Abstract
Recently, we and others have found that hyperfiltration-associated increase in biomechanical forces, namely, tensile stress and fluid flow shear stress (FFSS), can directly and distinctly alter podocyte structure and function. The ultrafiltrate flow over the major processes and cell body generates FFSS to podocytes. Our previous work suggests that the cyclooxygenase-2 (COX-2)-PGE2-PGE2 receptor 2 (EP2) axis plays an important role in mechanoperception of FFSS in podocytes. To address mechanotransduction of the perceived stimulus through EP2, cultured podocytes were exposed to FFSS (2 dyn/cm2) for 2 h. Total RNA from cells at the end of FFSS treatment, 2-h post-FFSS, and 24-h post-FFSS was used for whole exon array analysis. Differentially regulated genes (P < 0.01) were analyzed using bioinformatics tools Enrichr and Ingenuity Pathway Analysis to predict pathways/molecules. Candidate pathways were validated using Western blot analysis and then further confirmed to be resulting from a direct effect of PGE2 on podocytes. Results show that FFSS-induced mechanotransduction as well as exogenous PGE2 activate the Akt-GSK3β-β-catenin (Ser552) and MAPK/ERK but not the cAMP-PKA signal transduction cascades. These pathways are reportedly associated with FFSS-induced and EP2-mediated signaling in other epithelial cells as well. The current regimen for treating hyperfiltration-mediated injury largely depends on targeting the renin-angiotensin-aldosterone system. The present study identifies specific transduction mechanisms and provides novel information on the direct effect of FFSS on podocytes. These results suggest that targeting EP2-mediated signaling pathways holds therapeutic significance for delaying progression of chronic kidney disease secondary to hyperfiltration.
Keywords: exon array analysis, fluid flow shear stress, hyperfiltration, mechanotransduction, podocytes
INTRODUCTION
Normal glomerular filtration rate is important for maintaining homeostasis. A decrease in functional nephron mass results in adaptive changes in glomerular hemodynamics termed “hyperfiltration.” Glomerular hyperfiltration begins even before metabolic changes and clinical signs of chronic kidney disease (CKD) start to manifest. Hyperfiltration involves increased renal blood flow, glomerular capillary pressure (PGC), single-nephron glomerular filtration rate (SNGFR), filtration fraction, and decreased hydraulic conductivity associated with glomerular hypertrophy (6, 7). Traditionally, hyperfiltration-mediated injury has been studied using analysis of these hemodynamic parameters. However, several aspects of hyperfiltration remain poorly explained. These include the changes in podocyte structure/function and consequential loss of glomerular filtration barrier resulting in albuminuria. Therefore we and others have started reconsidering hyperfiltration in terms of biomechanical forces within the glomerulus (30, 31, 48, 54). We believe that investigating the effect of biomechanical forces on podocytes within the glomerulus may provide a better understanding of hyperfiltration-mediated kidney injury.
Localized in Bowman’s space, podocytes are exposed to mechanical forces, namely, tensile stress and fluid flow shear stress (FFSS; 16, 19, 51, 52). Glomerular capillary pressure (PGC) in the vascular compartment stretches the podocyte foot processes. This outwardly directed force working perpendicular to the direction of blood flow in the capillary generates tensile stress on the basolateral aspect of the podocytes that tightly cover the capillary on the outside through foot processes (16). Additionally, the flow of glomerular ultrafiltrate through Bowman’s space creates shear stress on the surface of the podocyte cell body. FFSS is believed to be largely exerted over the major processes and the soma of podocytes (19, 52). Thus PGC and SNGFR become the principal determinants of tensile stress and FFSS, respectively (15, 42, 54).
Recent work showed a 1.5- to 2.0-fold increase in the calculated FFSS over podocytes in unilaterally nephrectomized mice or rats (51). FFSS applied to podocytes in vitro resulted in an altered actin cytoskeleton, upregulation of cyclooxygenase-2 (COX-2), and increased secretion of prostaglandin E2 (PGE2; 52). We found that weak basal expression of prostanoid receptor PGE2 receptor 2 (EP2) in podocytes was upregulated by FFSS (53). Subsequent studies both in vitro and in vivo showed that FFSS resulted in increased expression of prostanoid receptor EP2 but not EP4 (50). Our previous work indicated that the COX-2-PGE2-EP2 axis plays a key role in mechanoperception, i.e., how podocytes perceive the force. The next logical step in this pursuit addresses mechanotransduction, i.e., how the podocyte converts mechanical stimulus into biochemical changes that elicit specific cellular responses.
Since PGE2 receptors are known to activate diverse signaling pathways, it is likely that FFSS affects a large number of proteins through the COX-2-PGE2-EP2 axis. Of the four PGE2 receptors, only EP2 activation results in nuclear translocation of activated β-catenin through five known intermediate mechanisms (Fig. 1). EP2 is a G protein-coupled receptor that activates the heterotrimeric (αβγ) Gs protein leading to 1) release of Gβγ complex, which activates phosphatidylinositol 3-kinase (PI3K)/Akt, which phosphorylates and inactivates GSK3β (49, 56), 2) binding of the Gα complex to axin and release of β-catenin from the axin-β-catenin-GSK3β complex, 3) generation of cAMP and activation of PKA, which activate β-catenin (23, 33, 57), 4) recruitment of β-arrestin-1 and phosphorylation of Src-kinase, which transactivates the EGF receptor (EGFR) signaling network of the PI3K/Akt, hepatocyte growth factor (HGF)/tyrosine-protein kinase Met (c-Met), and Ras/ERK pathways (10, 11, 41), and 5) activation of PI3K/Akt leading to activation of ERK (43). Studies suggest that phosphorylation at Ser552 or Ser675 induces β-catenin translocation to the nucleus with Akt preferentially phosphorylating β-catenin at Ser552 while PKA preferentially phosphorylates β-catenin at Ser675 (17, 21, 23, 55). A contemporaneous comprehensive analysis of several pathways appeared a suitable approach. Therefore, we planned to determine the effect of FFSS on genome-wide expression using exon array analysis with a secondary goal to explore the predicted signaling molecules and pathways and their intersection with known EP2-mediated signaling. To predict the signaling pathways, networks, and molecules, we used two bioinformatics tools, Enrichr and Ingenuity Pathway Analysis. Pathways activated by EP2 and those activated in other epithelial cells by FFSS were selected for further validation. Briefly, our results show that mechanotransduction in podocytes involves Akt-GSK3β-β-catenin (Ser552) and possibly MAPK/ERK signaling but not cAMP-PKA.
Fig. 1.
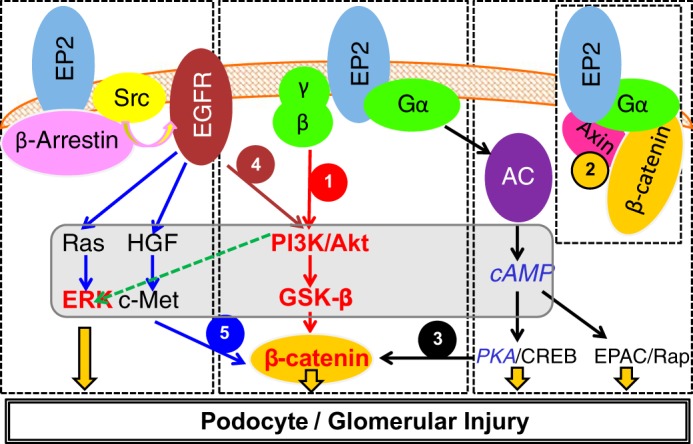
A compiled summary of signaling pathways activated by prostanoid receptor PGE2 receptor 2 (EP2) in osteocytes, neurodegenerative diseases, and colon cancer (see text for details). EP2 is a G protein-coupled receptor that activates the heterotrimeric Gs (αβγ) protein. β-Catenin can be recruited by EP2-mediated signaling by 1) phosphatidylinositol 3-kinase (PI3K)/Akt/GSK3β, which preferentially phosphorylates β-catenin at Ser552, 2) binding of the Gα complex to axin and release of β-catenin from the axin-β-catenin-GSK3β complex, 3) generation of cAMP by adenylate cyclase (AC) and activation of PKA, which preferentially phosphorylates β-catenin at Ser675, 4) recruitment by EP2 of β-arrestin 1 and phosphorylation of Src-kinase, which then transactivates the EGF receptor (EGFR) signaling network of PI3K/Akt, and 5) the hepatocyte growth factor (HGF)/tyrosine-protein kinase Met (c-Met) pathway. The study found Akt-GSK3β-β-catenin and ERK (shown in red and bold font), but not cAMP-PKA (shown in blue and italic font), to be important in mechanotransduction. CREB, cAMP-response element-binding protein; EPAC, exchange protein directly activated by cAMP; Rap, Rap GTP-binding protein.
MATERIALS AND METHODS
Cell Culture
These studies were performed using protocols approved by the Institutional Animal Care and Use Committee, Safety Subcommittee, and the R&D Committee at the Veterans Affairs Medical Center, Kansas City, MO. Conditionally immortalized female mouse podocyte line (kind gift from Dr. Peter Mundel) with thermosensitive tsA58 mutant T-antigen was used in these studies (47). Podocytes were propagated in RPMI 1640 with l-glutamine supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen, Carlsbad, CA) under permissive conditions with 10 U/ml of γ-interferon (Cell Sciences, Norwood, MA) and incubation at 33°C. Cells were transferred to nonpermissive conditions (37°C without γ-interferon) to induce differentiation. Three glass slides (25 × 75 × 1 mm; Fisher Scientific, Pittsburgh, PA) were maintained in one culture dish containing 12 ml of culture medium. The medium was changed 24 h before the application of FFSS. Differentiated podocytes were used for FFSS experiments on day 14.
Fluid Flow Shear Stress Application
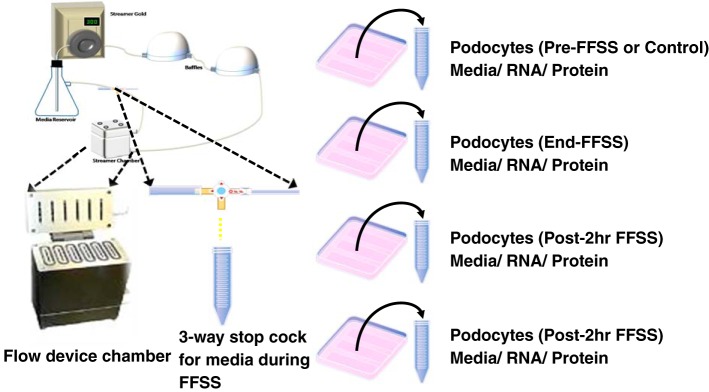
Fluid flow shear stress was applied to differentiated podocytes using a Flexcell Streamer Gold apparatus (Flexcell International, Hillsborough, NC) as described earlier (50). FFSS was applied at 2 dyn/cm2 (or 75 ml/min) for 2 h at 37°C with 5% CO2. Podocytes on a set of slides (untreated control group) were placed in the same incubator but were not exposed to FFSS. A three-way stopcock was attached to collect 1.5-ml aliquots of the medium at 0, 30, and 120 min during application of FFSS (Fig. 2). Collected medium was used for cAMP assay. Following FFSS treatment, podocytes on slides were returned to the original medium for recovery for up to 24 h in the incubator at 37°C under 5% CO2 humidified atmosphere. Samples obtained before exposing cells to FFSS, at the end of FFSS treatment, and at 2 and 24 h following FFSS treatment were termed Pre-FFSS (or Control), End-FFSS, Post-2 h FFSS, and Post-24 h FFSS, respectively. Pre-FFSS (Control), End-FFSS, and Post-2 h/24 h FFSS cells were harvested for analysis (Fig. 2). We chose post-2 h FFSS and post-24 h FFSS time points as a reflection of early and late changes based on published studies on podocytes and osteocytes (11, 50, 53, 61). In addition, podocytes were treated with PGE2 without application of FFSS and harvested at 0, 2, 5, 15, 60, and 120 min.
