleavage cycles. While fibrillarin localizes to the nucleus during interphase and is excluded from the nuclei undergoing mitosis, RNA pol I has a peak of concentration with the onset of mitosis in these early nuclear cycles (Figure 5B). Starting from n.c. 13 when the nucleolus forms, the dynamics of these two proteins overlap, and RNA pol I becomes enriched in the nuclei during interphase. However, although both fibrillarin and RNA pol I localize to the nucleolus, their involvement in the phase separation process appears to be different. After the nucleolus formation at n.c.13, the nucleoplasmic concentration of fibrillarin reaches a constant level, Ccr (Figure 5C), which is indicative of a phase separation occurring and reaching equilibrium. In contrast, RNA pol I does not reach an equilibrium concentration, and its nucleoplasmic levels increase toward the end of n.c. 13. In addition, the localization of RNA pol I to HANPs relative to the nucleolus is much weaker than fibrillarin (Figure 4C). These suggest that RNA pol I may not be a structural component of nucleolar phase separation, but instead may be recruited by other factors.
Although the changing concentration of certain critical components and posttranscriptional modifications like phosphorylation [42] may play broad roles in timing the formation of the nucleolus, our data suggest that seeding by rRNA may be the main mechanism regulating its temporal precision. A detectable nucleolus is not necessary for the initial transcription of rDNA. In mutants that lack rDNA however, although fibrillarin and RNA pol I can still form high concentration assemblies, the assembly process becomes variable and delayed. A similar loss in precision is observed when RNA pol I dependent transcription is reduced in embryos possessing normal rDNA, suggesting that it is the accumulation of rRNA transcripts, rather than the presence of the DNA itself, that drives nucleolar formation. By seeding the phase transition, this rRNA appears to dictate the spatiotemporal precision in the assembly, thereby turning an otherwise stochastic nucleation-limited process into a growth-limited event whose occurrence can be coordinated with other aspects of development.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for fly stocks. We thank all members of the E.F.W. and Schupbach laboratories, especially S. Little, B. He, and K. Doubrovinski for lively discussion. We thank S. Shvartsman, B. Machta, B. Bratton, A. Haji-Akbari, T. Schupbach, and R. Duronio for helpful comments. This work was supported in part by grant 5R37HD15587 from the National Institute of Child Health and Human Development (NICHD) to E.F.W. and Ruth Kirschstein National Research Service Award (NRSA) Postdoctoral Fellowship 1F32HD072653 from NICHD to S.A.B. E.F.W. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions: H.F., S.B., B.P. and E.W. designed the study, performed the experiments and analyzed the data. H.F. wrote the first draft of the manuscript. All authors participated in discussion of the data and in producing the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The Nucleolus under Stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. Available at: http://www.sciencedirect.com/science/article/pii/S1097276510007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. Available at: http://www.nature.com/nrm/journal/v8/n7/abs/nrm2184.html. [DOI] [PubMed] [Google Scholar]
- 3.Pederson T, Tsai RYL. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–6. doi: 10.1083/jcb.200812014. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2699146&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15635413. [DOI] [PubMed] [Google Scholar]
- 5.Scherl A. Functional Proteomic Analysis of Human Nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=133617&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. Directed Proteomic Analysis of the Human Nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0960982201006509. [DOI] [PubMed] [Google Scholar]
- 7.Tesařík J, Kopečný V, Plachot M, Mandelbaum J, Da Lage C, Fléchon JE. Nucleologenesis in the human embryo developing in vitro: Ultrastructural and autoradiographic analysis. Dev Biol. 1986;115:193–203. doi: 10.1016/0012-1606(86)90240-x. Available at: http://linkinghub.elsevier.com/retrieve/pii/001216068690240X. [DOI] [PubMed] [Google Scholar]
