Abstract
Trypanosoma cruzi, the protozoan parasite that causes Chagas disease in humans, has a complex life cycle that promotes survival in disparate environments. In each environment the parasite must fine-tune its metabolic pathways to divide and multiply. In the absence of recognizable transcriptional gene regulation, it is apparent that protein levels are determined by post-transcriptional mechanisms. Post-transcriptional gene control is influenced by RNA-binding proteins that target mRNAs in the cell’s cytoplasm. To initiate the study of post-transcriptional activities in T. cruzi, we studied this organism’s ortholog of RBP42, a trypanosomal RNA-binding protein. RBP42 was originally detected in Trypanosoma brucei and was shown to target a subset of mRNAs that encode proteins governing central-carbon metabolism. T. cruzi RBP42 structurally resembles T. brucei RBP42, sharing an NTF2 domain at its amino terminus and a single RNA-binding domain (specifically, the RNA recognition motif, or RRM), at its carboxy terminus. A phylogenetic analysis reveals that an NTF2 and a single RRM are distinguishing features of all RBP42 orthologs within the broad kinetoplastid grouping. T. cruzi RBP42 is expressed in all life cycle stages of the parasite as determined by immunoblot and immunofluorescence microscopy. In each case the protein is localized to the cytoplasm, indicating a role for T. cruzi RBP42 in post-transcriptional activities in all stages of the parasite life cycle. We speculate that RBP42 influences the dynamic metabolic pathways responsible for parasite infection and transmission.
Keywords: trypanosomes, Trypanosoma cruzi, mRNA binding proteins, Immunofluorescence, transgenic parasites, Chagas disease
INTRODUCTION
American trypanosomes, protozoan parasites that cause Chagas disease in humans, have a complex life cycle that requires morphological and metabolic changes controlled by complex gene expression programs (Jimenez 2014). In the absence of regulated transcription, trypanosome gene expression is regulated mainly at the post-transcriptional level (Araujo and Teixeira 2011). Post-transcriptional processes include mRNA maturation, localization, translation and decay.
RNA-binding proteins are key players in post-transcriptional events because they can coordinate translation of related mRNAs, much the way that DNA-binding proteins coordinate expression of related DNA sequences. Specific mRNAs can be grouped into RNA regulons, which encode functionally related proteins and rely on specific sets of RNA-binding proteins for their coordinated expression (Blackinton and Keene 2014; Keene 2007). Our work on the mRNA-binding protein RBP42 in the African trypanosome, Trypanosoma brucei, and work on other T. brucei mRNA-binding proteins including PUF9, suggests that RNA regulon networks are essential to these single-cell organisms (Archer et al. 2009; Das et al. 2015; Das et al. 2012; Kolev et al. 2014).
African and American trypanosomes are closely related organisms in terms of their genetic structure (El-Sayed et al. 2005). Their protein-coding genes are transcribed as multi-gene precursor mRNAs that mature into translatable mRNAs by the action of a very large collection of enzymes that separate and then stabilize individual protein coding molecules in the nucleus. mRNAs migrate to the cytoplasm where cytoplasmic mRNA-binding proteins direct and regulate their translation. Genomic and proteomic characterization of African and American trypanosomes reveal a common set of RNA-binding proteins that share not only their RNA-binding domains, but additional motifs that suggest, at least, similar functions for the RNA-binding proteins in the genetically similar, but clinically distinct, parasites (Romaniuk et al. 2016). Therefore, the function of specific proteins within the regulon may be phylogenetically conserved and possibly exploited for pharmaceutical intervention in the treatment of both T. cruzi and T. brucei.
The metabolically distinct forms of T. cruzi define the parasite’s life cycle stage and reflect its host environment (Tonelli et al. 2011). Parasites multiply as epimastigote forms in the midgut of Triatomine bugs, which serve as the insect vector for the parasite. Nutrient depletion leads to a metacyclogenesis (i.e., transformation into metacyclics) and parasite migration to the bug’s hindgut. Human infection occurs when the bug’s feces, laden with parasites, enter the body through skin-breaks or mucous membranes. Triatomines defecate on the skin and metacyclic parasites are rubbed or scratched into the bite wound. Other modes of transmission include oral ingestion of food contaminated with parasites or blood transfusion (Bonney 2014). Trypomastigotes enter human cells and multiply in the cell cytoplasm as amastigotes. Amastigotes fill their host cell cytoplasm and prepare to invade additional host cells by converting into trypomastigotes, which can survive extracellularly in the human host and insect’s blood meal.
T. brucei RBP42 is an essential protein in procyclic and bloodstream form parasites growing in culture, is located predominately in the parasite cytoplasm, near the ER and is associated with polysomes, suggesting a role for RBP42 in either mRNA translation, localization or stability. RBP42’s role as an important mRNA-binding protein is evidenced by our findings that the protein targets the coding region of mRNAs translated into proteins central to energy metabolism in the parasite form residing in its insect vector, Glossina flies (Das et al. 2012). Moreover, recent work from our laboratory shows that the protein profiles of many central carbon metabolism proteins are disrupted when bloodstream form parasites are depleted for RBP42 (our unpublished work). Finally, an exciting new study of T. brucei infection in the mouse model reveals that parasites residing the fat tissue of the mice have an increase in RBP42 mRNA compared to the parasites coursing through the bloodstream (Trindade et al. 2016).