Fig. 2.
Left: outline of the apparatus used for applying fluid flow shear stress (FFSS) to podocytes. The “fluid chamber” contains cells grown on six glass slides placed for FFSS studies, and a three-way stopcock is attached to collect media during application of FFSS. Right: experiment design for collecting supernatant culture medium, cells for RNA, and protein extraction at four different experimental time points (Pre-FFSS or Control, End-FFSS, Post-2 h FFSS, and Post-24 h FFSS).
Affymetrix GeneChip Mouse Exon 1.0 ST Array
Control and FFSS-treated podocytes were processed using the TRIzol/Phaselock protocol. Total RNA fraction was purified by RNA Clean-Up protocol using an RNeasy mini kit (no. 74104; Qiagen). RNA bound to the RNeasy column was treated with RNase-free DNase (no. 79254, Qiagen), eluted and quantitated using a Nanodrop-1000 spectrophotometer. Aliquots were analyzed on an Agilent 2100 Bioanalyzer using the RNA6000 LabChip Nano kit version II for quality assessment and quantification of RNA. The Affymetrix GeneChip Mouse Exon 1.0 ST Array (Thermo Fisher Scientific) with over 4.5 million unique 25-mer oligonucleotides constituting ~1.2 million probe sets, with ~4 probes per exon and ~40 probes per gene, was used. For gene-level expression analysis, multiple probes on different exons were summarized into a single gene-level expression data point. Two performance controls were incorporated into each array run: 1) target amplification and labeling control were incorporated by adding polyadenylated transcript controls from Bacillus subtilis using a Poly-A Control kit (no. 900433; Affymetrix) and 2) GeneChip hybridization was controlled by the addition 3′ Amplification Reagent Hybridization Controls (no. 900454; Affymetrix). Data were normalized using the Robust Multichip Average algorithm, which consisted of four steps: a background adjustment, quantile normalization, log 2 transformation, and final summarization. The data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE99988. Array data were analyzed using Partek Genomics Suite (http://www.partek.com/pgs). Two-way ANOVA with group and replicate effects were fitted to the model, and the confounding effects of replication were adjusted by the ANOVA model.
Kinases, Transcription Factors, and Protein-Protein Interaction Hubs Predicted Using Enrichr
Genes showing significant changes in expression compared with control (P < 0.01) were selected for input into Enrichr (http://amp.pharm.mssm.edu/Enrichr) to predict kinases, transcription factors, and protein-protein interaction hubs at P < 0.05 (8). The Kinase Enrichment Analysis (KEA) gene set library in Enrichr contains kinases and their known substrates as available in the Human Protein Reference Database (HPRD), PhosphoSite, PhosphoPOINT, Phospho.ELM, NetworKIN, and Molecular Interaction (MINT) databases. ChIP-x Enrichment Analysis (ChEA) is a gene set library in Enrichr that contains putative targets for transcription factors, based on reports profiling transcription factors binding to DNA in mammalian cells. The Protein-Protein Interaction Hubs gene set library is made from a human protein-protein interaction network.
Results for the group comparisons between End-, Post-2 h, or Post-24 h FFSS and the control groups were further investigated to identify the common kinases, transcription factors, and protein-protein hubs that were present across all three group comparisons. Biological significance of the results was then cross-referenced with published literature before laboratory validation.
Functional Annotation and Pathway and Network Analyses Using Ingenuity Pathway Analysis
To identify overrepresented canonical pathways, processes, and networks, the lists of differentially expressed genes (P < 0.01) within the three comparison sets (End-FFSS vs. Control; Post-2 h FFSS vs. Control; and Post-24 h FFSS vs. Control) were submitted for Ingenuity Pathway Analysis (IPA; Qiagen, Germantown, MD). Core analysis was performed by comparing the significant genes with the knowledge base within IPA, which comprises curated pathways within the program. IPA predicted significant biological functions and pathways (P < 0.05, Fisher’s exact test) affected in podocytes following exposure to FFSS. In addition, IPA was used to predict both upstream biological regulators and downstream effects on cellular and organismal biology. The settings for core analyses were as follows: Ingenuity Knowledge Base; “endogenous chemicals” included; “direct” and “indirect” relationships; molecules per pathway, 35; and networks per analysis, 25.
Enzyme Immunoassay to Determine Changes in Secreted and Intracellular cAMP
In a set of experiments, podocytes were incubated with isobutylmethylxanthine (IBMX; 0.5 µM, 15 min) to inhibit phosphodiesterase activity before applying FFSS. Levels of cAMP in media were measured using a cAMP EIA kit (no. 581001; Cayman Chemical, Ann Arbor, MI), collected at 0, 30, and 120 min, post-2 h, and post-24 h, and cell lysate was collected at end, post-2 h, and post-24 h following the manufacturer’s instructions.
Western Blotting to Determine Protein Phosphorylation of Kinases in Podocytes
Podocytes were lysed with radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors following application of FFSS. Total protein was determined using a DC Protein Assay Kit (Bio-Rad, Hercules, CA). Western blotting was performed as described previously (50, 53). Briefly, proteins were denatured in the sample buffer containing β-mercaptoethanol at 94°C for 5 min. Total protein (10 µg/lane) was electrophoresed by SDS-PAGE using a 10% Tris-glycine gel. Proteins were transferred to a polyvinylidene difluoride membrane, washed with PBST (0.1% Tween 20), and blocked using 5% nonfat milk powder. Antibodies to the following proteins were used at the dilutions indicated: Akt (cs-2938 at 1:1,000), phospho-Akt (Ser473) (cs-4060 at 1:2,000), GSK3β (cs-9315 at 1:1,000), phospho-GSK3β (Ser9) (cs-9336 at 1:1,000), β-catenin (cs-8480 at 1:1,000), phospho-β-catenin (Ser675, cs-4176; and Ser552, cs-5651; at 1:1,000), ERK1/2 (cs-4695 at 1:1,000), phospho-ERK 1/2 (Thr202/Tyr204) (cs-4370 at 1:2,000), p38 MAPK (cs-9212 at 1:1,000), phospho-p38 MAPK (Thr180/Tyr182) (cs-9215 at 1:1,000), PKA (cs-4782 1:1,000), and phospho-PKA (Thr197) (cs-4781 at 1:2,000) from Cell Signaling Technology, Danvers, MA, and β-actin (A5441 at 1:50,000; Sigma). After washing with PBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody to each primary antibody. Chemiluminescence (Pierce, Rockford, IL) reagent was used for detection on X-ray film. Developed X-ray films were imaged and analyzed using FluorChem using built-in AlphaEaseFC software (Alpha Innotech, San Leandro, CA). Samples from n = 4–6 replicates for each experimental treatment were analyzed.
Statistics
In addition to statistical software built within the bioinformatic tools Partek, Enrichr, and IPA, we used SPSS 23 statistical software for multiple group comparisons. The independent-samples Kruskal-Wallis test was used to compare cAMP levels and phosphorylated protein-to-total protein ratios on Western blots across multiple groups. A P value <0.05 was considered significant.
RESULTS
Exon Array Analysis Shows Altered Gene Expression in Podocytes Following Application of FFSS
The RNA integrity numbers for Control, End-FFSS, Post-2 h FFSS, and Post-24 h FFSS RNA samples were 9.1 ± 0.2, 9.4 ± 0.1, 9.2 ± 0.2, and 9.4 ± 0.1 (means ± SD), respectively. Two performance controls were incorporated into each array run: 1) polyadenylated transcript from B. subtilis (Poly-A Control kit, no. 900433; Affymetrix) as target amplification and biotin-labeling control and 2) 3′-Amplification Reagent (Hybridization Controls, no. 900454; Affymetrix) as a control for GeneChip hybridization. Hybridization controls were composed of biotin-labeled Escherichia coli transcripts (bioB, bioC, bioD, and cre), which were spiked at increasing concentrations into the hybridization cocktail. The results of quality controls are not shown here but are available at http://bioinformatics5.kumc.edu/mdms/login.php.
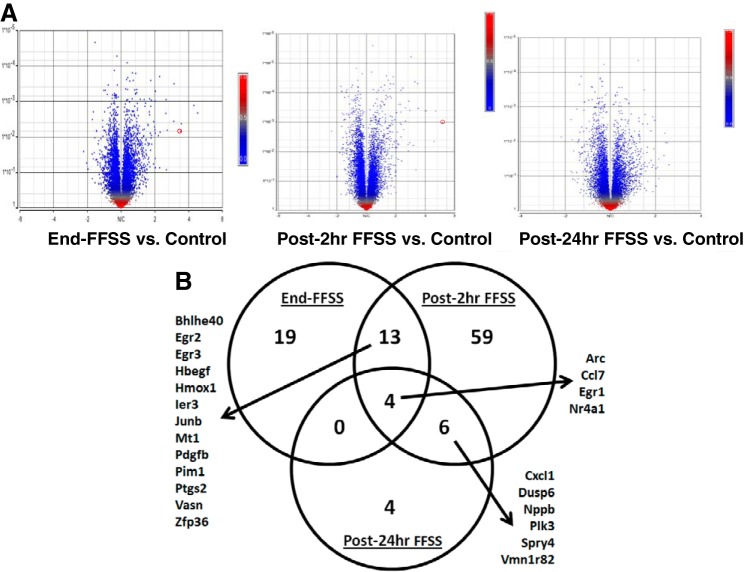
We examined raw data output containing 23,292 genes to determine the patterns of all gene expression in experimental groups using principal component analysis (PCA). The first three principal components summarized 58.9% of variation, where the number of the principal components was determined by scree plot. Our analysis adjusted for batch effects. Within each replicate (batch), Control, End-FFSS, Post-2 h FFSS, and Post-24 h FFSS were well separated in the first three principal components. The sample density histogram indicated little variation among treatment groups for gene expression intensity <3. Thus genes with mean expression intensity <3 and genes with no RefSeq ID were eliminated, and 17,494 genes remained in the study. The numbers of genes that were differentially regulated based on different breakdowns for fold change and P value are shown in Table 1. The Volcano plots show the distribution of all genes across fold change and P values (Fig. 3A). The genes that were differentially expressed at P < 0.05 and fold change >1.75 are shown in Fig. 3B and Table 2. The four genes common to all three experimental groups were activity-regulated cytoskeleton-associated protein (Arc), early growth response protein-1 (Egr1), C-C motif chemokine ligand 7 (Ccl7), and nuclear receptor subfamily 4 group A member 1 (Nr4a1).
