Abstract
The evolution of sequence-defined synthetic polymers made of building blocks beyond those compatible with polymerase enzymes or the ribosome has the potential to generate new classes of receptors, catalysts, and materials. Here we describe a ligase-mediated DNA-templated polymerization system and in vitro selection to evolve highly functionalized nucleic acid polymers (HFNAPs) made from 32 building blocks containing eight chemically diverse side-chains on a DNA backbone. Through iterated cycles of polymer translation, selection, and reverse translation, we discovered HFNAPs that bind PCSK9 and IL-6, two protein targets implicated in human diseases. Mutation and reselection of an active PCSK9-binding polymer yielded evolved polymers with high affinity (KD = 3 nM). This evolved polymer potently inhibited binding between PCSK9 and the LDL receptor. Structure-activity relationship studies revealed that specific side-chains at defined positions in the polymers are required for binding to their respective targets. Our findings expand the chemical space of evolvable polymers to include densely functionalized nucleic acids with diverse, researcher-defined chemical repertoires.
The gene-encoded synthesis and Darwinian selection of sequence-defined biopolymers are fundamental features of all known forms of life. These processes have been harnessed in the laboratory to evolve RNA1–3, DNA4–6, and polypeptides7–10 with a variety of binding and catalytic properties through iterated cycles of biopolymer translation, selection, replication, and mutation. The speed and effectiveness of the evolutionary process has inspired efforts to apply these principles to the much larger chemical space of synthetic polymers11. To date, however, the evolution of sequence-defined non-natural polymers in the laboratory has been limited to analogs of nucleic acids12–15 and polypeptides16 that can be synthesized by polymerases and ribosomes. Polymerases and ribosomes impose structural requirements on the building blocks that can be polymerized and thereby limit the diversity of synthetic polymers that are accessible to directed evolution. For example, polymerases use only mononucleotides as substrates, precluding the ability to encode a diverse set of codons and side chains. Known classes of polymerase-synthesized functional non-natural nucleic acid polymers, including those derived from non-natural sugar backbones17–20, uniform installation of hydrophobic21–28 or positively charged29–35 side-chains on nucleobases, or introduction of novel nucleobases among the four possibilities36–38, therefore have chemical diversities that are only modestly expanded beyond those of natural DNA and RNA, and fall short of the much more diverse chemical functionality present in proteins.
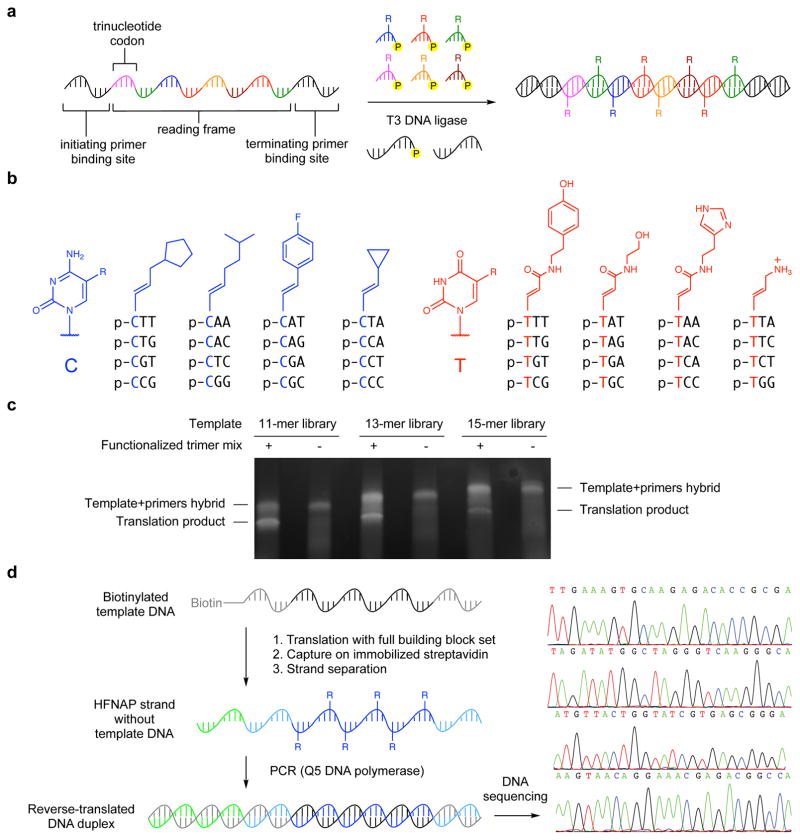
Previously we developed an in vitro system that uses DNA ligase to translate DNA sequences into sequence-defined highly functionalized nucleic acid polymers (HFNAPs) containing a wide range of side-chains chosen by the researcher39. In our original report, we showed that DNA sequences can be translated into HFNAPs using DNA ligase to catalyze the polymerization of up to 50 consecutive short, chemically functionalized oligonucleotide building blocks along a DNA template39 (Fig. 1a). We discovered that T4 DNA ligase accepts trinucleotide building blocks with a wide range of side-chains on the 5′ nucleobase, including side-chains at the C5 position of pyrimidine nucleobases that are both synthetically accessible and unlikely to disrupt Watson-Crick base pairing39.
Figure 1. Design and construction of the sequence-defined polymer library.
a, Reaction scheme for DNA ligase-mediated translation of DNA templates into sequence-defined highly functionalized nucleic acid polymers (HFNAPs). b, Structures of 5′-phosphorylated trinucleotide building blocks for HFNAP library synthesis. c, Translation of libraries of randomized DNA templates into HFNAPs that incorporate up to 15 consecutive functionalized trinucleotide building blocks. The translation reactions, as well as control reactions from which the trinucleotide building blocks were omitted, were analyzed by polyacrylamide gel electrophoresis on a non-denaturing 10% TBE gel and imaged by SYBR gold staining. d, A complete cycle of HFNAP translation, HFNAP strand isolation, and reverse translation back into DNA faithfully recovered sequence information from the original DNA templates. Left: experimental scheme; right: electrophoretic traces from Sanger sequencing. In control experiments in which the trinucleotide building blocks were omitted from the polymerization reactions, the PCR step did not generate any amplicons of the correct size.