These observations led us to investigate the expression and subcellular localization of RBP42 in T. cruzi. We were successful in epitope tagging and stably expressing the entire RBP42-encoding gene sequence in transgenic epimastigotes. The exogenous copy of RBP42 did not impair life cycle progression of epimastigotes, as the transgenic parasites were able to undergo metacyclogenesis and infect mammalian host cells. We show that RBP42 is produced all stages of the T. cruzi life cycle, indicating that post-transcriptional protein-mRNA interactions may contribute to the induction of quiescent and replicative states of the parasite’s life cycle. Therefore, further studies investigating the role of RBP42 in the metabolic changes during the T. cruzi life cycle may yield novel approaches in combating infection. Finally, our findings support a broad and possibly dynamic role for RBP42 in trypanosomatid biology.
MATERIALS AND METHODS
Trypanosome strains
Wild-type T. cruzi (Y strain) epimastigotes were grown in liver infusion tryptose (LIT) (Camargo 1964) medium, supplemented with 10% heat inactivated fetal bovine serum (FBS) at 27°C in a humidified chamber containing 5% CO2. The parasites were a kind gift from Silvia Moreno, at the University of Georgia. Metacyclic parasites were enriched by adding 10% fresh horse serum to stationary phase epimastigote cultures. Trypomastigotes and amastigotes were maintained in Vero and LLC-MK2 cells. Host cell lines were grown in DMEM supplemented with 10% FBS in a 37°C humidified chamber containing 5% CO2. Tc42 transgenic parasites contain an amino-terminal Ty1-tagged RBP42 followed by a G418 marker (neomycin resistance gene) inserted into a genomic ribosomal locus. TcGFP transgenic parasites contain a GFP followed by a G418 marker similarly inserted into a ribosomal locus. The transgenes were maintained in epimastigotes in LIT containing 120 µg/mL G418.
Plasmid constructions and transfections
T. cruzi RBP42 (Y strain) was amplified from genomic DNA using PCR primers corresponding to the termini of the TcCLB.509167.140 RBP42 coding region. Sequencing the Y strain-derived RBP42 gene indicated that it differed in 9 amino acid positions from the published TcCLB RBP42 gene. The sequence comparison is shown in Figure S1. An amino-terminal Ty1 tag was added to the RBP42 sequence by generating a PCR fragment encoding tandem Ty1 epitopes (EVHTNQDPLD), separated by a GT spacer, in which the first copy was preceded by an M in-frame with the natural second amino acid (L) of RBP42. Ty1-RBP42 was inserted between the XbaI and HindIII sites in the polylinker region of pTREX (Vazquez and Levin 1999. pTREX, a kind gift from Huan Huang, is derived from pRIBOTEX {Martinez-Calvillo, 1997 #1712). Cloning was done using the In-Fusion™ system (Clontech). Escherichia coli containing pTREX-GFP was a gift from Silvia Moreno (DaRocha et al. 2004). All plasmids used for transfections were prepared in our laboratory and quality control was done by direct DNA sequencing. The molecular weight of Ty1-RBP42 is 44 kDa. The untagged protein is 41.5 kDa.
Each construct was separately transfected into parasites using AMAXA Nuclefector™ II, program X-001, and the human T-cell Nuclefector™ kit (Lonza). Transfections were done using 100 µl of parasites (1×10^8/mL) obtained from a log-phase culture (1×10^7/mL) and 10 µg of plasmid DNA. Limiting dilution was used to generate two representative clonal cell lines, named Tc42 and TcGFP, which were used in the study. Parasites are described genetically in Table S1 and were selected in LIT and G418 (120 µg/mL). Transgenic organisms were obtained after 5–6 weeks of incubation.
Antibodies and Western analysis
Monoclonal anti-Ty1 antibody was purchased from Sloan Kettering Cancer Center (Bastin et al. 1996). Anti-RBP42 antibodies were generated in rabbits inoculated with recombinant T. brucei RBP42 (Das et al. 2012). Expression of RBP42 was determined by Western blot analysis of whole cell extracts prepared from parasites. Microscopic examination was initially used to assess purity of a particular life cycle stage in each sample; Stages were confirmed by characteristic positioning of nuclei and kinetoplastid in confocal microscopy. Protein extracts from 2 x10^6 cells, prepared by boiling in SDS-sample buffer, were separated on 10% PAGE-SDS and analyzed by immunoblotting using the ECLTM kit from Pierce. Anti-tubulin antibody was from Abcam.
Host cells
LLC-MK2 rhesus monkey kidney cells were obtained from ATCC, and cultured in DMEM medium with 10% FBS. Vero cells were from Silvia Moreno, at the University of Georgia. Vero cells were treated with 35 Gy gamma radiation to halt cell division. Neonatal rat cardiomyocytes were the gift of Maha Abdellatiff, at Rutgers-New Jersey Medical School.
Metacyclogenesis and host cell infections
Epimastigote cultures were aged to late log-phase growth (1×10^8/mL). Cultures (2 mL) were pelleted, resuspended in DMEM, and then used to infect Vero cells for 48 hrs in T-75 flasks, to produce intracellular amastigotes. Cultures were washed 3×with PBS and media was replaced with DMEM plus 10% fresh horse serum. After one week, the complement components in the horse serum had killed all epimastigotes. The trypomastigotes produced from the intracellular amastigotes where collected and used for analysis and for future infection experiments.
Immunofluorescence microscopy of epimastigotes and trypomastigotes
Mid-log phase Tc42 and TcGFP epimastigotes were washed twice with cold PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. Fixed cells were then washed twice with PBS and resuspended at 5×106 cells/mL in cold PBS. 200 µL of cells were spotted in the center of a pap-pen ring on poly-L-lysine coated slides and allowed to adhere for 20 min. Attached parasites were permeabilized with 0.2% Triton-X in PBS for 10 min and washed on-slide 5×with PBS. Fixed and permeabilized cells were blocked 1 h with 3% BSA, 10% goat serum in PBS and incubated 1 h at 37°C in primary antibody (monoclonal mouse α-Ty1, 1 µg/mL in 3% BSA in PBS). Isotype controls were blocked and incubated with mouse IgG at 1 µg/mL under the same conditions. Slides were washed with PBS (10 min, 5 times) and incubated in secondary antibody (Alexa Fluor® 594 α-mouse, 1:3000 in 3% BSA) for 1 h at 37°C. Parasites were then washed (10 min, 5 times) with PBS and mounted using Prolong® Gold Antifade Mountant with DAPI. Cells were imaged on a Nikon A1R confocal microscope.