Table 1.
Number of genes that met the statistical criteria using different cutoffs on Partek analysis
| P < 0.05 | P < 0.01 | P < 0.001 | P < 0.0001 | P < 0.05 and FC > 1.5 | P < 0.05 and FC > 1.75 | P < 0.05 and FC > 2 | P < 0.01 and FC > 1.5 | P < 0.01 and FC > 1.75 | P < 0.01 and FC > 2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| End-FFSS vs. Control | 1,276 | 320 | 48 | 3 | 88 | 38 | 17 | 40 | 28 | 15 |
| Post-2 h FFSS vs. Control | 2,444 | 895 | 175 | 32 | 196 | 83 | 41 | 137 | 64 | 35 |
| Post-24 h FFSS vs. Control | 1,208 | 308 | 49 | 2 | 72 | 20 | 7 | 29 | 8 | 2 |
Number of genes in each experimental group (End-FFSS, Post-2 h FFSS, and Post-24 h FFSS) that were different from the control group at P < 0.05, P < 0.01, P < 0.001, or P < 0.0001 is shown. Numbers of genes with fold changes (FC) >1.5, >1.75, and >2 at P < 0.05 and <0.01 are also shown. FFSS, fluid flow shear stress.
Fig. 3.
A: gene expression analysis presented as volcano plots shows the distribution of all genes across fold change and P values. Red circle denotes cyclooxygenase-2 (COX-2; Ptgs2) gene on the volcano plot. B: Venn diagram showing common genes between the three groups (End-FFSS vs. Control, Post-2 h FFSS vs. Control, and Post-24 h FFSS vs. Control) that were differentially expressed at P < 0.05 and fold change >1.75. FFSS, fluid flow shear stress.
Table 2.
Genes that were differentially expressed in podocytes following application of fluid flow shear stress in all experimental groups
| End-FFSS | Post-2 h FFSS | Post-24 h FFSS |
|---|---|---|
| Adm, Arc, Bhlhe40, Bmf, Btg2, Ccl7, Clec2d, Dusp2, Egr1, Egr2, Egr3, Fbxo32, Fos, Gadd45b, Gadd45g, Gpbar1, Hbegf, Hmox1, Ier2, Ier3, Junb, Krtap4-7, Mafb, Mt1, Nr4a1, Olfr1006, Olfr1511, Pdgfb, Pim1, Ptgs2, Scand3, Slc40a1, Snai1, Vasn, Vmn1r188, Zfp36 | 1810011O10Rik, Ahr, Angptl4, Arc, Atoh8, Bhlhe40, Ccl2, Ccl20, Ccl7, Celf2, Cxcl1, Cxcl10, Cxcl11, Dusp4, Dusp6, Egr1, Egr2, Egr3, Emp1, Fgf7, Fgfr1, Fosl1, Gjb3, Gjb4, Gm13154, Gm4952, Gpr146, Hbegf, Hk2, Hmga2, Hmox1, Hs3st1, Ier3, Il1rl1, Junb, Kcnk12, Klf5, Klhdc7a, Klk1, Lce1a2, Lce3b, Lgals3, Lif, Lrp8, Maff, Mt1, Mt2, Myc, Nfatc1, Nppb, Nqo1, Nr4a1, Nr4a2, Olfr1163, Olfr733, Pdgfb, Pim1, Plaur, Plk3, Pmepa1, Ppargc1a, Ptger4, Ptgs2, Runx1, Serpine1, Sesn1, Sgk1, Slc20a1, Slc7a11, Smad9, Spry4, Spsb1, Sult1c2, Tlr2, Tnfaip3, Tnfrsf12a, Tnfsf15, Uap1, Vasn, Vmn1r82, Zfp36 | Arc, Ccl7, Ciita, Cxcl1, Dusp6, Egr1, Gm4841, Nppb, Nr4a1, Olfr1197, Plk3, Spry4, Tnfsf10, Vmn1r82 |
The genes listed were differentially expressed at P < 0.05 and fold change >1.75 in podocytes following application of fluid flow shear stress (FFSS) in all experimental groups (End-FFSS vs. Control, Post-2 h FFSS vs. Control, and Post-24 h FFSS vs. Control).
Kinases, Transcription Factors, and Protein-Protein Interaction Hubs Predicted Using Enrichr in FFSS-Treated Podocytes
Control vs. experimental groups.
The differentially expressed genes from Control (P < 0.01) that were uploaded to Enrichr are available in Supplemental Material, Supplemental File A (supplemental material for this article is available online at the AJP-Renal Physiology website). The Kinase Enrichment Analysis (KEA) predicted 22, 28 and 7 kinases between Control and End-, Post-2 h, and Post-24 h FFSS (P < 0.05), respectively (Supplemental File B). The ChIP-x Enrichment Analysis (ChEA) predicted 189, 212, and 163 transcription factors between Control and End-, Post-2 h, and Post-24 h FFSS (P < 0.05), respectively (Supplemental File B). Protein-Protein Interaction Hubs (PPI Hubs) predicted 76, 131, and 72 hub proteins between Control and End-, Post-2 h, and Post-24 h FFSS (P < 0.05), respectively (Supplemental File B).
Molecules/pathways detected in all experimental groups.
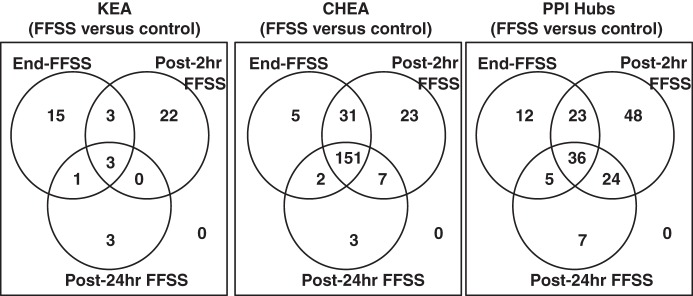
KEA, ChEA, and PPI were applied to identify similarities between End-, Post-2 h, and Post-24 h FFSS groups. Three kinases, 151 transcription factors, and 36 hub proteins were identified common to 3 experimental groups as shown in Table 3, and the overlap between the 3 experimental groups for KEA, ChEA, and PPI Hubs analysis is shown as a Venn diagram (Fig. 4). The KEA predicted GSK3β and ERK as potential kinases. Akt1 on PPI Hubs is upstream of GSK3β. The upstream and downstream molecules associated with MAPK/ERK on PPI Hubs included Raf1, mothers against decapentaplegic homolog 3 (SMAD3), and SMAD4. The activator protein-1 (AP-1) complex of c-Fos and c-Jun found on PPI Hubs is known to be regulated by ERK and GSK3β. EGFR is associated with Akt, MAPK, and JNK.
Table 3.
Predicted kinases on KEA, transcription factors on ChEA, and hub proteins on PPI Hubs on Enrichr analysis of gene sets that were differentially expressed in podocytes following application of fluid flow shear stress
| Predicted Kinases | Predicted Transcription Factors | Predicted Hub Proteins |
|---|---|---|
| GSK3β, MAPK1, MAPK3 | AR-19668381 (human), AR-20517297 (human), CCND1-20090754 (mouse), CEBPB-20176806 (mouse), CHD1-19587682 (mouse), CLOCK-20551151 (human), CNOT3-19339689 (mouse), CREB1-20920259 (mouse), CREM-20920259 (mouse), CTCF-18555785 (mouse), CTNNB1-20460455 (human), CUX1-19635798 (human), DACH1-20351289 (human), DNAJC2-21179169 (human), E2F1-17053090 (human), E2F1-18555785 (mouse), E2F1-21310950 (human), E2F4-17652178 (human), E2F4-21247883 (human), EGR1-20690147 (human), ELF1-20517297 (human), ELK1-19687146 (human), EOMES-20176728 (mouse), EOMES-21245162 (human), EP300-20729851 (mouse), EP300-21415370 (mouse), ERG-20887958 (mouse), ESR1-17901129 (mouse), ESR1-21235772 (human), ESRRB-18555785 (mouse), EST1-17652178 (human), ETS1-20019798 (human), FLI1-20887958 (mouse), FLI1-21571218 (human), FOXA2-19822575 (human), FOXP2-21765815 (mouse), FOXP3-21729870 (human), GABP-19822575 (human), GATA1-19941827 (mouse), GATA1-21571218 (human), GATA2-19941826 (human), GATA2-20887958 (mouse), GATA2-21571218 (human), GATA2-21666600 (human), GATA3-20176728 (mouse), GATA4-21415370 (mouse), GFI1B-20887958 (mouse), HNF4A-19761587 (human), HNF4A-19822575 (human), HOXB4-20404135 (mouse), JARID1A-20064375 (mouse), JUN-21703547 (human), KDM5B-21448134 (mouse), KLF1-20508144 (mouse), KLF4-18358816 (mouse), KLF4-18555785 (mouse), KLF4-19030024 (mouse), LMO2-20887958 (mouse), MEF2A-21415370 (mouse), MEIS1-20887958 (mouse), MITF-21258399 (human), MTF2-20144788 (mouse), MYB-21317192 (mouse), MYC-18358816 (mouse), MYC-19030024 (mouse), MYC-19079543 (mouse), MYC-19915707 (human), MYC-20876797 (human), MYCN-18555785 (mouse), NACC1-18358816 (mouse), NANOG-16153702 (human), NANOG-16518401 (mouse), NANOG-18347094 (mouse), NANOG-18358816 (mouse), NANOG-18555785 (mouse), NANOG-18692474 (mouse), NANOG-21062744 (human), NFE2L2-20460467 (mouse), NR0B1-18358816 (mouse), NR1I2-20693526 (mouse), NRF2-20460467 (mouse), PADI4-21655091 (human), PAX3-FKHR-20663909 (human), PDX1-19855005 (mouse), PHF8-20622854 (human), POU5F1-16518401 (mouse), POU5F1-18347094 (mouse), POU5F1-18358816 (mouse), POU5F1-18692474 (mouse), PPARD-21283829 (human), PPARG-20176806 (mouse), PPARG-20887899 (mouse), PRDM14-21183938 (mouse), RAD21-21589869 (mouse), RCOR3-21632747 (mouse), REST-18959480 (mouse), REST-19997604 (mouse), REST-21632747 (mouse), RNF2-16625203 (mouse), RUNX1-20887958 (mouse), RUNX1-21571218 (human), SALL4-18804426 (mouse), SALL4-18804426_ESC (mouse), SCL-21571218 (human), SETDB1-19884255 (mouse), SETDB1-19884257 (mouse), SFPI1-20887958 (mouse), SIN3A-21632747 (mouse), SIN3B-21632747 (mouse), SMAD-19615063 (human), SMAD2-18955504 (human), SMAD3-18955504 (human), SMAD3-21741376 (human), SMAD4-21741376 (human), SMARCA4-20176728 (mouse), SOX17-20123909 (mouse), SOX2-16153702 (human), SOX2-18358816 (mouse), SOX2-18692474 (mouse), SOX2-20726797 (human), SOX2-21211035 (human), SPI1-20176806 (mouse), SRF-21415370 (mouse), STAT1-20625510 (human), STAT4-19710469 (mouse), SUZ12-18692474 (mouse), SUZ12-18974828 (mouse), SUZ12-20075857 (mouse), TAL1-20566737 (mouse), TAL1-20887958 (mouse), TBX3-20139965 (mouse), TBX5-21415370 (mouse), TCF3-18347094 (mouse), TCF3-18467660 (mouse), TCF3-18692474 (mouse), TCF4-18268006 (human), TCFAP2C-20176728 (mouse), TCFCP2L1-18555785 (mouse), TET1-21451524 (mouse), TFAP2C-20629094 (human), TFEB-21752829 (human), THAP11-20581084 (mouse), TRIM28-19339689 (mouse), TRP63-18441228 (mouse), WT1-20215353 (mouse), YAP1-20516196 (mouse), YY1-21170310 (mouse), ZFP281-18358816 (mouse), ZFP281-18757296 (mouse), ZFP42-18358816 (mouse), ZFX-18555785 (mouse) | Akt1, ATXN1, CDK1, CDK2, CREBBP, EGFR, EP300, FOS, GSK3B, HDAC1, HSP90AA1, IKBKB, JUN, MAPK1, MAPK3, MAPK8, MYC, NR3C1, PPP2CA, PRKCA, PRKCB, PRKDC, RAC1, RAF1, RELA, RPS6KA3, SFN, SMAD2, SMAD3, SMAD4, STAT1, STAT3, SUMO1, TP53, TRAF6, UBC |