This artificial translation system allows researchers, in principle, to mimic and even expand the chemical repertoire of protein building blocks in an evolvable synthetic polymer system. The broad chemical scope of HFNAPs gives them the potential to adopt unique folding and functional properties distinct from those of known natural or non-natural nucleic acid polymers. The original HFNAP system proved unable to support the evolution of functional polymers, however, likely because of the limited diversity provided by its eight-codon genetic code and the long, flexible linkers present in the building blocks. In this study, we designed a new HFNAP “genetic code”, translation system, and in vitro selection system that overcomes these challenges, then applied the resulting HFNAP evolution system to generate sequence-defined synthetic polymers that binds two protein targets of biomedical interest.
Results
Our new genetic code was designed to offer a high degree of both codon and side-chain diversity to evolving polymers (Fig. 1b). We included the maximum number of different trinucleotides containing a 5′ pyrimidine (all 32 possible YNN combinations, where Y = C or T and N = A, C, G, or T) as codons. The corresponding 32 building blocks were each linked to one of eight side-chains (four codons per side-chain) that include hydrophobic, aliphatic, aromatic, halogenated, polar, and charged groups, several of which are not found among proteinogenic amino acid side-chains. The linkers between side-chains and nucleobases were redesigned compared with our original genetic code with limited conformational flexibility in order to increase the likelihood that the polymer backbone, nucleobases, and side-chains would cooperatively adopt defined folded structures. We assigned each side-chain to a set of four codons that collectively contained the same balance of A/T versus C/G bases following the side-chain-functionalized 5′ pyrimidine base.
We improved DNA-templated polymerization (artificial “translation”) reactions by screening ligase enzymes and adjusting polymerization conditions. We found that subjecting translation reactions to a slow (0.01 °C/s) temperature ramp to 4 °C before initiating ligation with T3 DNA ligase substantially improved yields of full-length HFNAP from libraries of DNA templates containing random coding regions of 45 nt, which encoded the incorporation of 15 consecutive side-chain-functionalized trinucleotide building blocks of mixed sequence (Fig. 1c and Supplementary Fig. 1). To test the ability of translated polymers to be “reverse-translated” back into DNA, thereby enabling iterated cycles of translation, selection, reverse translation, and PCR amplification, we performed polymerization on four templates that each encoded the incorporation of eight different building blocks and collectively covered all 32 building blocks, and then subjected the resulting HFNAP products, separated from template DNA, to reverse translation in a PCR reaction using Q5 DNA polymerase39 (Fig. 1d). One of the PCR primers binds a 3′-overhang (Fig. 1d, green) present in the HFNAP products but absent in template DNA, precluding the amplification of any contaminating template DNA. DNA sequencing of the resulting PCR products showed that the original sequence information in the templates was faithfully recovered (Fig. 1d), indicating that both translation from DNA to HFNAPs and reverse translation from HFNAPs to their encoding DNA occur with high sequence fidelity using this set of building blocks.
These observations are qualitatively consistent with results from Hili and coworkers, who reported fidelities ranging from 95.1% to 98.4% per codon for ligase-mediated DNA-templated polymerization of functionalized pentanucleotides40. Perfect fidelity is not expected for a ligase-mediated polymerization, which lacks proofreading mechanisms, but we reasoned that the level of fidelity in our system may be sufficient to support iterated selection for functional polymers, consistent with our previous mock selection results39. Modest levels of mutations may also confer a benefit to the selection, as reported by Benner and coworkers for selections of aptamers containing novel nucleobases37.
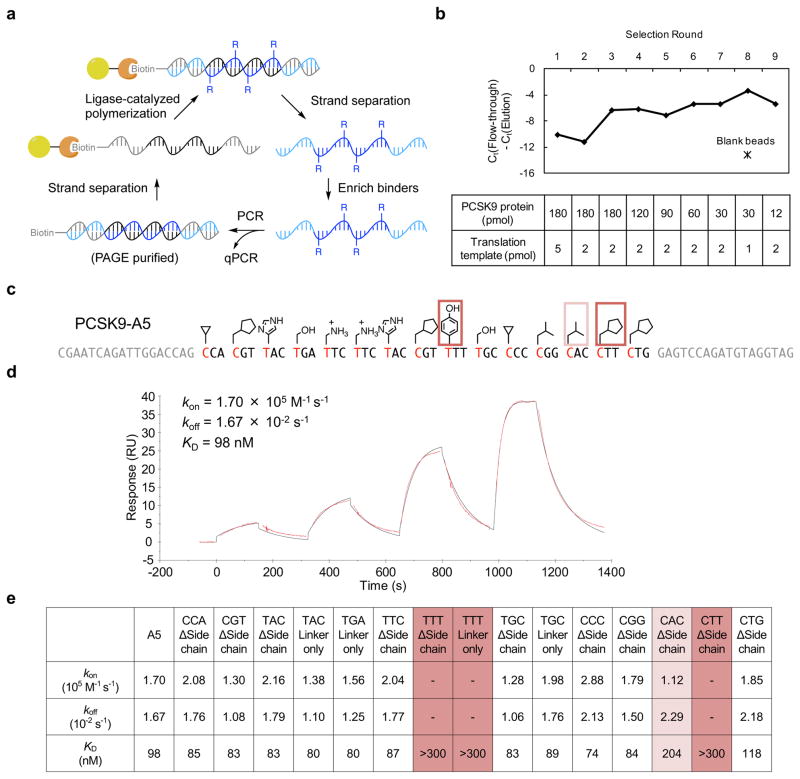
Encouraged by these developments, we generated a library of HFNAPs containing 15 consecutive building blocks drawn from the set of 32 (theoretical polymer library space = 3 × 1022; average HFNAP molecular weight = 28 kDa) and subjected the resulting library (starting quantity = 3 × 1012 molecules) to iterated rounds of in vitro selection for binding to PCSK9 protein, a target implicated in low-density lipoprotein (LDL) metabolism and cardiovascular disease41–43 (Fig. 2a). The HFNAP library was incubated with PCSK9 protein immobilized on agarose beads. After washing the beads with buffer, the bound HFNAPs were eluted by boiling the beads in buffer containing detergent. The surviving HFNAPs were reverse translated in a PCR reaction using Q5 DNA polymerase, and the resulting double-stranded DNAs were purified by PAGE. The non-template strands were removed by alkaline denaturation, and the streptavidin-bound template strands were translated back into HFNAPs by ligase-catalyzed DNA-templated polymerization for the next round of PCSK9-binding selection. We note that the ability of HFNAPs to be directly reverse-translated by a DNA polymerase, though not an absolute requirement for iterated selection as demonstrated by the use of display methods to evolve nucleic acids17,44, provides a practical and high-fidelity way to complete a selection cycle.
