Significance
Crops carrying broad-spectrum resistance loci provide an effective strategy for controlling infectious disease. Despite their importance, few broad-spectrum resistance loci have been reported, and the underlying mechanisms controlling the trait remain largely unknown. This report describes the identification of a gene, called “bsr-k1,” conferring broad-spectrum resistance and demonstrates that the encoded protein regulates immunity-related genes. Loss of function of BSR-K1 in rice leads to enhanced broad-spectrum resistance to two serious rice diseases with no major penalty on yield. This report provides insights into broad-spectrum resistance and offers an efficient strategy to breeding durably resistant rice.
Keywords: blast disease, broad-spectrum resistance, innate immunity, TPR protein, rice
Abstract
Crops carrying broad-spectrum resistance loci provide an effective strategy for controlling infectious disease because these loci typically confer resistance to diverse races of a pathogen or even multiple species of pathogens. Despite their importance, only a few crop broad-spectrum resistance loci have been reported. Here, we report the identification and characterization of the rice bsr-k1 (broad-spectrum resistance Kitaake-1) mutant, which confers broad-spectrum resistance against Magnaporthe oryzae and Xanthomonas oryzae pv oryzae with no major penalty on key agronomic traits. Map-based cloning reveals that Bsr-k1 encodes a tetratricopeptide repeats (TPRs)-containing protein, which binds to mRNAs of multiple OsPAL (OsPAL1–7) genes and promotes their turnover. Loss of function of the Bsr-k1 gene leads to accumulation of OsPAL1–7 mRNAs in the bsr-k1 mutant. Furthermore, overexpression of OsPAL1 in wild-type rice TP309 confers resistance to M. oryzae, supporting the role of OsPAL1. Our discovery of the bsr-k1 allele constitutes a significant conceptual advancement and provides a valuable tool for breeding broad-spectrum resistant rice.
Plant disease causes devastating yield losses in crop production, threatening global food security (1, 2). Repeated over-application of pesticides to control plant diseases has polluted many environments around the globe (3, 4). Crop genetic improvement for resistance is the most economical and environmentally friendly approach to preventing disease outbreaks (3, 4). Although many resistance (R) genes conferring race-specific resistance have been deployed in plant breeding, their resistance often remains effective for only a few years, presumably due to strong selection pressure for evolution of virulent races (5, 6). In contrast, the broad-spectrum resistance controlled by multiple genes or quantitative trait loci (QTLs) is often durable and therefore more effective for disease management (5, 6).
To date, nine plant genes conferring broad-spectrum resistance have been isolated: Arabidopsis RPW8.2 (7), rice Xa21, STV11, pi21, Pigm, and bsr-d1 (8–12), wheat Lr34 and Yr36 (13, 14), and barley mlo (15). Only a few of these genes have their molecular mechanisms of action elucidated. Arabidopsis RPW8.2 is specifically targeted to the plant extrahaustorial membrane adjacent to fungal haustorium and activates the salicylic acid (SA)-dependent defense response, leading to broad-spectrum resistance against diverse races of powdery mildew (16). Rice Xa21 encoding a receptor kinase phosphorylates downstream genes to trigger a series of defense responses, bringing about broad-spectrum resistance to Xanthomonas oryzae pv oryzae (Xoo) (17). Rice STV11 encodes a sulfotransferase catalyzing the conversion of SA to sulfonated SA, conferring broad-spectrum resistance to rice stripe virus (9). Rice bsr-d1 reduces the expression of H2O2-degrading enzymes through the binding of a repressive MYB transcription factor (MYBS1) to the bsr-d1 promoter, generating broad-spectrum resistance to Magnaporthe oryzae (12). Wheat Yr36 encodes a kinase-START protein, which phosphorylates the thylakoid-associated ascorbate peroxidase in chloroplast, resulting in broad-spectrum resistance to diverse races of stripe rust (18).
In some cases, close linkage to undesirable agricultural traits has hindered the use of broad-spectrum resistance genes to effectively control diseases of elite cultivars. For example, wheat Lr34 lines produce less grain than lines without Lr34 (19). The barley recessive mlo mutant displays early senescence-like leaf chlorosis (20). The rice pi21 gene is tightly linked with a gene causing inferior grain quality (10). Thus, there is a need to identify novel genes conferring broad-spectrum resistance for integration into crop genomes without penalizing major superior agricultural traits.