Enrichr analysis of genes sets that were differentially expressed (P < 0.01) in podocytes following application of fluid flow shear stress. KEA, Kinase Enrichment Analysis; ChEA, ChIP-x Enrichment Analysis; PPI Hubs, Protein-Protein Interaction Hubs. Transcription factors: AR, androgen receptor; CCND1, cyclin-D1; CEBPB, CCAAT/enhancer-binding protein-β; CHD1, chromodomain-helicase-DNA-binding protein-1; CLOCK, basic helix-loop-helix-PAS transcription factor; CNOT3, C-C chemokine receptor type 4 (CCR4)-NOT transcription complex subunit 3; CREB1, cAMP-response element-binding protein-1; CREM, cAMP-response element modulator; CTCF, CCCTC-binding factor; CTNNB1, β-catenin; CUX1, cut-like homeobox 1; DACH1, Dachshund homolog 1; DNAJC2, dnaJ homolog subfamily C member 2; E2F, E2 factor; EGR1, early growth response protein-1; ELF1, E74-like factor 1; ELK1, erythroblast transformation-specific (ETS) domain-containing protein-1; EOMES, eomesodermin; EP300, E1A-associated protein p300; ERG, ETS-related gene; ESR1, estrogen receptor 1; ESRRB, estrogen-related receptor-β; EST1, telomerase-activating protein-1; ETS1, E26 transformation-specific sequence-1; FLI1, friend leukemia integration 1; FOX, forkhead box protein; GABP, GA-binding protein; GATA1-4, GATA-binding factor 1–4; GFI1B, zinc finger protein Gfi-1b; HNF4A, hepatocyte nuclear factor 4α; HOXB4, homeobox protein Hox-B4; JARID1A, lysine-specific demethylase 5A; KDM5B, lysine-specific demethylase 5B; KLF, Krueppel-like factor; LMO2, LIM domain only 2; MEF2A, myocyte-specific enhancer factor 2A; MEIS1, homeobox protein Meis1; MITF, microphthalmia-associated transcription factor; MTF2, metal-response element-binding transcription factor 2; MYB, Myb proto-oncogene protein; MYC, Myc proto-oncogene protein; MYCN, N-myc proto-oncogene protein; NACC1, nucleus accumbens-associated protein-1; NANOG, homeobox protein Nanog; NFE2L2, nuclear factor (erythroid-derived 2)-like 2; NR0B1, nuclear receptor subfamily 0 group B member 1; NR1I2, nuclear receptor subfamily 1 group I member 2; NRF2, nuclear factor E2-related factor 2; PADI4, protein-arginine deiminase type 4; PAX3-FKHR, paired box protein Pax-3-FKHR protein; PDX1, pancreas/duodenum homeobox protein-1; PHF8, plant homeodomain finger protein-8; POU5F1, POU domain, class 5 transcription factor 1; PPARD and PPARG, peroxisome proliferator-activated receptor-δ and -γ, respectively; PRDM14, PR domain zinc finger protein-14; RAD21, double-strand-break repair protein rad21 homolog; RCOR3, repressor element-1 silencing transcription factor (REST) corepressor 3; RNF2, E3 ubiquitin-protein ligase RING2; RUNX1, runt-related transcription factor 1; SALL4, Sal-like protein-4; SCL, stem cell leukemia transcription factor; SETDB1, histone-lysine N-methyltransferase Setdb1; SFPI1, transcription factor PU.1; SIN, paired amphipathic helix protein; SMAD, mothers against decapentaplegic homolog; SMARCA4, transcription activator BRG1; SOX, SRY-related high-mobility group (HMG) box transcription factor; SPI1, transcription factor PU.1; SRF, serum response factor; SUZ12, polycomb protein Suz12; TAL1, T cell acute lymphocytic leukemia protein-1; TBX, T-box transcription factor; TCF3 and TCF4, transcription factor 3 and 4, respectively; TCFAP2C, transcription factor AP-2γ; TCFCP2L1, transcription factor CP2-like protein-1; TET1, methylcytosine dioxygenase Tet1; TFAP2C, transcription factor AP-2γ; TFEB, transcription factor EB; THAP11, THAP domain-containing protein-11; TRIM28, transcription intermediary factor 1β; TRP63, tumor protein 63; WT1, Wilms tumor protein homolog; YAP1, Yes-associated protein-1; YY1, transcriptional repressor protein Yy1; ZFP281, zinc finger protein; ZFX, zinc finger X-chromosomal protein. Hub proteins: ATXN1, ataxin-1; CDK, cyclin-dependent kinase; CREBBP, CREB-binding protein; EGFR, EGF receptor; HDAC1, histone deacetylase 1; HSP90AA1, heat shock protein Hsp 90α; IKBKB, inhibitor of NF-κB kinase subunit-β; NR3C1, glucocorticoid receptor; PPP2CA, serine/threonine-protein phosphatase 2A catalytic subunit α-isoform; PRKCA and PRKCB, protein kinase C α- and β-type, respectively; PRKDC, DNA-dependent protein kinase catalytic subunit; RAC1, Ras-related C3 botulinum toxin substrate 1; RAF1, RAF proto-oncogene serine/threonine-protein kinase; RELA, transcription factor p65; RPS6KA3, ribosomal protein S6 kinase-α3; SFN, 14-3-3 protein-σ; SUMO1, small ubiquitin-related modifier 1; TP53, cellular tumor antigen p53; TRAF6, TNF receptor-associated factor 6; UBC, polyubiquitin-C.
Fig. 4.
Results of Kinase Enrichment Analysis (KEA), ChIP-x Enrichment Analysis (ChEA), and Protein-Protein Interaction Hubs (PPI Hubs) predicted by Enrichr analysis based on genes that were differentially expressed (P < 0.01) in each experimental group compared with the control group, i.e., End-FFSS vs. Control, Post-2 h FFSS vs. Control, and Post-24 h FFSS vs. Control, shown as Venn diagrams. Left: KEA shows three kinases common to all experimental groups compared with the control group. Middle: ChEA shows 151 transcription factors present in all the experimental groups compared with the control group. Right: PPI Hubs shows 36 hub proteins shared by all experimental groups compared with the control group. FFSS, fluid flow shear stress.
Thus, using KEA, ChEA, and PPI Hubs and the predicted EP2-mediated signaling, we identified Akt-GSK3β-β-catenin and MAPK/ERK as potential signaling pathways in FFSS. PPI Hubs also identified other serine/threonine kinases such as cyclin-dependent kinases (CDK1 and CDK2), protein kinase C (PRKCA and PRKCB), and ribosomal S6 kinase (RPS6KA3) and a possible role for histone acetylation suggested by cAMP-response element-binding protein (CREB)-binding protein (CREBBP), E1A-associated protein p300 (EP300), histone deacetylase 1 (HDAC1), and MYC.
Pathways and Networks Predicted Using IPA in Podocytes Following Application of FFSS
Canonical pathway analysis.
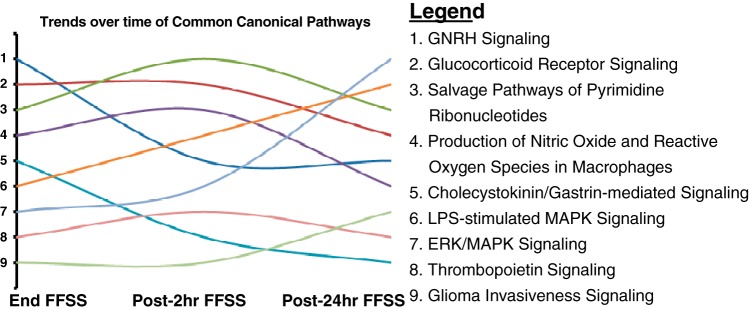
IPA identified 70, 167, and 32 canonical pathways that were significantly altered (P < 0.05) at End-, Post-2 h, and Post-24 h FFSS, respectively (Supplemental File C). The IPA also suggested nine canonical pathways that showed changes in all three groups (Table 4). In addition, rank order distribution showed that “MAPK/ERK signaling” and “LPS-stimulated MAPK signaling” were trending upward while “GNRH signaling” and “cholecystokinin/gastrin-mediated signaling” (where GNRH is gonadotropin-releasing hormone) were trending down (Fig. 5). Additionally, FFSS was also associated with 1) “production of nitric oxide (NO) and reactive oxygen species in macrophages” and 2) “salvage pathways of pyrimidine ribonucleotides.”
Table 4.
