Abstract
The opportunistic pathogen Pseudomonas aeruginosa colonizes the lungs of susceptible individuals by deploying virulence factors targeting host defenses. The secreted factor Cif (cystic fibrosis transmembrane conductance regulator inhibitory factor) dysregulates the endocytic recycling of CFTR and thus reduces CFTR abundance in host epithelial membranes. We have postulated that the decrease in ion secretion mediated by Cif would slow mucociliary transport and decrease bacterial clearance from the lungs. To test this hypothesis, we explored the effects of Cif in cultured epithelia and in the lungs of mice. We developed a strategy to interpret the “hurricane-like” motions observed in reconstituted cultures and identified a Cif-mediated decrease in the velocity of mucus transport in vitro. Presence of Cif also increased the number of bacteria recovered at two time points in an acute mouse model of pneumonia caused by P. aeruginosa. Furthermore, recent work has demonstrated an inverse correlation between the airway concentrations of Cif and 15-epi-lipoxin A4, a proresolving lipid mediator important in host defense and the resolution of pathogen-initiated inflammation. Here, we observe elevated levels of 15-epi-lipoxin A4 in the lungs of mice infected with a strain of P. aeruginosa that expresses only an inactive form of cif compared with those mice infected with wild-type P. aeruginosa. Together these data support the inclusion of Cif on the list of virulence factors that assist P. aeruginosa in colonizing and damaging the airways of compromised patients. Furthermore, this study establishes techniques that enable our groups to explore the underlying mechanisms of Cif effects during respiratory infection.
Keywords: opportunistic pathogens, gram-negative bacteria, host-pathogen interactions, α/β-hydrolase, airway clearance, mucociliary transport, pro-resolution mediators
INTRODUCTION
The simple act of breathing repeatedly exposes the lung epithelium to airborne microbes. Because the respiratory tract represents a potentially attractive niche for colonization, it is advantageous for the system to possess multiple mechanisms to prevent infection. The mucociliary elevator, which relies on the hydration generated by CFTR-mediated ion secretion (16), removes many inhaled pathogens. Those that bypass the mucociliary transport barrier confront both innate and acquired immune responses, coordinated by signals that control inflammation and resolution (3).
Pathogenic bacteria deploy a wide array of virulence factors that subvert host responses, facilitating infections and exacerbating their severity (14a, 28). Additionally, some invaders exploit preexisting weaknesses in host defenses. One such opportunistic pathogen is Pseudomonas aeruginosa, which can cause pneumonia in otherwise healthy people but most frequently infects patients with underlying disorders such as cystic fibrosis (CF), chronic obstructive pulmonary disease, non-CF bronchiectasis, or conditions requiring mechanical ventilation. In each case, P. aeruginosa infections are linked to poorer prognoses (8, 18–20).
Virulence factors used by P. aeruginosa, including elastase, pyocyanin, and the toxins injected by the type III secretion system, contribute to infection (14a). P. aeruginosa also secretes an epoxide hydrolase first identified as a trigger for degradation of the CF transmembrane conductance regulator (CFTR) in cultured epithelial cells (4, 15, 24). Because mutation of CFTR debilitates mucociliary clearance in CF patients, the ability of the CFTR inhibitory factor (Cif) to decrease surface expression of CFTR implies that Cif may dysregulate mucociliary clearance and thus alter the rate of bacterial clearance in the airway. Cif also targets endogenous epoxide molecules involved in inflammation and resolution (9, 12), potentially disrupting clearance through a second pathway. In this study, we employ reconstituted human bronchial epithelia and a mouse model of acute pneumonia to directly test the hypothesis that Cif can impair the in vivo host airway defenses that target P. aeruginosa.
MATERIALS AND METHODS
Bacterial culture.
The optical density of overnight cultures of P. aeruginosa grown in LB broth was quantified at λ = 600 nm (OD600). Where specified, racemic styrene oxide at 1 mM was added to overnight cultures to induce cif expression. Cultures were then pelleted, washed two times with PBS, and resuspended in PBS at equal optical density.
Mucociliary transport assays.
Fully differentiated primary human bronchial epithelial cells from healthy donors were cultured at an air-liquid interface on 6.5-mm Transwell filters (MucilAir; Epithelix) and maintained in serum-free MucilAir medium in a 5% CO2-95% air incubator at 37°C.
PA14 or isogenic PA14 Δcif preincubated (3 h) in serum-free MucilAir medium, or buffer control, were mixed with fluorescent polystyrene beads [red-orange (565/580 nm), 15 µm FluoSpheres; Molecular Probes] in 0.1% DMSO and incubated for 5 min at room temperature. At time 0, the bacteria/bead suspension was applied to the apical side of four donor-independent sets of healthy MucilAir epithelia maintained at 37°C and 5% CO2-95% air on a Tokai Hit heated microscope stage. Bead movement was visualized by a Nikon LiveScan Swept Field Confocal Microscope with a 561-nm laser line and a ×10 objective. Movies were acquired for 5 min at 10-min intervals over 30 min (820-μm2 field, 24 frames/min). Beads (≥15) were selected from still images of each movie at minute 12 of the experiment and tracked over five frames. Bead velocity at each time point was calculated using the Nikon NIS Elements 2.2 Research Duo software with particle tracking module.
A random-intercept mixed-effect linear model was applied to log2-transformed velocities, with the experimental treatments as the fixed effect and donor identification as a random effect (package “nlme” in Rv3.3.1). Fixed-effect differences and P values between pairs of treatment groups were derived from the full linear model that includes both random and fixed effects.
Immunoblots.