Figure 2. Selection of PCSK9-binding polymers from a random HFNAP library.
a, Overview of translation, selection, and reverse translation scheme. b, PCSK9 binding selection progress. The HFNAP pool’s bulk affinity to PCSK9-coated beads was assessed by quantifying the amount of HFNAP in the flow-through versus the elution at each round of selection by quantitative PCR. Higher values in the graph indicate higher ratios of polymers that bound to immobilized PCSK9 and were eluted relative to polymers that flowed through the immobilized PCSK9. c, Sequence and side-chain structure of selected polymer PCSK9-A5. Side-chains essential for binding activity are boxed. d, SPR sensogram characterizing binding kinetics between surface-immobilized PCSK9-A5 polymer and the target PCSK9 protein. The concentrations of injected PCSK9 were 10, 30, 100, and 300 nM. The observed sensogram is shown in red and the fitted curve with the kinetic parameters listed is shown in black. e, Kinetic parameters for binding of PCSK9-A5 or its side-chain-deficient variants to PCSK9 protein, as measured by SPR. For the variants “TTT ΔSide chain”, “TTT Linker only”, and “CTT ΔSide chain”, no SPR signal was observed at highest analyte concentration tested (300 nM PCSK9).
As the iterated rounds of selection progressed, the fraction of HFNAP that was retained on PCSK9-linked beads generally increased, consistent with enrichment of PCSK9-binding polymers, even though we steadily elevated selection stringency by decreasing the amount of PCSK9 protein (Fig. 2b). At the eighth round of selection, the polymer population was retained by PCSK9 protein-conjugated agarose beads with an efficiency of approximately 10%. In contrast, less than 0.1% of the same polymer population was retained on agarose beads not conjugated to any protein (Fig. 2b), suggesting that the ability of the selected polymers to bind PCSK9-linked beads arose from their ability to bind PCSK9, rather than agarose beads.
Results from high-throughput DNA sequencing after nine rounds of selection indicated that the HFNAP pool had strongly converged to just seven sequence families containing conserved sub-sequences suggestive of common binding motifs (Supplementary Fig. 2). Sequences within the same family were likely descendants of a single parental polymer derived through mutations that accumulated through the selection process, as evidenced by high levels of homology. We individually synthesized the seven most highly enriched HFNAPs by ligase-mediated DNA-templated polymerization and tested their retention on immobilized PCSK9 or immobilized thrombin, an unrelated protein. Five of the seven tested polymers exhibited substantial apparent binding activity to immobilized PCSK9 beyond any apparent binding to immobilized thrombin. We also assayed unfunctionalized DNA sequences (lacking any side-chains) corresponding to these seven HFNAPs and observed no evident PCSK9-binding activity (Supplementary Fig. 2). Together, these results suggest that the HFNAP populations emerging from nine rounds of in vitro selection converged on a small number of polymer families that bind immobilized PCSK9 in a manner dependent on the polymers’ side-chains.
We characterized in depth PCSK9-A5, the polymer with the highest apparent PCSK9 binding activity (Fig. 2c). We synthesized biotinylated PCSK9-A5 by templated translation and confirmed its binding affinity for PCSK9 (dissociation constant KD = 98 nM) by surface plasmon resonance (SPR) (Fig. 2d). To probe the role of the side-chain functional groups, we synthesized side-chain mutants of PCSK9-A5 in which all instances of each building block were replaced by the corresponding trinucleotide either lacking any side-chain or containing a linker but missing the side-chain’s functional group (Supplementary Fig. 3). The removal of a single phenol side-chain in codon 9 or a single cyclopentyl side-chain in codon 14 completely abolished the binding between PCSK9-A5 and its target (Fig. 2c and 2e, dark red). Furthermore, the removal of the isopentyl side-chain at codon 13 resulted in an approximately two-fold reduction in affinity (Fig. 2c and 2e, light red). The individual removal of other side-chains had less significant effects on binding affinity.
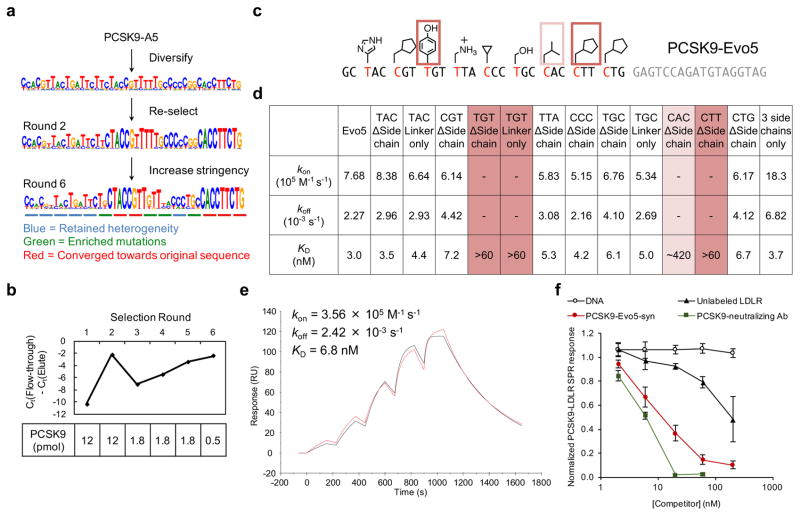
Given the vast sequence space of the HFNAP library (3 × 1022 possible polymers), evolution would likely generate polymer variants with improved activity over those polymers present in the initial population of 3 × 1012 HFNAP molecules. To evolve the PCSK9-A5 polymer into variants with improved PCSK9 affinity, we synthesized a library of mutated PCSK9-A5 templates containing 79% identity and 21% diversity (79:21 at the pyrimidine-only first position and 79:7:7:7 at the second and third positions of each codon) for each nucleotide in the variable region (Fig. 3a). We subjected the resulting mutated PCSK9-A5 library to six additional iterated cycles of translation and selection for PCSK9 binding (Supplementary Fig. 4) After just one round of enrichment at a stringency level comparable to that of the last round of our initial selection, the mutated PCSK9-A5 library exhibited bulk affinity for PCSK9-conjugated beads (Fig. 3b). High-throughput DNA sequencing revealed that the mutant population after a second round of translation and selection began to converge toward the sequence of PCSK9-A5 at many positions (Fig. 3a). In subsequent rounds, we reduced the amount of immobilized PCSK9 to further increase selection stringency. Four additional rounds of translation and selection resulted in steadily improved retention of the polymer population on immobilized PCSK9 (Fig. 3b).