Nine common canonical pathways identified by Ingenuity Pathway Analysis based on genes that were differentially expressed in FFSS-treated podocytes at End-FFSS, Post-2 h FFSS, and Post-24 h FFSS compared with control
|
P Value |
|||
|---|---|---|---|
| Common Canonical Pathways on IPA | End-FFSS | Post-2 h FFSS | Post-24 h FFSS |
| Cholecystokinin/gastrin-mediated signaling | 0.010 | 0.030 | 0.044 |
| MAPK/ERK signaling | 0.033 | 0.006 | 0.003 |
| Glioma invasiveness signaling | 0.036 | 0.048 | 0.034 |
| Glucocorticoid receptor signaling | 0.007 | <0.001 | 0.019 |
| GNRH signaling | 0.001 | 0.001 | 0.027 |
| LPS-stimulated MAPK signaling | 0.014 | <0.001 | 0.013 |
| Production of nitric oxide and reactive oxygen species in macrophages | 0.009 | <0.001 | 0.029 |
| Salvage pathways of pyrimidine ribonucleotides | 0.007 | <0.001 | 0.013 |
| Thrombopoietin signaling | 0.033 | 0.015 | 0.037 |
Ingenuity Pathway Analysis (IPA) based on genes that were differentially expressed (P < 0.01) in fluid flow shear stress (FFSS)-treated podocytes compared with control. GNRH, gonadotropin-releasing hormone.
Fig. 5.
Ingenuity Pathway Analysis outlined as trends for the nine canonical pathways that were altered in all experimental groups based on differential gene expression (P < 0.01) in each experimental group compared with control (End-FFSS vs. Control, Post-2 h FFSS vs. Control, and Post-24 h FFSS vs. Control). The rank order distribution shown in Table 4 suggests 1) an upward trend of MAPK/ERK signaling and LPS-stimulated MAPK signaling with time and 2) a downward trend of gonadotropin-releasing hormone (GNRH) signaling and cholecystokinin/gastrin-mediated signaling.
Network analysis.
IPA identified 22, 45, and 19 networks at End-, Post-2 h, and Post-24 h FFSS, respectively (Supplemental File D). IPA also detected three networks that were shared by all three groups, namely, 1) “lipid metabolism, molecular transport, small molecule biochemistry,” 2) “developmental disorder, hereditary disorder, metabolic disease,” and 3) “hereditary disorder, neurological disease, organismal injury and abnormalities.”
Upstream regulator analysis.
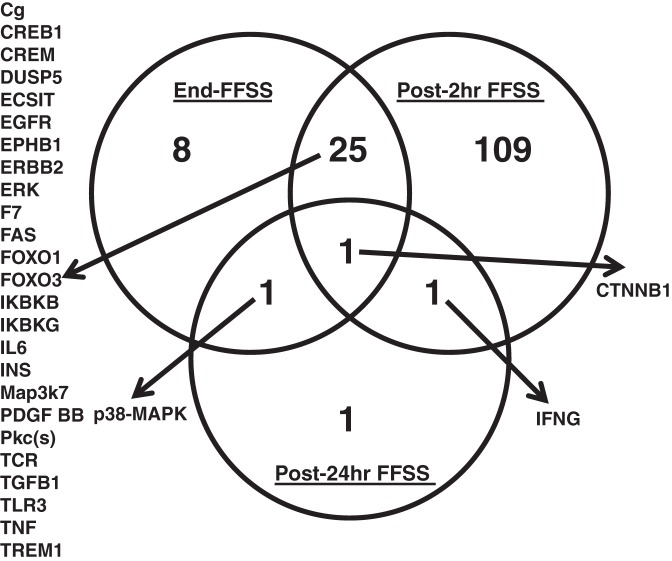
The upstream regulators included transcription regulators, kinases, cytokines, transmembrane receptors, growth factors, transporters, etc., at End-, Post-2 h, and Post-24 h FFSS. IPA identified 35, 136, and 4 upstream regulators at End-, Post-2 h, and Post-24 h FFSS, respectively. These data are shown in Supplemental File E. The upstream regulators detected in all three groups are shown in Fig. 6. The transcription factor β-catenin was present in all three groups. Transcription factor β-catenin would be relevant to the MAPK/ERK and LPS-Stimulated MAPK Signaling identified in the nine canonical pathways (Table 4).
Fig. 6.
Ingenuity Pathway Analysis (IPA) using differentially expressed genes (P < 0.01) in experimental groups compared with control (End-FFSS vs. Control, Post-2 h FFSS vs. Control, and Post-24 h FFSS vs. Control). Predicted by IPA, the Venn diagram shows the overlap in upstream regulators in experimental groups. Upstream regulators include transcription regulators, kinases, cytokines, transmembrane receptors, growth factors, transporters, etc. CREB1, cAMP-response element-binding protein-1; CREM, cAMP-response element modulator; CTNNB1, β-catenin; DUSP5, dual-specificity protein phosphatase 5; ECSIT, evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial; EGFR, EGF receptor; EPHB1, ephrin type B receptor 1; ERBB2, receptor tyrosine-protein kinase erbB-2; F7, coagulation factor VII; FAS, tumor necrosis factor receptor superfamily member 6; FOXO, forkhead box protein O; IFNG, interferon-γ ; IKBKB and IKBKG, inhibitor of NF-κB kinase subunit-β and -γ, respectively; INS, insulin; PDGF BB, PDGF β-polypeptide b; TCR, T cell receptor; TGFB1, transforming growth factor-β1; TLR3, Toll-like receptor 3; TREM1, triggering receptor expressed on myeloid cells 1.
Thus a large data set analysis approach using bioinformatics tools Enrichr and IPA identified candidate kinases, transcription factors, and protein-protein interaction hubs, canonical pathways, networks, and upstream regulators that may be involved in mechanotransduction in podocytes exposed to FFSS. To validate some of the pathways from the numerous possibilities presented by these analyses, we targeted those reportedly altered by FFSS or those intersecting with EP2-mediated signaling in other epithelial cells, e.g., osteocytes and renal tubular cells (Fig. 1).
Fluid Flow Shear Stress Does Not Increase cAMP Generation in Podocytes
There was no detectable cAMP in the medium at onset (0 min), during FFSS (30 min), or at the end of FFSS (120 min) (<3 pmol/ml; data not shown). There was no significant change in cAMP levels secreted into the medium or intracellular cAMP in podocytes treated with FFSS.
Levels of cAMP in the medium from podocytes exposed to FFSS were measured before FFSS and during recovery at Post-2 h and at Post-24 h following FFSS (n = 4). Levels of cAMP in the medium at Post-2 h (295.7 ± 92.7 pmol) and Post-24 h (267.8 ± 77.7 pmol, means ± SD) were not significantly changed compared with Pre-FFSS (334.3 ± 64.5 pmol, P = 0.50).
Intracellular cAMP levels in End-FFSS (2.38 ± 0.39), Post-2 h (1.53 ± 0.40), and Post-24 h FFSS (2.03 ± 0.22) were not changed compared with Control cAMP (2.38 ± 0.46 pmol/μg DNA, P = 0.054). In contrast to the anticipated increase, a decrease in cAMP was observed at Post-2 h FFSS that was statistically insignificant.
Western Blot Analysis Demonstrated Changes in Phosphorylation of Kinases in Podocytes Following FFSS Treatment
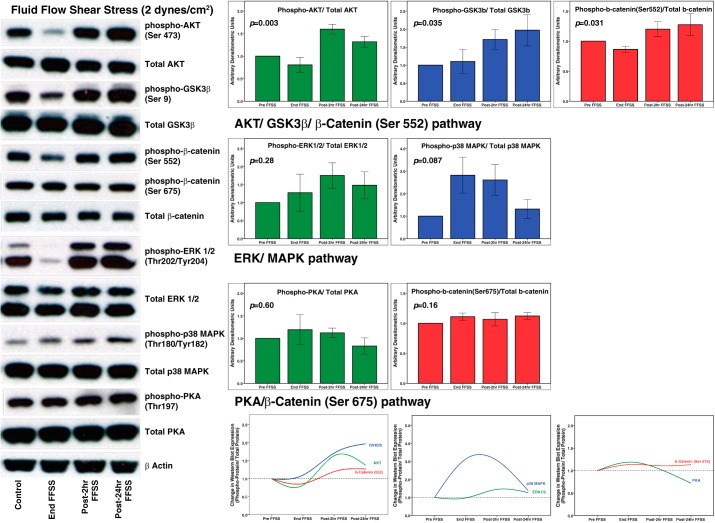
Figure 7 shows representative results of Western blotting for phosphorylation of Akt, GSK3β, β-catenin, ERK1/2, p38 MAPK, and PKA proteins in podocytes at End-FFSS, Post-2 h FFSS, and Post-24 h FFSS (n = 4–6). Changes in the phosphorylation of these proteins over the 24-h period following application of FFSS compared with Control are shown as bar graphs (Fig. 7). The change over time as a group on Kruskal-Wallis test was significant for phospho-Akt (P = 0.003), phospho-GSK3β (P = 0.035), and phospho-β-catenin (Ser552) (P = 0.031) suggesting that the Akt-GSK3β-β-catenin pathway is stimulated by FFSS. Although phospho-ERK (P = 0.280) and phospho-p38 MAPK (P = 0.087) showed change over time, it did not reach statistical significance. There was no change in phospho-PKA (P = 0.599) and phospho-β-catenin (Ser675) (P = 0.164). These results are in line with the molecules identified by KEA, ChEA, protein-protein hub interaction, and EP2-mediated signaling analyses.
Fig. 7.
Western blotting shows that fluid flow shear stress (FFSS) increases expression of key proteins of signaling pathways predicted by Ingenuity Pathway Analysis and Enrichr. These include phospho-Akt (Ser473), phospho-GSK3β (Ser9), phospho-β-catenin (Ser552), phospho-β-catenin (Ser675), phospho-ERK 1/2 (Thr202/Tyr204), phospho-p38 MAPK (Thr180/Tyr182), and phospho-PKA (Thr197). Cultured podocytes were exposed to FFSS at 2 dyn/cm2 for 2 h. Control groups were maintained identically in the medium without FFSS. Total protein lysates at end, post-2 h, and post-24 h were analyzed by SDS-PAGE followed by Western blotting. β-Actin was used as the loading control. Bar graphs show fold change (experimental/control) in density ratios of phosphorylated protein/total protein. Images of representative immunoblots are shown at left, and the trend over the 24-h period is shown at bottom. Results are presented as means ± SE of n = 4–6 replicate experiments.