- 8.Kopečný V, Fléchon JE, Camous S, Fulka J. Nucleologenesis and the onset of transcription in the eight-cell bovine embryo: Fine-structural autoradiographic study. Mol Reprod Dev. 1989;1:79–90. doi: 10.1002/mrd.1080010202. Available at: http://dx.doi.org/10.1002/mrd.1080010202. [DOI] [PubMed] [Google Scholar]
- 9.Geuskens M, Alexandre H. Ultrastructural and autoradiographic studies of nucleolar development and rDNA transcription in preimplantation mouse embryos. Cell Differ. 1984;14:125–134. doi: 10.1016/0045-6039(84)90037-x. Available at: http://linkinghub.elsevier.com/retrieve/pii/004560398490037X. [DOI] [PubMed] [Google Scholar]
- 10.Korčeková D, Gombitová A, Raška I, Cmarko D, Lanctôt C. Nucleologenesis in the Caenorhabditis elegans Embryo. PLoS One. 2012;7:e40290. doi: 10.1371/journal.pone.0040290. Available at: http://dx.plos.org/10.1371/journal.pone.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verheggen C, Le Panse S, Almouzni G, Hernandez-Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis. J Cell Biol. 1998;142:1167–80. doi: 10.1083/jcb.142.5.1167. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2149348&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasano Y, Hokii Y, Inoue K, Sakamoto H, Ushida C, Fujiwara T. Distribution of U3 small nucleolar RNA and fibrillarin during early embryogenesis in Caenorhabditis elegans. Biochimie. 2008;90:898–907. doi: 10.1016/j.biochi.2008.02.001. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0300908408000345. [DOI] [PubMed] [Google Scholar]
- 13.Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H. Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma. 1985;92:330–336. doi: 10.1007/BF00327463. Available at: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=3902398&retmode=ref&cmd=prlinks\npapers2://publication/uuid/8525C68D-C5CB-4539-A0BB-14E784081C4D. [DOI] [PubMed] [Google Scholar]
- 14.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. Available at: http://www.pnas.org/cgi/doi/10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyman Aa, Weber Ca, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25288112. [DOI] [PubMed] [Google Scholar]
- 16.Haji-Akbari A, DeFever RS, Sarupria S, Debenedetti PG. Suppression of sub-surface freezing in free-standing thin films of a coarse-grained model of water. Phys Chem Chem Phys. 2014;16:25916–25927. doi: 10.1039/c4cp03948c. Available at: http://xlink.rsc.org/?DOI=C4CP03948C. [DOI] [PubMed] [Google Scholar]
- 17.Haji-Akbari A, Debenedetti PG. Direct calculation of ice homogeneous nucleation rate for a molecular model of water. Proc Natl Acad Sci. 2015;112:10582–10588. doi: 10.1073/pnas.1509267112. Available at: http://arxiv.org/abs/1505.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson KA, Borisy GG. Kinetic analysis of microtubule self-assembly in vitro. J Mol Biol. 1977;117:1–31. doi: 10.1016/0022-2836(77)90020-1. Available at: http://linkinghub.elsevier.com/retrieve/pii/0022283677900201. [DOI] [PubMed] [Google Scholar]
- 19.Grob A, Colleran C, McStay B. Construction of syntheticnucleoli in human cells reveals how a major functional nuclear domain isformed and propagated through cell division. Genes Dev. 2014;28:220–230. doi: 10.1101/gad.234591.113. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3923965&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21240286. [DOI] [PubMed] [Google Scholar]
- 21.Salzler HR, Tatomer DC, Malek PY, McDaniel SL, Orlando AN, Marzluff WF, Duronio RJ. A Sequence in the Drosophila H3-H4 Promoter Triggers Histone Locus Body Assembly and Biosynthesis of Replication-Coupled Histone mRNAs. Dev Cell. 2013;24:623–634. doi: 10.1016/j.devcel.2013.02.014. Available at: http://linkinghub.elsevier.com/retrieve/pii/S1534580713001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. Available at: http://www.genesdev.org/cgi/doi/10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- 23.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. [Accessed October 22, 2014];EMBO J. 1989 8:4015–24. doi: 10.1002/j.1460-2075.1989.tb08584.x. Available at: www.ncbi.nlm.nih.gov/pubmed/401576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann BP. Nucleolus-organizing regions in salivary gland chromosomes of Drosophila melanogaster. Zeitschrift für Zellforsch und Mikroskopische Anat. 1938;28:1–11. Available at: http://dx.doi.org/10.1007/BF00368209. [Google Scholar]