Here we report the rice bsr-k1 (broad-spectrum resistance Kitaake-1) mutant that displays broad-spectrum resistance against M. oryzae and Xoo and maintains major agronomic traits. Map-based cloning reveals that Bsr-k1 encodes a tetratricopeptide repeats (TPRs)-containing protein. Disruption of Bsr-k1 enhances resistance to diverse isolates of M. oryzae and Xoo. The BSR-K1 protein binds to mRNAs of the defense-related OsPAL genes, promoting their turnover. Loss of function of BSR-K1 decreases mRNA turnover, resulting in OsPAL mRNA accumulation in the bsr-k1 mutant. Collectively, our study reveals the mechanism by which BSR-K1 regulates the immune response and provides a tool for breeding rice with broad-spectrum resistance.
Results
The Rice bsr-k1 Mutant Confers Enhanced Resistance Against Diverse Races of M. oryzae and Xoo.
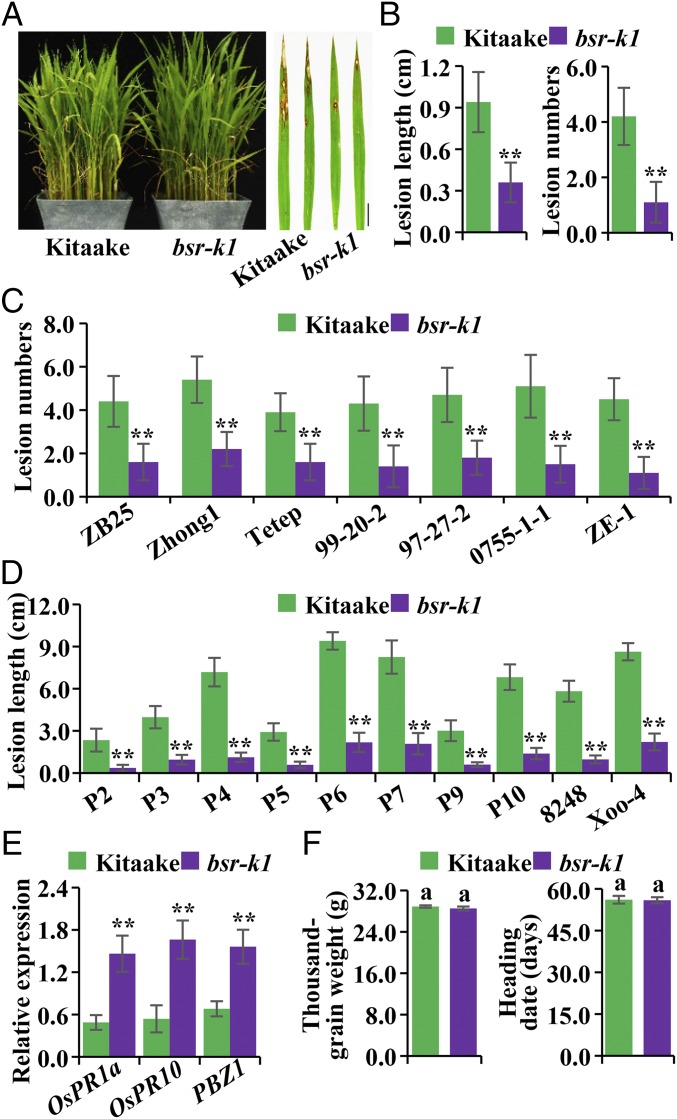
To identify genes involved in rice blast resistance, we generated a mutant population containing ∼50,000 lines in the model rice Kitaake cultivar through ethylmethane sulfonate (EMS)-mediated mutagenesis. We then inoculated these lines with mixed spores derived from seven Kitaake-compatible M. oryzae isolates (ZB25, Zhong1, Tetep, 99-20-2, 97-27-2, 0755-1-1, and ZE-1) in the field. We obtained five mutant lines showing enhanced disease resistance compared with wild-type Kitaake. Of these, the bsr-k1 mutant exhibits significantly smaller lesion length and fewer lesions than Kitaake (Fig. 1 A and B). We validated the smaller lesion size and lower fungal biomass accumulation in bsr-k1 (Fig. S1A) using a punch inoculation method (21). We then tested the seven Kitaake-compatible blast isolates individually in field inoculation. We found that the lesion numbers were dramatically reduced in bsr-k1 with all isolates (Fig. 1C). For example, bsr-k1 yields 1.6 ± 0.8 lesions, while Kitaake has 4.4 ± 1.2, when inoculated with isolate ZB25 (Fig. 1C). To assess if bsr-k1 enhances resistance to other pathogens, we inoculated bsr-k1 and Kitaake with 10 Xoo isolates (P2–P7, P9, P10, 8248, and Xoo-4) that are commonly used to test for broad-spectrum resistance to rice bacterial blight (22). We found that bsr-k1 confers enhanced resistance to all 10 Xoo isolates (Fig. 1D).