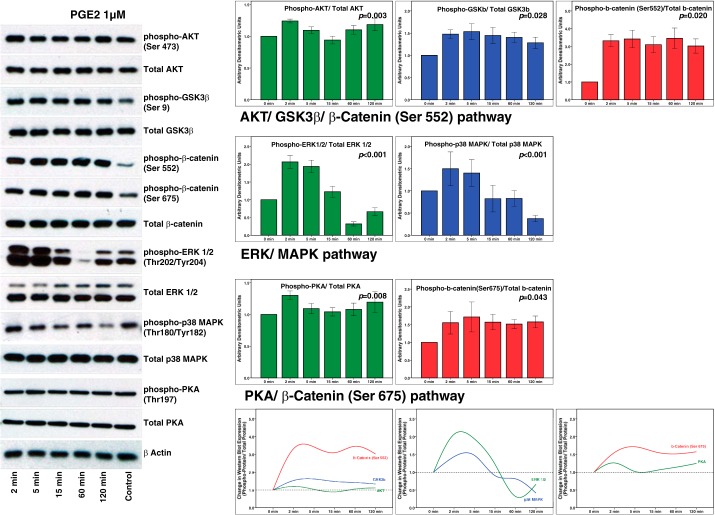
Figure 8 shows results of Western blotting to determine changes in the phosphorylation of Akt, GSK3β, β-catenin, ERK1/2, p38 MAPK, and PKA proteins at 0, 2, 5, 15, 60, and 120 min in podocytes treated with 1 µM PGE2 (n = 4–6). Changes in phosphorylation of these proteins over 120 min following incubation with PGE2 are shown as bar graphs. The change over time as a group on Kruskal-Wallis test was significant for phospho-Akt (P = <0.001), phospho-GSK3β (P = 0.002), and phospho-β-catenin (Ser552) (P = 0.004) suggesting that the Akt-GSK3β-β-catenin pathway is stimulated using PGE2. Similarly, phospho-ERK (P < 0.001), phospho-p38 MAPK (P = 0.001), phospho-PKA (P = 0.007), and phospho-β-catenin (Ser675) (P = 0.004) changed over time suggesting that MAPK/ERK and PKA pathways are recruited by PGE2.
Fig. 8.
Western blot analysis shows that exogenously added PGE2 induces changes similar to those caused by fluid flow shear stress. These include results for phospho-Akt (Ser473), phospho-GSK3β (Ser9), phospho-β-catenin (Ser552), phospho-β-catenin (Ser675), phospho-ERK 1/2 (Thr202/Tyr204), phospho-p38 MAPK (Thr180/Tyr182), and phospho-PKA (Thr197). Cultured podocytes were exposed to PGE2 (1 µM) for 0, 2, 5, 15, 60, and 120 min. Total protein in cell lysates was analyzed by SDS-PAGE followed by Western blotting. β-Actin was used as the loading control. Bar graphs show fold change as ratios (experimental/control) of density of phosphorylated protein/total protein. Images of representative immunoblots are shown at left, and the trend over the 24-h period is shown at bottom. Results are presented as means ± SE of n = 4–6 replicate experiments.
DISCUSSION
Hyperfiltration-mediated glomerular injury has been implicated in progression of CKD, but the underlying mechanism is not well understood. Our studies address the mechanism of injury from hyperfiltration in terms of changes in hyperfiltration-associated biomechanical forces within the glomerulus rather than conventional hemodynamic parameters. Cellular response to biomechanical forces can be understood through the processes involved in mechanoperception and mechanotransduction. The signaling events involved in mechanotransduction in FFSS-treated podocytes are not known. The present study addressed the changes in the molecules and signaling networks that may contribute to mechanotransduction in podocytes exposed to FFSS.
Podocytes, a significant structural component of the glomerular filtration barrier, are vulnerable to hyperfiltration-associated biomechanical forces, namely, tensile stress and FFSS. We have shown that podocytes are mechanoresponsive (50, 52). In vitro application of tensile stress causes formation of actin-rich centers and radial stress fibers (14, 16, 36) while FFSS disrupts actin stress fibers with the formation of a cortical actin ring (19, 52). Mechanoperception in cells is associated with changes in NO, ATP, Ca2+, and PGE2 as elements of initial cellular effects of mechanical forces (27, 29, 46, 60). We found that the COX-2-PGE2-EP2 axis plays an important role in mechanoperception in podocytes, although the contribution of other pathways remains open for investigation. We also found that in vitro FFSS results in the upregulation of COX-2 and increased secretion of PGE2 (52) and the increased expression of prostanoid receptor EP2, but not EP4 (53), and COX-2, but not COX-1, in podocytes (50). Results of the present work show that podocytes exposed to FFSS invoke more than one mechanism for mechanotransduction. These include Akt-GSK3β-β-catenin and possibly MAPK/ERK signaling, but not cAMP-PKA.
We adopted a large data set analytics approach to identify multiple molecules, signaling pathways, and networks recruited in podocytes exposed to FFSS. However, such an approach is also fraught with false positive signals. Therefore we used two independent bioinformatics tools, Enrichr and IPA, to identify molecules that were consistently identified using both programs. IPA and Enrichr have been used by others to identify innate and adaptive immune systems in Parkinson’s disease, Alzheimer’s disease, and multiple sclerosis (20). Enrichr has also been used to identify pathways in Alzheimer’s disease, hearing impairment, and thrombosis (12, 26, 32). Initial data, facilitated by a distinct distribution of signals, were refined to obtain fewer targets common to all groups, i.e., End-FFSS and at 2 and 24 h after FFSS. The PCA and volcano plots also showed that the gene sets were well separated and expression profile was not skewed (Fig. 3). Additionally, Fig. 5 shows that analyses at multiple time points also enabled a convenient detection of temporal changes in signaling pathways.
Table 1 shows the number of genes at different degrees of stringency used for analysis. We selected P < 0.01 as a cutoff for analysis as bioinformatics tools are considered to yield robust results when ~500 differentially expressed genes are entered for analysis. Among the genes that respond early, we have previously reported an upregulation of COX-2 gene immediately after FFSS and its return to baseline by 24 h after FFSS (50). The present array analysis also showed upregulated COX-2 (Ptgs2) gene at End- and Post-2 h FFSS but not at Post-24 h FFSS (Fig. 3A). Several other genes besides that of COX-2 were immediately upregulated in cells treated with FFSS. Ccl7, Nr4a1, Arc, and Egr1 (Fig. 3B) belong to immediate early genes showing rapid activation in response to a stimulus. Ccl7 is a secreted cytokine while Arc and Egr1 proteins are important for neuronal plasticity (3, 39). Nr4a1 can be induced by various stimuli including prostaglandins, growth factors, calcium, fluid shear stress, etc. (1, 37). Arc was identified to be critical in the dynamic changes of foot processes in podocytes in the puromycin aminonucleoside model of glomerulopathy (38). Nr4a1 increases significantly in models of diabetic nephropathy, while transforming growth factor-β1 (TGFβ1)-ERK1/2-Egr-1 increases in podocytes treated with high glucose (40, 62). Thus FFSS appears to alter the expression of several genes in addition to that of COX-2, which we had identified previously.
Protein phosphorylation being among the most prominent cellular responses, we looked for changes in kinases in FFSS-treated podocytes. A total of 49 kinases, ~9% of 540 known mouse kinases, were identified by KEA at each time point (Supplemental File B; 4). Thus the response to FFSS involves specific kinases and not a global change in kinases. Among the most noticeable kinases in KEA, identification of GSK3β was further supported by PPI that included Akt1 and GSK3β (Table 3 and Fig. 4). In parallel, β-catenin was identified in Enrichr ChEA and IPA upstream regulators (Table 3 and Fig. 6). Thus the Akt-GSK3β-β-catenin signaling pathway is important in podocyte response to FFSS. Others have shown upregulation of β-catenin in podocytes under pathological conditions and abrogation of kidney injury following β-catenin gene deletion (13, 22, 58).
Studies on the effect of FFSS on epithelial cells from other tissues support our findings. Osteocytes, embedded in mineralized bone matrix, are terminally differentiated epithelial cells that express podoplanin as podocytes do. Cytoplasmic processes (dendrites) from osteocytes traverse within the canaliculi of mature bone and connect with other osteocytes and with cells on bone surface through adhesion molecules and gap junctions. Mechanistically, activation of the Wnt/β-catenin signaling pathway is a physiological response to mechanical loading in the bone. Inactivation of GSK3β is required for β-catenin activation. Active GSK3β phosphorylates β-catenin at Ser31/33/Thr41 before ubiquitination and degradation. PGE2 secretion appears to be an important upstream event in Wnt/β-catenin activation in osteocytes (2, 27, 28, 45, 59). PGE2 engages EP2 followed by GSK3β phosphorylation and inactivation resulting in increased intracellular β-catenin that translocates to the nucleus where it binds to cofactors T cell factor (Tcf) and lymphoid enhancer-binding factor (LEF) and regulates the transcription of genes including that of COX-2 in osteocytes (2, 27, 28, 45, 59). Thus FFSS-induced release of PGE2 activates EP2 in osteocytes leading to activation of the Akt-GSK3β-β-catenin pathway (2, 9, 27, 28, 45, 59). Of the four PGE2 receptors (EP1–EP4), only EP2 activation results in nuclear translocation of activated β-catenin. Figure 7 shows Western blotting data that validate the predicted signaling pathway Akt-GSK3β-β-catenin (Ser552) in podocytes following exposure to FFSS. This EP2-mediated signaling pathway is also relevant in mediating the effect of FFSS on osteocytes. These findings are further supported by the experiments to determine the direct effect of PGE2 on podocytes shown in Fig. 8.
Additional kinases identified using KEA and PPI included ERK1/2 (Table 3 and Fig. 4) and the MAPK/ERK canonical signaling pathway by IPA (Table 4 and Fig. 5). MAPK/ERK signaling pathway identified by us is relevant to the effect of FFSS. Although P values were not significant, changes visualized using Western blotting (Fig. 7) support the role for predicted p38 MAPK/ERK signaling pathway in podocytes following exposure to FFSS. Furthermore, these changes could be replicated by exogenously adding PGE2 to podocytes (Fig. 8).
FFSS causes similar activation of MAPK/ERK signaling in tubular cells. As in podocytes, tensile stress and FFSS induce distinct effects on the tubular segment of the nephron. Increased tubular urine flow caused by diuretics or extracellular isotonic volume expansion causes circumferential stretch and FFSS on tubular cells. Circumferential stretch applied to cortical collecting duct decreases PGE2 secretion while FFSS increases PGE2 (5, 35). FFSS, but not circumferential stretch, upregulates COX-2, neutral sphingomyelinase, endothelin, phospho-ERK, and phospho-p38 in cortical collecting duct cells (5, 18, 34, 35).
Increased PGE2 has been associated with the activation of cAMP-PKA, but neither Enrichr analysis nor IPA indicated upregulation of this pathway in FFSS-treated podocytes (25). Further analysis did not detect increased cAMP levels in the media or cells, nor did we detect increased phosphorylation of PKA after FFSS (Fig. 7). However, we did observe increased phosphorylation of PKA following incubation of podocytes with PGE2 (Fig. 8). This could be due to differential responses of EP2 and EP4 receptors, both being Gs-coupled membrane-localized G protein-coupled receptors that share 30% homology and activate adenylate cyclase. Under normal conditions, podocytes express predominantly EP4 and very little EP2, which is induced in cells treated with FFSS (53). Our results do not support a significant role of the cAMP-PKA pathway in transducing the effects of FFSS in podocytes. It is possible that the changes in cAMP-PKA occur in a very early response that is being missed in our experimental design or that the cAMP-PKA pathway is recruited only at much greater FFSS than 2 dyn/cm2 that elicited all the observed changes in podocytes. The higher FFSS (8–16 dyn/cm2) required to activate the Akt-GSK3β-β-catenin pathway in osteocytes does indeed seem to result in a slight upregulation of the cAMP-PKA pathway (28).
