Abstract
Resolvin conjugates in tissue regeneration (RCTRs) are new chemical signals that accelerate resolution of inflammation, infection, and tissue regeneration. Herein, using liquid chromatography–tandem mass spectrometry–based metabololipidomics, we identified RCTRs in human spleen, lymph node, bone marrow, and brain. In human spleen incubated with Staphylococcus aureus, endogenous RCTRs were increased along with conversion of deuterium-labeled docosahexaenoic acid, conferring pathway activation. Physical and biological properties of endogenous RCTRs were matched with those prepared by total organic synthesis. The complete stereochemical assignment of bioactive RCTR1 is 8R-glutathionyl-7S,17S-dihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid, RCTR2 is 8R-cysteinylglycinyl-7S,17S-dihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid, and RCTR3 is 8R-cysteinyl-7S,17S-dihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid. These stereochemically defined RCTRs stimulated human macrophage phagocytosis, efferocytosis, and planaria tissue generation. Proteome profiling demonstrated that RCTRs regulated both proinflammatory and anti-inflammatory cytokines with human macrophages. In microfluidic chambers, the three RCTRs limited human polymorphonuclear cell migration. In hind-limb ischemia-reperfusion–initiated organ injury, both RCTR2 and RCTR3 reduced polymorphonuclear cell infiltration into lungs. In infectious peritonitis, RCTR1 shortened the resolution intervals. Each RCTR (1 nmol/L) accelerated planaria tissue regeneration by approximately 0.5 days, with direct comparison to both maresin and protectin CTRs. Together, these results identify a new bioactive RCTR (ie, RCTR3) in human tissues and establish the complete stereochemistry and rank-order potencies of three RCTRs in vivo. Moreover, RCTR1, RCTR2, and RCTR3 each exert potent anti-inflammatory and proresolving actions with human leukocytes.
Inflammation, although protective in host defense,1 when uncontrolled is associated with many widely occurring diseases. In addition to the well-appreciated chronic inflammatory diseases, such as arthritis and periodontal disease,2 many other diseases in humans are also known to be associated with a chronic state of inflammation. These include cardiovascular disease,3, 4 neurodegenerative diseases,5 metabolic syndrome, obesity,6 aging, and even poor nutrition.7 There are numerous chemical mediators that are involved in the initiation of the acute inflammatory response to microbial invasion that stimulate recruitment of leukocytes to sites of inflammation from the vasculature.1, 8 These include eicosanoids (prostaglandins and leukotrienes) and complement components as well as many cytokines, chemokines, and vasoactive mediators.9
By investigating the resolution of self-limiting inflammation and bacterial infections, this laboratory uncovered a novel superfamily of proresolving mediators that are biosynthesized de novo from essential fatty acids.2, 10, 11, 12, 13 These new chemical structures are potent in evoking the cardinal signs of resolution of inflammation2 (namely, they each limit further neutrophil infiltration, counterregulate proinflammatory mediators, and stimulate macrophage phagocytosis of cell debris and microbes). Their families were coined the resolvins, protectins, and maresins to distinguish their unique biosynthetic pathways and structures.2 Each member of these families carries potent proresolving actions that are evoked by specific receptors. The roles of these receptors were validated in a wide range of animal experimental models of diseases, emphasizing critical roles of controlling leukocyte trafficking and their responses that contribute to disease pathologies and infectious inflammation.14 Given their potent actions with isolated human cells, in vivo pharmacologic and physiological actions in animal models, and endogenous biosynthesis in human cells and tissues,12 the superfamily of proresolving lipid-derived mediators is collectively coined the specialized proresolving mediators (SPMs).2, 12 Because many of the anti-inflammatory agents used in the clinic today can be immune suppressive, the hunt for new approaches to control excess inflammation has highlighted the endogenous resolution mediators and mechanisms for new therapeutic approaches because they are not immunosuppressive.12, 15
Tissue injury, inflammation, and infection place demands on the system to regenerate damaged tissue.9 Recently, maresin 1 was found to stimulate tissue regeneration, in addition to its proresolving actions in inflammation,16 indicating that resolution-phase mediators can also directly affect tissue regeneration in primordial model organisms. Along these lines, focusing on the resolution of infectious inflammation and the cardinal signs of resolution led to the identification of three novel proresolving and tissue-regenerative biosynthetic pathways and mediators in each pathway that are potent new sulfido-conjugated chemical mediators.17, 18 These peptide-containing conjugates with docosahexaenoic acid (DHA)–derived backbones encompass the distinguishing chemical features in the biosynthesis and structures of the maresins, protectins, and resolvins.18 They include maresin conjugates in tissue regeneration (MCTRs), protectin conjugates in tissue regeneration (PCTRs), and resolvin conjugates in tissue regeneration (RCTRs). Each pathway is biosynthesized via pivotal allylic epoxide–containing intermediates. Because mass spectrometry–based identification can give only the basic structures without stereochemistry when deduced from biological sources,19, 20 it is imperative to establish the complete stereochemical assignments of the new RCTRs via matching by total organic synthesis with materials of known stereochemistry to confirm their structures and potent bioactions. Recently, we established the complete stereochemistry of MCTR1, MCTR2, and MCTR321 as well as PCTR1.22 Herein, we addressed the complete stereochemical assignments of the newest family of proresolving mediators found to stimulate tissue regeneration (ie, RCTRs) and established their production in human tissues.
Materials and Methods
Human Organs, Cell Isolation, and Mice
Human brain, spleen, lymph nodes, and bone marrow were purchased from the Cooperative Human Tissue Network, National Cancer Institute (Eastern Division, Philadelphia, PA). For lipid mediator (LM) profiling, they were weighed, and 1 mL methanol-containing 500 pg internal standard mixture was added to each incubation.
Human neutrophils were isolated from whole blood that was collected from healthy volunteers, as approved by the Partners Human Research Committee (protocol 1999P001297). Peripheral blood neutrophils were isolated by density gradient. To induce apoptosis, polymorphonuclear neutrophils (PMNs) were cultured overnight with phosphate-buffered saline (PBS) and labeled with 10 ng/mL bisbenzimide H 33258 (Sigma-Aldrich, St. Louis, MO).
Human monocytes were isolated from deidentified leukopacks obtained from Children's Hospital Blood Bank (Boston, MA) with the use of Ficoll-Histopaque 1077-1 (Sigma-Aldrich). Macrophage differentiation was induced with 10 ng/mL human granulocyte-macrophage colony-stimulating factor (R&D Systems, Minneapolis, MN) for 6 days in RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen, Grand Island, NY), 2 mmol/L l-glutamine (Lonza, Basel, Switzerland), and penicillin-streptomycin (Lonza).
Animal experimental procedures were approved by the Standing Committee on Animals of Brigham and Women's Hospital (protocol no. 2016N000145) and complied with institutional guidelines, the USDA Animal Welfare Act, Public Health Service Policy on Humane Care and Use of Laboratory Animals (https://grants.nih.gov/grants/olaw/references/phspol.htm), and the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals (https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf). FVB male mice (6 to 8 weeks old; Charles River Laboratories, Wilmington, MA) were fed ad libitum Laboratory Rodent Diet 20-5058 (Lab Diet; Purina Mills, Great Summit, MO).
Peritonitis experiments were conducted as done previously by Dalli et al.18 Briefly, Escherichia coli (serotype O6:K2:H1) was cultured in Luria-Bertani broth and harvested at midlog phase (OD 600 nm, approximately 0.5 absorbance units). Mice were inoculated with E. coli (105 colony-forming units/mouse), and 12 hours later, they were administered with RCTR1, RCTR2, RCTR3 (100 ng/mouse) or vehicle via i.p. injection. At indicated intervals, mice were euthanized and exudate was collected with 5 mL of PBS without calcium or magnesium. Leukocyte populations were determined using flow cytometry and fluorescent-labeled antibodies, including anti-mouse F4/80 (clone BM8; 17-4801-82; eBioscience, San Diego, CA), anti-mouse CD11b (clone M1/70; 25-0112-81; eBioscience), anti-mouse Ly-6C (clone HK14; 128005; BioLegend, Dedham, MA), and anti-mouse Ly-6G (clone 1A8; 127607; BioLegend). Macrophages, monocytes, and PMNs were identified as F4/80+CD11b+, F4/80−CD11b+LY-6C+, and F4/80−CD11b+LY-6G+, respectively. Resolution indexes were calculated as in Bannenberg et al.23
LM Metabololipidomics
Before sample extraction, ice-cold methanol–containing deuterium (d)-labeled d4-leukotriene (LT)B4, d4-5S–hydroxyeicosatetraenoic acid, d4–prostaglandin E2, d5-resolvin D2, d5-LTC4, d5-LTD4, and d2-PCTR3 internal standards (500 pg each) were added to facilitate quantification. All samples were kept at -20°C for 45 minutes to allow protein precipitation and then spun at 800 × g for 5 minutes. Products were extracted using solid-phase extraction with C18 columns (100 mg/3 mL isolute; Biotage, Charlotte, NC). Samples were rapidly acidified to pH = 3.5 just before loading onto solid-phase extraction columns and being washed with 4 mL of water, pH = 7.4. Next, columns were washed with 5 mL of hexane. Methyl formate fractions (3 mL) were then collected for LM profiling using liquid chromatography–tandem mass spectrometry (LC-MS-MS) in the negative ion mode.24 For profiling of cysteinyl leukotrienes and DHA-sulfido conjugates, methanol fractions (5 mL) were collected and LC-MS-MS was performed in the positive ion mode. Then, samples were concentrated with a continuous flow of nitrogen to avoid oxidation using a manifold apparatus (TurboVap LV; Biotage). Samples were suspended in 50 μL methanol/water 1:1 mixture and were analyzed by a liquid chromatography–tandem mass spectrometry system, QTrap 5500 (AB Sciex, Framingham, MA) equipped with a Shimadzu LC-20AD high-performance liquid chromatograph (Shimadzu, Tokyo, Japan). A Poroshell 120 EC-18 column (100 mm × 4.6 mm × 2.7 μm; Agilent Technologies, Santa Clara, CA) was kept in a column oven maintained at 50°C, and sulfido lipid mediators were eluted with a gradient of methanol/water/acetic acid from 55:45:0.1 (v/v/v) to 100:0:0.1 at 0.5 mL/minute flow rate.
To monitor and quantify the levels of targeted LM, a multiple reaction monitoring method was devised with signature ion fragments for each molecule. Molecules were identified using published criteria, including retention times and at least six diagnostic ions. Calibration curves were obtained using synthetic and authentic LM mixtures, including d5-LTC4, d5-LTD4, d2-PCTR3, LTC4, LTD4, LTE4, MCTR1, MCTR2, MCTR3, PCTR1, PCTR2, PCTR3, RCTR1, RCTR2, and RCTR3 at 4, 10, 20, 40, 100, 200, 400, and 1000 pg. Linear calibration curves for each compound were obtained with r2 values of 0.98 to 0.99. Peak areas of the multiple reaction monitoring transitions were used for quantification. Each RCTR configuration prepared by total organic synthesis was confirmed by nuclear magnetic resonance imaging.25 Structural integrity of all synthetic compounds was verified via LC-MS-MS using established criteria17, 26 before each experiment.
Proteome Profiler Arrays
Human granulocyte-macrophage colony-stimulating factor differentiated macrophages were incubated with 10 nmol/L RCTR1, RCTR2, RCTR3, MCTR1, MCTR2, MCTR3, or vehicle (containing 0.1% ethanol) for 15 minutes at 37°C, then with 100 ng serum-treated zymosan (STZ), and finally incubated at 37°C, in 5% CO2 for 24 hours. Cell-free supernatants were collected and proteome profiler array was processed according to the manufacturer's instructions (ARY022; R&D Systems). Densitometric analysis of dot blots was performed using ImageJ software version 1.50i (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Human Organ Incubations
Nine human spleens (1.5 g each) were incubated in PBS at 37°C for 60 minutes, followed by incubation in the presence or absence of Staphylococcus aureus (107 colony-forming units), 60 minutes. In separate sets of experiments, three human spleens (approximately 3 g each) from healthy donors were obtained and incubated in PBS at 37°C for 1 hour, followed by incubation with 1 μmol/L of d5-DHA in the presence or absence of S. aureus (107 colony-forming units), 60 minutes, 37°C. All incubations were stopped with addition of cold methanol (2× that of the incubation volume) and taken for solid-phase extraction.