Fig. 1.
Phenotypic characterization of the bsr-k1 allele. (A) Field inoculation of bsr-k1 and wild-type Kitaake plants. Shown are photographs of representative plants and leaves taken 10 d postinoculation (dpi) with spore mixtures of seven Kitaake-compatible blast isolates. (Scale bar: 1 cm.) (B) Lesion lengths and numbers (mean ± SD, n > 30) after inoculation with seven Kitaake-compatible blast isolates. (C) Lesion numbers (mean ± SD, n > 30) after inoculation with seven individual blast isolates (names are given under bars). (D) Lesion lengths measured 14 dpi with 10 individual Xoo isolates (names are given below the bars). (E) Comparison of defense gene expression levels (mean ± SEM, n = 3) in bsr-k1 and Kitaake. This experiment was repeated twice with similar results. (F) Comparison of thousand-grain weights and heading dates. Error bars represent mean ± SD (n > 30). The same letter above bars indicates same statistical group (P > 0.05, Tukey’s multiple-comparison test). Asterisks indicate statistical significance (**P ≤ 0.01, t test).
Because the immune response in plants is usually accompanied by the induction of defense-related genes, such as OsPR1a (Os07g0129200), OsPR10 (Os12g0555200), and PBZ1 (Os12g0555500) (21), we measured the expression of these three genes in bsr-k1 and Kitaake plants using qRT-PCR. We observed that expression levels of these three defense-related genes are moderately elevated (two- to threefold) in bsr-k1 compared with Kitaake plants (Fig. 1E). Together, these results demonstrate that the bsr-k1 mutant confers broad-spectrum resistance to M. oryzae and Xoo compared with Kitaake.
The bsr-k1 Mutant Maintains Key Agronomic Traits.
We next monitored several key agronomic traits in the bsr-k1 mutant to assess its application potential in rice breeding. We examined bsr-k1 and Kitaake plants for their yield-related traits (in Lingshui, Hainan, China), including thousand-grain weight, grain length and width, panicle length, grain numbers per panicle, grain yield per plant, and other traits, including tiller numbers and heading date. Few differences were observed between bsr-k1 and Kitaake (Fig. 1F and Fig. S1B). For example, the thousand-grain weights of bsr-k1 (28.52 ± 0.38 g) and Kitaake (28.89 ± 0.23 g) are similar, and the average heading dates of bsr-k1 (55.9 ± 1.1 d) and Kitaake (56.1 ± 1.4 d) are comparable (Fig. 1F). These results suggest that the bsr-k1 allele does not penalize major agronomic traits.
Map-Based Cloning Identified the Causative Mutation of bsr-k1.