Figure 3. Evolution of an improved PCSK9-binding polymer.
a, Evolution scheme and DNA sequencing results of the diversification and iterated selection of PCSK9-A5 variants with increased PCSK9 binding activity. b, Affinity maturation of the diversified PCSK9-A5 pool. The evolving polymer pool’s bulk affinity to immobilized PCSK9 was assessed by quantifying the amount of HFNAP in the flow-through and the elution at each round of selection by quantitative PCR. c, Sequence and side-chain structure of the resulting PCSK9-Evo5 polymer. Side chains essential for binding activity are boxed. d, Kinetic parameters for binding of PCSK9-Evo5 or its side-chain-deficient variants to PCSK9 protein, as measured by SPR. For the variants “TGT ΔSide chain”, “TGT Linker only”, and “CTT ΔSide chain”, no SPR signal was observed at the highest analyte concentration tested (60 nM PCSK9). For the variant “CAC ΔSide chain”, the binding interaction fits a two-state reaction kinetic model with KD ≈ 420 nM. Representative sensograms are provided in Supplementary Fig. 5. e, SPR sensogram characterizing binding kinetics between PCSK9-Evo5-syn and surface-immobilized PCSK9 protein. The concentrations of injected PCSK9-Evo5-syn were 1.8, 6, 18, 60, and 180 nM. The observed sensogram is shown in red and the fitted curve with the kinetic parameters listed is shown in black. f, SPR response on an LDLR-coated surface produced by flowing PCSK9 in the presence of either PCSK9-Evo5-syn, unfunctionalized DNA of identical sequence to PCSK9-Evo5-syn, unlabeled LDLR, or a known PCSK9-neutralizing monoclonal antibody. The SPR response shown is normalized to the response in experiments without any competitor (defined as an SPR response of 1). Error bars represent s.d. (n = 3). Representative raw sensograms are provided in Supplementary Fig. 11.
High-throughput sequencing revealed new consensus codons at four out of 15 positions within the population of evolved polymers (Fig. 3a). Among these four positions, codons 6, 10, and 12 evolved a different side-chain compared with that of PCSK9-A5, while codon 9 evolved a different codon (TGT instead of TTT) encoding the same phenol side-chain at this position. In addition, five other codons converged to the original sequence in PCSK9-A5, and six other codon positions, mostly near the 5′ end, retained sequence heterogeneity introduced during mutation, suggesting that these positions do not strongly contribute to binding activity (Fig. 3a). The three side-chain functional groups that were shown to be crucial to the PCSK9-binding activity of PCSK9-A5 were all maintained in the consensus sequence of the evolved polymer population.
We synthesized a biotinylated, truncated HFNAP (designated PCSK9-Evo5; Fig. 3c) retaining only the evolved consensus sequence and the 3′ constant region, and measured its affinity for PCSK9 to be KD = 3.0 nM (Fig. 3d and Supplementary Fig. 5), representing a 33-fold increase over that of PCSK9-A5. In contrast, a similarly truncated version of PCSK9-A5 (designated PCSK9-A5Trunc) did not substantially improve affinity (KD = 68 nM) over that of full-length PCSK9-A5 (KD = 98 nM) (Supplementary Fig. 6). Additional truncation of PCSK9-Evo5 from the 3′-end resulted in complete loss of affinity (Supplementary Fig. 6). Similar to the structure-activity relationships observed for PCSK9-A5, the removal of a single phenol or cyclopentyl side-chain from PCSK9-Evo5 abolished its affinity to PCSK9 protein (Fig. 3d), and the removal of the isopentyl side-chain severely impaired target binding (the binding interaction approximately fits a two-state reaction kinetic model with KD ≈ 420 nM; Fig. 3d and Supplementary Fig. 5). The individual removal of other side-chains had less significant effects on binding affinity; indeed an HFNAP containing only three side chains (phenol at position 9, isopentyl at position 21, and cyclopentyl at position 24) and the rest of the Evo5 sequence as unfunctionalized DNA maintains strong binding to PCSK9 (KD = 3.7 nM; Fig. 3d and Supplementary Fig. 5). Together, these results establish the evolution (iterated selection with intervening mutation and replication) of a sequence-defined synthetic polymer with improved target affinity.
To gain further insight into the secondary structures of PCSK9-binding HFNAPs and the affinity improvement of PCSK9-Evo5 over PCSK9-A5, we subjected the DNA sequences of PCSK9-Evo5 and PCSK9-A5Trunc to secondary structure prediction using mFold45. The 3′ region, conserved between PCSK9-A5Trunc and PCSK9-Evo5, was predicted to form a stem-loop structure containing a bulge. The 5′ region, which includes all the consensus mutations leading to PCSK9-Evo5, contained significantly less (in the case of PCSK9-A5Trunc) or no (in the case of PCSK9-Evo5) predicted secondary structure based on Watson-Crick base-pairing (Supplementary Fig. 7). Consistent with the observed side-chain structure-activity relationships shown in Figure 3d, these calculations further suggest crucial contributions of side chains, which are not captured in the secondary structure model, to HFNAP folding and target-binding activity.