Real-Time Imaging Phagocytosis of E. Coli
Human macrophages were plated onto 8-well chamber slides (5 × 104 cells per well) overnight before the experiments. Chamber slides were kept in a Stage Top Incubation system for microscopes equipped with a built-in digital gas mixer and temperature regulator (TOKAI HIT model INUF-K14; Tokyo, Japan). Cells were treated with RvD1, RCTR1, RCTR2, RCTR3 (10 nmol/L), or vehicle control (containing 0.1% ethanol) for 15 minutes at 37°C. E. coli serotype O6:K2:H1 was cultured in Luria-Bertani broth, collected at midlog phase (OD600 nm ≈ 0.5; 0.5 × 109 colony-forming units/mL), followed by labeling with Baclight Green (B35000; Molecular Probes, Waltham, MA). E. coli was added at a ratio of 50:1 (E. coli/macrophage) to initiate phagocytosis. Fluorescent images were then recorded every 10 minutes for 60 or 150 minutes (37°C) with a Keyence BZ-9000 (BIOREVO) inverted fluorescence phase-contrast microscope (40× objective) equipped with a monochrome/color-switching camera using BZ-II Viewer software version 2.2 (Keyence, Woburn, MA). Six separate experiments were performed. In each experiment, three fields (×40) per condition (per well) were recorded. Green fluorescence intensity was quantified using a BZ-II Analyzer (Keyence).27
Efferocytosis
Human macrophages were plated onto 96-well plates (5 × 104 cells per well) and incubated with RCTR1, RCTR2, RCTR3, or RvD1 (0.1 pmol/L to 10 nmol/L) or vehicle for 15 minutes at 37°C, pH = 7.45. Fluorescently labeled apoptotic neutrophils were then added at a ratio of 5:1 (PMN/macrophage) for 60 minutes. Extracellular fluorescence was quenched with Trypan blue 0.026%. Cells were washed with PBS, and fluorescence was recorded using a fluorescent plate reader (excitation/emission, 350/461 nm).17
Hind-Limb Ischemia/Reperfusion in Mice
Mice were anesthetized by i.p. injection of a mixture of xylazine (80 mg/kg) and ketamine (10 mg/kg). Bilateral hind-limb ischemia was initiated using tourniquets placed on each limb.28 Mice were subjected to ischemia for 1 hour, tourniquets were removed, and reperfusion ensued for 2 hours. Ten minutes before reperfusion, vehicle (saline containing 0.1% ethanol), RCTR1, RCTR2, or RCTR3 (500 ng) was administered intravenously. Mice were then euthanized, and lungs were collected for quantification of PMN infiltration and for LC-MS-MS metabololipidomics. PMNs were quantified by myeloperoxidase levels. Briefly, lungs were homogenized and centrifuged, supernatants were collected, and tissue levels of myeloperoxidase were determined with myeloperoxidase enzyme-linked immunosorbent assay (mouse myeloperoxidase Duo Set; DY3667; R&D Systems).
Real-Time Neutrophil Chemotaxis
Human neutrophils were isolated, as described, and suspended in Hanks' balanced salt solution (Gibco Life Technologies, Gaithersburg, MD). PMNs (3 × 106 cells/mL) were placed at ibidi chemotaxis chambers and treated per manufacturer's instructions and as described earlier29 (μ-Slide Chemotaxis; catalog number 80326; ibidi cells in focus; ibidi, Planegg, Germany). PMNs were incubated with vehicle (Hanks' balanced salt solution containing 0.1% ethanol), RCTR1, RCTR2, or RCTR3 at 10 nmol/L for 10 minutes at 37°C. Gradients of LTB4 were formed in the chemotaxis chamber connected to the main channel where neutrophils were placed. Neutrophil migration in the gradient and their responses of different conjugates were recorded every 30 seconds over a 30-minute period with a digital monochrome/color-switching camera using BZ-II Viewer software. Cell migration was analyzed using the cell-tracking function in ImageJ software, and tracks were analyzed using ibidi Chemotaxis and Migration Tool. Chemotaxis indexes were calculated with the following formula: chemotaxis indexes = yend/accumulated distance.
At least 20 cells were tracked per donor, with four donors per tested compound. Migration directness was calculated with the ratio between euclidian/accumulated distance.30
Planaria Tissue Regeneration
Tissue regeneration was assessed, as previously described.18 In brief, planaria (Dugesia japonica) were kept in water (Poland Spring, Poland, ME) at 20°C. All animals were starved for 7 days before surgery. Head resection was performed (postocular surgical injury), and planaria were then placed in water that contained RCTR1, RCTR2, or RCTR3 at 1 to 100 nmol/L or vehicle control. Rate of tissue regeneration was monitored for 6 days. Blastema regeneration was quantified with ImageJ software. Tissue regeneration index analysis was used to normalize the total area of regenerated tissue and the postocular width, where tissue regeneration index = total area of regenerated tissue/postocular width.
Statistical Analysis
Significance was determined with the use of t-test when comparing two experimental groups. One- or two-way analysis of variance with a post Dunnett's multiple comparisons test was used when more than two experimental groups were analyzed. Analysis was conducted with GraphPad Prism version 6 (GraphPad Software, La Jolla, CA). Statistical significance was set at P < 0.05.
Results
Human Tissues Produce RCTR1, RCTR2, and RCTR3: Stereochemical Assignments
To identify the RCTRs in vivo and establish their stereochemistry, RCTRs were first obtained from human tissues. These included brain, spleen, lymph nodes, and bone marrow (Table 1 and Figure 1, A and B) that were selected because these tissues are enriched with the precursor DHA and SPM.13, 26, 31, 32 Next, the physical properties of these endogenous materials were matched with RCTR prepared by total organic synthesis25 using established criteria19 (Figure 1C). These criteria include retention times in the multiple reaction monitoring chromatographs, fragmentation patterns in the MS-MS spectra, and at least six diagnostic ions in the MS-MS spectra.26 The diagnostic ions in the mass spectra were identical between both endogenous and synthetic RCTRs. In addition to RCTR1 and RCTR2, identified in the study by Dalli et al,17 material that appeared to be consistent with RCTR3 was found, raising the question of whether RCTR3 carries bioactivity. This matching approach was required because the amounts produced in vivo were routinely <100 μg that would be needed for direct determination of the double bond geometries using nuclear magnetic resonance imaging and purified mediators. This approach enables us to establish the complete stereochemistry of the resolvins, protectins, and maresins.2
Table 1.
Human Endogenous Bioactive Sulfido-Conjugate Metabolome: Direct Comparison of CTRs to Leukotrienes
| Sulfido conjugates | Brain, pg (n = 5) | Spleen, pg (n = 7) | Lymph node, pg (n = 6) | Bone marrow, pg (n = 4) |
|---|---|---|---|---|
| MCTR1 | 5.1 ± 2.1 | 10.0 ± 2.5 | 4.5 ± 2.0 | 5.9 ± 2.4 |
| MCTR2 | 2.6 ± 1.2 | 7.9 ± 1.7 | 1.6 ± 1.1 | - |
| MCTR3 | 0.6 ± 0.3 | - | 0.8 ± 0.2 | - |
| PCTR1 | 1.7 ± 0.8 | 1.1 ± 0.8 | 1.2 ± 0.5 | - |
| PCTR2 | 0.8 ± 0.4 | - | 0.3 ± 0.2 | - |
| PCTR3 | 1.0 ± 0.4 | - | 1.3 ± 0.5 | - |
| RCTR1 | - | 394.4 ± 112.4 | - | 182.4 ± 56.4 |
| RCTR2 | - | 160.5 ± 71.1 | - | 91.8 ± 41.7 |
| RCTR3 | 4.3 ± 1.9 | 62.3 ± 27.9 | 5.0 ± 1.7 | 24.3 ± 3.0 |
| LTC4 | 3.8 ± 1.6 | 139.9 ± 68.3 | 4.8 ± 2.8 | 36.5 ± 23.7 |
| LTD4 | 0.6 ± 0.1 | 77.4 ± 57.9 | 2.9 ± 2.1 | 2.0 ± 0.6 |
| LTE4 | 5.7 ± 3.6 | 100.4 ± 39.4 | 2.9 ± 0.9 | 2.1 ± 1.0 |
Human brain, spleen, lymph nodes, and bone marrow were matched with synthetic material obtained by total organic synthesis. Levels of specialized proresolving lipid mediator sulfido conjugates in different human organs are expressed as means ± SEM pg/500 mg of tissue and pg/4 mL of bone marrow.
-, limit of detection approximately 0.1 pg; LT, leukotriene.
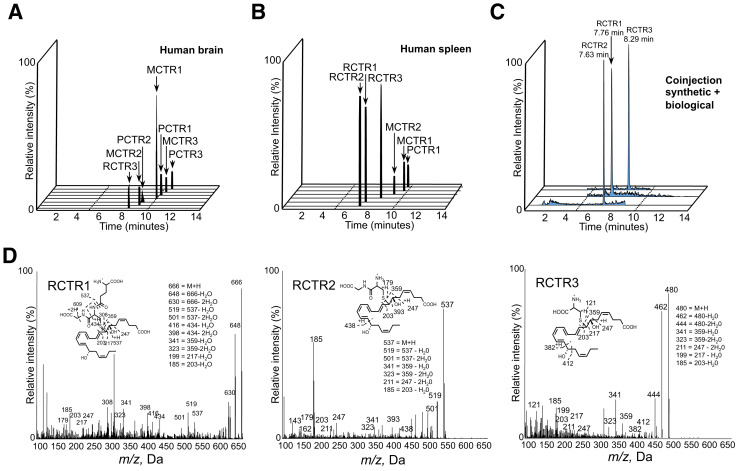
Figure 1.
Stereochemistry of endogenous human RCTR1, RCTR2, and RCTR3. Gently minced human brain and spleen tissue sections were extracted, and levels of sulfido conjugates were assessed by lipid mediator metabololipidomics liquid chromatography–tandem mass spectrometry (MS-MS; Materials and Methods). A: Human brain identification of endogenous sulfido-conjugated products MCTR1, MCTR2, MCTR3, PCTR1, PCTR2, PCTR3, and RCTR3; representative multiple reaction monitoring (MRM) chromatograms from five human donors (Materials and Methods). B: Human spleen identification of endogenous MCTR1, MCTR2, PCTR1, RCTR1, RCTR2, and RCTR3; representative MRM chromatograms from seven human donors. C: Coinjections of synthetic and biological materials from human spleens. D: Human spleen identification of RCTR1, RCTR2, and RCTR3; representative MS-MS spectra. m/z, mass/charge ratio.
RCTR1 gave a mass/charge ratio (m/z) 666 = M + H, m/z 648 = 666-H2O, m/z 630 = 666-2H2O, m/z 519 = 537-H2O, m/z 501 = 537-2H2O, m/z 416 = 434-H2O, m/z 398 = 434-2H2O, m/z 341 = 359-H2O, m/z 323 = 359-2H2O, m/z 199 = 217-H2O, and m/z 185 = 203-H2O (Figure 1D). RCTR2 gave m/z 537 = M + H, m/z 519 = 537-H2O, m/z 501 = 537-2H2O, m/z 341 = 359-H2O, m/z 323 = 359-2H2O, m/z 211 = 247-2H2O, and m/z 185 = 203-H2O (Figure 1D). RCTR3 gave m/z 480 = M + H, m/z 462 = 480-H2O, m/z 444 = 480-2H2O, m/z 341 = 359-H2O, m/z 323 = 359-2H2O, m/z 211 = 247-2H2O, m/z 199 = 217-H2O, and m/z 185 = 203-H2O (Figure 1D). Quantification was achieved using the multiple reaction monitoring transitions of parent ion and characteristic daughter ion, and linear regression with standard curve for each LM (Materials and Methods). In addition, coinjection of synthetic and endogenous RCTR1, RCTR2, and RCTR3 demonstrated coelution at 7.76 minutes for RCTR1, at 7.63 minutes for RCTR2, and at 8.29 minutes for RCTR3 (Figure 1C). These results confirmed the stereochemistry of endogenous RCTR1 and RCTR2 and documented formation of RCTR3 in human tissues.