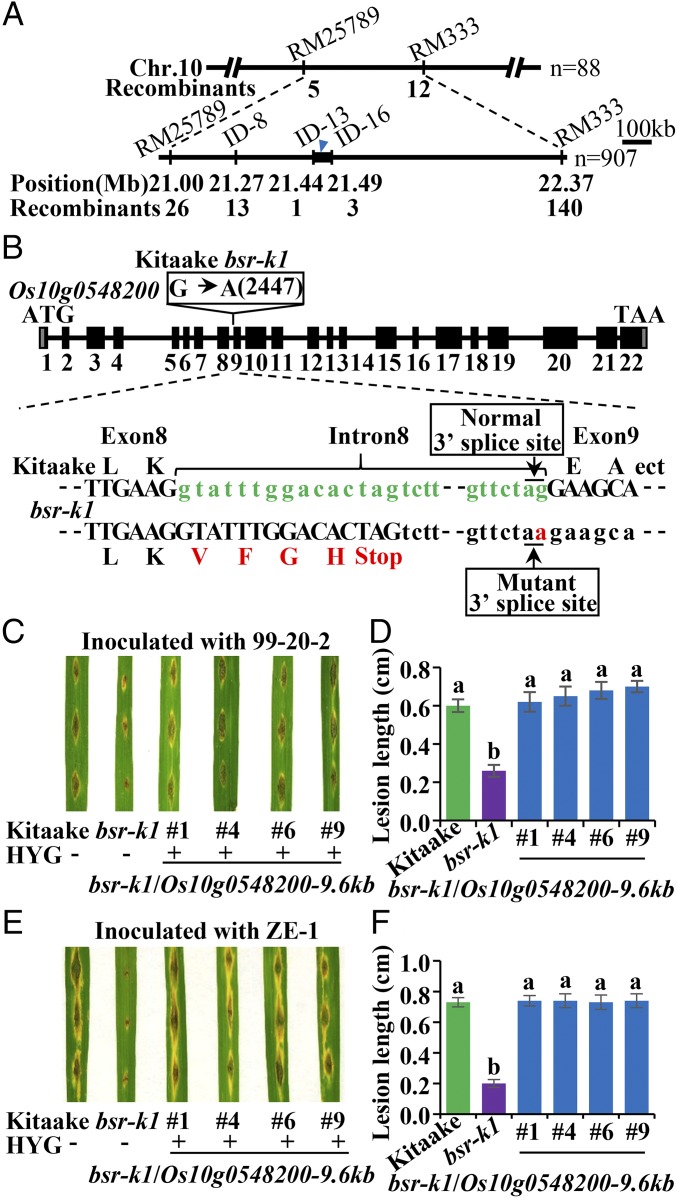
To identify the bsr-k1 locus, we generated three F2 populations by crossing the resistant bsr-k1 mutant with susceptible cultivars Kitaake and Joden, separately. Genetic analyses showed that the bsr-k1 resistance phenotype was controlled by a single recessive locus (Dataset S1). We selected 995 F2 plants from Joden × bsr-k1 that exhibited high-level resistance against blast isolate 99-20-2 and determined the resistance level in each F3 progeny (Dataset S1). By using 88 resistant F2 individuals, we first mapped the bsr-k1 locus to a 1.37-Mb interval between markers RM25789 and RM333 on chromosome 10 (Fig. 2A). Analysis of another 907 resistant F2 individuals located it to a 50-kb region between insertion–deletion markers ID13 and ID16 (Fig. 2A) containing 11 predicted genes (Dataset S2). We then sequenced the genomic DNA sample bulked from 25 resistant BC2F4 individuals and Kitaake control DNA separately using whole-genome resequencing. In the 50-kb region, we identified a single nucleotide substitution (G2447A) located at the 3′ splice site of intron 8 in gene Os10g0548200 (Fig. 2B and Fig. S2A). Sequencing of cDNA confirms that bsr-k1 produces a variant Os10g0548200 transcript with a 71-bp insertion derived from intron 8, causing a premature termination (Fig. S2 B–D).
Fig. 2.