Samples were separated by SDS-PAGE using a 12% (vol/vol) polyacrylamide gel and transferred to a PVDF membrane. After blocking, blots were incubated with a Cif-specific rabbit polyclonal antibody validated previously (9) in 50 mM Tris, pH 8.5, 100 mM NaCl containing 0.1% (vol/vol) Tween 20, 5% (wt/vol) BSA, and 0.02% (wt/vol) NaN3. Cif antibody in Figs. 2A, 2C, and 3A was detected with a goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (Bio-Rad). Cif antibody in Fig. 3D was detected with goat anti-rabbit IgG secondary antibody conjugated with alkaline phosphatase (Sigma).
Fig. 2.
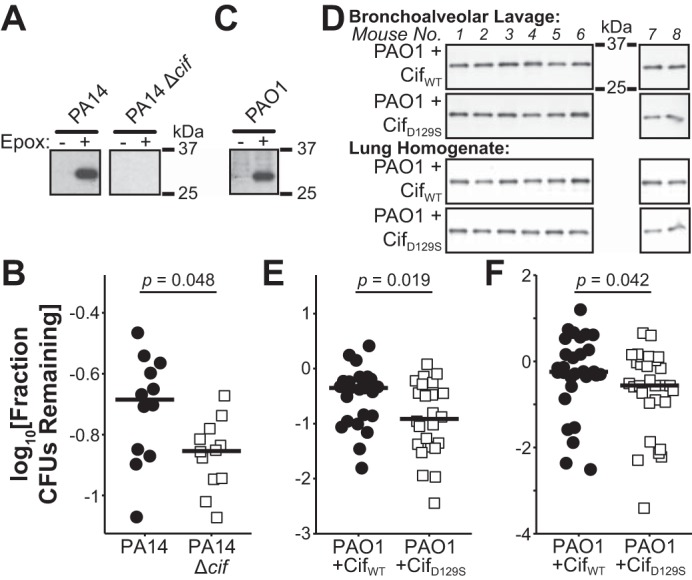
Cif increases the number of colony-forming units (CFUs) recovered from mice in an acute pneumonia model. A: immunoblots for Cif from PA14 and PA14 Δcif strains grown with or without an epoxide inducer. B: ratio of recovered CFUs at 6 h postinfection to input CFUs from bronchoalveolar lavage fluid (BALF) of mice treated with epoxide-induced PA14 or PA14 Δcif (n = 2 experiments, 6 mice for each treatment group/experiment). C: immunoblots for Cif from strain PAO1 grown with or without an epoxide inducer. D: immunoblots from one experimental set of mice for Cif in BALF (E) and lung homogenate (LH) (F) samples. Blots generated at the same time and processed in parallel. E and F: ratio of recovered CFUs at 6 h postinfection to input CFUs from BALF (E, n = 3 experiments, 4–8 mice for each treatment group/experiment) or LH (F, n = 4 experiments, 4–8 mice for each treatment group/experiment) of mice treated with PAO1 in combination with either exogenous, purified CifWT, or CifD129S protein. A and C: panels derived from a single blot. B, E, and F: black bars, medians; significance calculated using a one-tailed exact Wilcoxon-Mann-Whitney test from the R package “coin.”
Fig. 3.
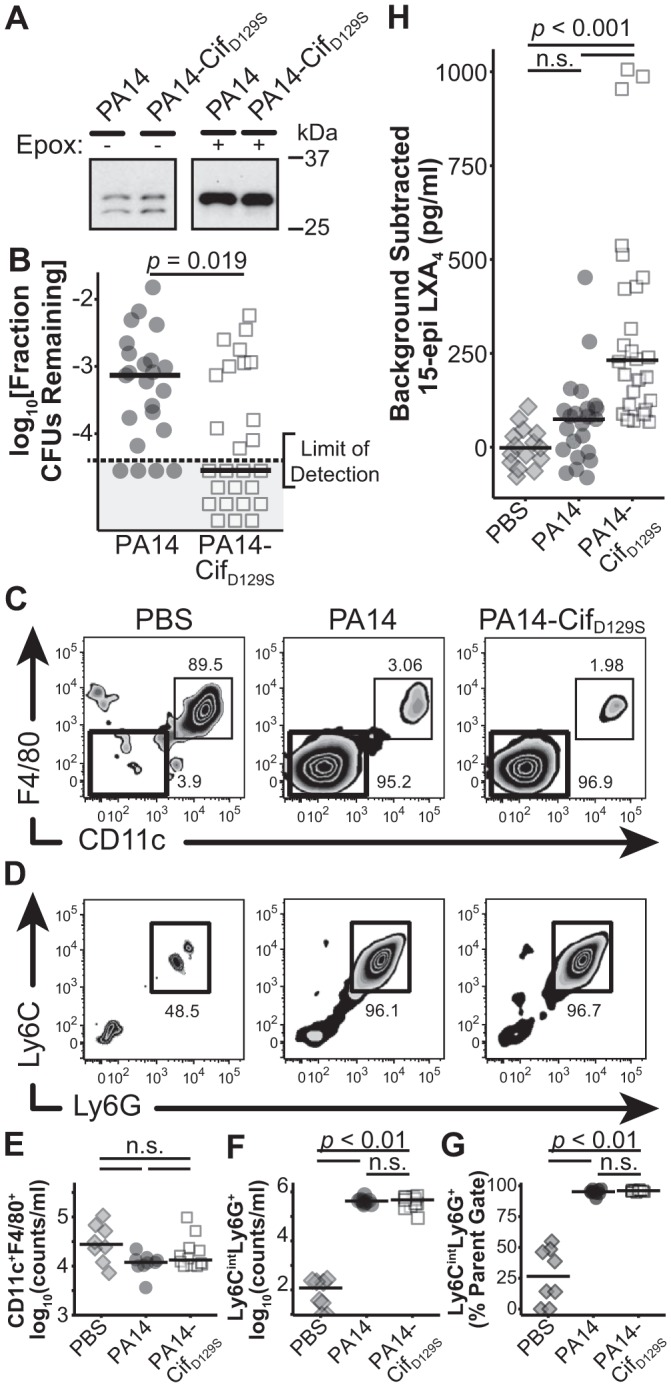
Removal of Cif catalytic activity decreases recovered CFU counts and increases the amount of 15-epi-lipoxin A4 (LXA4) detected in an acute pneumonia model. A: immunoblot of Cif from PA14 and PA14-CifD129S in the absence (1:4 sample dilution) and presence (1:20 sample dilution) of an epoxide inducer. Blots generated at the same time and processed in parallel. B: ratio of recovered CFUs at 24 h postinfection to input CFUs from LH of mice treated with PA14 or PA14-CifD129S (n = 4 experiments, 4–6 mice for each treatment group/experiment; significance calculated using a 1-tailed exact Wilcoxon-Mann-Whitney test from the R package coin.). The gray box indicates those mice where no CFUs were detected. C: representative plots from BALF samples gated on live CD45+CD11b+ markers. Thin-lined black boxes indicate the CD11c+F4/80+ macrophage population, with the percent of parent gate indicated above the black box; thick-lined black boxes indicate the F4/80−CD11c− population displayed in D. D: representative plots from BALF samples gated on live CD45+CD11b+CD11c−F4/80− markers. Black inset boxes indicate the Ly6G+Ly6Cint neutrophil population, with the percent of parent gate indicated below the black box. E: counts of live CD45+CD11b+CD11c−F4/80− macrophages/ml BALF. Not significant (ns), P > 0.05. F: counts of live CD45+CD11b+CD11c−F4/80−Ly6G+Ly6Cint neutrophils/ml BALF. G: percent of parent gates (live