Sulfido-conjugated lipid mediator levels, including MCTRs, PCTRs, and RCTRs, were identified and quantified in human brain, spleen, lymph nodes, and bone marrow (Table 1). Demographics for each subject are given in Supplemental Table S1. LC-MS-MS–based LM profiling in the positive ion mode that targeted molecules containing a DHA backbone and carrying a glutathionyl, cysteinylglycinyl, or a cysteinyl group to capture the three families of sulfide conjugates gave seven distinct chromatographic peaks obtained from human brain and lymph nodes, including MCTR1, MCTR2, MCTR3, PCTR1, PCTR2, PCTR3,21 and RCTR3 (Figure 1A and Table 1). From human spleen tissue, MCTR1, MCTR2, PCTR1, and RCTR1, RCTR2, and RCTR3 were identified (Figure 1B). Human bone marrow gave RCTR1, RCTR2, and RCTR3, which were present together with MCTR1. Cysteinyl leukotrienes were present in all human tissues examined (Table 1). In human spleen and bone marrow, the highest levels of DHA-derived conjugates were RCTR family, specifically RCTR1. These results indicate that RCTRs were produced in select human tissues with distinct profiles.
After matching of the physical properties of synthetic stereochemically defined materials with those of endogenously produced RCTR from both human (Figure 1) and murine (Supplemental Figure S1 and Supplemental Table S2) systems, the production of RCTRs was examined in tissue slices of human spleen (Materials and Methods). Staphylococcus aureus incubations with spleen increased all three RCTRs (Table 2). In addition, when human spleen tissues were incubated with S. aureus and d5-labeled DHA, the RCTR and resolvin pathway markers, including d5-17-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid and d5-7-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid, were increased after S. aureus incubation (Table 3). In mouse infectious exudates, the addition of DHA significantly increased MCTRs, PCTRs, and RCTR, but not cysteinyl leukotrienes (Supplemental Figure S1 and Supplemental Table S2). Together, these results indicate that RCTR biosynthetic machinery/enzymes were present in spleen and infectious exudates and were activated by bacteria to initiate RCTR biosynthesis.
Table 2.
RCTR Formation by Human Spleen
| RCTR | Spleen, pg | Spleen + Staphylococcus aureus, pg |
|---|---|---|
| 1 | 106.9 | 206.6 |
| 2 | 7.6 | 16.1 |
| 3 | 12.3 | 14.5 |
Human spleen tissue was incubated in phosphate-buffered saline at 37°C for 1 hour, followed by 1-hour incubation in the presence or absence of 107 colony-forming units Staphylococcus aureus. Results are representative of nine human donors, expressed as pg/1.5 g spleen tissue.
Table 3.
Conversion of d5-DHA to Resolvin Precursors in Human Spleen
| Variable | Spleen, pg | +Staphylococcus aureus, pg |
|---|---|---|
| d5-17-HDHA | 50.4 | 109.2 |
| d5-14-HDHA | 29.3 | 104.5 |
| d5-7-HDHA | 5.6 | 11.0 |
Human spleen tissue was preincubated in phosphate-buffered saline at 37°C for 1 hour, followed by 1-hour incubation with 1 μmol/L d5-DHA in the presence or absence of 107 colony-forming units Staphylococcus aureus. Results are representative of three human donors, expressed as pg/3 g spleen tissue.
d5-DHA, deuterium-labeled docosahexaenoic acid; HDHA, hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid.
RCTRs Stimulate Phagocytosis and Efferocytosis with Human Macrophages
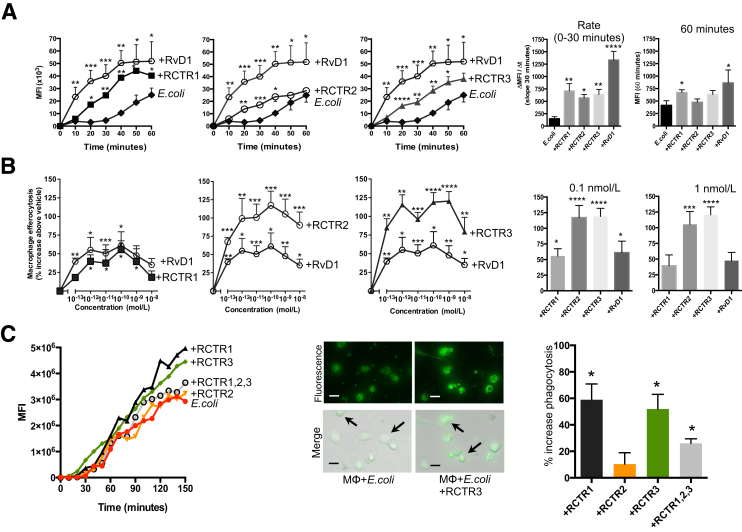
Phagocytosis and clearance of cellular debris, such as apoptotic PMNs, known as efferocytosis, is critical in clearance of infections and progression to resolution.33 In this regard, biologically derived RCTR1 and RCTR2 display potent actions in efferocytosis and phagocytosis of E. coli by human macrophages.17 Next, it was addressed whether the stereochemically defined RCTR1 and RCTR2 carry these bioactions, and also assessed whether RCTR3 carries these proresolving actions. Using real-time imaging microscopy, the kinetics of E. coli phagocytosis were investigated by human macrophages. RvD1 was used for direct comparison with each RCTR. At 10 nmol/L, each RCTR significantly increased phagocytosis of fluorescently labeled E. coli as early as 20 minutes after addition compared with E. coli alone (Figure 2A). Among the three RCTRs tested, RCTR1 gave the highest rate of E. coli phagocytosis from 0 to 30 minutes (Δmean fluorescence intensity/Δtime, approximately 750 mean fluorescence intensity/minute) and E. coli ingestion at 60 minutes (mean fluorescence intensity, approximately 40,000), followed by RCTR3 and RCTR2.
Figure 2.
Stereochemically defined RCTR1, RCTR2, and RCTR3 stimulate phagocytosis and efferocytosis with human macrophages. Differentiated human monocyte–derived macrophages (MΦ) collected from healthy donors were plated (5 × 104 cells per well). A: Macrophages were treated with 10 nmol/L of RvD1, RCTR1, RCTR2, RCTR3, or vehicle for 15 minutes at 37°C. Real-time phagocytosis was performed using Baclight green-labeled Escherichia coli with a ratio of 1:50, macrophages/E. coli. In each experiment, three fields (×20) per condition (per well) were recorded. Data are expressed as mean fluorescence intensity (MFI). Bar graphs represent the slopes at 0 to 30 minutes and maximum MFI at 60 minutes. B: Macrophages (5 × 104 cells per well) were plated and incubated with 0.1 pmol/L to 10 nmol/L of RCTR1, RCTR2, RCTR3, RvD1, or vehicle for 15 minutes at 37°C. Fluorescently labeled apoptotic neutrophils (Materials and Methods) were added at a ratio of 1:5 macrophages/apoptotic polymorphonuclear cells for 60 minutes. Then, extracellular fluorescence was quenched with Trypan blue, 0.026%. Cells were washed with phosphate-buffered saline, and fluorescence was recorded using a fluorescent plate reader (excitation/emission, 350/461 nm) and quantified as percentage increases above vehicle. C: Macrophages (5 × 104 cells per well) were plated onto 8-well chamber slides and incubated with RCTR1, RCTR2, RCTR3 individually or all three together (10 nmol/L each; denoted all RCTRs). Real-time phagocytosis was performed as described in A. Time course from one representative donor. Representative images of macrophages from E. coli or E. coli + RCTR incubations after 150 minutes of phagocytosis; fluorescence and overlay of fluorescence and bright-field images are shown. Arrows indicate macrophages with ingested fluorescent E. coli. Results are percentage increase in phagocytosis above E. coli + vehicle, at 150 minutes. Data are expressed as means ± SEM (A–C). n = 6 (A and B); n = 3 (C). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus E. coli + vehicle (t-test). Scale bars = 20 μm (C).
Each of the RCTRs dose dependently stimulated phagocytosis of human apoptotic neutrophils (ie, efferocytosis) compared with RvD1, where RCTR2 and RCTR3 displayed essentially equal potency at concentrations as low as 0.1 pmol/L to 10 nmol/L, with approximately 80% to 120% increase in efferocytosis when compared with vehicle. At 0.1 and 1 nmol/L, RCTR1 and RvD1 were less potent, thus establishing a rank-order potency of RCTR3 ≥ RCTR2 > RCTR1 (Figure 2B). Although each of the RCTRs proved to be statistically significant and potent activators of macrophage phagocytosis (eg, efferocytosis and E. coli phagocytosis), when added together they did not further enhance phagocytosis (Figure 2C). These results indicated that RCTR1, RCTR2, and RCTR3 are potent agonists of both phagocytosis and efferocytosis. They also demonstrated the stereoselectivity of these actions.
RCTRs Regulate Select Inflammation-Associated Proteins with Human Macrophages
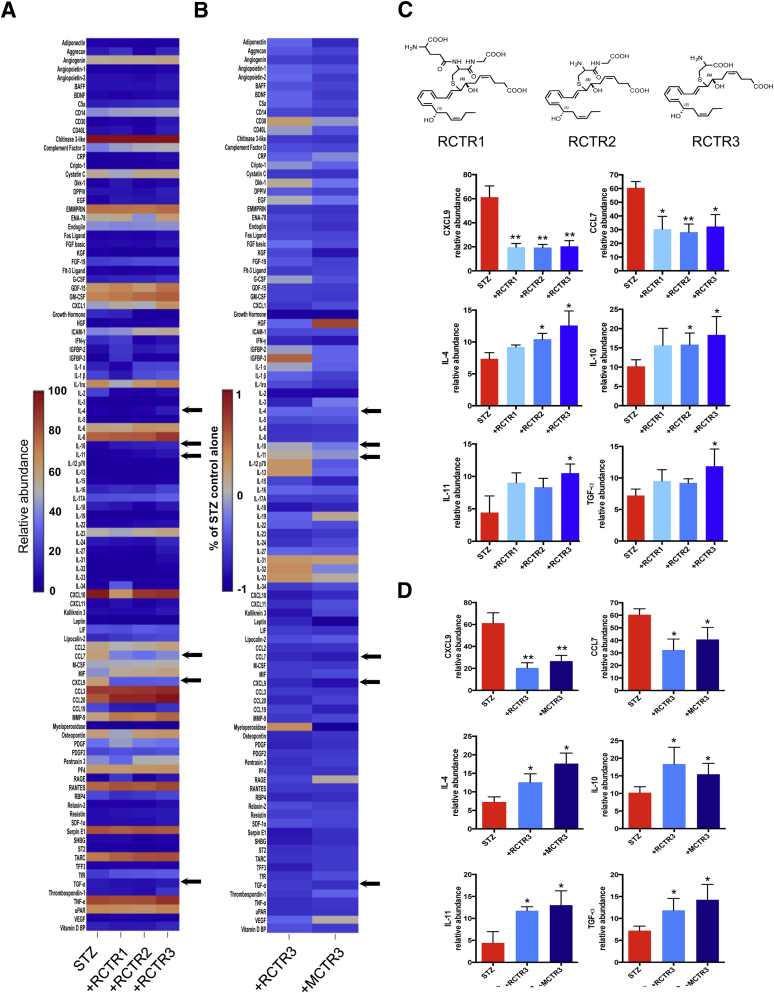
Next, proteome profiling was performed to examine whether RCTRs regulate cytokines and inflammation-related proteins produced by human macrophages during phagocytosis. Macrophages were challenged with 100 ng STZ for 24 hours in the presence or absence of each RCTR (Figure 3A) or MCTR (Figure 3B and Supplemental Figure S2), each at 10 nmol/L. Cell-free supernatants were collected for proteome profiling. STZ alone stimulated release of a panel of inflammation-related proteins, including cytokines and chemokines. RCTR1, RCTR2, or RCTR3 incubated together with STZ each significantly down-regulated select proteins, including inflammatory chemokines CXCL9 and chemokine (C-C motif) ligand 7 compared with STZ alone (Figure 3, A and C). In addition, RCTR2 and RCTR3 statistically significantly up-regulated IL-4 and IL-10 (Figure 3), which are known to be anti-inflammatory.34 RCTR3 also increased IL-11 and transforming growth factor (TGF)-α (Figure 3, A and C). TGF-α is known to promote wound healing.35 IL-11 is also known to have anti-inflammatory properties.36, 37
Figure 3.