Positional cloning of bsr-k1. (A) Fine mapping of the bsr-k1 locus. The molecular markers and the number of recombinants are indicated. (B) Structure of the Bsr-k1 gene and the mutation in bsr-k1. Filled boxes indicate exons (numbered 1–22) of Os10g0548200. The change from G to A in Os10g0548200 in bsr-k1 is indicated in the open box above exon 9. Nucleotide sequences between exon 8 and exon 9 in Kitaake and bsr-k1 with deduced amino acid sequences are shown below. The substitution in bsr-k1 (in red) abolishes the 3′ splice site of intron 8, resulting in premature termination of translation. (C–F) Blast-resistance test of Bsr-k1–complemented lines. (C and E) Photographs of four independent representative lines at 7 dpi with blast fungal isolates 99-20-2 (C) and ZE-1 (E). (D and F) Lesion lengths (mean ± SD, n > 10) of the complemented lines, bsr-k1, and Kitaake inoculated with blast isolates 99-20-2 (D) and ZE-1 (F). Different letters above bars indicate significant differences (P < 0.05, Tukey’s test). This punch inoculation was repeated twice with similar results.
To confirm that the bsr-k1 resistance phenotype was caused by this mutation, we introduced the Os10g0548200 genomic DNA (9.6 kb) into bsr-k1. All 15 independent, complemented lines carrying wild-type Os10g0548200 reverted to susceptibility to all seven blast isolates. We chose four of the complemented lines for further investigation. Punch inoculation with M. oryzae isolates 99-20-2 and ZE-1 revealed that the lesions in these complemented lines were similar in size to those in Kitaake whereas the bsr-k1 mutant was resistant (Fig. 2 C–F). These complemented lines also show susceptibility to Xoo similar to Kitaake (Fig. S3A). We then generated two independent Os10g0548200-KO lines in Kitaake using the CRISPR/Cas9 approach (Fig. S3 B and C) and performed inoculation with M. oryzae and Xoo. These KO lines show enhanced resistance, displaying smaller lesion size, less M. oryzae DNA accumulation, and lower Xoo population than Kitaake (Fig. S3 D–F). In addition, plants overexpressing Os10g0548200 show much larger lesions compared with Kitaake (Fig. S3 G–J). Taken together, these results demonstrate that the Os10g0548200 gene hosts the bsr-k1 mutation where a single nucleotide change results in enhanced resistance.
Bsr-K1 Is Constitutively Expressed and Encodes a Cytoplasm-Localized TRP.