CD45+CD11b+CD11c−F4/80−) composed of neutrophils (Ly6G+Ly6Cint). E-G: n = 2 experiments, 4–6 mice for each treatment group/experiment; F and G: ns, P ≥ 0.2. H: background-subtracted levels of 15-epi LXA4 detected in BALF from mice treated with PBS, PA14, or PA14-CifD129S (n = 4 experiments, 4–6 mice for each treatment group/experiment; ns, P ≥ 0.2.). E–H: black bars, medians; significance calculated using a Kruskal-Wallis Rank Sum test followed by a multiple-comparisons test in the R package “pgirmess.”
Protein purification.
Recombinant Cif or hydrolysis-deficient mutant CifD129S protein was purified by nickel-affinity chromatography as described previously (2). Protein was concentrated (Amicon Ultra 10K NMWL), dialyzed into GIBCO Dulbecco’s phosphate-buffered saline (DPBS), and diluted to 10 mg/ml.
Mouse acute pneumonia model.
The protocol for animal infection was approved by Dartmouth’s Institutional Animal Care and Use Committee, in accordance with Association for Assessment and Accreditation of Laboratory Animal Care guidelines. All efforts were made to minimize animal suffering.
Mouse experiments were completed as described previously (27), with modification. Purchased male C57BL/6J mice 8–12 wk old (Jackson Laboratories) were inoculated with DPBS-resuspended OD600-normalized cultures of P. aeruginosa or buffer control via oropharyngeal aspiration following anesthesia with isoflourane. Mice in the PAO1 treatment groups received two doses of 50 μl PAO1 with Cif protein [5 × 107 colony-forming units (CFUs); 1 mg protein]. Mice in the PA14 treatment groups received one dose of 40 μl (1 × 106 CFUs) of P. aeruginosa PA14, isogenic PA14 Δcif, or isogenic PA14-CifD129S. At 6 or 24 h after infection, mice were anesthetized and euthanized with isofluorane followed by cervical dislocation. Tracheas were cannulated, and lungs were lavaged using DPBS. Bronchoalveolar lavage fluid (BALF) samples were immediately put on ice, and aliquots were snap-frozen neat, in methanol, or in Laemmli sample buffer. Lungs were excised and immediately placed in 1 ml of cold DPBS followed by homogenization, and frozen as above. Exact CFUs contained in the input and recovered samples were determined by plating serial dilutions of the bacterial suspensions on Pseudomonas Isolation Agar (Difco) followed by incubation at 37°C for 24 h. Each point on a single plot represents an independent animal.
Construction of PA14-CifD129S.
CifD129S with 1,000 bp of 5′- and 3′-genomic flanking sequences was cloned in the plasmid pMQ30 using Gibson assembly (NEB). Plasmids were transformed to E. coli S17/λpir strain and selected with 10 μg/ml gentamycin. Plamids were conjugated in P. aeruginosa strain PA14 for integration at the endogenous cif locus, as previously described (14, 15, 23). Integration and plasmid excision were verified through PCR and sequencing. No growth differences were observed between the new strain and the published strains.
15-epi-lipoxin A4 ELISA.
BALF samples snap-frozen in 3 vol of methanol were stored at −80°C until solid-phase extraction and quantification of 15-epi-lipoxin A4 (15-epi LXA4) by ELISA (Neogen) as previously described (5). Each point on the plot represents an independent animal.
Neutrophil and macrophage quantification.