RCTRs regulate select proteome profiles of inflammation-associated proteins with human macrophages: Direct comparison of RCTR3 and MCTR3. Proteome Profiler cytokine array was performed using human macrophages (2 × 106 cells). Macrophages were incubated with 10 nmol/L RCTR1, RCTR2, RCTR3, MCTR3, or vehicle (containing 0.1% ethanol) for 15 minutes at 37°C, then with 100 ng of serum-treated zymosan (STZ) and incubated at 37°C, with 5% CO2 for 24 hours. A: Results are expressed in a heat map as relative abundance (Materials and Methods). B: Results are expressed in a heat map as fold-change comparing RCTR3 or MCTR3 with STZ. Arrows in A and B indicate proteins shown in C and D. C: Top panels: Structures of RCTR1, RCTR2, and RCTR3. C and D: Levels of CXCL9, chemokine (C-C motif) ligand (CCL) 7, IL-4, IL-10, IL-11, and transforming growth factor (TGF)-α were determined by densitometry analysis of dot blots. Data are expressed as the means ± SEM (C and D). n = 3 separate experiments with three separate healthy donors (A–D). ∗P < 0.05, ∗∗P < 0.01 versus STZ (t-test). BAFF, B-cell activating factor; BDNF, brain-derived neurotrophic factor; CRP, C-reactive protein; DPPIV, dipeptidyl-peptidase-4; EGF, epidermal growth factor; FGF, fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GDF, growth and differentiation factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; HGF, hepatocyte growth factor; ICAM, intercellular adhesion molecule; IFN, interferon; IGFBP, insulin growth factor binding protein; LIF, leukemia inhibitory factor; M-CSF, macrophage colony-stimulating factor; MIF, macrophage migration inhibitory factor; MMP, matrix metallopeptidase; PDGF, platelet-derived growth factor; RAGE, receptor for advanced glycation end products; RANTES, regulated on activation normal T cell expressed and secreted; RBP, retinol binding protein; SDF, stromal cell–derived factor; SHBG, sex hormone–binding globulin; TARC, thymus and activation regulated chemokine; TFF, trefoil factor family; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
In comparison to RCTRs, when each of the MCTR1, MCTR2, or MCTR3 was incubated with STZ-challenged macrophages, they each gave statistically significant down-regulation of CXCL9 and chemokine (C-C motif) ligand 7 lower than those levels produced in the presence of STZ alone (Supplemental Figure S2). Each of the three MCTRs gave essentially equal potency in up-regulating IL-4, IL-10, IL-11, and TGF-α. In these experiments, RCTR3 and MCTR3 gave comparable potencies in regulating these select cytokines and growth factor (Figure 3D). Taken together, STZ challenge heightened proinflammatory chemokine levels that were regulated by both RCTRs and MCTRs (Figure 3 and Supplemental Figure S2), which each dampened specific proinflammatory and increased anti-inflammatory cytokines. These results indicate that, within the RCTR family, RCTR3 was the most potent in stimulating specific cytokines, giving the rank-order potency of RCTR3 > RCTR2 > RCTR1.
Direct Action of RCTRs on Human PMNs Limits Chemotaxis
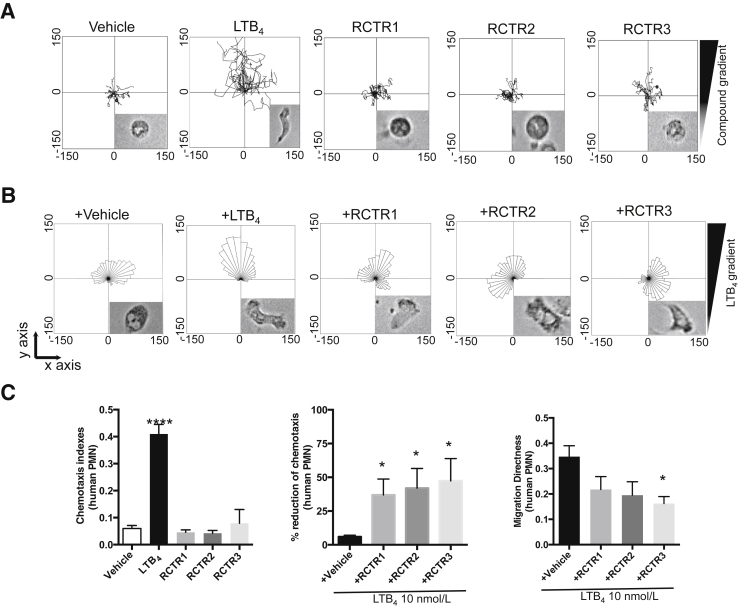
Proresolving mediators limit PMN infiltration, leading to the cessation of their recruitment to the site of inflammation.2 Therefore, it was next tested whether the new RCTRs have direct actions on human PMNs. To address this, the actions of RCTRs were first examined in regulating PMN chemotaxis in real time at the single-cell level using chemotaxis chambers that provide defined spatiotemporal concentration gradients (Materials and Methods). LTB4, a potent PMN chemoattractant,38 was used for direct comparison. Isolated PMNs gave round morphology in the absence of a stimulus gradient (Figure 4A). In the presence of an LTB4 gradient, cells changed shape to a polarized morphology and migrated toward LTB4. RCTR1, RCTR2, or RCTR3 did not significantly increase chemotaxis when added alone above that of vehicle control (Figure 4A). Because PMN apoptosis accelerates resolution, RCTRs were tested and it was found that at 10 nmol/L they each did not alter human PMN apoptosis at 24 and 48 hours (Supplemental Figure S3).
Figure 4.
RCTR1, RCTR2, and RCTR3 each reduces human neutrophil chemotaxis. Neutrophils were isolated from whole blood (Materials and Methods) of healthy volunteers, and 6 μL was placed on ibidi microfluidics chambers at a concentration of 3 × 106 cells/mL. A: Chemotaxis toward 10 nmol/L of leukotriene (LT) B4, RCTR1, RCTR2, RCTR3, or vehicle [Hanks' balanced salt solution (HBSS) + 0.1% ethanol] was monitored under a light microscope in real time over 30 minutes. Representative trajectory paths are shown from a representative donor. Insets: Morphology of neutrophils after exposure to vehicle, LTB4, or RCTR 1 to 3. B: Cells were exposed to 10 nmol/L of RCTR1, RCTR2, RCTR3, or vehicle (HBSS + 0.1% ethanol) for 10 minutes. Chemotaxis toward an LTB4 gradient (10 nmol/L) was monitored in real time over 30 minutes. Rose plots of migrating cells are shown from a representative donor. C: Results are chemotaxis indexes for human neutrophils with 10 nmol/L of LTB4, RCTR1, RCTR2, RCTR3, or vehicle. Percentage reduction of LTB4-initiated neutrophil chemotaxis at 30 minutes. Results are polymorphonuclear cell (PMN) migration directness toward LTB4 gradient in the absence or presence of RCTR1, RCTR2, or RCTR3 (10 nmol/L). P values are given using one-tailed paired t-test (middle panel) or one-way analysis of variance with Tukey's multiple comparison post-test (left and right panel). Data are expressed as means ± SEM (C). n = 4 (A–C). ∗P < 0.05 ∗∗∗∗P < 0.0001 versus vehicle.
Next, it was investigated whether RCTRs can limit LTB4-initiated chemotaxis. Each RCTR (10 nmol/L) was injected into the microfluidic chambers containing human PMNs, followed by injection of LTB4 to form a gradient of the chemoattractant. Migrating PMNs within the chemotactic gradient were recorded in real time by phase-contrast microscopy and reported in rose plots (Figure 4B). RCTR1, RCTR2, and RCTR3 each statistically significantly reduced LTB4-initiated chemotaxis by 37%, 42%, and 47%, respectively (Figure 4C). In addition, the directionality of neutrophil migration was calculated for LTB4 and each of the RCTRs (Materials and Methods) (Figure 4C). Exposure of neutrophils to LTB4 gradient initiated migration directness. Incubation of RCTR3 with neutrophil statistically significantly reduced LTB4-initiated chemotaxis. RCTR1 and RCTR2 did not reach statistically significant differences for reducing LTB4-initiated chemotaxis. These results demonstrate, at the single-cell level, that each RCTR at 10 nmol/L reduced directional migration of human PMNs toward LTB4 gradient.
RCTRs Reduce Ischemia/Reperfusion-Initiated PMN-Mediated Lung Injury
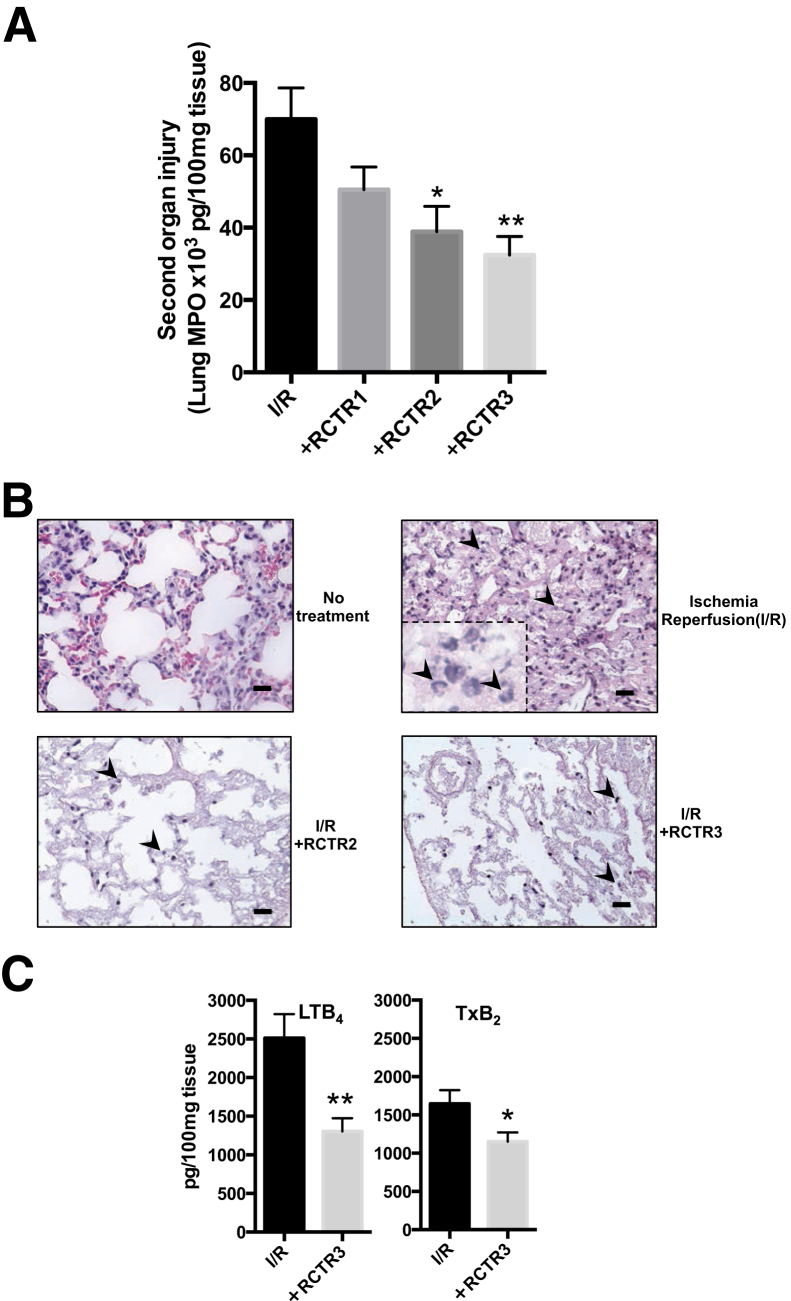
Because RCTRs limited chemotaxis with isolated human PMNs, the in vivo actions of RCTRs in PMN-mediated remote organ injury initiated by ischemia/reperfusion (I/R) were next examined. Surgical clamping procedures are well appreciated to lead to aberrant PMN activation via vessel occlusion that causes remote organ injury, contributing to longer hospitalization.39 A hind-limb I/R (tourniquet occlusion) model of second organ lung injury, an established murine model of operating room surgical insults in humans,28 was used. Ischemia was initiated by 1-hour occlusion of hind limbs, followed by 2-hour reperfusion that caused PMN infiltration into the lungs compared with no treatment (Figure 5, A and B). The i.v. administration of RCTR1, RCTR2, or RCTR3 (500 ng each) 10 minutes before reperfusion led to reduction of PMN infiltration into lungs compared with I/R alone. RCTR3 was the most potent, giving approximately 50% reduction, followed by RCTR2 (approximately 40% reduction) and RCTR1 (approximately 30% reduction) (Figure 5A).