IFA of amastigotes
LLC-MK2 cells were seeded on 22×22 mm coverslips in 6 well plates and infected with Tc42, TcGFP, or both transgenic trypomastigotes. Four days post-infection cells were washed once with PBS and fixed with 4% PFA in PBS at 4°C for 4 hr. Cells were then washed 6×10 min with PBS and permeabilized 20 min with 0.2% Triton-X in PBS and washed (5 min, 3 times). Fixed and permeabilized cells were blocked overnight at 4°C and incubated in primary antibody (monoclonal mouse α-Ty1, 1 µg/mL; mouse IgG, 1 µg/mL) for 48 h at 4°C. Cells were then washed (10 min, 5 times) with PBS and incubated in secondary antibody (Alexa Fluor® 594 α-mouse, 1:3000; Alexa Fluor® 680 Phalloidin, according to manufacturer’s protocol) for 1.5 h. Parasites were washed (10 min, 5 times) with PBS and mounted using Prolong® Gold Antifade Mountant with DAPI. Cells were imaged on a Nikon A1R confocal microscope.
Protein alignments
RBP42 alignments were performed using Clustal Omega (Goujon et al. 2010; Sievers et al. 2011)Sievers et al. 2011) and prepared in GeneDoc (Nicholas et al. 1997).
RESULTS AND DISCUSSION
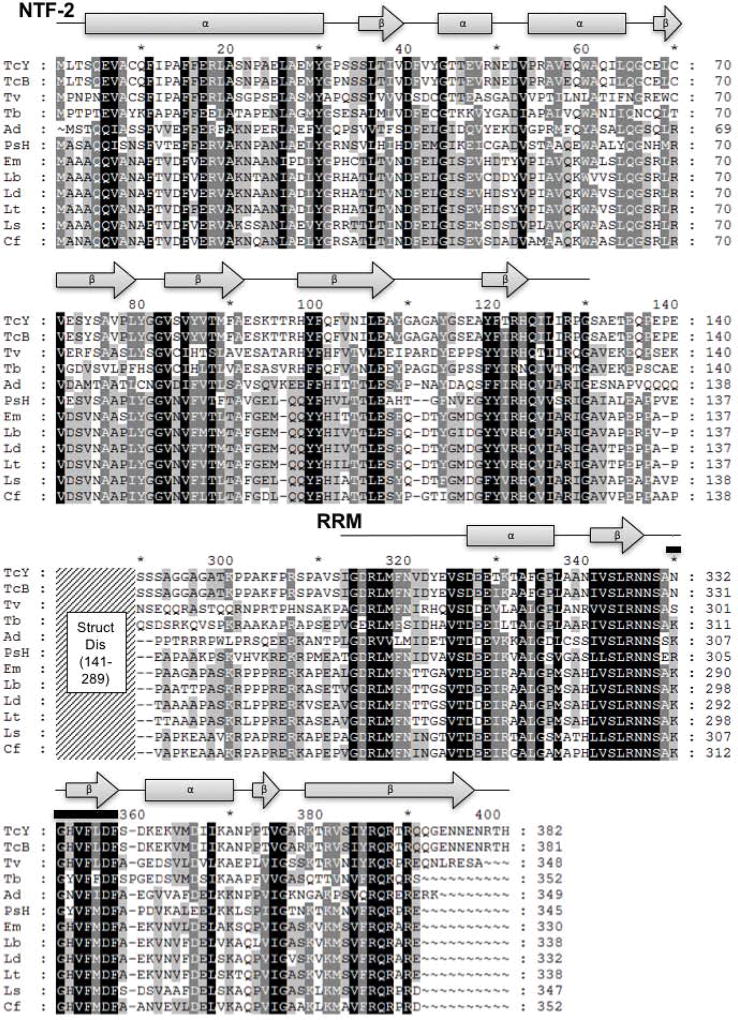
We began our study of the RNA-binding protein RBP42 in T. cruzi by cloning and sequencing the homolog of RBP42 that is present in the Y strain, as the Y strain is extensively used for molecular genetic analysis of T. cruzi gene expression (Lander et al. 2015). Moreover, this strain progresses through the complete T. cruzi life cycle in culture, including intracellular amastigote development. Using the published Cl Brener RBP42 sequence, we amplified the Y strain RBP42 using PCR primers derived from the first four and last four amino acids present at the amino and carboxy terminal sequences of Cl Brener RBP42. As expected, the Y strain RBP42 is extremely homologous, but not identical, to the Cl Brener RBP42 protein. The comparison between the two is shown in Figure S1. We established the conserved domain structure among all kinetoplastid RBP42 proteins by generating an extensive sequence comparison (Figure 1). RBP42s share two striking features: an NTF2 domain and a single RRM domain. Using this information in our Cl Brener and Y strain comparison, we observe that of the 11 total amino acid differences, three are charge-conserved amino acid differences, and nine are non-conserved. The nine non-conserved amino acid differences predominately lie outside of the conserved NTF2 and RRM domains. Specifically, six are in the disordered region and two of the remaining three differences are in hyper-variant positions (Figure 1). As Chagas disease is caused by several different strains of T. cruzi (Bern 2015; Rassi et al. 2010), differences in protein sequences may contribute to different courses of infection.