In the large volume of data analyzed, IPA also detected “production of nitric oxide and reactive oxygen species in macrophages” and “salvage pathways of pyrimidine ribonucleotides,” which we have not yet explored. However, given the relevance of NO, ATP, and Ca2+ in mechanoperception, these pathways may provide important targets in podocytes (29, 46, 60). Friedrich et al. (19), using laminar flow at 0.25–1.75 dyn/cm2, found that broadband tyrosine kinase inhibitors such as genistein and AG 82 (tyrphostin A25) caused podocyte loss but not specific inhibitors such as PP2 (src family) or PD153035 (EGF receptor) suggesting that tyrosine kinases may have a role in FFSS. We detected only a few tyrosine kinases [Bruton's tyrosine kinase (BTK), receptor tyrosine-protein kinase erbB-4 (ERBB4), FGF receptor 2 (FGFR2), and JAK2 in these studies; Supplemental File B]. Huang et al. (24) applied rotational fluid movement with maximum FFSS of 10 dyn/cm2. They demonstrated increased phospholipase D (PLD) activity, c-Src phosphorylation, and mTOR activation suggesting the importance of the c-Src/PLD1/mTOR pathway in podocytes exposed to FFSS. Recently, Rinschen et al. (44) suggested transcriptional coactivator Yes-associated protein (YAP) involvement in mechanotransduction in podocytes. This transcriptional factor was also present among our 151 common transcriptional factors (Table 3).
In summary, the present results show that mechanotransduction in podocytes involves Akt-GSK3β-β-catenin with a likely role of MAPK/ERK signaling, but not cAMP-PKA signaling. Our earlier work demonstrated that the COX-2-PGE2-EP2 axis is involved in mechanoperception, and the present work shows that predicted signaling pathways in mechanotransduction also intersect with known EP2-mediated signaling pathways (Fig. 1). Our results also indicate a likely role of additional molecules, pathways, and networks in mechanotransduction that are open to the research community for investigation. These novel findings on the effects of FFSS would not only lead to a better understanding of the mechanism of hyperfiltration-mediated glomerular injury but also be clinically relevant. Currently, renin-angiotensin-aldosterone system blockers are used to decrease capillary pressure as the main approach to delay progression of CKD. Little progress has been made past these blockers to ameliorate hyperfiltration-mediated injury. Investigations into biomechanical forces in the glomerulus may translate into identification of new targets for therapeutic interventions. We believe that EP2 is a potential target for developing new interventions to mitigate hyperfiltration-mediated injury.
GRANTS
The array analysis was performed at the Kansas Intellectual and Developmental Disabilities Research Center Core (University of Kansas Medical Center) supported by the Smith Intellectual and Developmental Disabilities Research Center (National Institute of Child Health and Human Development Grant U54-HD-090216) and “Molecular Regulation of Cell Development and Differentiation” (Centers of Biomedical Research Excellence Grant 5P20GM104936-10). This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant R01-DK-107490 (T. Srivastava), Department of Veterans Affairs Grant VA BX001037 (V. J. Savin), NIDDK Grant 1R01-DK-064969 (E. T. McCarthy), funds from the Midwest Biomedical Research Foundation (V. J. Savin and M. Sharma), and The Sam and Helen Kaplan Research Fund in Pediatric Nephrology (U. S. Alon and T. Srivastava).
DISCLAIMERS
The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.S., H.D., U.S.A., R.E.G., A.E.-M., E.T.M., R.S., M.L.J., V.J.S., and M.S. conceived and designed research; T.S., J.Z., R.S.D., and M.S. performed experiments; T.S., H.D., D.P.H., R.S.D., and M.S. analyzed data; T.S., H.D., D.P.H., U.S.A., R.E.G., A.E.-M., E.T.M., R.S., M.L.J., V.J.S., and M.S. interpreted results of experiments; T.S., R.S.D., and M.S. prepared figures; T.S. drafted manuscript; T.S., H.D., D.P.H., U.S.A., R.E.G., J.Z., R.S.D., A.E.-M., E.T.M., R.S., M.L.J., V.J.S., and M.S. edited and revised manuscript; T.S., H.D., D.P.H., U.S.A., R.E.G., J.Z., R.S.D., A.E.-M., E.T.M., R.S., M.L.J., V.J.S., and M.S. approved final version of manuscript.
Supplemental Data
REFERENCES
- 1.Bandoh S, Tsukada T, Maruyama K, Ohkura N, Yamaguchi K. Mechanical agitation induces gene expression of NOR-1 and its closely related orphan nuclear receptors in leukemic cell lines. Leukemia 11: 1453–1458, 1997. doi: 10.1038/sj.leu.2400800. [DOI] [PubMed] [Google Scholar]
- 2.Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone 42: 606–615, 2008. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci 28: 11760–11767, 2008. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci USA 101: 11707–11712, 2004. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrisoza-Gaytan R, Liu Y, Flores D, Else C, Lee HG, Rhodes G, Sandoval RM, Kleyman TR, Lee FY, Molitoris B, Satlin LM, Rohatgi R. Effects of biomechanical forces on signaling in the cortical collecting duct (CCD). Am J Physiol Renal Physiol 307: F195–F204, 2014. doi: 10.1152/ajprenal.00634.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celsi G, Larsson L, Seri I, Savin V, Aperia A. Glomerular adaptation in uninephrectomized young rats. Pediatr Nephrol 3: 280–285, 1989. doi: 10.1007/BF00858530. [DOI] [PubMed] [Google Scholar]
- 7.Celsi G, Savin J, Henter JI, Sohtell M. The contribution of ultrafiltration pressure for glomerular hyperfiltration in young nephrectomized rats. Acta Physiol Scand 141: 483–487, 1991. doi: 10.1111/j.1748-1716.1991.tb09109.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128, 2013. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherian PP, Cheng B, Gu S, Sprague E, Bonewald LF, Jiang JX. Effects of mechanical strain on the function of gap junctions in osteocytes are mediated through the prostaglandin EP2 receptor. J Biol Chem 278: 43146–43156, 2003. doi: 10.1074/jbc.M302993200. [DOI] [PubMed] [Google Scholar]
- 10.Chun KS, Lao HC, Trempus CS, Okada M, Langenbach R. The prostaglandin receptor EP2 activates multiple signaling pathways and β-arrestin1 complex formation during mouse skin papilloma development. Carcinogenesis 30: 1620–1627, 2009. doi: 10.1093/carcin/bgp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun KS, Lao HC, Langenbach R. The prostaglandin E2 receptor, EP2, stimulates keratinocyte proliferation in mouse skin by G protein-dependent and β-arrestin1-dependent signaling pathways. J Biol Chem 285: 39672–39681, 2010. doi: 10.1074/jbc.M110.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong W, Meng X, Li J, Zhang Q, Chen F, Liu W, Wang Y, Cheng S, Yao X, Yan J, Kim S, Saykin AJ, Liang H, Shen L; Alzheimer’s Disease Neuroimaging Initiative . Genome-wide network-based pathway analysis of CSF t-tau/Aβ1-42 ratio in the ADNI cohort. BMC Genomics 18: 421, 2017. doi: 10.1186/s12864-017-3798-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai C, Stolz DB, Kiss LP, Monga SP, Holzman LB, Liu Y. Wnt/beta-catenin signaling promotes podocyte dysfunction and albuminuria. J Am Soc Nephrol 20: 1997–2008, 2009. doi: 10.1681/ASN.2009010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durvasula RV, Petermann AT, Hiromura K, Blonski M, Pippin J, Mundel P, Pichler R, Griffin S, Couser WG, Shankland SJ. Activation of a local tissue angiotensin system in podocytes by mechanical strain. Kidney Int 65: 30–39, 2004. doi: 10.1111/j.1523-1755.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- 15.Endlich K, Kliewe F, Endlich N. Stressed podocytes-mechanical forces, sensors, signaling and response. Pflugers Arch 469: 937–949, 2017. doi: 10.1007/s00424-017-2025-8. [DOI] [PubMed] [Google Scholar]
- 16.Endlich N, Kress KR, Reiser J, Uttenweiler D, Kriz W, Mundel P, Endlich K. Podocytes respond to mechanical stress in vitro. J Am Soc Nephrol 12: 413–422, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, Lu Z. Phosphorylation of β-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem 282: 11221–11229, 2007. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores D, Liu Y, Liu W, Satlin LM, Rohatgi R. Flow-induced prostaglandin E2 release regulates Na and K transport in the collecting duct. Am J Physiol Renal Physiol 303: F632–F638, 2012. doi: 10.1152/ajprenal.00169.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich C, Endlich N, Kriz W, Endlich K. Podocytes are sensitive to fluid shear stress in vitro. Am J Physiol Renal Physiol 291: F856–F865, 2006. doi: 10.1152/ajprenal.00196.2005. [DOI] [PubMed] [Google Scholar]
- 20.Gagliano SA, Pouget JG, Hardy J, Knight J, Barnes MR, Ryten M, Weale ME. Genomics implicates adaptive and innate immunity in Alzheimer’s and Parkinson’s diseases. Ann Clin Transl Neurol 3: 924–933, 2016. doi: 10.1002/acn3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirisina R, Porter-Westpfahl KS, Hembree M, Johnson T, Wiedemann LM, Barrett TA, Hood L, Wu H, Li L. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet 39: 189–198, 2007. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikkilä E, Juhila J, Lassila M, Messing M, Perälä N, Lehtonen E, Lehtonen S, Sjef Verbeek J, Holthofer H. β-Catenin mediates adriamycin-induced albuminuria and podocyte injury in adult mouse kidneys. Nephrol Dial Transplant 25: 2437–2446, 2010. doi: 10.1093/ndt/gfq076. [DOI] [PubMed] [Google Scholar]