Figure 5.
RCTR2 and RCTR3 limit reflow organ injury and reduce production of leukotriene (LT) B4 and thromboxane (TxB2). Hind-limb ischemia-reperfusion (I/R) was induced by applying tourniquets to the hind limbs of 6- to 8-week-old male FVB mice. After 50 minutes, RCTR1, RCTR2, or RCTR3, at 500 ng/mouse i.v., was each directly compared with vehicle (saline containing 0.1% ethanol), followed by removal of tourniquets; reperfusion ensued for 2 hours. Then, animals were sacrificed, and lungs were collected. A: Myeloperoxidase (MPO) levels were assessed by enzyme-linked immunosorbent assay. B: Hematoxylin and eosin histology sections of mouse lungs; arrowheads indicate infiltrated neutrophils. C: Homogenized lungs were extracted and eicosanoids were identified and quantified using lipid mediator metabololipidomics (Materials and Methods). Data are expressed as means ± SEM (A and C). n = 6 to 7 (A–C). ∗P < 0.05, ∗∗P < 0.01 versus I/R alone (one-way analysis of variance). Scale bars = 20 μm (B). Original magnification, ×40 (B, main images); ×100 (B, inset).
Using targeted lipid mediator metabololipidomics, it was assessed whether RCTRs regulated proinflammatory eicosanoids after reperfusion. The i.v. administration of RCTR3 led to a statistically significant reduction of both LTB4 (>45%) and thromboxane (approximately 30%) (Figure 5C) in lung tissues. Together, these results demonstrate that RCTR3 in vivo controlled PMN-mediated lung injury, reduced proinflammatory LM, and exerted organ protection, as illustrated herein with hind-limb occlusion-initiated reflow lung injury. RCTR3 proved to be the most potent of the RCTR family, with the rank-order potency RCTR3 >> RCTR2 >>> RCTR1.
RCTRs Accelerate Resolution of Infectious Inflammation
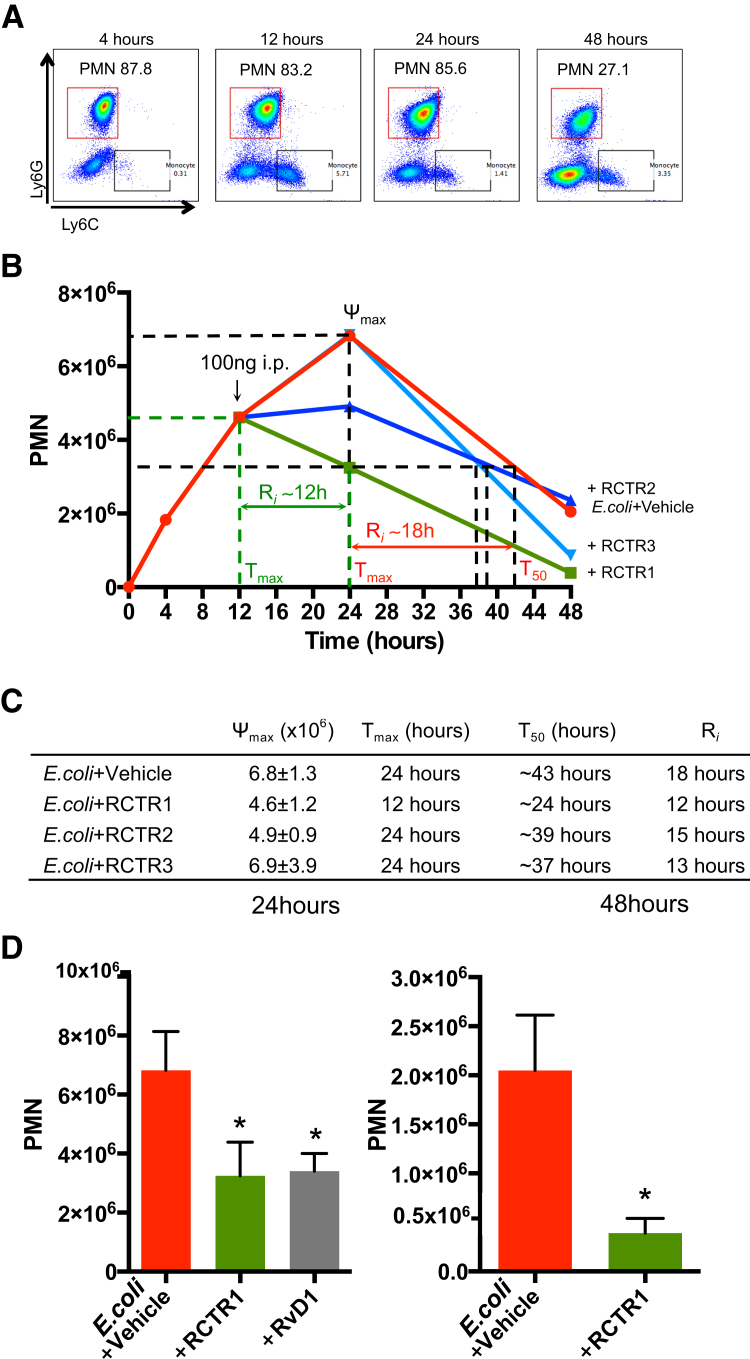
Given that RCTRs each display potent actions in enhancing phagocytosis (Figure 2), it was tested whether each of the RCTRs also stimulate resolution in vivo in murine infections. Quantification of PMNs with flow cytometer (Figure 6A) in the exudates of E. coli (1 × 105 colony-forming units/mouse) i.p. inoculated mice gave a self-resolving inflammatory response, with maximal neutrophil infiltration at approximately 24 hours that subsequently declined; there was a calculated T50 of approximately 43 hours and a resolution interval of approximately 18 hours. The i.p. injection of RCTR1 at 12 hours after E. coli inoculation significantly reduced neutrophil infiltration at 24 hours and shortened the resolution interval from approximately 18 hours to approximately 12 hours. Neither RCTR2 nor RCTR3 gave a statistically significant difference in PMN numbers at 24 hours; their resolution interval values were approximately 27 hours for RCTR2 and approximately 13 hours for RCTR3 (Figure 6, B and C). At 24 hours, RCTR1 and RvD1 were essentially equally potent in reducing PMN infiltration (Figure 6D). By comparison, RCTR1 was most potent of all three RCTRs in accelerating resolution of E. coli infection (Figure 6D). Thus, activation of the RCTR and D-series resolvin pathways produced molecules that equally accelerated resolution of infectious inflammation.
Figure 6.
RCTR1 accelerates resolution of Escherichia coli infection. FVB mice (males, 6 weeks old) were inoculated with E. coli (105 colony-forming units). RvD1, RCTR1, RCTR2, RCTR3 (100 ng each), or vehicle was given by i.p. injection 12 hours after E. coli inoculation, and peritoneal exudates were collected at 24 and 48 hours. Total leukocytes were enumerated using light microscopy, and polymorphonuclear cell (PMN) numbers were determined by flow cytometry. A: Representative flow cytometry dot plots of phosphatidylethanolamine-Ly6G and fluorescein isothiocyanate–Ly6C. PMNs (Ly6G+Ly6C−) are denoted in red squares, and monocytes (Ly6C+Ly6G−) are denoted in black squares. For gating strategy, see Materials and Methods. B: Time courses of PMN numbers. C: Resolution indexes. D: Results are PMN numbers at 24 and 48 hours. Data are expressed as means ± SEM (D). n = 3 to 4 mice per group (A–D). ∗P < 0.05 versus E. coli + vehicle (unpaired t-test).
Planaria Tissue Regeneration: Comparison of RCTRs, PCTRs, and MCTRs
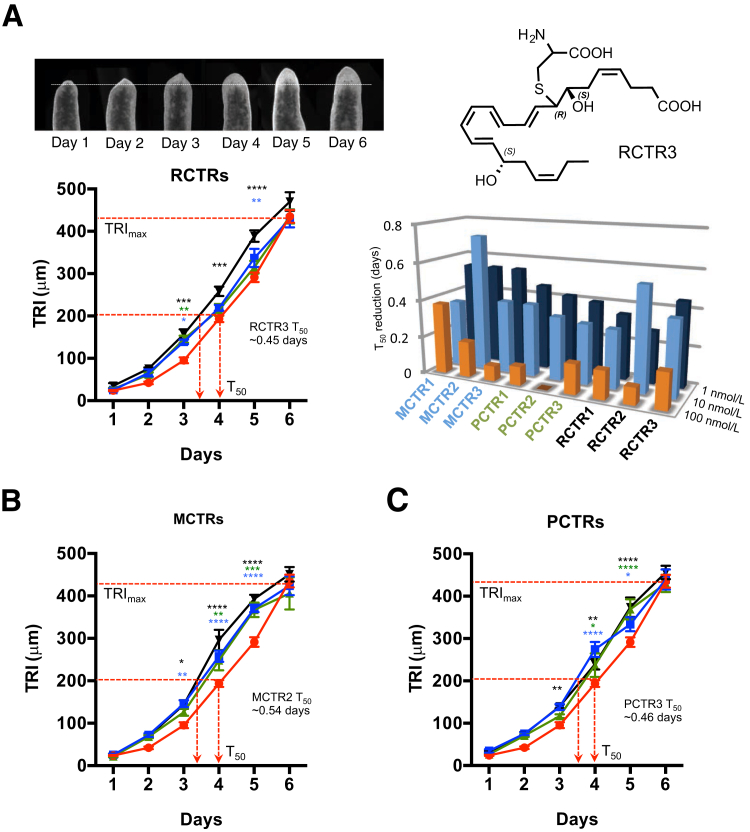
Planaria undergo regeneration via evolutionarily conserved pathways, making it an ideal system to test the tissue-regenerative actions of vertebrate-derived molecules.40 Isolated biologically derived RCTR1 and RCTR2 accelerated planaria tissue regeneration.17 Thus, it was required herein to assess the tissue regenerative action of stereochemically defined RCTR1 and RCTR2 prepared by total organic synthesis, and also determine whether RCTR3 shared this regenerative action. All nine stereochemically defined CTRs (ie, MCTRs, PCTRs, and RCTRs) were compared directly. Using defined tissue regeneration indexes17, 18 after head resection–injury, planaria gave a maximum tissue regeneration index of approximately 400 μm at 6 days. Tissue regeneration index50, representing 50% regeneration (approximately 200 μm), and T50, representing the time point when regeneration reaches 50% maximum (4 days), were also defined. Incubation of planaria with the RCTRs (1 to 100 nmol/L) accelerated tissue regeneration, shortening T50. Among all three RCTRs, RCTR3 was the most effective at 1 nmol/L, shortening T50 approximately 0.45 days, whereas RCTR2 was the most potent at 10 nmol/L, reducing T50 approximately 0.55 days (Figure 7A). Also, for each RCTR, 1 and 10 nmol/L proved to be more potent than 100 nmol/L. Incubations of injured planaria with each stereochemically defined MCTR (Figure 7B) or PCTR (Figure 7C) also led to acceleration in tissue regeneration with reductions in T50. Of the MCTR family, MCTR2 appeared to be the most potent at 10 nmol/L. In comparison, PCTR1, RCTR2, and RCTR3 gave essentially equal potency at 1 and 10 nmol/L. Taken together, these results demonstrated that MCTR, PCTR, and RCTR families each stimulated planaria tissue regeneration after surgical injury. The functions, stereochemistry, and rank-order potencies of RCTRs are summarized in Figure 8.
Figure 7.