Figure 1. Kinetoplastid RBP42 homologues consist of an amino-terminal NTF2-like domain and a carboxy-terminal RRM domain.
Shown are the eight secondary structures that help define an NTF2-like domain and six secondary structures consistent with a RRM-like domain; filled gray rectangles represent predicted T. cruzi Y strain RBP42 α-helices, filled gray arrows represent predicted T. cruzi Y strain RBP42 β-strands, and black lines represent additional amino acids included in the domains. Within the T. cruzi Y strain RRM, the RNP1 RNA-binding signature sequence motif K/R-G-F/Y-G/A-F/Y-V/I/L-X-F/Y is found as NGHVFLDF (residues 350-357, amino acids are overlined); this sequence is highly conserved amongst the RBP42 homologues. The middle portion of the alignment, residues 141-289 represented by the diagonally-lined region, was omitted from the figure as this region is predicted to be structurally disordered (Struct Dis) and reveals less similarity among these sequences. Amino acid sequences are represented in single letter amino acid code; intensity of gray scale shading from black to no shading indicates degree of identity ranging from identical or highly similar to limited or no similarity. Secondary structure prediction based on the T. cruzi Y strain RBP42 amino acid sequence was done using the Phyre2 web portal (Kelley et al. 2015). I-TASSER, PSIPRED, and JPred all returned similar results to the Phyre2 prediction (Roy et al. 2012) (Drozdetskiy et al. 2015)
(Buchan et al. 2013) (Zhang 2009). Gene annotations are: TcY, T. cruzi Y strain; TcB, T. cruzi CL Brener Esmeraldo-like (TcCLB.509167.140); Tv, T. vivax Y486 (TvY486_0603830); Tb, T. brucei TREU927 (Tb927.6.4440); Ad, Angomonas deanei (gi: 528258120, gb: EPY37436.1); PsH, Phytomonas sp. isolate Hart1 (gi: 594148441, emb: CCW68155.1); Em, Endotrypanum monterogeii strain LV88 (EMOLV88_300036900); Lb, Leishmania braziliensis MHOM/BR/75/M2903 (LBRM2903_300037500); Ld, Leishmania donovani BPK282A1 (LdBPK_303130.1); Lt, Leishmania tropica L590 (LTRL590_300039500); Ls, Leptomonas seymouri ATCC 30220 (Lsey_0105_0110); Cf, Crithidia fasciculata strain Cf-Cl (CFAC1_260055900). NTF, nuclear transport factor; RRM, RNA recognition motif.
The conservation of both the NTF2 and RRM domains in RBP42 orthologs across diverse groups of trypanosomatidae indicates that this two-domain modular structure, linked by a flexible region, is a central feature of this protein. Although an NTF2 domain is usually associated with nuclear-cytoplasmic transport, recent structural work demonstrated that an NTF2-like domain contributes to RNA binding when it is linked to an RRM in mRNA binding protein Mex67 in budding yeast (Aibara et al. 2015; Hieronymus and Silver 2003). Interestingly, in the case of both the Mex67 and RBP42 proteins, their RRMs are possibly weak version of RRMs, as MEX67 lacks the typical RNP1 and RNP2 motifs and RBP42 lacks the RNP2 and typical beta-subfold that initiates the βαββαβ fold characteristic of RRMs. Taken together, these observations suggest that RBP42 may use both its RRM and NTF2 domains for binding of target mRNAs identified in our TbRBP42 study (Das et al. 2012). As Tb927.11.6440, a hypothetical RNA binding protein, may interact with RBP42 in T. brucei, it is further possible that a heterocomplex makes RNA-binding contacts via the various protein domains (Gazestani et al. 2017).
Our bioinformatic analysis revealed that trypanosome RBP42 is widely dispersed through multiple species of these flagellated protozoa. RBP42 is found in parasites from all three groups of human disease-causing parasites, namely American Trypanosomes (T. cruzi), African trypanosomes (T. vivax, T. brucei) and Leishmania (L. braziliensi, L. donovani and L. tropica). In addition, in the case of T. brucei and T. cruzi, phosphoproteomic analysis reveals that there are phosphorylated forms of RBP42, although the dynamic patterns of these modifications are unknown (Marchini et al. 2011; Nett et al. 2009; Urbaniak et al. 2013). We identified RBP42 sequences in the digenetic trypanosomatids within the Endotrypanum genus (E. monterogeii strain LV88). RBP42 has been maintained in Trypanosomatida that reside exclusively in insects (Crithidia fasciculate, Leptomonas seymouri) and plants (Phytomonas sp. isolate Hart). Finally, we observed that even the presence of an endosymbiont bacteria, which occurs in Angomonas deanei, did not eliminate the apparent need for RBP42 in trypanosomes.