To further characterize PCSK9-Evo5, we executed the multi-milligram-scale total synthesis of a variant of PCSK9-Evo5 (PCSK9-Evo5-syn), which has an additional 3′ inverted dT base for exonuclease resistance46, using standard phosphoramidite chemistry on solid support (Supplementary Fig. 8). We planned to install all side-chain-functionalized building blocks entirely through corresponding side-chain-functionalized phosphoramidite reagents, with the nucleophilic functional groups protected with base-labile protecting groups that would be removed under standard oligonucleotide deprotection conditions. Although most of the side-chain-functionalized nucleoside phosphoramidites were readily synthesized (Supplementary Methods), the imidazole-bearing thymidine reagent proved difficult to prepare. We therefore synthesized PCSK9-Evo5-syn on solid phase with an activated ester (NHS-carboxy-dT) in place of the imidazole-functionalized thymidine, and coupled histamine to that position in the bead-bound polymer chain to install the imidazole side-chain before global deprotection and cleavage from solid support with ammonium hydroxide (Supplementary Fig. 8). The identity of PCSK9-Evo5-syn (approximately 2 mg from a one-micromole solid-phase synthesis, ~13% overall yield) was confirmed by high-resolution mass spectrometry (Supplementary Fig. 9). High-affinity binding (KD = 6.8 nM) between totally synthetic PCSK9-Evo5-syn and biotinylated Avi-tagged PCSK9 was confirmed by SPR (Fig. 3e). In addition, binding of Alexa Fluor 647-labeled Evo5 at low nM concentrations to PCSK9 was confirmed in an electrophoretic mobility shift assay (EMSA) (Supplementary Fig. 10).
PCSK9 regulates cholesterol metabolism by binding the LDL receptor (LDLR) and promoting the lysosomal degradation of LDLR 42. We tested the ability of PCSK9-Evo5-syn to disrupt PCSK9-LDLR binding in an SPR assay. PCSK9-Evo5-syn dose-dependently reduced binding of PCSK9 to surface-immobilized LDLR (Fig. 3f and Supplementary Fig. 11). The potency of PCSK9-Evo5-syn inhibition of PCSK9-LDLR binding (IC50 = ~9 nM) was similar to that of a known PCSK9-neutralizing monoclonal antibody (Fig. 3f and Supplementary Fig. 11). In contrast, unfunctionalized DNA of the same sequence as PCSK9-Evo5-syn produced no inhibitory effect (Fig. 3f and Supplementary Fig. 11), consistent with the necessity of the side chains implicated in PCSK9 binding (Fig. 2c–e and Fig. 3c–d). We also tested the affinity of surface-immobilized PCSK9-Evo5 to different PCSK9 protein constructs and found that a truncated PCSK9 variant lacking the prodomain exhibited no apparent binding to PCSK9-Evo5 (Supplementary Fig. 12), implicating the PCSK9 prodomain, known to be involved in a secondary binding interface between PCSK9 and LDLR47, in mediating the interaction of PCSK9 with PCSK9-Evo5.
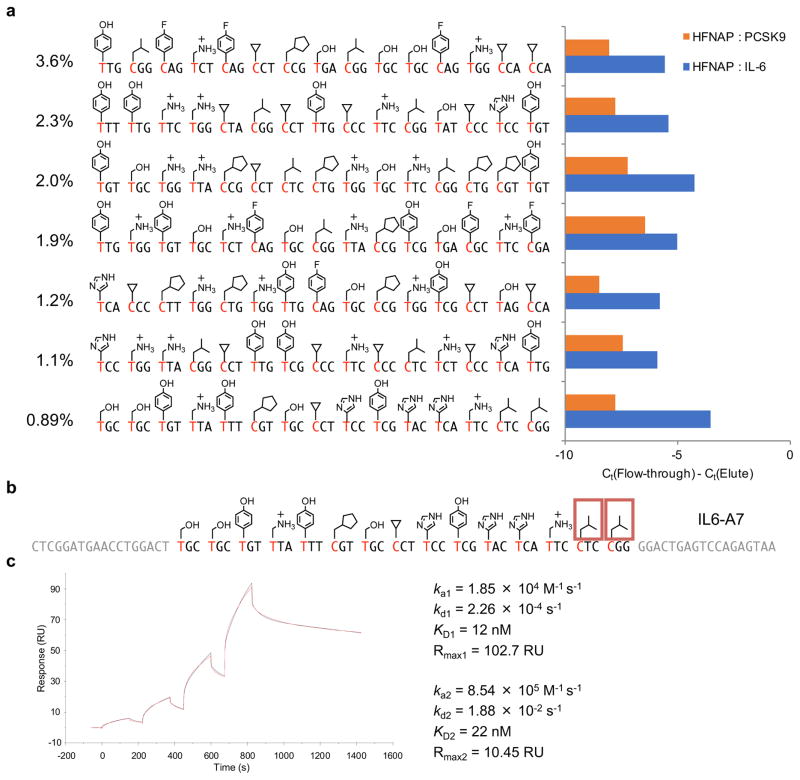
To test the generality of our polymer evolution system and to investigate the potential of this new class of polymers to evolve receptors to different proteins, we performed a separate selection for HFNAPs that bind a protein unrelated to PCSK9. We chose human interleukin-6 (IL-6), a key cytokine involved in inflammation and the target of many drugs and drug candidates48, including modified DNA aptamers24,25. After seven iterated cycles of translation, selection for binding to immobilized IL-6 protein, reverse translation, and amplification, the most abundant sequence accounted for 3.6% of the population (Fig. 4a). The top seven HFNAPs were individually synthesized and assayed for binding to immobilized IL-6. Based on its high apparent binding activity to immobilized IL-6, but not to immobilized PCSK9 (Fig. 4a), the HFNAP IL6-A7 (Fig. 4b) was chosen for further characterization. Binding of biotinylated IL6-A7 to the target IL-6 protein was confirmed by SPR. Although the binding kinetics of IL6-A7 to IL-6 protein did not conform to a classical one-to-one binding model, a phenomenon often observed in aptamer-protein binding18,24, fitting to a heterogeneous ligand model resulted in an apparent affinity of KD = 12 nM for the major component and KD = 22 nM for the minor component (Fig. 4c).