Acceleration of tissue regeneration in planaria: Direct comparisons of RCTRs, PCTRs, and MCTRs. Anterior portions of brown planaria were surgically removed, and remaining planaria were exposed to vehicle or increasing concentrations (1 to 100 nmol/L) of compounds. A: Images (top row) are planaria head regeneration from 1 to 6 days. The dashed white line indicates head resection at day 0 and head regeneration by day 6. Bottom left panel: Tissue regeneration index (TRI) of RCTR1 (blue), RCTR2 (green), RCTR3 (black) at 1 nmol/L each or vehicle alone (red). Bottom right panel: Direct comparison of T50 of nine DHA-derived conjugates. B: TRI of MCTR1 (blue), MCTR2 (green), or MCTR3 (black). C: TRI of PCTR1 (blue), PCTR2 (green), or PCTR3 (black) at 1 nmol/L each or vehicle alone (red) were assessed for 6 days. Data are expressed as means ± SEM (A–C). n = 8 planaria/group from two independent experiments (A–C). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus vehicle alone (two-way analysis of variance with Dunnett's multiple comparisons test).
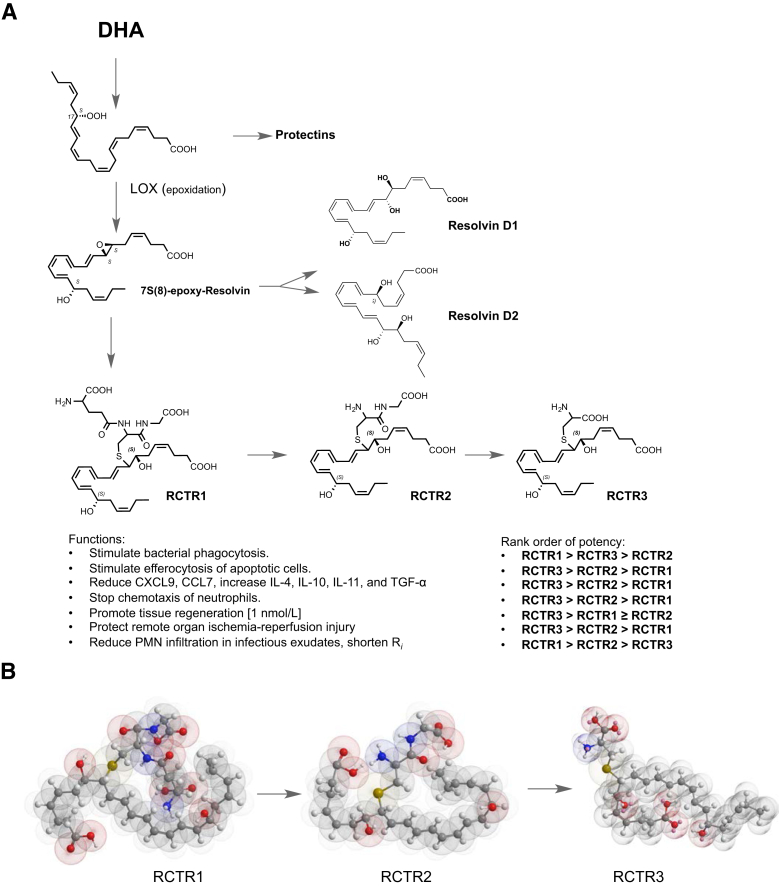
Figure 8.
Biosynthesis, stereochemistry, and RCTR functions. A: DHA is converted via lipoxygenase to the 17-hydroperoxy intermediate to produce protectins2 or can undergo a second lipoxygenation (namely, hydrogen abstraction with insertion of molecular oxygen) at the carbon-7 position, yielding 7S,17S-dihydro(peroxy)-4Z, 9E,11E,13Z,15E,19Z-docosahexaenoic acid, which is enzymatically converted to an allylic epoxide, followed by production of RCTR1.17 This glutathionyl conjugate can be converted to RCTR2 and subsequently to RCTR3. Complete stereochemistry of each bioactive RCTR is determined herein and is as shown. Functions of RCTRs in human phagocytes, mouse, and planaria were determined, and rank-order potencies were established. B: Structures in three-dimensional configuration using energy minimization (ChemBio3D Ultra 14.0; PerkinElmer, Cambridge, MA) for RCTR1, RCTR2, and RCTR3. Each gave distinct overall three-dimensional structures that may relate to their separate actions and rank orders in potencies.
Discussion
Herein, the complete stereochemistry of three novel members of the resolvin family that are peptide-lipid conjugates, which carry proresolving actions and stimulate tissue regeneration, were determined. Namely, endogenous RCTR1, RCTR2, and RCTR3 were matched to physical properties of synthetic RCTRs, establishing the bioactions and stereochemistry of the RCTR pathway (Figure 8). RCTRs were produced in human tissues and organs, and their levels increased in human spleen on exposure to S. aureus. Using stereochemically defined RCTRs, their rank-order potencies in enhancing proresolving actions defining the resolution program of acute inflammation were examined.41 These actions include stimulating human macrophage phagocytosis and efferocytosis of apoptotic PMNs, limiting human PMN chemotaxis in vitro and PMN infiltration in mice, and regulating specific lipid mediators and inflammation-associated cytokines, as well as promoting planaria tissue regeneration. Regulation of surgically injured planaria was selected to assess RCTR functions because the regeneration process in these primitive organisms is likely to be conserved, as are the structures of the SPM2 and RCTRs.
Sulfido-conjugated lipid mediators are produced in human lymphoid organs. For example, MCTRs and PCTRs are formed in lymph nodes at concentrations (0.5 to 4.5 pmol/L) commensurate with their known bioactive concentration ranges.17, 18, 21, 22 RCTR production and biosynthetic pathways in lymphoid organs are likely regulated in vivo during bacterial infections, as exemplified by the results obtained with S. aureus. SPM production has been documented in bone marrow.32, 42 For example, lipoxins, RvD1, RvE1, and PD1 are present in rat bone marrow, and their levels are higher in ovariectomized compared with sham-operated animals.32 Also, RvD1 and RvD2 are produced in mouse bone marrow, and increase with parathyroid hormone stimulation.42 Our results are the first to demonstrate that human bone marrow produces sulfido conjugates, including MCTRs, PCTRs, and RCTRs (Table 1). Along these lines, a lymphoid tissue, PLA2G2D, was identified as a resolving sPLA2. It is present in both lymph nodes and bone marrow, reducing hypersensitivity in skin inflammation via mobilizing DHA to produce proresolving lipid mediators in lymph nodes, including protectin D1 and resolvin D1.43 These earlier findings are in line with our present results indicating that biosynthetic pathways for SPM and DHA-derived sulfido-conjugate branches for MCTRs, PCTRs, and RCTRs are active in lymphoid tissues (spleen, lymph node, and bone marrow). Along these lines, SPMs, including RvD1, RvD2, and MaR1, prevent naïve CD4+ T-cell differentiation into types 1 and 17 helper T cells, as well as enhance generation and function of Foxp3(+) regulatory T cells.44 In addition, RvD1 promotes differentiation of IgG-producing B cells and inhibits the differentiation of IgE-producing B cells without being immune suppressive.45 Whether RCTRs also carry functions in adaptive immunity remained to be determined.
Proteome profiling of STZ-treated macrophages indicated that RCTR1, RCTR2, and RCTR3 as well as MCTR1, MCTR2, and MCTR3 each down-regulated CXCL9 and chemokine (C-C motif) ligand 7 with essentially equal potencies (Figure 3 and Supplemental Figure S2). CXCL9 (alias monokine induced by γ interferon) is a T-cell chemoattractant, which is induced by interferon-γ.46 RvD1 treatment leads to decrease in CXCL9 levels in inflamed human visceral adipose tissue.47 In HIV patients, infiltrated mucosal macrophages showed increased CXCL9 associated with their low phagocytic activity.48 Herein, RCTR3 significantly up-regulated IL-10, as well as IL-4, IL-11, and TGF-α (Figure 3B). IL-4 promotes M2a macrophage, whereas IL-10 promotes M2c phenotype,49 which are proresolving phenotypes.50 Hence, RCTRs reduced cytokines that stimulate M1 differentiation (eg, interferon-γ–CXCL9 axis) and up-regulated cytokines stimulating M2 differentiation (eg, IL-4 and IL-10). Also, TGF-α increases keratinocyte migration,51 and these cells promote wound healing.52 IL-11, up-regulated by RCTR3 (Figure 3C), is immunoregulatory and organ protective in collagen-induced arthritis,37 liver ischemia-reperfusion,36 and experimental autoimmune encephalomyelitis.53 Taken together, these results obtained with isolated human macrophages demonstrated that RCTRs at picomolar to low nanomolar range stimulated phagocytosis and efferocytosis (Figure 2) as well as regulated specific cytokines (Figure 3) that control macrophage phenotypes and display tissue-protective functions. These functions are likely to be relevant in the biosynthesis and actions of RCTRs in human spleen, lymph nodes, and bone marrow, where they are produced in response to bacteria.
Neutrophils (PMNs) are critical in host defense and in mediating collateral tissue damage when their congregation is excessive.1, 54 With isolated human PMNs, RCTRs at 10 nmol/L each directly stopped LTB4-initiated PMN chemotaxis, as demonstrated at the single-cell level in real time using chemotaxis chambers (Figure 4). By comparison, select SPMs share this action. For example, RvD1 (10 nmol/L) stops PMN migration toward IL-8 gradient, whereas its precursor DHA, at the same concentrations, does not share this action.55 Eicosapentaenoic acid–derived RvE1 and RvE2 (10 nmol/L each) also stop PMN chemotaxis and stimulate rapid shape change using single-cell monitoring with microfluidic chamber.56 RvD2, lipoxin A4, and lipoxin B4 also shared this potent action, reducing directional PMN migration toward IL-8.30 Temporal LM profiling during PMN swarming establishes LM class switching [ie, LTB4, prostaglandin D2, and prostaglandin E2 are initially released during PMN swarming toward zymosan particles, whereas lipoxin A4 levels significantly increase as cells reach a density at congregation to trigger an approximately 20-fold increase in stop signals (ie, lipoxin A4, RvE3, and PD1)].57 Injection of lipoxin A4 into microfluidic chambers stops PMN swarming. These findings, together with our present results, indicate that RCTRs and SPMs display potent and direct actions on human PMNs, controlling directional migration and swarming in response to inflammatory stimuli, such as cytokine, lipid mediator, and microbial particles. Temporal cessation of PMN infiltration is a key cardinal sign of resolution of inflammation.2, 12, 15 Hence, as determined herein, RCTRs each carry this key resolution function with isolated human PMNs.
Along with the direct actions of RCTRs on isolated human PMNs, protective actions of RCTRs in vivo in PMN-mediated I/R-initiated acute lung injury are also reported (Figure 5). LTB4 is a potent PMN activator in acute inflammation and injury from I/R. Both BLT1 (an LTB4 receptor) expression and LTB4 biosynthesis are critical for PMN infiltration in reperfusion-initiated remote organ injury, as demonstrated using BLT1 transgenic and 5-lipoxygenase–deficient mice.28 Herein, RCTR3 limited PMN infiltration into lungs after I/R and reduced LTB4 levels by approximately 50%. These organ-protective actions of RCTRs are shared by MCTR1 and MCTR2.18 Therefore, RCTR3 limited PMN infiltration into lungs after I/R injury, at least in part via reducing LTB4.
Figure 8A illustrates the proposed biosynthetic pathway and the complete stereochemistry, functions, and rank-order potencies of RCTR1, RCTR2, and RCTR3. Resolvins (RvD1 and RvD2) and RCTRs share the common precursor DHA and an epoxide intermediate. DHA is first converted via 17-lipoxygenation to a hydroperoxy-containing intermediate that undergoes a second lipoxygenation to insert a molecular oxygen at the carbon-7 position, yielding 7S,17S-dihydro(peroxy)-4Z, 9E,11E,13Z,15E,19Z-docosahexaenoic acid, which is enzymatically converted to an allylic epoxide, 7S(8)-epoxy-resolvin. This epoxide is converted to either resolvins (ie, RvD1 and RvD2) or RCTR1 with the complete stereochemistry of 8R-glutathionyl 7S,17S-dihydroxy-4Z, 9E,11E,13Z,15E,19Z-docosahexaenoic acid. This glutathionyl conjugate is next transformed enzymatically to RCTR2, which is then subsequently converted to bioactive RCTR3 with the complete stereochemistry of 8R-cysteinyl,7S,17S-dihydroxy-4Z, 9E,11E,13Z,15E,19Z-docosahexaenoic acid, which proved to be a potent bioactive molecule.