- 25.McCain J, Danzy L, Hamdi A, Dellafosse O, DiMario P. Tracking nucleolar dynamics with GFP-Nopp140 during Drosophila oogenesis and embryogenesis. Cell Tissue Res. 2006;323:105–15. doi: 10.1007/s00441-005-0044-9. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16158326. [DOI] [PubMed] [Google Scholar]
- 26.Edgar B. Parameters controlling transcriptional activation during early drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. Available at: http://linkinghub.elsevier.com/retrieve/pii/0092867486900097. [DOI] [PubMed] [Google Scholar]
- 27.Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. Available at: http://emboj.embopress.org/content/18/9/2610.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groβhans J, Müller HAJ, Wieschaus E. Control of Cleavage Cycles in Drosophila Embryos by frühstart. Dev Cell. 2003;5:285–294. doi: 10.1016/s1534-5807(03)00208-9. Available at: http://linkinghub.elsevier.com/retrieve/pii/S1534580703002089. [DOI] [PubMed] [Google Scholar]
- 29.Misteli T, Phair RD. No Title. Nature. 2000;404:604–609. doi: 10.1038/35007077. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10766243. [DOI] [PubMed] [Google Scholar]
- 30.McKnight SL, Miller OL. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. Available at: http://www.ncbi.nlm.nih.gov/pubmed/822943. [DOI] [PubMed] [Google Scholar]
- 31.Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. Available at: http://www.molbiolcell.org/cgi/doi/10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0168952512000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern. 1973;3:610–621. Available at: http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=4309314. [Google Scholar]
- 34.Blythe SA, Wieschaus EF. Zygotic Genome Activation Triggers the DNA Replication Checkpoint at the Midblastula Transition. Cell. 2015;160:1169–1181. doi: 10.1016/j.cell.2015.01.050. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0092867415001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CF, Brangwynne CP, Gharakhani J, Hyman Aa, Jülicher F. Spatial Organization of the Cell Cytoplasm by Position-Dependent Phase Separation. Phys Rev Lett. 2013;111:088101. doi: 10.1103/PhysRevLett.111.088101. Available at: http://link.aps.org/doi/10.1103/PhysRevLett.111.088101. [DOI] [PubMed] [Google Scholar]
- 36.Zwicker D, Decker M, Jaensch S, Hyman Aa, Julicher F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc Natl Acad Sci. 2014;111:E2636–E2645. doi: 10.1073/pnas.1404855111. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24979791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber SC, Brangwynne CP. Getting {RNA} and Protein in Phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. Available at: http://www.sciencedirect.com/science/article/pii/S0092867412006344. [DOI] [PubMed] [Google Scholar]
- 38.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. Available at: http://www.pnas.org/content/112/38/E5237.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-Mediated H3.3 Incorporation Is Required for DNA Replication and Ribosomal RNA Transcription in the Mouse Zygote. Dev Cell. 2014;30:268–279. doi: 10.1016/j.devcel.2014.06.022. Available at: http://linkinghub.elsevier.com/retrieve/pii/S1534580714004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verheggen C, Almouzni G, Hernandez-Verdun D. The ribosomal RNA processing machinery is recruited to the nucleolar domain before RNA polymerase I during Xenopus laevis development. J Cell Biol. 2000;149:293–306. doi: 10.1083/jcb.149.2.293. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2175160&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dundr M, Misteli T, Olson MOJ. The Dynamics of Postmitotic Reassembly of the Nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. Available at: http://www.jcb.org/cgi/doi/10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2:189–194. doi: 10.4161/nucl.2.3.16246. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3149879&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