- 23.Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase stabilizes β-catenin through inhibition of its ubiquitination. Mol Cell Biol 25: 9063–9072, 2005. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C, Bruggeman LA, Hydo LM, Miller RT. Shear stress induces cell apoptosis via a c-Src-phospholipase D-mTOR signaling pathway in cultured podocytes. Exp Cell Res 318: 1075–1085, 2012. doi: 10.1016/j.yexcr.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain M, Javeed A, Ashraf M, Yuzhu H, Mukhtar MM. Multilevel pharmacological manipulation of adenosine-prostaglandin E2/cAMP nexus in the tumor microenvironment: a ‘two hit’ therapeutic opportunity. Pharmacol Res 73: 8–19, 2013. doi: 10.1016/j.phrs.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Jha PK, Vijay A, Sahu A, Ashraf MZ. Comprehensive Gene expression meta-analysis and integrated bioinformatic approaches reveal shared signatures between thrombosis and myeloproliferative disorders. Sci Rep 6: 37099, 2016. doi: 10.1038/srep37099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamel MA, Picconi JL, Lara-Castillo N, Johnson ML. Activation of β-catenin signaling in MLO-Y4 osteocytic cells versus 2T3 osteoblastic cells by fluid flow shear stress and PGE2: implications for the study of mechanosensation in bone. Bone 47: 872–881, 2010. doi: 10.1016/j.bone.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitase Y, Barragan L, Qing H, Kondoh S, Jiang JX, Johnson ML, Bonewald LF. Mechanical induction of PGE2 in osteocytes blocks glucocorticoid-induced apoptosis through both the β-catenin and PKA pathways. J Bone Miner Res 25: 2657–2668, 2010. doi: 10.1002/jbmr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinnulend J, Semeins CM, Ajubi NE, Nijweide PJ, Burger EH. Pulsating fluid flow increases nitric oxide (NO) synthesis by osteocytes but not periosteal fibroblasts: correlation with prostaglandin upregulation. Biochem Biophys Res Commun 217: 640–648, 1995. doi: 10.1006/bbrc.1995.2822. [DOI] [PubMed] [Google Scholar]
- 30.Kriz W, Lemley KV. A potential role for mechanical forces in the detachment of podocytes and the progression of CKD. J Am Soc Nephrol 26: 258–269, 2015. doi: 10.1681/ASN.2014030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kriz W, Lemley KV. Potential relevance of shear stress for slit diaphragm and podocyte function. Kidney Int 91: 1283–1286, 2017. doi: 10.1016/j.kint.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 32.Lebeko K, Manyisa N, Chimusa ER, Mulder N, Dandara C, Wonkam A. A genomic and protein-protein interaction analyses of nonsyndromic hearing impairment in cameroon using targeted genomic enrichment and massively parallel sequencing. OMICS 21: 90–99, 2017. doi: 10.1089/omi.2016.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leone V, di Palma A, Ricchi P, Acquaviva F, Giannouli M, Di Prisco AM, Iuliano F, Acquaviva AM. PGE2 inhibits apoptosis in human adenocarcinoma Caco-2 cell line through Ras-PI3K association and cAMP-dependent kinase A activation. Am J Physiol Gastrointest Liver Physiol 293: G673–G681, 2007. doi: 10.1152/ajpgi.00584.2006. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Flores D, Carrisoza-Gaytán R, Rohatgi R. Biomechanical regulation of cyclooxygenase-2 in the renal collecting duct. Am J Physiol Renal Physiol 306: F214–F223, 2014. doi: 10.1152/ajprenal.00327.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Flores D, Carrisoza-Gaytán R, Rohatgi R. Cholesterol affects flow-stimulated cyclooxygenase-2 expression and prostanoid secretion in the cortical collecting duct. Am J Physiol Renal Physiol 308: F1229–F1237, 2015. doi: 10.1152/ajprenal.00635.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martineau LC, McVeigh LI, Jasmin BJ, Kennedy CR. p38 MAP kinase mediates mechanically induced COX-2 and PG EP4 receptor expression in podocytes: implications for the actin cytoskeleton. Am J Physiol Renal Physiol 286: F693–F701, 2004. doi: 10.1152/ajprenal.00331.2003. [DOI] [PubMed] [Google Scholar]
- 37.Maxwell MA, Muscat GEO. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal 4: e002, 2006. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao J, Fan Q, Cui Q, Zhang H, Chen L, Wang S, Guan N, Guan Y, Ding J. Newly identified cytoskeletal components are associated with dynamic changes of podocyte foot processes. Nephrol Dial Transplant 24: 3297–3305, 2009. doi: 10.1093/ndt/gfp338. [DOI] [PubMed] [Google Scholar]
- 39.Minatohara K, Akiyoshi M, Okuno H. Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol Neurosci 8: 78, 2016. doi: 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa D, Eguchi J, Wada J, Terami N, Hatanaka T, Tachibana H, Nakatsuka A, Horiguchi CS, Nishii N, Makino H. Nuclear hormone receptor expression in mouse kidney and renal cell lines. PLoS One 9: e85594, 2014. doi: 10.1371/journal.pone.0085594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pai R, Nakamura T, Moon WS, Tarnawski AS. Prostaglandins promote colon cancer cell invasion; signaling by cross-talk between two distinct growth factor receptors. FASEB J 17: 1640–1647, 2003. doi: 10.1096/fj.02-1011com. [DOI] [PubMed] [Google Scholar]
- 42.Pichler Sekulic S, Sekulic M. Rheological influence upon the glomerular podocyte and resultant mechanotransduction. Kidney Blood Press Res 40: 176–187, 2015. doi: 10.1159/000368493. [DOI] [PubMed] [Google Scholar]
- 43.Pozzi A, Yan X, Macias-Perez I, Wei S, Hata AN, Breyer RM, Morrow JD, Capdevila JH. Colon carcinoma cell growth is associated with prostaglandin E2/EP4 receptor-evoked ERK activation. J Biol Chem 279: 29797–29804, 2004. doi: 10.1074/jbc.M313989200. [DOI] [PubMed] [Google Scholar]
- 44.Rinschen MM, Grahammer F, Hoppe AK, Kohli P, Hagmann H, Kretz O, Bertsch S, Höhne M, Göbel H, Bartram MP, Gandhirajan RK, Krüger M, Brinkkoetter PT, Huber TB, Kann M, Wickström SA, Benzing T, Schermer B. YAP-mediated mechanotransduction determines the podocyte’s response to damage. Sci Signal 10: eaaf8165, 2017. doi: 10.1126/scisignal.aaf8165. [DOI] [PubMed] [Google Scholar]
- 45.Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, Cullen DM, Zhao W, Li C, Kharode Y, Sauter L, Babij P, Brown EL, Hill AA, Akhter MP, Johnson ML, Recker RR, Komm BS, Bex FJ. Wnt/β-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem 281: 31720–31728, 2006. doi: 10.1074/jbc.M602308200. [DOI] [PubMed] [Google Scholar]
- 46.Saunders MM, You J, Zhou Z, Li Z, Yellowley CE, Kunze EL, Jacobs CR, Donahue HJ. Fluid flow-induced prostaglandin E2 response of osteoblastic ROS 17/2.8 cells is gap junction-mediated and independent of cytosolic calcium. Bone 32: 350–356, 2003. doi: 10.1016/S8756-3282(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 47.Shankland SJ, Pippin JW, Reiser J, Mundel P. Podocytes in culture: past, present, and future. Kidney Int 72: 26–36, 2007. doi: 10.1038/sj.ki.5002291. [DOI] [PubMed] [Google Scholar]
- 48.Sharma M, Sharma R, McCarthy ET, Savin VJ, Srivastava T. Hyperfiltration-associated biomechanical forces in glomerular injury and response: potential role for eicosanoids. Prostaglandins Other Lipid Mediat 2017: S1098-8823(17)30008-4, 2017 10.1016/j.prostaglandins.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng H, Shao J, Washington MK, DuBois RN. Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J Biol Chem 276: 18075–18081, 2001. doi: 10.1074/jbc.M009689200. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava T, Alon US, Cudmore PA, Tarakji B, Kats A, Garola RE, Duncan RS, McCarthy ET, Sharma R, Johnson ML, Bonewald LF, El-Meanawy A, Savin VJ, Sharma M. Cyclooxygenase-2, prostaglandin E2, and prostanoid receptor EP2 in fluid flow shear stress-mediated injury in the solitary kidney. Am J Physiol Renal Physiol 307: F1323–F1333, 2014. doi: 10.1152/ajprenal.00335.2014. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava T, Celsi GE, Sharma M, Dai H, McCarthy ET, Ruiz M, Cudmore PA, Alon US, Sharma R, Savin VA. Fluid flow shear stress over podocytes is increased in the solitary kidney. Nephrol Dial Transplant 29: 65–72, 2014. doi: 10.1093/ndt/gft387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava T, McCarthy ET, Sharma R, Cudmore PA, Sharma M, Johnson ML, Bonewald LF. Prostaglandin E(2) is crucial in the response of podocytes to fluid flow shear stress. J Cell Commun Signal 4: 79–90, 2010. doi: 10.1007/s12079-010-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava T, McCarthy ET, Sharma R, Kats A, Carlton CG, Alon US, Cudmore PA, El-Meanawy A, Sharma M. Fluid flow shear stress upregulates prostanoid receptor EP2 but not EP4 in murine podocytes. Prostaglandins Other Lipid Mediat 104-105: 49–57, 2013. doi: 10.1016/j.prostaglandins.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava T, Thiagarajan G, Alon US, Sharma R, El-Meanawy A, McCarthy ET, Savin VJ, Sharma M. Role of biomechanical forces in hyperfiltration-mediated glomerular injury in congenital anomalies of the kidney and urinary tract. Nephrol Dial Transplant 32: 759–765, 2017. doi: 10.1093/ndt/gfw430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem 281: 9971–9976, 2006. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- 56.Tessner TG, Muhale F, Riehl TE, Anant S, Stenson WF. Prostaglandin E2 reduces radiation-induced epithelial apoptosis through a mechanism involving AKT activation and bax translocation. J Clin Invest 114: 1676–1685, 2004. doi: 10.1172/JCI22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426, 1999. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 58.Wang D, Dai C, Li Y, Liu Y. Canonical Wnt/β-catenin signaling mediates transforming growth factor-β1-driven podocyte injury and proteinuria. Kidney Int 80: 1159–1169, 2011. doi: 10.1038/ki.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia X, Batra N, Shi Q, Bonewald LF, Sprague E, Jiang JX. Prostaglandin promotion of osteocyte gap junction function through transcriptional regulation of connexin 43 by glycogen synthase kinase 3/β-catenin signaling. Mol Cell Biol 30: 206–219, 2010. doi: 10.1128/MCB.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You J, Jacobs CR, Steinberg TH, Donahue HJ. P2Y purinoceptors are responsible for oscillatory fluid flow-induced intracellular calcium mobilization in osteoblastic cells. J Biol Chem 277: 48724–48729, 2002. doi: 10.1074/jbc.M209245200. [DOI] [PubMed] [Google Scholar]
- 61.Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, Zhao S, Harris M, Harris SE, Feng JQ, Bonewald LF. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol 26: 4539–4552, 2006. doi: 10.1128/MCB.02120-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Li H, Hao J, Zhou Y, Liu W. High glucose increases Cdk5 activity in podocytes via transforming growth factor-β1 signaling pathway. Exp Cell Res 326: 219–229, 2014. doi: 10.1016/j.yexcr.2014.04.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.