With isolated human macrophages in vitro, RvD1 appeared to be more potent than RCTRs in stimulating phagocytosis of E. coli (Figure 2A), whereas RCTR2 and RCTR3 were more potent in enhancing macrophage efferocytosis (Figure 2B). These results emphasize the importance of chemical structures and stereochemical biosynthesis of each product. These could dictate their functions and potencies in activating proresolving leukocyte responses, possibly via interacting with distinct receptors and downstream signaling, because the three-dimensional models of RCTR1, RCTR2, and CRTR3 suggest that they are each spatially distinct (Figure 8B). RCTR1 and RvD1 both shorten resolution intervals during infection (Figure 6), which is a determinant of initiation and temporal clearance in contained sites of inflammation.23 The ability of RCTR1 and RvD1 to share this vital function implicates pathway redundancies in these mediators that accelerate resolution. It is likely that RvD1 and RCTR pathways are operating in differential cellular targets and/or with different temporal biosynthesis in target organs. The complete stereochemical assignments of these bioactive RCTRs herein permit future studies that are required to document their endogenous production in target organs, investigate the cellular and molecular targets of each RCTR, and assess their relationships with D-series resolvin pathways in governing resolution and tissue regeneration.
Cysteinyl leukotrienes are well-known sulfido-conjugated leukotrienes that are biosynthesized from arachidonic acid [via a 5,6-epoxide intermediate (ie, LTA4)], including LTC4, LTD4, and LTE4. LTC4 and LTD4 are potent bronchoconstrictors and increase vascular permeability.58 LTE4 was earlier considered essentially inactive. In comparison, sulfido-conjugated RCTRs are biosynthesized from docosahexaenoic acid via a 7,8-epoxide intermediate (Figure 8). Among them, RCTR1 was the most potent in enhancing bacterial phagocytosis by human macrophages and accelerating infectious infection. RCTR3 appeared to be the most potent in stimulating efferocytosis, regulating inflammation-related proteins, stopping PMN chemotaxis and PMN-mediated acute lung injury, and promoting planaria tissue regeneration (Figure 8). Along these lines, MCTR3 is also the most potent of three MCTRs in stimulating macrophage phagocytosis in vitro and in vivo.59 These findings are in sharp contrast to the diminished potencies and biological actions of LTE4 compared with LTC4 and LTD4 in human tissues and experimental animal models.60
Together, these results establish the complete and unique stereochemistry of the individual bioactive RCTRs and their production in human organs. The rank-order potencies of RCTR1, RCTR2, and RCTR3 for their anti-inflammatory and proresolving actions on isolated human phagocytes, as well as in vivo in mice and planaria tissue regeneration, were also determined. Thus, our findings provide new targets and pathways to actively stimulate resolution of inflammation, clear infection, and enhance tissue regeneration.
Acknowledgments
We thank Mary H. Small for assistance with manuscript preparation, and Molly C. Jenssen and Justin T. English (Charles N. Serhan laboratory) for technical assistance.
Footnotes
Supported in part by NIH grant P01GM095467 (C.N.S.).
The Rous-Whipple Award is given by the American Society for Investigative Pathology (ASIP) to a senior pathologist with a distinguished career in experimental pathology research and continued productivity at the time of the award. This article stems from the ASIP Rous-Whipple Award Lecture: “Decoding Novel Resolution Mediators and Mechanisms in Infectious Inflammation and Tissue Regeneration,” presented by C.N.S. at the annual meeting of the American Society for Investigative Pathology in San Diego, CA, April 21 to 25, 2018.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.01.004.
Supplemental Data
RCTRs are produced in mice challenged with Escherichia coli exudate. FVB mice were challenged with 5 × 105E. coli for 24 hours. Then, lavage was performed with 5 mL of phosphate-buffered saline+/+ and was incubated with 30 μmol/L of DHA. Methanol fractions were analyzed and quantified for all sulfido conjugates. Results are shown as pie charts of sulfido-conjugated specialized proresolving mediators and leukotriene profiles in exudates incubated without (left panel) or with (right panel) 30 μmol/L DHA. n = 5. LT, leukotriene.
MCTRs regulate selected inflammatory cytokines with human macrophages. Proteome Profiler cytokine array was performed using human macrophages (2 × 106 cells). Macrophages were incubated with 10 nmol/L MCTR1, MCTR2, MCTR3, or vehicle (containing 0.1% ethanol) for 15 minutes at 37°C, then with 100 ng of serum-treated zymosan (STZ) and incubated at 37°C, with 5% CO2 for 24 hours. A: Results are expressed in a heat map as relative abundance to reference spots. Arrows indicate proteins shown in B. B: Levels of CXCL9, chemokine (C-C motif) ligand (CCL) 7, IL-4, IL-10, IL-11, and transforming growth factor (TGF)-α were determined by densitometry analysis of dot blots. Data are expressed as the means ± SEM from three separate donors (B). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus STZ (t-test). BAFF, B-cell activating factor; BDNF, brain-derived neurotrophic factor; CRP, C-reactive protein; DPPIV, dipeptidyl-peptidase-4; EGF, epidermal growth factor; FGF, fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GDF, growth and differentiation factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; HGF, hepatocyte growth factor; ICAM, intercellular adhesion molecule; IFN, interferon; IGFBP, insulin growth factor binding protein; LIF, leukemia inhibitory factor; M-CSF, macrophage colony-stimulating factor; MIF, macrophage migration inhibitory factor; MMP, matrix metallopeptidase; PDGF, platelet-derived growth factor; RAGE, receptor for advanced glycation end products; RANTES, regulated on activation normal T cell expressed and secreted; RBP, retinol binding protein; SDF, stromal cell–derived factor; SHBG, sex hormone–binding globulin; TARC, thymus and activation regulated chemokine; TFF, trefoil factor family; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
RCTRs do not alter survival of human neutrophils. Human polymorphonuclear cells (PMNs) were incubated for either 24 or 48 hours in phosphate-buffered saline (PBS) with calcium and magnesium (pH = 7.2, 37°C, 5% CO2), in vehicle PBS with calcium and magnesium containing 0.1% ethanol, or with the addition of 10 nmol/L RCTR1, RCTR2, RCTR3, or RvD1. Cell viability was assessed using flow cytometry with surface staining for annexin V and propidium iodide (PI; V13242; Invitrogen). Annexin+, PI+ represents apoptotic PMN populations. There were no statistically significant differences in apoptotic populations between all groups (two-way analysis of variance with Tukey's multiple comparison test). Data are expressed as means ± SEM. n = 3 healthy volunteers.
References
- 1.Cotran R.S., Kumar V., Collins T. ed 6. WB Saunders Co; Philadelphia: 1999. Robbins Pathologic Basis of Disease; p. 1425. [Google Scholar]
- 2.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridker P.M., Everett B.M., Thuren T., MacFadyen J.G., Chang W.H., Ballantyne C., Fonseca F., Nicolau J., Koenig W., Anker S.D., Kastelein J.J.P., Cornel J.H., Pais P., Pella D., Genest J., Cifkova R., Lorenzatti A., Forster T., Kobalava Z., Vida-Simiti L., Flather M., Shimokawa H., Ogawa H., Dellborg M., Rossi P.R.F., Troquay R.P.T., Libby P., Glynn R.J. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 4.Libby P., Tabas I., Fredman G., Fisher E.A. Inflammation and its resolution as determinants of acute coronary syndromes. Circ Res. 2014;114:1867–1879. doi: 10.1161/CIRCRESAHA.114.302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiala M., Terrando N., Dalli J. Specialized pro-resolving mediators from Omega-3 fatty acids improve amyloid-beta phagocytosis and regulate inflammation in patients with minor cognitive impairment. J Alzheimers Dis. 2015;48:293–301. doi: 10.3233/JAD-150367. [DOI] [PubMed] [Google Scholar]
- 6.Claria J., Lopez-Vicario C., Rius B., Titos E. Pro-resolving actions of SPM in adipose tissue biology. Mol Aspects Med. 2017;58:83–92. doi: 10.1016/j.mam.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Calder P.C. Marine omega-3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim Biophys Acta. 2015;1851:469–484. doi: 10.1016/j.bbalip.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Serhan C.N., Ward P.A., Gilroy D.W. Cambridge University Press; New York: 2010. Fundamentals of Inflammation. [Google Scholar]
- 9.Majno G., Joris I. ed 2. Oxford University Press; New York: 2004. Cells, Tissues, and Disease: Principles of General Pathology. [Google Scholar]
- 10.Serhan C.N., Clish C.B., Brannon J., Colgan S.P., Chiang N., Gronert K. Novel functional sets of lipid-derived mediators with antiinflammatory actions generated from omega-3 fatty acids via cyclooxygenase 2-nonsteroidal antiinflammatory drugs and transcellular processing. J Exp Med. 2000;192:1197–1204. doi: 10.1084/jem.192.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan C.N., Hong S., Gronert K., Colgan S.P., Devchand P.R., Mirick G., Moussignac R.-L. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter pro-inflammation signals. J Exp Med. 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serhan C.N. Discovery of specialized pro-resolving mediators marks the dawn of resolution physiology and pharmacology. Mol Aspects Med. 2017;58:1–11. doi: 10.1016/j.mam.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong S., Gronert K., Devchand P., Moussignac R.-L., Serhan C.N. Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood and glial cells: autacoids in anti-inflammation. J Biol Chem. 2003;278:14677–14687. doi: 10.1074/jbc.M300218200. [DOI] [PubMed] [Google Scholar]
- 14.Chiang N., Serhan C.N. Structural elucidation and physiologic functions of specialized pro-resolving mediators and their receptors. Mol Aspects Med. 2017;58:114–129. doi: 10.1016/j.mam.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perretti M., Cooper D., Dalli J., Norling L.V. Immune resolution mechanisms in inflammatory arthritis. Nat Rev Rheumatol. 2017;13:87–99. doi: 10.1038/nrrheum.2016.193. [DOI] [PubMed] [Google Scholar]
- 16.Serhan C.N., Dalli J., Karamnov S., Choi A., Park C.K., Xu Z.Z., Ji R.R., Zhu M., Petasis N.A. Macrophage pro-resolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB J. 2012;26:1755–1765. doi: 10.1096/fj.11-201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalli J., Ramon S., Norris P.C., Colas R.A., Serhan C.N. Novel proresolving and tissue-regenerative resolvin and protectin sulfido-conjugated pathways. FASEB J. 2015;29:2120–2136. doi: 10.1096/fj.14-268441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalli J., Chiang N., Serhan C.N. Identification of 14-series sulfido-conjugated mediators that promote resolution of infection and organ protection. Proc Natl Acad Sci U S A. 2014;111:E4753–E4761. doi: 10.1073/pnas.1415006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R., Chiang N., Oh S.F., Serhan C.N. Metabolomics-lipidomics of eicosanoids and docosanoids generated by phagocytes. Curr Protoc Immunol. 2011;95:14.26.1–14.26.26. doi: 10.1002/0471142735.im1426s95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serhan C.N., Petasis N.A. Resolvins and protectins in inflammation-resolution. Chem Rev. 2011;111:5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalli J., Vlasakov I., Riley I.R., Rodriguez A.R., Spur B.W., Petasis N.A., Chiang N., Serhan C.N. Maresin conjugates in tissue regeneration biosynthesis enzymes in human macrophages. Proc Natl Acad Sci U S A. 2016;113:12232–12237. doi: 10.1073/pnas.1607003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramon S., Dalli J., Sanger J.M., Winkler J.W., Aursnes M., Tungen J.E., Hansen T.V., Serhan C.N. The Protectin PCTR1 is produced by human M2 macrophages and enhances resolution of infectious inflammation. Am J Pathol. 2016;186:962–973. doi: 10.1016/j.ajpath.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bannenberg G.L., Chiang N., Ariel A., Arita M., Tjonahen E., Gotlinger K.H., Hong S., Serhan C.N. Molecular circuits of resolution: formation and actions of resolvins and protectins. J Immunol. 2005;174:4345–4355. doi: 10.4049/jimmunol.174.7.4345. [DOI] [PubMed] [Google Scholar]