Cells within the BALF were pelleted, resuspended in buffer [DPBS supplemented with 2 mM EDTA and 0.2% (wt/vol) BSA], and aliquoted in a 96-well round-bottom plate. Samples were washed two times in buffer and Fc receptor blocked with anti-CD16 and anti-CD32. The suspensions were stained with antibodies against CD45-APC-Cy7 (30-F11), CD11b-PerCP (M1/70), CD11c-APC (N418), F4/80-FITC (BM8), Ly6C-PE (HK1.4), and Ly6G-PE-Cy7 (1A8) (BioLegend). Cell counts were acquired on a Miltenyl MACS Quant10 Analyzer, and the data were analyzed with FlowJo (version 9.8.1). Neutrophils were defined as live CD45+CD11b+CD11c−F4/80−Ly6CintLy6G+ cells, and macrophages were defined as live CD45+CD11b+CD11c+F4/80+ cells. Only a fraction of each sample was tested, and absolute numbers were corrected for dilution and volume.
Figure generation and statistics.
Plots were generated with the R (version 3.3.1) package “ggplot2.” Microscopy images were generated with ImageJ (version 1.51j). All figures were assembled using Adobe Illustrator (version CS6). Statistical significance was calculated using R as described in individual legends in Figs. 1–4.
RESULTS AND DISCUSSION
Cif decreases mucociliary transport.
We have previously demonstrated that Cif reduces apical CFTR (4, 15, 24), an effect that is predicted to reduce ion transport and thus reduces both the airway surface liquid volume and mucociliary transport (MCT). To test whether Cif reduces MCT, we employed reconstituted human bronchial epithelia, which secrete mucus and transport it through ciliary motion. We applied fluorescently labeled beads mixed with either P. aeruginosa wild-type (WT) strain PA14, an isogenic deletion construct PA14 Δcif, or a buffer control to the apical side of the epithelial cell cultures (9, 15). The beads that become trapped in the mucus allow us to monitor the motions of the mucus by videomicroscopy. Movies 5 min in duration were recorded beginning at time points 0, 10, and 20 min after treatment [Supplemental Movies (Supplemental data for this article may be found on the journal website.)], as shown in the timeline in Fig. 1A.
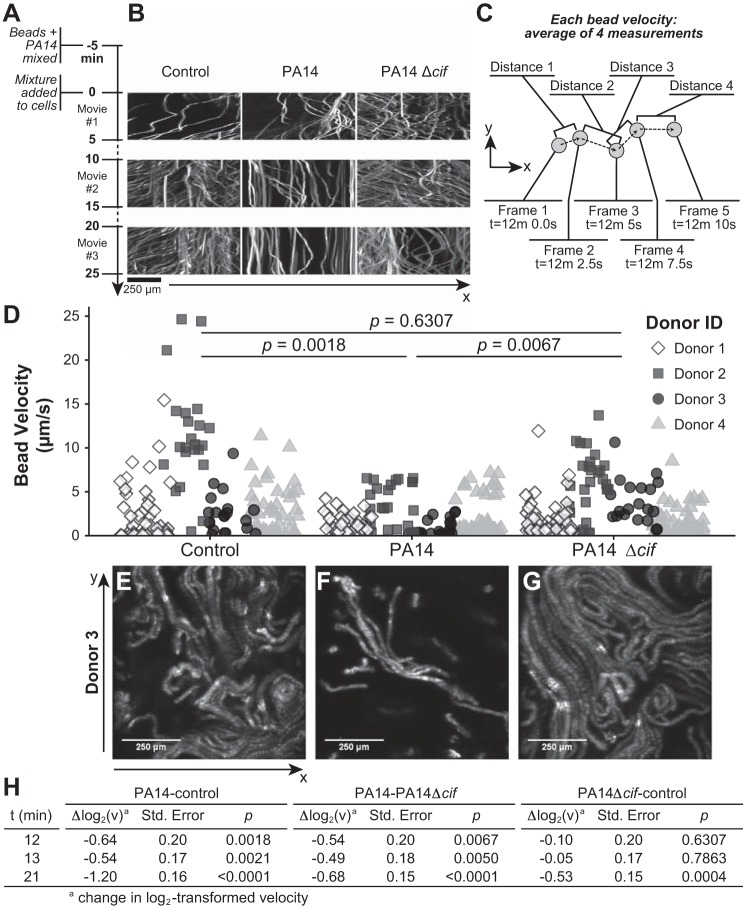
Fig. 1.
Deletion of Cif (cystic fibrosis transmembrane conductance regulator inhibitory factor) slows PA14 inhibition of mucociliary transport (MCT). A: timeline of the experiment. B: view of bead locations in one positional axis (horizontal axis) over time (vertical axis) from the epithelium of donor 3. C: diagram of the velocity calculation for each bead shown as a point in D. D: quantification of individual bead velocities from each condition between time point 12 min 0.0 s and time point 12 min 10.0 s, stratified by donor ID. P values indicate significance derived from log2 transformation and modeling as described in the text and in H. E: maximum intensity projection of the movie from the control condition between the 10- and 15-min marks from donor 3. F: projection from the same donor and capture as E, where PA14 was added to the apical side of the culture dish. G: projection from the same donor and capture as E, where PA14 Δcif was added to the apical side of the culture dish. H: results of a random-intercept mixed-effect linear model used to assess the magnitude and statistical significance of differences in log2-transformed bead velocities associated with treatment with vehicle control, PA14, or PA14 Δcif.
As observed in previous MCT studies, a single epithelial cell culture dish displayed a range of bead speeds and directions (11, 22). To explore gross differences, we rendered the movies as volumes, with time as the vertical axis (Fig. 1B). In this view, motion can be read as movement parallel to the horizontal axis in Fig. 1B. Beads that remain stationary through the duration of the movie appear as vertical lines. This view focused our attention on the 10- through 15-min captures. This time period not only displayed the greatest qualitative difference between our experimental conditions, but parallels earlier studies into the effects of P. aeruginosa on CFTR levels (24).