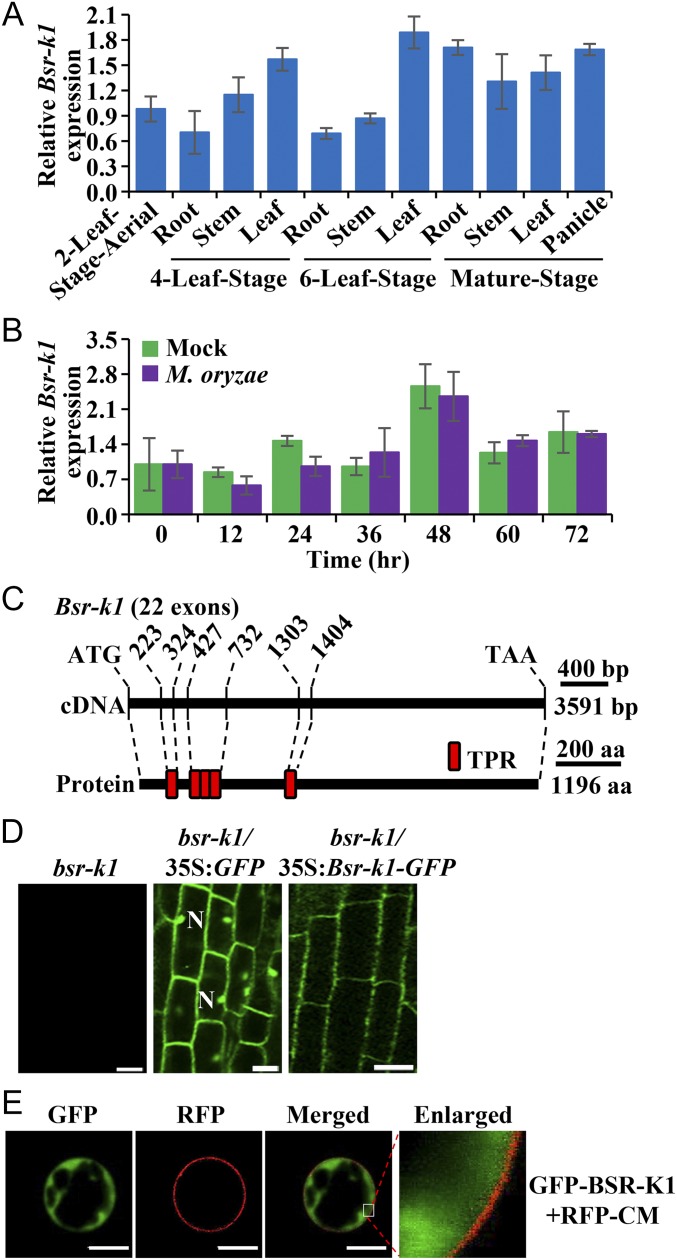
To investigate the expression pattern of Bsr-k1, we sampled roots, stems, leaves, and panicles of Kitaake plants at the four- and six-leaf and mature stages and performed qRT-PCR to measure RNA levels. We found that Bsr-k1 is expressed across all stages and in various tissues but predominantly in leaves (Fig. 3A). To determine whether Bsr-k1 is induced by M. oryzae infection, we assessed the Bsr-k1 RNA level in Kitaake before and after M. oryzae inoculation and found that Bsr-k1 RNA levels remain similar in mock- and M. oryzae-inoculated samples (Fig. 3B). These results suggest that Bsr-k1 is constitutively expressed in rice and is not induced by M. oryzae infection.
Fig. 3.
Expression pattern of Bsr-k1 and subcellular localization of BSR-K1. (A) Bsr-k1 RNA expression pattern. (B) Bsr-k1 RNA levels (mean ± SEM, n = 3) post M. oryzae or mock inoculation. This experiment was repeated twice with similar results. (C) Schematic structure of BSR-K1. The nucleotide numbers above the cDNA indicate the locations of the five TPR motifs. The TPR motifs are represented as red boxes. (D) Subcellular localization of BSR-K1 in rice root tips. Photographs of representative root tips were taken under confocal laser-scanning microscopy. N, nucleus. (Scale bars: 20 μm.) (E) Subcellular localization of BSR-K1 in rice protoplasts. The white square in the merged image is enlarged and shown at the right. CM, mCherry-cytomembrane. (Scale bars: 5 μm.)
The Bsr-k1 ORF encodes a protein of 1,196 amino acids containing five tandem TPRs (Fig. 3C). To determine the subcellular localization of the BSR-K1 protein, we used confocal laser-scanning microscopy to analyze the roots of plants carrying a functional GFP-tagged BSR-K1 protein, which complemented the bsr-k1 mutant (Fig. S3 G–J), and found that its GFP signal is excluded from the nucleus, whereas the GFP protein alone is present in both the cytoplasm and nucleus (Fig. 3D). We validated this result by expressing the GFP-BSR-K1 protein in Nicotiana benthamiana (Fig. S4) and in rice protoplasts, where the GFP-BSR-K1 protein was detected only in the cytoplasm but not in the nucleus or cytomembrane (visualized by the mCherry-cytomembrane marker, RFP-CM, in Fig. 3E). Together, these results suggest that BSR-K1 functions in the cytoplasm.
BSR-K1 Binds to OsPAL1 mRNA in Rice.