Figure 4. Characterization of IL-6-binding HFNAPs selected from a random library.
a, Retention of individual selection-enriched HFNAPs on immobilized IL-6 (target; blue bars) or immobilized PCSK9 (non-target; red bars). The percentages of each sequence in the pool after seven rounds of selection are listed to the left. The experiment was performed as a screen to identify hits for further characterization, and was not performed in replicates. b, Sequence and side-chain structure of IL6-A7. Side-chains essential for binding activity are boxed. c, SPR sensogram characterizing binding kinetics between biotinylated IL6-A7 and its target IL-6 protein. The concentrations of injected IL-6 were 10, 30, 100, and 300 nM. The observed sensogram is shown in red and the fitted curve with the kinetic parameters listed is shown in black.
The binding kinetics and affinity of IL6-A7 to E. coli-expressed IL-6 (used in the selection) and to human HEK293 cell-expressed IL-6 protein were comparable (Supplementary Fig. 13). Next we measured by SPR the IL-6 affinity of IL6-A7 side-chain mutants in which all instances of a trimer building block were replaced by the corresponding trinucleotide lacking any side-chain. The mutants missing isopentyl side-chain in either codon 14 or codon 15 exhibited severely impaired binding (Supplementary Fig. 13), implicating these two side chains as key determinants of IL-6 binding activity in IL6-A7.
Discussion
We used a ligase-mediated DNA-templated polymerization system and in vitro selection to evolve HFNAPs, nucleic acid polymers that are densely functionalized with chemically diverse side-chains. HFNAPs that bind PCSK9 and IL-6 were selected from random polymer libraries. Through diversification and reselection, we evolved an improved PCSK9-binding HFNAP (Evo5) with KD = 3 nM. We characterized structure-activity relationships within this polymer, revealing side chains at specific positions that are critical to target-binding activity. Evo5 potently inhibits binding between PCSK9 and the LDL receptor.
Collectively, these findings represent to our knowledge the first laboratory evolution of functional, genetically encoded sequence-defined synthetic polymers without the constraints imposed by polymerases or ribosomes. The DNA-templated, ligase-based translation system developed here supports many rounds of iterated selection of polymers with diverse side-chains, including side-chains that mimic—and extend beyond—the repertoire of amino acid side-chains found in proteins. Both the PCSK9-binding and IL-6-binding polymers generated in this system exhibit position-dependent and side-chain dependent structure-activity relationships resembling those of proteins. Finally, we note that the PCSK9-binding polymers generated in this work are strongly dependent on the presence of multiple side-chains with different physical properties, consistent with the importance of chemical diversity to the functional potential of these polymers.
Recently, Gawande and coworkers performed selections for PCSK9 aptamers from modified DNA libraries in which all instances of one or both pyrimidines (C and/or T) were replaced by side-chain-functionalized variants27. High-affinity aptamers with dissociation constants similar to those of FDA-approved anti-PCSK9 monoclonal antibodies (evolocumab, KD = 8.0 pM49, and alirocumab, KD = 0.58 nM50) were enriched from doubly modified libraries in which hydrophobic or phenolic side chains were present on 50% of the nucleobases on average. Aptamers enriched from singly modified libraries (25% hydrophobic side chains on average) were less potent (KD ≥ 100 pM), while libraries containing hydrophilic side chains or consisting of unmodified DNA did not produce aptamers with KD ≤ 30 nM. Consistent with their findings, the highest affinity binders from our HFNAP library, which contains a roughly equal mix of hydrophilic and hydrophobic side chains installed at 33% total frequency, has KD = 3 nM to PCSK9. We note, though, that different modifications may be suitable for other applications, as demonstrated by DNA-based catalysts functionalized with nitrogen nucleophiles as side-chains29–33,35. Therefore, the diverse, balanced set of side-chains in HFNAPs, similar to the natural repertoire of proteins, may be more versatile in other settings.
The ligase-based polymerization method allows straightforward redesign of the genetic code of the polymer, as we exploited to expand the sequence and structural diversity of the polymers used in this work compared with those of our original system39. This feature also enables researchers to generate and select HFNAPs with side-chains tailored toward specific applications, as recently demonstrated by Hili and coworkers for scaffolding peptides on a DNA template51. Moreover, the side-chain flexibility of this polymer evolution system raises the possibility of performing parallel evolution experiments with libraries of different side-chain compositions to shed light on the fundamental relationship between the structure of the building blocks in a genetic code and the evolutionary potential of the resulting polymers.
Methods
Additional experimental procedures and characterization data are provided in the Supplementary Information.
Synthesis of HFNAP by templated translation via DNA ligase-mediated polymerization
DNA template [up to 10 pmol, either in solution or immobilized on MyOne Streptavidin C1 magnetic beads (ThermoFisher Scientific)], polymerization initiation and termination primers (1.5 equivalents each relative to template), functionalized trinucleotide building blocks (10 equivalents relative to template for each occurrence of the corresponding codon) and 10x T4 RNA ligase reaction buffer (New England Biolabs; 1 μL) were mixed in a total volume of 8 μL in a PCR tube. The mixture was subjected to the following temperature program on a thermocycler: 95 °C for 10 sec; 65 °C for 4 min; a ramp from 65 °C to 4 °C at 0.1 °C per 10 s. To the PCR tube were added 1 μL of 10 mM ATP and 1 μL of T3 DNA ligase (New England Biolabs). The reaction was incubated at 4 °C for 12 h and then at 16 °C for 2 h.