Because we predicted Cif would decrease MCT, it was crucial to assess all beads, not just those in rapid motion. By selecting beads from a still image at minute 12, we eliminated the risk of preferentially selecting fast-moving particles. The positions of 15–25 selected beads were tracked in the x–y plane over the following four consecutive frames, each captured 2.5 s apart (Fig. 1C). We converted the four distances traveled between frames into velocity measurements and plotted the average velocity of each bead in Fig. 1D. A number of factors, including positional differences in the z-axis and directional changes between captured frames, may alter the observed velocities for individual beads. Averaging generated a velocity that more accurately approximates each bead’s true velocity within a specified window of time.
Figure 1, E, F, and G, shows the additive superposition of the two-dimensional images captured between the 10- and 15-min time points for the reconstituted epithelia of one donor, following incubation with either buffer, PA14, or PA14 Δcif, respectively. In this depiction, each particle appears as a two-dimensional trajectory. As mentioned above, we expected the nonlinear movements of the beads embedded in the mucus. Figure 1D shows bead velocity as a function of both donor and experimental conditions. In the control group, the range of velocities varies by donor, indicating that each donor displays different velocity set points as previously described (22, 26). However, we can glean some qualitative information by comparing the treatment groups within each donor set. For example, the beads incubated with buffer and tracked on the epithelium from donor 3 (Fig. 1D) display a range of velocities between 0 and 10 μm/s. When incubated with PA14, the fastest beads only travel 2.5 μm/s. In contrast, when incubated with PA14 Δcif, the range of bead velocities approximates that of control cells, values much faster than those seen with WT PA14.
To quantify and assess the significance of the differences among the treatment groups, we employed a mixed-effect linear model. After log2 transformation of the data for a normal distribution, this method of analysis allowed us to compare across the treatment regimes while accounting for the contribution of interdonor differences. At time points in Supplemental Movies 2 (10–15 min) and 3 (20–25 min), exposure to PA14 significantly reduced bead velocity relative to control treatment or to treatment with PA14 Δcif (Fig. 1, D and H). Treatment with PA14 Δcif did not cause a significant change in bead velocity compared with control during Supplemental Movie 2. By Supplemental Movie 3, PA14 Δcif significantly slowed bead velocity, although WT PA14 caused a greater decrease (Fig. 1H). Based on these data, Cif decreases MCT in vitro and thus may also reduce the clearance of P. aeruginosa from the host.
Cif decreases bacterial clearance in vivo.
To explore the effect of Cif on clearance of P. aeruginosa in vivo, we employed a mouse pneumonia model in which P. aeruginosa was aspirated in the lungs of mice and clearance was assessed by counting CFUs recovered in BALF and lung homogenate (LH). Our initial experiment employed the bacterial strains used in the MCT experiments, PA14 and PA14 Δcif. To normalize across experiments and groups, we calculated the remaining CFU fractions by dividing counts of recovered CFUs by the input CFU count per mouse. Although no significant differences were detected between the treatment groups at 6 h postinfection, the BALF samples displayed a trend (P = 0.20), with mice treated with PA14 retaining a greater fraction of input CFUs in the BALF (median = 0.10, interquartile range = 0.04–0.13) than BALF from mice treated with PA14 Δcif (median = 0.04, interquartile range = 0.03–0.04).
We hypothesized that, by priming the production of Cif by P. aeruginosa, we could increase the effect of Cif on the system. Exposing P. aeruginosa to an epoxide inducer increases bacterial synthesis of Cif (Fig. 2A). Mice inoculated with epoxide-exposed PA14 generated significantly higher CFU counts than mice inoculated with epoxide-exposed PA14 Δcif (Fig. 2B). A difference of this magnitude corresponds to a decrease in virulence for P. aeruginosa strains deficient in phenazine production, a well-characterized virulence factor (21).
Despite the decrease in clearance mediated by Cif, we were unable to detect Cif protein in the BALF from mice infected with PA14. Because BALF samples from human CF patients contained Cif at levels detectable by immunoblot (9), we assessed what effect a detectable dose of Cif has on bacterial clearance. For these studies, we employed purified Cif protein and the P. aeruginosa strain PAO1. Although PAO1 produces Cif when exposed to an industrial epoxide (Fig. 2C), PAO1 does not induce a Cif-mediated decrease of CFTR (24). PAO1 has the additional advantage of being less virulent than PA14 (17, 25), which we predicted would allow us to better identify the effect of Cif on clearance. To the PAO1 inoculum, we added either purified CifWT protein or the catalytically inactive CifD129S protein (2). At 6 h postinoculation, we could detect the protein in both the BALF and LH of the inoculated mice (Fig. 2D). The presence of CifWT increases the number of recovered CFUs in both the BALF and LH compared with the inoculum supplemented with the catalytically dead mutant CifD192S, and the use of exogenous Cif increased the magnitude of the effect compared with the earlier experiments (Fig. 2, D and E). Thus, Cif disrupts host clearance of P. aeruginosa.
Cif alters clearance and 15-epi LXA4 concentrations in vivo.