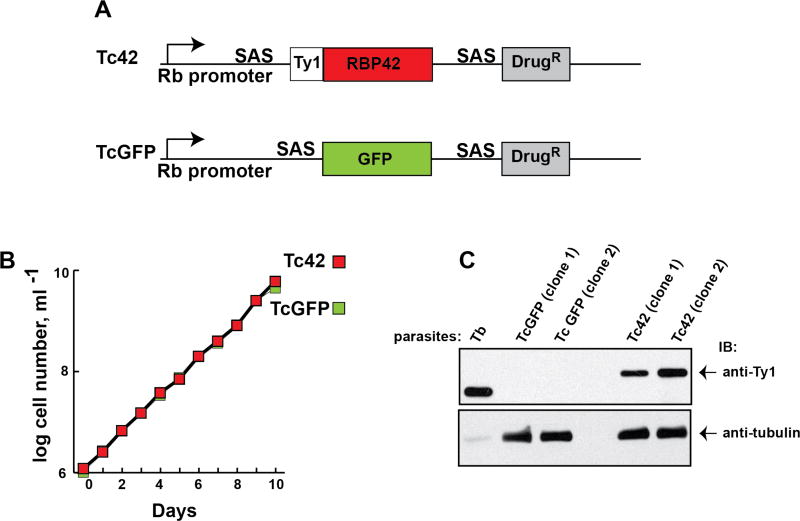
To assess the subcellular localization of RBP42 in epimastigotes, the form that can be transfected with transgenes, we developed a cell line that contained an exogenous and homologous copy of RBP42 that was tagged with the small, non-obtrusive Ty1-epitope tag. To not interfere with the RNA-binding potential of the exogenous protein, we placed the Ty1-tag in-frame at the amino terminus of RBP42. Thus, the carboxy-terminal RRM was likely unperturbed. As a control, we produced a GFP-reporter transgenic cell line, using the identical pTREX expression vector and G418-selection plasmid that was used to generate the Ty1-RBP42 cell line. The plasmid components that drove the transgenic genes in the Tc42 and TcGFP parasites are diagramed in Figure 2, panel A. Parasites containing Ty1-RBP42 exhibited a normal growth phenotype (Figure 2, panel B). We determined the specificity of the anti-Ty1 antibody and the expression of the RBP42 transgene using Western blot analysis (Figure 2, panel C). As expected, anti-Ty1 antibody selectively detected the Ty1-RBP42 protein and did not cross-hybridize with any other proteins in the parasites that were constitutively expressed or induced as a result of the pTREX backbone plasmid or drug selection scheme. As an additional control, we determined the expression levels of RBP42 in the Tc42 cell line by Western blot analysis. Anti-RBP42 antibody, produced in rabbits challenged with recombinant T. brucei RBP42 protein, recognizes T. cruzi RBP42 less well than it recognizes T. brucei RBP42, as expected since the proteins are highly similar but not identical. Using our heterologous antibody we were able to detect endogenous RBP42 in wild-type and transgenic T. cruzi epimastigotes via Western blot analysis. RBP42 appears as a single band in this analysis, although it is likely that there are post-translationally modified forms of the protein in some trypanosomatids (Figure 3, panel A) (Urbaniak et al. 2013). Tc42 parasites contain two distinct RBP42 proteins, an endogenous protein and the one derived from the Ty1-tagged RBP42 transgene. Interestingly, Ty1-tagged RBP42 expression levels are comparable with endogenous RBP42 levels in the Tc42 cells.
Figure 2. Transgenic T. cruzi epimastigotes express Ty1-tagged RBP42.
Panel A: Schematic of relevant regions of Tc42 and TcGFP constructs derived from pTREX. The ribosomal RNA promoter (Rb promoter) and splice acceptor sequences (SAS) are indicated. The SAS contains the HX1 fragment (from TcP2beta) with its polypyrimidine tract and AG splice-leader acceptor site. The Ty1-epitope tag (Ty1), protein coding region (TcRBP42 in blue and GFP in green) and G418 drug resistance marker (DrugR) are indicated. (RBP is the abbreviation for RNA-binding protein and 42 clarifies that RBP42 is the 42nd identified RNA-binding protein in trypanosomatids.)
Panel B: Growth curves of transgenic T. cruzi epimastigotes containing Tc42 or TcGFP constructs. To keep parasites in log phase growth, cultures were diluted to 1×10^6 cells/mL when their density reached 1×10^7 cells/mL. Direct microscopic observation using a hemocytometer was done for all cell counts. Results of a single clone are shown; however, similar growth curves were observed for additional clones.
Panel C: Immunoblot analysis of total soluble protein from transgenic cell lines shows the Ty1-tagged RPB42 is present in Tc42 parasites and absent in TcGFP parasites. Protein from T. brucei (Tb) parasites expressing Ty1-tagged RBP42 was used as a positive control for Ty1 antibody specificity. Protein loading of T. cruzi extracts was confirmed using anti-tubulin. To easily detect T. brucei RBP42, protein concentrations were adjusted. Antibodies are indicated on the right.
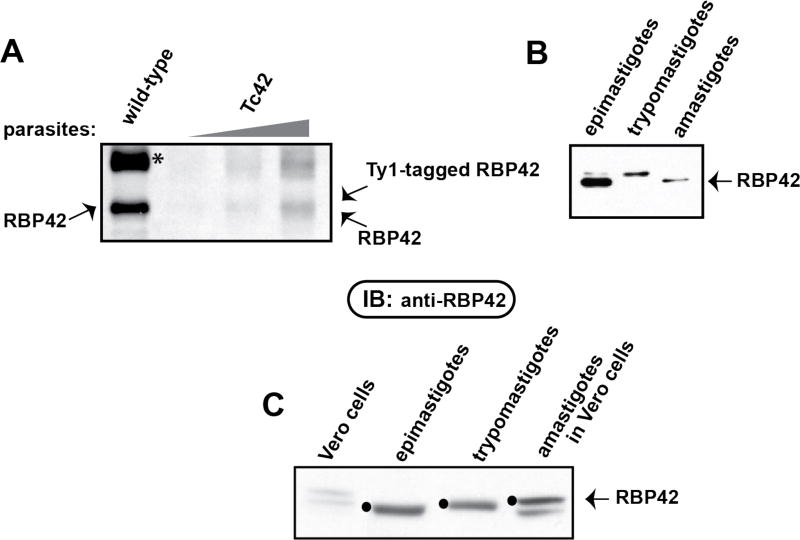
Figure 3. Expression level of Ty1-tagged RBP42 is similar to endogenous RBP42 in the epimastigote form of transgenic parasites.