Selections of HFNAP that bind protein targets
Selection bait was prepared by immobilizing recombinant protein onto AminoLink Plus aldehyde-functionalized agarose resin via reductive amination with a MicroLink Protein Coupling Kit (ThermoFisher Scientific). Loading was 1 mg PCSK9 protein (ACROBiosystems) per mL resin for the initial PCSK9 binder selection and the first two rounds of PCSK9 binder re-selection; 150 μg PCSK9 per mL resin for rounds 3–5 of the re-selection; 40 μg protein per mL resin for round 6 of the re-selection; and 250 μg IL-6 protein (PeproTech) per mL resin throughout the IL-6 binder selection.
To initiate the selection, primer extension was performed with a biotinylated primer on 5 pmol of the sense strand randomized DNA library (Integrated DNA Technologies or TriLink BioTechnologies) with Klenow (exo-) polymerase (New England Biolabs). Biotinylated species was captured on streptavidin magnetic beads, which were then washed three times with 20 mM NaOH and then twice with 1x T4 RNA ligase reaction buffer. The bead-immobilized template strand library was then translated in a ligase-mediated polymerization to produce HFNAPs. The beads were suspended in 20mM NaOH to denature the HFNAP-template hybrids. HFNAP strands in the supernatant were cleaned up with a MinElute column (Qiagen).
The HFNAP library was added to DPBS (with calcium and magnesium; Lonza) supplemented with BSA (0.1 mg/ml final) and Tween-20 (0.01% final), and then incubated with PCSK9 resin in a micro-spin filtration column (Pierce) at room temperature for 1 h on a rotor. (The amounts of resin-bound protein used in each round of the PCSK9 selection are indicated in Fig. 2b. Throughout the IL-6 selection, 240 pmol of immobilized IL-6 protein was used in each round.) The flow-through was collected by centrifugation at 1000 g into an Eppendorf tube. The beads were washed three times with 50 μL each of DPBS. The column was cut open, and the beads were collected by centrifugation into an Eppendorf tube and then incubated in 50 μL of lithium dodecyl sulfate (LDS) loading buffer (Life Technologies) at 95 °C for 15 min. After cooling, HFNAP strands were isolated from the mixture by cleaning up with a QiaQuick column (Qiagen), eluting the HFNAP into 50 μL of water.
Samples of 1 μL each from the flow-through, the three washes, and the elution were quantified by qPCR (20 μL reaction volume) using the iTaq Supermix (Bio-rad). The number of cycles for the qPCR curve of the elution sample to reach the end of exponential growth was used as the number of cycles for the preparative PCR (400 μL reaction volume split into 8 × 50 μL) of the selection elution pool (20 μL) with Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs), using a biotinylated primer for the strand that will serve as translation template. The PCR product was cleaned up with a MinElute column and PAGE purified on a non-denaturing 10% TBE gel. A portion (indicated in Fig. 2b) of the dsDNA product was captured on streptavidin magnetic beads to initiate the next round of selection.
Surface plasmon resonance (SPR) assays
All SPR assays were performed at 25 °C on a Biacore X100 or Biacore T200 (GE Healthcare Life Sciences). Binding kinetics between enzymatically synthesized biotinylated HFNAPs and unlabeled recombinant proteins were measured using single-cycle kinetics with the Biotin CAPture kit (GE Life Sciences) using 0.9x HBS-EP buffer (GE Life Sciences) at a flow rate of 30 μL/min. The injected PCSK9 concentration ranged from 10 to 300 nM for PCSK9-A5 and its variants, or from 2 to 60 nM for PCSK9-Evo5 and its variants. The injected IL-6 concentration ranged from 10 to 300 nM.
Binding kinetics between chemically synthesized PCSK9-Evo5-syn and biotinylated Avi-tagged PCSK9 (ACROBiosystems) were measured using single-cycle kinetics on a Series S SA chip (GE Life Sciences) using 0.9x HBS-EP buffer at a flow rate of 30 μL/min. The injected PCSK9-Evo5-syn concentration ranged from 1.8 to 180 nM.
Binding of PCSK9 on surface-immobilized LDLR in the presence of various competing agents was measured on a Series S SA chip using 10 mM HEPES, 150 mM NaCl, 0.1 mM CaCl2, 0.005% Tween-20, pH 7.5 as bulk buffer at a flow rate of 10 μL/min. The injected solutions contained 20 nM PCSK9 and various competing agents ranging from 2 to 200 nM.
Data availability
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the authors on request.
Supplementary Material
Acknowledgments
This work was supported by the DARPA Fold Fx program (N66001-14-2-4053), the NIH R01 EB022376 (formerly R01 GM065400) and R35 GM118062, and the Howard Hughes Medical Institute. Z.C. was partially supported by the Y. Kishi Graduate Prize in Chemistry and Chemical Biology sponsored by the Eisai Corporation. The authors wish to thank Dr. Jia Niu, Dr. Ryan Hili, Dr. Juan Pablo Maianti, Dr. Dmitry L. Usanov, and Alix Chan for helpful discussions. We thank the Center for Macromolecular Interactions in the Department of Biological Chemistry and Molecular Pharmacology at Harvard Medical School for access to Biacore T200 SPR instrument, and thank Dr. Matt Blome, Dr. Brian Lang, and Dr. Kelly Arnett for technical assistance on SPR experiments. We thank the Harvard FAS Small Molecule Mass Spectrometry facility for access to ESI-MS instruments and thank Dr. Sunia A. Trauger for technical assistance.
Footnotes
Author Contributions
Z.C. and D.R.L. conceived and designed the study. Z.C., P.A.L., A.P.B., and J.C.C. performed the experiments. Z.C., P.A.L. and D.R.L. wrote the manuscript.