To test the effects of Cif’s catalytic activity on bacterial clearance without the use of exogenous protein, we generated a new strain of P. aeruginosa. We introduced a chromosomal mutation in PA14 that results in the production of only the catalytically inactive mutant CifD129S protein by PA14. The protein CifD129S is expressed by the new PA14-CifD129S strain, and exposure to an epoxide inducer increases expression to the same degree as in the WT strain (Fig. 3A). Additionally, because our recently published results linked Cif’s catalytic activity to a reduction of proresolution lipid mediators (9), we moved our time point to 24 h postinfection. This time point captures both a slice of the resolution phase (1) and bacterial clearance at a later interval. At 24 h, mice inoculated with PA14 retain a higher fraction of input CFUs in the LH samples compared with mice inoculated with PA14-CifD129S (Fig. 3B), confirming the ability of Cif to reduce bacterial clearance from mouse lungs, as well as the effect’s persistence at the 24-h time point.
In addition to CFUs, we also examined the effect of Cif on innate immune cell infiltration and 15-epi LXA4 concentrations in the BALF of these mice (Fig. 3, C–H). Whereas no significant differences in BALF macrophage counts were evident at 24 h (Fig. 3, C and E), mice inoculated with either PA14 or PA14-CifD129S displayed significantly higher numbers of neutrophils compared with the PBS control but no difference when compared with each other (Fig. 3, D, F, and G). Only mice treated with PA14-CifD129S showed elevated 15-epi LXA4 concentrations (Fig. 3H). As we predicted, mice treated with PA14 did not exhibit elevated levels of 15-epi LXA4, consistent with Cif-mediated hydrolysis of a key upstream signal, 14,15-epoxyeicosatrienoic acid (9). Taken together, our results suggest that removal of Cif’s catalytic activity changes the dynamics of the interaction between P. aeruginosa and multiple host defense mechanisms in the lung, leading to the observed decrease in bacterial clearance.
Conclusions.
In summary, the data implicate Cif as a virulence factor in which catalytic activity manipulates the host-pathogen interaction for the benefit of P. aeruginosa (Fig. 4). We originally identified Cif as a factor that can reduce overall CFTR ion-channel activity in vitro, with likely implications for airway colonization (15). In this study, we present the first direct evidence that P. aeruginosa can deploy Cif to decrease bacterial clearance in vivo, potentially through dysregulation of both MCT and 15-epi LXA4 signaling. These findings underline the clinical relevance of Cif established by previous studies of patient samples: sputum contains Cif mRNA, serum harbors Cif-specific antibodies, and BALF concentrations of Cif correlate with reduced airway function (9, 10).
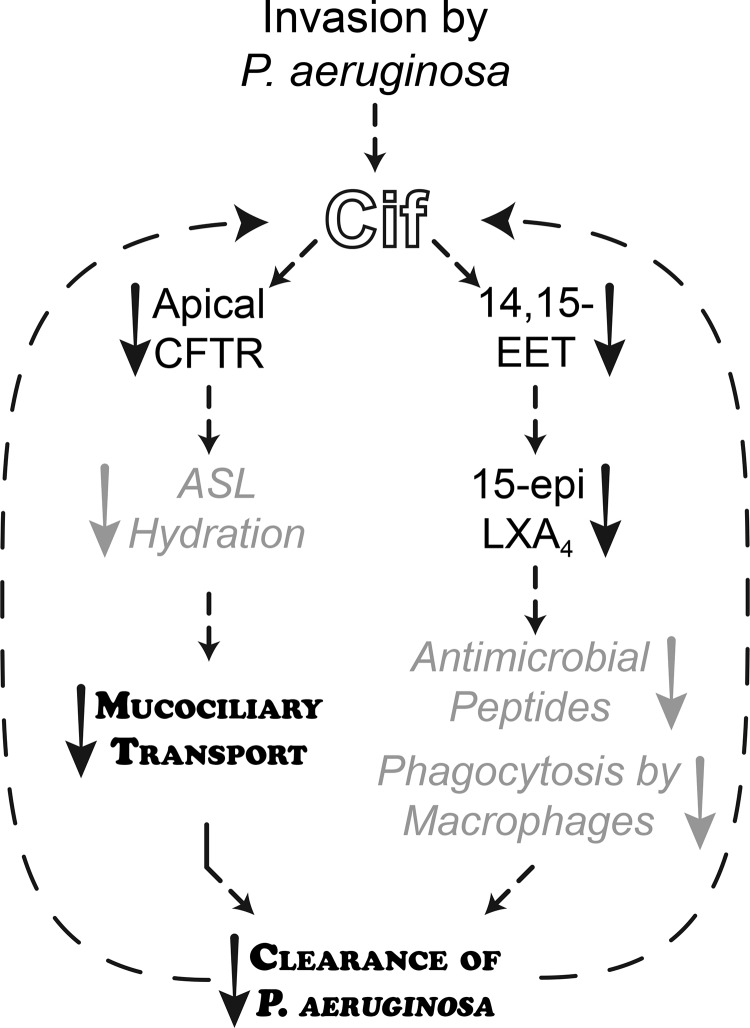
Fig. 4.
Cif decreases host clearance of P. aeruginosa. As we have shown previously, Cif causes a decrease in cystic fibrosis transmembrane conductance regulator (CFTR) abundance at the apical membrane of bronchial epithelial cells that in turn decreases ion secretion. This likely dehydrates the airway surface liquid (ASL) and decreases MCT. The effect of Cif on MCT is notably rapid; like other virulence factors, the decrease in MCT may derive from a direct effect on cilia although a rapid CFTR-driven MCT effect has been seen with other stimuli (7). In addition to the effect of Cif effect on CFTR, our previous work shows that Cif hydrolyzes 14,15-epoxyeicosatrienoic acid (14,15-EET). Hydrolysis of 14,15-EET decreases the synthesis of 15-epi LXA4, a proresolving lipid mediator that enhances phagocytosis by macrophages and may increase production of antimicrobial peptides (3). Together, the effects of Cif on MCT and 15-epi LXA4 decrease clearance of P. aeruginosa in the acute pneumonia model, likely increasing the severity or duration of the infection. Finally, because cif expression persists in CF clinical isolates (9, 10), reduced clearance is likely to yield continued exposure to Cif. Key: Standard black text, published work; gray arrows and italic text, proposed mechanisms (3, 7); bold, small capital text, effects demonstrated in this work.