Panel A: Immunoblotting using anti-RBP42 antibody detected comparable levels of endogenous and Ty1-RBP42 in wild-type and Tc42 transgenic parasites. Asterisk (*) indicates a cross-reacting protein routinely detected with this antibody, prompting us to perform IFA assays using the highly specific anti-Ty1 antibody.
Panel B: Immunoblot analysis of total soluble protein from Tc42 transgenic parasites at the epimastigote, intracellular amastigote and trypomastigote stages of the T. cruzi life cycle. Parasite life cycle stages were identified by microscopy and each culture was determined to be a ~99% pure population. Epimastigotes were transformed into metacyclics, which were used to infect Vero cells. Intracellular amastigotes were harvested at day 4 post-infection. Trypomastigotes developed from the intracellular parasites by day 10 and were collected by concentrating the culture media. Equal amounts to total protein were loaded into each gel lane.
Panel C: As in Panel B, except that here pre-infection Vero cell extracts are included as a negative control. RBP42 is indicated by filled black circles.
Using established methods, we were able to generate metacyclic parasites from the Tc42 epimastigotes that were able to infect mammalian cells. The infected mammalian cells, whether they were LLC-MK2 or Vero cells, supported the transformation of metacyclics into amastigotes that eventually filled the host cell cytoplasm. Amastigotes differentiated into trypomastigotes and were released from host cells. The released trypomastigotes were able to continue the infection cycle by infecting new mammalian host cells. We assayed for RBP42 expression in these life cycle stages using Western blot analysis (Figure 3, panel B) and immunofluorescent imaging (Figures 4 and 5). Western blot analysis demonstrated that RBP42 is expressed throughout the T. cruzi life cycle in wild-type parasites. The cross-hybridizing proteins in the epimastigote and amastigote samples likely indicate proteolytically clipped protein; thus, the relative amounts of RBP42 in each life cycle stage, as well as any potentially different post-translational modifications, can be assessed in our assays.
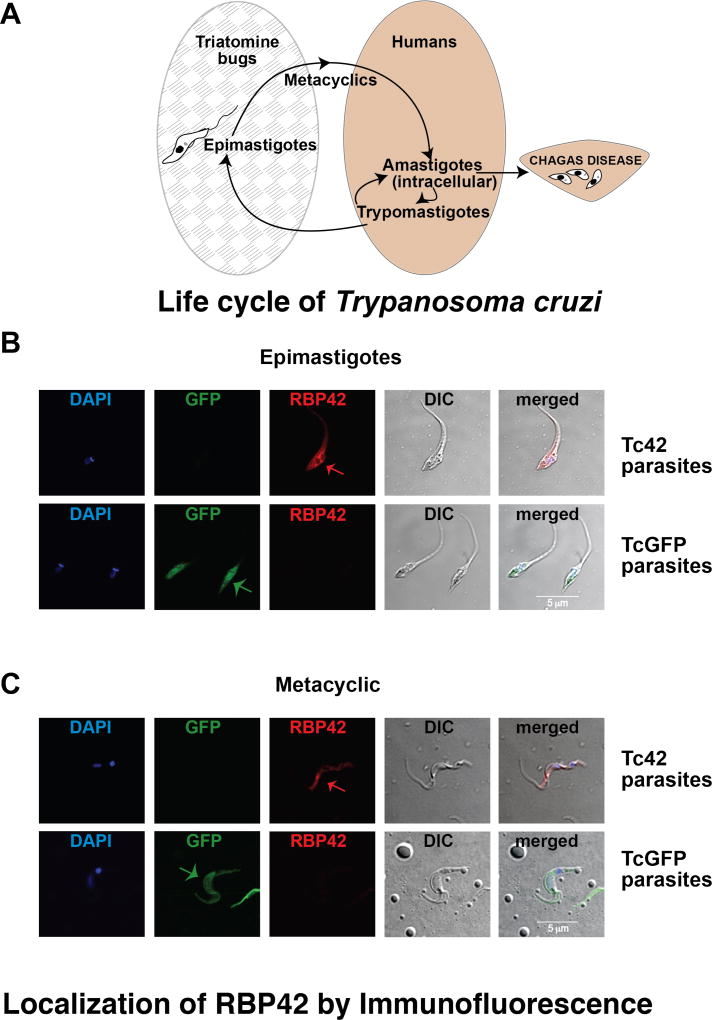
Figure 4. RBP42 is normally expressed in multiple stages of the parasite life cycle.
Panel A: Diagram of T. cruzi cycling through its insect vector (Triatomine bugs; grey patterned ellipse) and mammalian host (Human; beige ellipse). Amastigotes thrive in human tissues, leading to Chagas disease.
Panel B and C: Immunolocalization of RBP42 in epimastigote and metacyclic stages of the parasite life cycle. Nuclear and kinetoplastid staining is blue (DAPI); GFP fluorescence is green (GFP); RBP42 staining is red and detected by anti-Ty1 antibody (RBP42); differential interference contrast (DIC) is shown as a composite image (Merge). Arrows indicate staining of RBP42 and GFP fluorescence.