- 24.English J.T., Norris P.C., Hodges R.R., Dartt D.A., Serhan C.N. Identification and profiling of specialized pro-resolving mediators in human tears by lipid mediator metabolomics. Prostaglandins Leukot Essent Fatty Acids. 2017;117:17–27. doi: 10.1016/j.plefa.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez A.R., Spur B.W. First total synthesis of pro-resolving and tissue-regenerative resolvin sulfido-conjugates. Tetrahedron Lett. 2017;58:1662–1668. [Google Scholar]
- 26.Colas R.A., Shinohara M., Dalli J., Chiang N., Serhan C.N. Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am J Physiol Cell Physiol. 2014;307:C39–C54. doi: 10.1152/ajpcell.00024.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang N., de la Rosa X., Libreros S., Serhan C.N. Novel resolvin D2 receptor axis in infectious inflammation. J Immunol. 2017;198:842–851. doi: 10.4049/jimmunol.1601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang N., Gronert K., Clish C.B., O'Brien J.A., Freeman M.W., Serhan C.N. Leukotriene B4 receptor transgenic mice reveal novel protective roles for lipoxins and aspirin-triggered lipoxins in reperfusion. J Clin Invest. 1999;104:309–316. doi: 10.1172/JCI7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malet-Engra G., Viaud J., Ysebaert L., Farce M., Lafouresse F., Laurent G., Gaits-Iacovoni F., Scita G., Dupre L. CIP4 controls CCL19-driven cell steering and chemotaxis in chronic lymphocytic leukemia. Cancer Res. 2013;73:3412–3424. doi: 10.1158/0008-5472.CAN-12-3564. [DOI] [PubMed] [Google Scholar]
- 30.Norling L.V., Headland S.E., Dalli J., Arnardottir H.H., Haworth O., Jones H.R., Irimia D., Serhan C.N., Perretti M. Proresolving and cartilage-protective actions of resolvin D1 in inflammatory arthritis. JCI Insight. 2016;1:e85922. doi: 10.1172/jci.insight.85922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salem N., Jr., Vandal M., Calon F. The benefit of docosahexaenoic acid for the adult brain in aging and dementia. Prostaglandins Leukot Essent Fatty Acids. 2015;92:15–22. doi: 10.1016/j.plefa.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen R.C., Gotlinger K.H., Serhan C.N., Kruger M.C. Identification of inflammatory and proresolving lipid mediators in bone marrow and their lipidomic profiles with ovariectomy and omega-3 intake. Am J Hematol. 2008;83:437–445. doi: 10.1002/ajh.21170. [DOI] [PubMed] [Google Scholar]
- 33.Serhan C.N., Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 34.Chomarat P., Vannier E., Dechanet J., Rissoan M.C., Banchereau J., Dinarello C.A., Miossec P. Balance of IL-1 receptor antagonist/IL-1 beta in rheumatoid synovium and its regulation by IL-4 and IL-10. J Immunol. 1995;154:1432–1439. [PubMed] [Google Scholar]
- 35.Eming S.A., Wynn T.A., Martin P. Inflammation and metabolism in tissue repair and regeneration. Science. 2017;356:1026–1030. doi: 10.1126/science.aam7928. [DOI] [PubMed] [Google Scholar]
- 36.Yu J., Feng Z., Tan L., Pu L., Kong L. Interleukin-11 protects mouse liver from warm ischemia/reperfusion (WI/Rp) injury. Clin Res Hepatol Gastroenterol. 2016;40:562–570. doi: 10.1016/j.clinre.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Walmsley M., Butler D.M., Marinova-Mutafchieva L., Feldmann M. An anti-inflammatory role for interleukin-11 in established murine collagen-induced arthritis. Immunology. 1998;95:31–37. doi: 10.1046/j.1365-2567.1998.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malawista S.E., de Boisfleury Chevance A., van Damme J., Serhan C.N. Tonic inhibition of chemotaxis in human plasma. Proc Natl Acad Sci U S A. 2008;105:17949–17954. doi: 10.1073/pnas.0802572105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eltzschig H.K., Eckle T. Ischemia and reperfusion: from mechanism to translation. Nat Med. 2011;17:1391–1401. doi: 10.1038/nm.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott S.A., Sanchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:301–326. doi: 10.1002/wdev.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwab J.M., Chiang N., Arita M., Serhan C.N. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447:869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCauley L.K., Dalli J., Koh A.J., Chiang N., Serhan C.N. Cutting edge: parathyroid hormone facilitates macrophage efferocytosis in bone marrow via proresolving mediators resolvin D1 and resolvin D2. J Immunol. 2014;193:26–29. doi: 10.4049/jimmunol.1301945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miki Y., Yamamoto K., Taketomi Y., Sato H., Shimo K., Kobayashi T., Ishikawa Y., Ishii T., Nakanishi H., Ikeda K., Taguchi R., Kabashima K., Arita M., Arai H., Lambeau G., Bollinger J.M., Hara S., Gelb M.H., Murakami M. Lymphoid tissue phospholipase A2 group IID resolves contact hypersensitivity by driving antiinflammatory lipid mediators. J Exp Med. 2013;210:1217–1234. doi: 10.1084/jem.20121887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiurchiu V., Leuti A., Dalli J., Jacobsson A., Battistini L., Maccarrone M., Serhan C.N. Proresolving lipid mediators resolvin D1, resolvin D2, and maresin 1 are critical in modulating T cell responses. Sci Transl Med. 2016;8:353ra111. doi: 10.1126/scitranslmed.aaf7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim N., Ramon S., Thatcher T.H., Woeller C.F., Sime P.J., Phipps R.P. Specialized proresolving mediators (SPMs) inhibit human B-cell IgE production. Eur J Immunol. 2016;46:81–91. doi: 10.1002/eji.201545673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasperini S., Marchi M., Calzetti F., Laudanna C., Vicentini L., Olsen H., Murphy M., Liao F., Farber J., Cassatella M.A. Gene expression and production of the monokine induced by IFN-gamma (MIG), IFN-inducible T cell alpha chemoattractant (I-TAC), and IFN-gamma-inducible protein-10 (IP-10) chemokines by human neutrophils. J Immunol. 1999;162:4928–4937. [PubMed] [Google Scholar]
- 47.Titos E., Rius B., Lopez-Vicario C., Alcaraz-Quiles J., Garcia-Alonso V., Lopategi A., Dalli J., Lozano J.J., Arroyo V., Delgado S., Serhan C.N., Claria J. Signaling and immunoresolving actions of resolvin D1 in inflamed human visceral adipose tissue. J Immunol. 2016;197:3360–3370. doi: 10.4049/jimmunol.1502522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allers K., Fehr M., Conrad K., Epple H.J., Schurmann D., Geelhaar-Karsch A., Schinnerling K., Moos V., Schneider T. Macrophages accumulate in the gut mucosa of untreated HIV-infected patients. J Infect Dis. 2014;209:739–748. doi: 10.1093/infdis/jit547. [DOI] [PubMed] [Google Scholar]
- 49.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Dalli J., Serhan C.N. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120:e60–e72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Fan J., Chen M., Li W., Woodley D.T. Transforming growth factor-alpha: a major human serum factor that promotes human keratinocyte migration. J Invest Dermatol. 2006;126:2096–2105. doi: 10.1038/sj.jid.5700350. [DOI] [PubMed] [Google Scholar]
- 52.Werner S., Krieg T., Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 53.Gurfein B.T., Zhang Y., Lopez C.B., Argaw A.T., Zameer A., Moran T.M., John G.R. IL-11 regulates autoimmune demyelination. J Immunol. 2009;183:4229–4240. doi: 10.4049/jimmunol.0900622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delano M.J., Ward P.A. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Invest. 2016;126:23–31. doi: 10.1172/JCI82224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasuga K., Yang R., Porter T.F., Agrawal N., Petasis N.A., Irimia D., Toner M., Serhan C.N. Rapid appearance of resolvin precursors in inflammatory exudates: novel mechanisms in resolution. J Immunol. 2008;181:8677–8687. doi: 10.4049/jimmunol.181.12.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh S.F., Dona M., Fredman G., Krishnamoorthy S., Irimia D., Serhan C.N. Resolvin E2 formation and impact in inflammation resolution. J Immunol. 2012;188:4527–4534. doi: 10.4049/jimmunol.1103652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reátegui E., Jalali F., Khankhel A.H., Wong E., Cho H., Lee J., Serhan C.N., Dalli J., Elliott H., Irimia D. Microscale arrays for the profiling of start and stop signals coordinating human-neutrophil swarming. Nat Biomed Eng. 2017;1 doi: 10.1038/s41551-017-0094. 0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu Rev Pharmacol Toxicol. 2009;49:123–150. doi: 10.1146/annurev.pharmtox.011008.145616. [DOI] [PubMed] [Google Scholar]
- 59.Dalli J., Sanger J.M., Rodriguez A.R., Chiang N., Spur B.W., Serhan C.N. Identification and actions of a novel third maresin conjugate in tissue regeneration: MCTR3. PLoS One. 2016;11:e0149319. doi: 10.1371/journal.pone.0149319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samuelsson B. Role of basic science in the development of new medicines: examples from the eicosanoid field. J Biol Chem. 2012;287:10070–10080. doi: 10.1074/jbc.X112.351437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RCTRs are produced in mice challenged with Escherichia coli exudate. FVB mice were challenged with 5 × 105E. coli for 24 hours. Then, lavage was performed with 5 mL of phosphate-buffered saline+/+ and was incubated with 30 μmol/L of DHA. Methanol fractions were analyzed and quantified for all sulfido conjugates. Results are shown as pie charts of sulfido-conjugated specialized proresolving mediators and leukotriene profiles in exudates incubated without (left panel) or with (right panel) 30 μmol/L DHA. n = 5. LT, leukotriene.
MCTRs regulate selected inflammatory cytokines with human macrophages. Proteome Profiler cytokine array was performed using human macrophages (2 × 106 cells). Macrophages were incubated with 10 nmol/L MCTR1, MCTR2, MCTR3, or vehicle (containing 0.1% ethanol) for 15 minutes at 37°C, then with 100 ng of serum-treated zymosan (STZ) and incubated at 37°C, with 5% CO2 for 24 hours. A: Results are expressed in a heat map as relative abundance to reference spots. Arrows indicate proteins shown in B. B: Levels of CXCL9, chemokine (C-C motif) ligand (CCL) 7, IL-4, IL-10, IL-11, and transforming growth factor (TGF)-α were determined by densitometry analysis of dot blots. Data are expressed as the means ± SEM from three separate donors (B). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus STZ (t-test). BAFF, B-cell activating factor; BDNF, brain-derived neurotrophic factor; CRP, C-reactive protein; DPPIV, dipeptidyl-peptidase-4; EGF, epidermal growth factor; FGF, fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GDF, growth and differentiation factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; HGF, hepatocyte growth factor; ICAM, intercellular adhesion molecule; IFN, interferon; IGFBP, insulin growth factor binding protein; LIF, leukemia inhibitory factor; M-CSF, macrophage colony-stimulating factor; MIF, macrophage migration inhibitory factor; MMP, matrix metallopeptidase; PDGF, platelet-derived growth factor; RAGE, receptor for advanced glycation end products; RANTES, regulated on activation normal T cell expressed and secreted; RBP, retinol binding protein; SDF, stromal cell–derived factor; SHBG, sex hormone–binding globulin; TARC, thymus and activation regulated chemokine; TFF, trefoil factor family; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
RCTRs do not alter survival of human neutrophils. Human polymorphonuclear cells (PMNs) were incubated for either 24 or 48 hours in phosphate-buffered saline (PBS) with calcium and magnesium (pH = 7.2, 37°C, 5% CO2), in vehicle PBS with calcium and magnesium containing 0.1% ethanol, or with the addition of 10 nmol/L RCTR1, RCTR2, RCTR3, or RvD1. Cell viability was assessed using flow cytometry with surface staining for annexin V and propidium iodide (PI; V13242; Invitrogen). Annexin+, PI+ represents apoptotic PMN populations. There were no statistically significant differences in apoptotic populations between all groups (two-way analysis of variance with Tukey's multiple comparison test). Data are expressed as means ± SEM. n = 3 healthy volunteers.