We have now streamlined the tools necessary to delve into the mechanistic underpinnings of Cif-disrupted mucociliary transport and 15-epi LXA4 production (Fig. 4). To quantify MCT, we developed methods of analysis to account for the inter- and intraculture variation and non-Gaussian properties of the data (Fig. 1), which could be more broadly applied to other MCT datasets. The acute pneumonia model will function as a platform to test the mechanism and timeline underlying Cif’s dysregulation of inflammatory and resolution processes (Fig. 4). The techniques outlined above, combined with our work developing inhibitors of Cif hydrolysis (13) and identifying alternative substrates (12), provide us with the tools to systematically interrogate Cif’s molecular mechanisms and to explore the interplay of ion flux, mucus transport, and inflammatory signaling in vivo.
GRANTS
This work was supported by National Institutes of Health Grants R01-AI-091699, R01-HL-074175, P20-GM-113132, P30-GM-106394, T32-GM-008704, R01-HL-123771, T32-AI-060525, P01-GM-095467, and U24-AI-118656, CFF (STANTO15R0), Gilead Research Scholars in Cystic Fibrosis, and the Munck-Pfefferkorn Fund.
DISCLOSURES
D.R.M. and C.D.B. are coinventors of Cif inhibitor intellectual property held by Dartmouth. B.D.L. is an inventor on patents (pro-resolving mediators) licensed by Brigham and Women’s Hospital for clinical development. E.D. is currently employed by Agilent Technologies.
AUTHOR CONTRIBUTIONS
K.L.H., E.D., S.M.-M., B.A.F., C.D.B., J.M.B., B.A.S., D.A.H., and D.R.M. conceived and designed research; K.L.H., E.D., S.M.-M., T.B.S., and B.A.F. performed experiments; K.L.H., E.D., T.H.H., T.B.S., and B.A.F. analyzed data; K.L.H., T.B.S., B.A.F., J.M.B., B.D.L., B.A.S., D.A.H., and D.R.M. interpreted results of experiments; K.L.H. prepared figures; K.L.H. and D.R.M. drafted manuscript; K.L.H., E.D., S.M.-M., T.H.H., T.B.S., B.A.F., J.M.B., B.D.L., B.A.S., D.A.H., and D.R.M. edited and revised manuscript; K.L.H., E.D., S.M.-M., T.H.H., T.B.S., B.A.F., C.D.B., J.M.B., B.D.L., B.A.S., D.A.H., and D.R.M. approved final version of manuscript.
Supplemental Data
Buffer-Treated Reconstituted Human Epithelia at 0 minutes
Buffer-Treated Reconstituted Human Epithelia at 10 minutes
Buffer-Treated Reconstituted Human Epithelia at 20 minutes
PA14-Treated Reconstituted Human Epithelia at 0 minutes
PA14-Treated Reconstituted Human Epithelia at 10 minutes
PA14-Treated Reconstituted Human Epithelia at 20 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 0 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 10 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 20 minutes
ACKNOWLEDGMENTS
We thank Drs. Brent Berwin, Robert Cramer, Joshua Obar, George O’Toole, and Scott Gerber for discussions and suggestions.
REFERENCES
- 1.Abdulnour RE, Sham HP, Douda DN, Colas RA, Dalli J, Bai Y, Ai X, Serhan CN, Levy BD. Aspirin-triggered resolvin D1 is produced during self-resolving gram-negative bacterial pneumonia and regulates host immune responses for the resolution of lung inflammation. Mucosal Immunol 9: 1278–1287, 2016. doi: 10.1038/mi.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahl CD, Hvorecny KL, Bomberger JM, Stanton BA, Hammock BD, Morisseau C, Madden DR. Inhibiting an epoxide hydrolase virulence factor from Pseudomonas aeruginosa protects CFTR. Angew Chem Int Ed Engl 54: 9881–9885, 2015. doi: 10.1002/anie.201503983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basil MC, Levy BD. Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nat Rev Immunol 16: 51–67, 2016. doi: 10.1038/nri.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomberger JM, Ye S, Maceachran DP, Koeppen K, Barnaby RL, O’Toole GA, Stanton BA. A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS Pathog 7: e1001325, 2011. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang N, Takano T, Clish CB, Petasis NA, Tai HH, Serhan CN. Aspirin-triggered 15-epi-lipoxin A4 (ATL) generation by human leukocytes and murine peritonitis exudates: development of a specific 15-epi-LXA4 ELISA. J Pharmacol Exp Ther 287: 779–790, 1998. [PubMed] [Google Scholar]
- 7.Clunes LA, Davies CM, Coakley RD, Aleksandrov AA, Henderson AG, Zeman KL, Worthington EN, Gentzsch M, Kreda SM, Cholon D, Bennett WD, Riordan JR, Boucher RC, Tarran R. Cigarette smoke exposure induces CFTR internalization and insolubility, leading to airway surface liquid dehydration. FASEB J 26: 533–545, 2012. doi: 10.1096/fj.11-192377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cystic Fibrosis Foundation Patient Registry Annual Data Report 2015 (online). https://www.cff.org/UploadedFiles/research/ClinicalResearch/Patient-Registry-Report-2009.pdf.