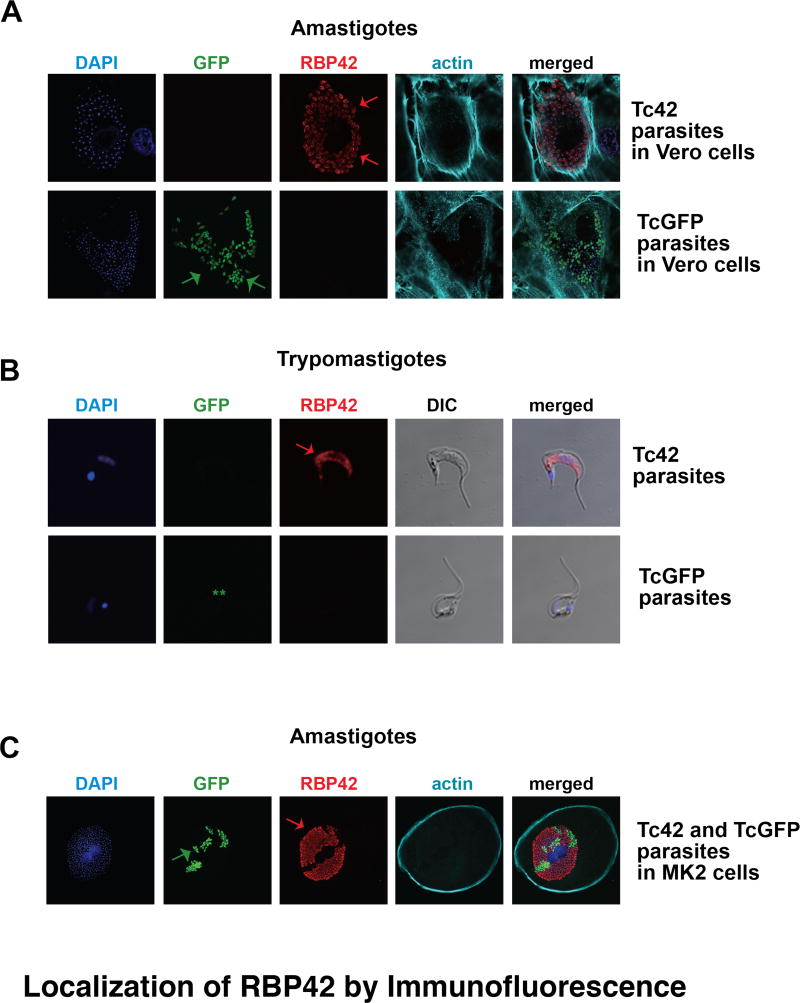
Figure 5. Immunolocalization of RBP42 in amastigote and trypomastigote stages of the parasite life cycle.
Panel A and B: Immunolocalization of RBP42 in amastigote and trypomastigote stages of the parasite life cycle. Staining is as described in Figure 4, with the addition of phalloidin staining to visualize host cell periphery. In Panel B, the two asterisks (**) indicate that the GFP signal was lost in these parasites. Arrows indicate staining of RBP42 and GFP fluorescence.
Panel C: LLC-MCK cells were co-infected with similar numbers of Tc42 and TcGFP trypomastigotes, which then differentiated into amastigotes. Staining is as described in Panel A and B.
As predicted from our Western blot analyses, we were able to visualize Ty1-tagged RBP42 in parasites in all life cycle stages (Figure 4). Epimastigotes from the Tc42 cell line did not fluoresce for GFP, but did fluoresce for Ty1-RBP42, as shown in the Epi-Ty1 image (Figure 4, panel B). As a positive control, we observed GFP fluorescence in the TcGFP parasites. As a negative control for RBP42 staining, we did not observe red fluorescence in the TcGFP parasites. In the Tc42 parasites, fluorescence was localized to the cell cytoplasm and was punctate, whereas the GFP fluorescence was homogenously dispersed throughout the cytoplasm. We did not observe RBP42-dependent staining in the microtubule-containing structures, such as the plasma membrane and the flagellum. Staining intensity was concentrated peri-nuclearly, similar to the staining patterns seen in African Trypanosomes. This was expected, as the cytoplasmic-directing domains of TbRBP42 are most likely shared by the TcRBP42 protein.
We used this staining protocol to determine if Ty1-RBP42 is also expressed in metacyclic stages of the parasite. Immunofluorescence imaging indicated that metacyclic parasites expressed RBP42. Lack of colocalization of the RBP42 signal with DAPI indicates the protein is cytoplasmic.
In order to determine if Ty1-RBP42 was expressed in the intracellular stages, amastigotes were visualized in Vero cells, as well as in LLC-MK2 cells, using confocal microscopy. Ty1-RBP42 was present in all amastigotes within Vero cells. Vero cells were visualized using Phalloidin 680 (actin-specific fungal toxin conjugated to a fluorophore) to mark the cell periphery, and DAPI was used to mark the individual amastigotes. Interestingly, the Ty1-RBP42 transgene persisted in parasites without drug selection, which was not the case for the GFP reporter transgene. Specifically, not all of the DAPI-stained amastigotes fluoresced green on the GFP channel. The selective maintenance of RBP42, and not GFP, is consistent with a function for RBP42, but not GFP, in parasites.
Lastly, we assessed the expression of Ty1-RBP42 as parasites transitioned from amastigotes to trypomastigotes. Trypomastigotes are non-dividing forms whose function is to continue the parasite’s life cycle in the mammalian host. We were surprised to discover that Ty1-RBP42 protein was observed in this life cycle stage, as this stage is often viewed as metabolically quiescent and might not utilize cytoplasmic RNA-binding proteins as extensively as do other life cycle stages. In addition, we anticipate that Ty1-RBP42 expression is regulated by the same (yet unknown) mechanisms that control endogenous RBP42, as trypanosomes rely on post-transcriptional reactions to modulate intracellular protein levels.
In summary, we have detected RBP42 throughout the life cycle of the harmful human parasite T. cruzi. We confirmed the findings by others that GFP is an excellent reporter gene to follow transgenic parasites in the epimastigote, metacyclic and amastigote phases of the parasite life cycle (DaRocha et al. 2004). We have shown that RBP42 is expressed throughout the parasite life cycle, and, at the level of resolution afforded by our confocal microscopy, continues to reside in the cytoplasm. As is the case for some RNA-binding proteins, as well as for NTF2-containing proteins, shuttling between the cytoplasm and nucleus often occurs as part of a stress response. Now that we have the ability to efficiently detect and follow Ty1-RBP42 in T. cruzi, we can begin to dissect its contribution to environmental signals encountered by the parasite during its life cycle.