Competing Financial Interests
The authors have filed patent applications on aspects of this work.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 4.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 6.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 7.Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci U S A. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H. In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414:405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi J, et al. cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucleic Acids Res. 2009;37:e108. doi: 10.1093/nar/gkp514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brudno Y, Liu DR. Recent Progress Toward the Templated Synthesis and Directed Evolution of Sequence-Defined Synthetic Polymers. Chem Biol. 2009;16:265–276. doi: 10.1016/j.chembiol.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaput JC, Yu H, Zhang S. The Emerging World of Synthetic Genetics. Chem Biol. 2012;19:1360–1371. doi: 10.1016/j.chembiol.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr Opin Chem Biol. 2012;16:245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- 14.Pinheiro VB, Holliger P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 2014;32:321–328. doi: 10.1016/j.tibtech.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenstein M. Nucleoside Triphosphates — Building Blocks for the Modification of Nucleic Acids. Molecules. 2012;17:13569–13591. doi: 10.3390/molecules171113569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers JM, Suga H. Discovering functional, non-proteinogenic amino acid containing, peptides using genetic code reprogramming. Org Biomol Chem. 2015;13:9353–9363. doi: 10.1039/c5ob01336d. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 18.Pinheiro VB, et al. Synthetic Genetic Polymers Capable of Heredity and Evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor AI, et al. Catalysts from synthetic genetic polymers. Nature. 2014;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn MR, Chaput JC. Reverse Transcription of Threose Nucleic Acid by a Naturally Occurring DNA Polymerase. ChemBioChem. 2016;17:1804–1808. doi: 10.1002/cbic.201600338. [DOI] [PubMed] [Google Scholar]
- 21.Vaught JD, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–4151. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 22.Gold L, et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies DR, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A. 2012;109:19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, et al. Chemically-Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking its Interaction with Interleukin-6 Receptor. J Biol Chem. 2014;289:8706–8719. doi: 10.1074/jbc.M113.532580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelinas AD, et al. Crystal Structure of Interleukin-6 in Complex with a Modified Nucleic Acid Ligand. J Biol Chem. 2014;289:8720–8734. doi: 10.1074/jbc.M113.532697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imaizumi Y, et al. Efficacy of Base-Modification on Target Binding of Small Molecule DNA Aptamers. J Am Chem Soc. 2013;135:9412–9419. doi: 10.1021/ja4012222. [DOI] [PubMed] [Google Scholar]
- 27.Gawande BN, et al. Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci U S A. 2017;114:2898–2903. doi: 10.1073/pnas.1615475114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolle F, Brändle GM, Matzner D, Mayer G. A Versatile Approach Towards Nucleobase-Modified Aptamers. Angew Chem Int Ed. 2015;54:10971–10974. doi: 10.1002/anie.201503652. [DOI] [PubMed] [Google Scholar]
- 29.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 30.Perrin DM, Garestier T, Hélène C. Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucleosides Nucleotides. 1999;18:377–391. doi: 10.1080/15257779908043083. [DOI] [PubMed] [Google Scholar]
- 31.Perrin DM, Garestier T, Hélène C. Bridging the Gap between Proteins and Nucleic Acids: A Metal-Independent RNAseA Mimic with Two Protein-Like Functionalities. J Am Chem Soc. 2001;123:1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]
- 32.Lermer L, Roupioz Y, Ting R, Perrin DM. Toward an RNaseA mimic: A DNAzyme with imidazoles and cationic amines. J Am Chem Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- 33.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+) Nucleic Acids Res. 2009;37:1638–1649. doi: 10.1093/nar/gkn1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoji A, Kuwahara M, Ozaki H, Sawai H. Modified DNA Aptamer That Binds the (R)-Isomer of a Thalidomide Derivative with High Enantioselectivity. J Am Chem Soc. 2007;129:1456–1464. doi: 10.1021/ja067098n. [DOI] [PubMed] [Google Scholar]
- 35.Sidorov AV, Grasby JA, Williams DM. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 2004;32:1591–1601. doi: 10.1093/nar/gkh326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sefah K, et al. In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci U S A. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, et al. Evolution of functional six-nucleotide DNA. J Am Chem Soc. 2015;137:6734–6737. doi: 10.1021/jacs.5b02251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 39.Hili R, Niu J, Liu DR. DNA ligase-mediated translation of DNA into densely functionalized nucleic acid polymers. J Am Chem Soc. 2013;135:98–101. doi: 10.1021/ja311331m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei Y, Kong D, Hili R. A High-Fidelity Codon Set for the T4 DNA Ligase-Catalyzed Polymerization of Modified Oligonucleotides. ACS Comb Sci. 2015;17:716–721. doi: 10.1021/acscombsci.5b00119. [DOI] [PubMed] [Google Scholar]
- 41.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence Variations in PCSK9, Low LDL, and Protection against Coronary Heart Disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 42.Lambert G, Charlton F, Rye KA, Piper DE. Molecular basis of PCSK9 function. Atherosclerosis. 2009;203:1–7. doi: 10.1016/j.atherosclerosis.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Stein EA, et al. Effect of a Monoclonal Antibody to PCSK9 on LDL Cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 44.Brudno Y, Birnbaum ME, Kleiner RE, Liu DR. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat Chem Biol. 2010;6:148–155. doi: 10.1038/nchembio.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortigão JFR, et al. Antisense Effect of Oligodeoxynucleotides with Inverted Terminal Internucleotidic Linkages: A Minimal Modification Protecting against Nucleolytic Degradation. Antisense Res Dev. 1992;2:129–146. doi: 10.1089/ard.1992.2.129. [DOI] [PubMed] [Google Scholar]
- 47.Lo Surdo P, et al. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep. 2011;12:1300–1305. doi: 10.1038/embor.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 49.Gibbs JP, et al. Impact of Target-Mediated Elimination on the Dose and Regimen of Evolocumab, a Human Monoclonal Antibody Against Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) J Clin Pharmacol. 2017;57:616–626. doi: 10.1002/jcph.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kühnast S, et al. Alirocumab inhibits atherosclerosis, improves the plaque morphology, and enhances the effects of a statin. J Lipid Res. 2014;55:2103–2112. doi: 10.1194/jlr.M051326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo C, Watkins CP, Hili R. Sequence-Defined Scaffolding of Peptides on Nucleic Acid Polymers. J Am Chem Soc. 2015;137:11191–11196. doi: 10.1021/jacs.5b07675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the authors on request.