- 9.Flitter BA, Hvorecny KL, Ono E, Eddens T, Yang J, Kwak DH, Bahl CD, Hampton TH, Morisseau C, Hammock BD, Liu X, Lee JS, Kolls JK, Levy BD, Madden DR, Bomberger JM. Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators. Proc Natl Acad Sci USA 114: 136–141, 2017. doi: 10.1073/pnas.1610242114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gifford AH, Willger SD, Dolben EL, Moulton LA, Dorman DB, Bean H, Hill JE, Hampton TH, Ashare A, Hogan DA. Use of a multiplex transcript method for analysis of Pseudomonas aeruginosa gene expression profiles in the cystic fibrosis lung. Infect Immun 84: 2995–3006, 2016. doi: 10.1128/IAI.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoegger MJ, Awadalla M, Namati E, Itani OA, Fischer AJ, Tucker AJ, Adam RJ, McLennan G, Hoffman EA, Stoltz DA, Welsh MJ. Assessing mucociliary transport of single particles in vivo shows variable speed and preference for the ventral trachea in newborn pigs. Proc Natl Acad Sci USA 111: 2355–2360, 2014. doi: 10.1073/pnas.1323633111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hvorecny KL, Bahl CD, Kitamura S, Lee KSS, Hammock BD, Morisseau C, Madden DR. Active-site flexibility and substrate specificity in a bacterial virulence factor: Crystallographic snapshots of an epoxide hydrolase. Structure 25: 697–707.e4, 2017. doi: 10.1016/j.str.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura S, Hvorecny KL, Niu J, Hammock BD, Madden DR, Morisseau C. Rational design of potent and selective inhibitors of an epoxide hydrolase virulence factor from Pseudomonas aeruginosa. J Med Chem 59: 4790–4799, 2016. doi: 10.1021/acs.jmedchem.6b00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchma SL, Connolly JP, O’Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol 187: 1441–1454, 2005. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Le Berre R, Nguyen S, Nowak E, Kipnis E, Pierre M, Quenee L, Ader F, Lancel S, Courcol R, Guery BP, Faure K; Pyopneumagen Group . Relative contribution of three main virulence factors in Pseudomonas aeruginosa pneumonia. Crit Care Med 39: 2113–2120, 2011. doi: 10.1097/CCM.0b013e31821e899f. [DOI] [PubMed] [Google Scholar]
- 15.MacEachran DP, Ye S, Bomberger JM, Hogan DA, Swiatecka-Urban A, Stanton BA, O’Toole GA. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect Immun 75: 3902–3912, 2007. doi: 10.1128/IAI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mall MA. Role of cilia, mucus, and airway surface liquid in mucociliary dysfunction: lessons from mouse models. J Aerosol Med Pulm Drug Deliv 21: 13–24, 2008. doi: 10.1089/jamp.2007.0659. [DOI] [PubMed] [Google Scholar]
- 17.Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 6: e29113, 2011. doi: 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy TF. The many faces of Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Clin Infect Dis 47: 1534–1536, 2008. doi: 10.1086/593187. [DOI] [PubMed] [Google Scholar]
- 19.Pasteur MC, Bilton D, Hill AT; British Thoracic Society Bronchiectasis non-CF Guideline Group . British Thoracic Society guideline for non-CF bronchiectasis. Thorax 65, Suppl 1: i1–i58, 2010. doi: 10.1136/thx.2010.136119. [DOI] [PubMed] [Google Scholar]
- 20.Ramírez-Estrada S, Borgatta B, Rello J. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infect Drug Resist 9: 7–18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Recinos DA, Sekedat MD, Hernandez A, Cohen TS, Sakhtah H, Prince AS, Price-Whelan A, Dietrich LEP. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc Natl Acad Sci USA 109: 19420–19425, 2012. doi: 10.1073/pnas.1213901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sears PR, Yin W-N, Ostrowski LE. Continuous mucociliary transport by primary human airway epithelial cells in vitro. Am J Physiol Lung Cell Mol Physiol 309: L99–L108, 2015. doi: 10.1152/ajplung.00024.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72: 5027–5036, 2006. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swiatecka-Urban A, Moreau-Marquis S, Maceachran DP, Connolly JP, Stanton CR, Su JR, Barnaby R, O’toole GA, Stanton BA. Pseudomonas aeruginosa inhibits endocytic recycling of CFTR in polarized human airway epithelial cells. Am J Physiol Cell Physiol 290: C862–C872, 2006. doi: 10.1152/ajpcell.00108.2005. [DOI] [PubMed] [Google Scholar]
- 25.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA 96: 715–720, 1999. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanner A. Clinical aspects of mucociliary transport. Am Rev Respir Dis 116: 73–125, 1977. [DOI] [PubMed] [Google Scholar]
- 27.Wargo MJ, Gross MJ, Rajamani S, Allard JL, Lundblad LKA, Allen GB, Vasil ML, Leclair LW, Hogan DA. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am J Respir Crit Care Med 184: 345–354, 2011. doi: 10.1164/rccm.201103-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb SAR, Kahler CM. Bench-to-bedside review: Bacterial virulence and subversion of host defences. Crit Care 12: 234, 2008. doi: 10.1186/cc7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Buffer-Treated Reconstituted Human Epithelia at 0 minutes
Buffer-Treated Reconstituted Human Epithelia at 10 minutes
Buffer-Treated Reconstituted Human Epithelia at 20 minutes
PA14-Treated Reconstituted Human Epithelia at 0 minutes
PA14-Treated Reconstituted Human Epithelia at 10 minutes
PA14-Treated Reconstituted Human Epithelia at 20 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 0 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 10 minutes
PA14 delta-cif-Treated Reconstituted Human Epithelia at 20 minutes