Supplementary Material
Acknowledgments
We thank David Engman, Cheryl Olson, Silvia Moreno and Melina Galizzi for their patience and advice in helping us set up the T. cruzi system in our laboratory. We are indebted to Eliseo Eugenin for help with the confocal microscopy.
References
- Aibara S, Valkov E, Lamers M, Stewart M. Domain organization within the nuclear export factor Mex67:Mtr2 generates an extended mRNA binding surface. Nucleic Acids Res. 2015;43(3):1927–36. doi: 10.1093/nar/gkv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo PR, Teixeira SM. Regulatory elements involved in the post-transcriptional control of stage-specific gene expression in Trypanosoma cruzi : a review. Mem Inst Oswaldo Cruz. 2011;106(3):257–66. doi: 10.1590/s0074-02762011000300002. [DOI] [PubMed] [Google Scholar]
- Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS pathogens. 2009;5(8):e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77(2):235–9. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bern C. Chagas' Disease. N Engl J Med. 2015;373(5):456–66. doi: 10.1056/NEJMra1410150. [DOI] [PubMed] [Google Scholar]
- Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney KM. Chagas disease in the 21st century: a public health success or an emerging threat? Parasite. 2014;21:11. doi: 10.1051/parasite/2014012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41(Web Server issue):W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo EP. Growth and Differentiation in Trypanosoma Cruzi IOrigin of Metacyclic Trypanosomes in Liquid Media. Rev Inst Med Trop Sao Paulo. 1964;6:93–100. [PubMed] [Google Scholar]
- DaRocha WD, Silva RA, Bartholomeu DC, Pires SF, Freitas JM, Macedo AM, Vazquez MP, Levin MJ, Teixeira SM. Expression of exogenous genes in Trypanosoma cruzi : improving vectors and electroporation protocols. Parasitol Res. 2004;92(2):113–20. doi: 10.1007/s00436-003-1004-5. [DOI] [PubMed] [Google Scholar]
- Das A, Bellofatto V, Rosenfeld J, Carrington M, Romero-Zaliz R, del Val C, Estévez AM. High throughput sequencing analysis of Trypanosoma brucei DRBD3/PTB1-bound mRNAs. Mol Biochem Parasitol. 2015;199(1–2):1–4. doi: 10.1016/j.molbiopara.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, Estevez AM, Bellofatto V. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA. 2012;18(11):1968–83. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389–94. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–9. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Gazestani VH, Yip CW, Nikpour N, Berghuis N, Salavati R. TrypsNetDB: An integrated framework for the functional characterization of trypanosomatid proteins. PLoS Negl Trop Dis. 2017;11(2):e0005368. doi: 10.1371/journal.pntd.0005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33(2):155–61. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Jimenez V. Dealing with environmental challenges: mechanisms of adaptation in Trypanosoma cruzi. Res Microbiol. 2014;165(3):155–65. doi: 10.1016/j.resmic.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ullu E, Tschudi C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell Microbiol. 2014;16(4):482–9. doi: 10.1111/cmi.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander N, Li ZH, Niyogi S, Docampo R. CRISPR/Cas9-Induced Disruption of Paraflagellar Rod Protein 1 and 2 Genes in Trypanosoma cruzi Reveals Their Role in Flagellar Attachment. MBio. 2015;6(4):e01012. doi: 10.1128/mBio.01012-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini FK, de Godoy LM, Rampazzo RC, Pavoni DP, Probst CM, Gnad F, Mann M, Krieger MA. Profiling the Trypanosoma cruzi phosphoproteome. PLoS One. 2011;6(9):e25381. doi: 10.1371/journal.pone.0025381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett IR, Martin DM, Miranda-Saavedra D, Lamont D, Barber JD, Mehlert A, Ferguson MA. The phosphoproteome of bloodstream form Trypanosoma brucei causative agent of African sleeping sickness. Mol Cell Proteomics. 2009;8(7):1527–38. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K, Nicholas H, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14. [Google Scholar]
- Rassi A, Jr, Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375(9723):1388–402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- Romaniuk MA, Cervini G, Cassola A. Regulation of RNA binding proteins in trypanosomatid protozoan parasites. World J Biol Chem. 2016;7(1):146–57. doi: 10.4331/wjbc.v7.i1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Yang J, Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012;40(Web Server issue):W471–7. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli RR, Augusto Lda S, Castilho BA, Schenkman S. Protein synthesis attenuation by phosphorylation of eIF2alpha is required for the differentiation of Trypanosoma cruzi into infective forms. PLoS One. 2011;6(11):e27904. doi: 10.1371/journal.pone.0027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S, Rijo-Ferreira F, Carvalho T, Pinto-Neves D, Guegan F, Aresta-Branco F, Bento F, Young SA, Pinto A, Van Den Abbeele J, Ribeiro RM, Dias S, Smith TK, Figueiredo LM. Trypanosoma brucei Parasites Occupy and Functionally Adapt to the Adipose Tissue in Mice. Cell Host Microbe. 2016;19(6):837–48. doi: 10.1016/j.chom.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DM, Ferguson MA. Global Quantitative SILAC Phosphoproteomics Reveals Differential Phosphorylation Is Widespread between the Procyclic and Bloodstream Form Lifecycle Stages of Trypanosoma brucei. Journal of proteome research. 2013;12(5):2233–44. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez MP, Levin MJ. Functional analysis of the intergenic regions of TcP2beta gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene. 1999;239(2):217–25. doi: 10.1016/s0378-1119(99)00386-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;(77 Suppl 9):100–13. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.