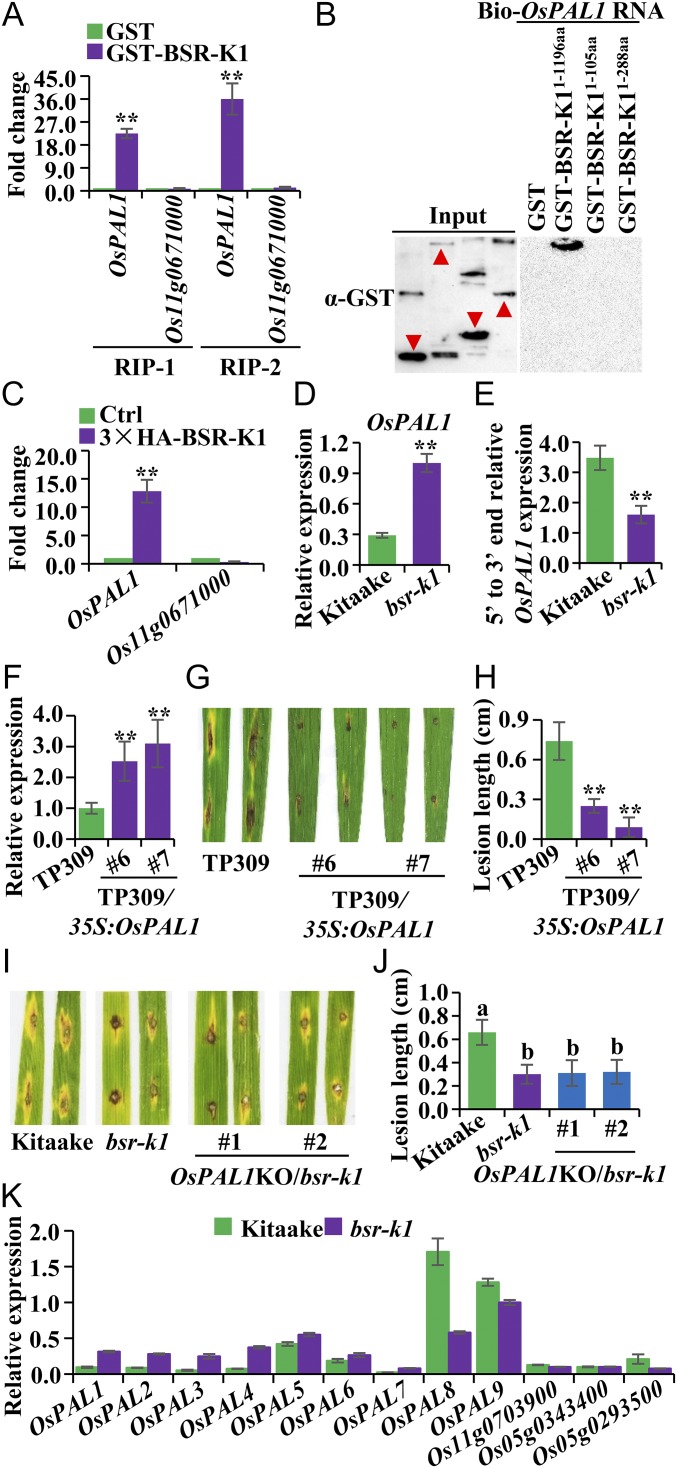
TPR-containing proteins have been reported to regulate mRNA metabolism (23–25). We hypothesized that BSR-K1, containing TPRs, may associate selectively with specific RNA transcripts. To address this, we performed two independent experiments of in vitro RNA immunoprecipitation (RIP) assays by separately coincubating GST and recombinant GST-BSR-K1 protein with rice total RNA (Fig. S5 A and B) and subsequently purifying GST, followed by high-throughput sequencing to identify BSR-K1–associated mRNAs. RNA electrophoresis analysis revealed that GST-BSR-K1, but not GST alone, is able to bind to RNA in vitro (Fig. S5C). By comparing the RNAs coprecipitated with GST-BSR-K1 (RIP) and GST (mock), we identified 82 and 110 RNA fragments from these two RIP assays, respectively, that were specifically and significantly coprecipitated by the GST-BSR-K1 protein (Dataset S3 A and B); 40 of the fragments were found in both assays (Dataset S3C). Interestingly, OsPAL1 RNA fragments are highly represented in both RIP assays (Dataset S3C). Previous studies have shown up-regulation of PAL1 genes, which results in enhanced immunity upon pathogen infection (26, 27). We then performed qRT-PCR to validate the enrichment of OsPAL1 RNA in the RIP precipitates (Fig. 4A). Further analysis showed that biotinylated OsPAL1 RNA was able to pull down the full-length BSR-K1 (GST-BSR-K11–1,196 aa) protein but not two truncated, nonfunctional BSR-K1 proteins, GST-BSR-K1–105 aa and GST-BSR-K11–288 aa (Fig. 4B). Together, these results demonstrate direct binding of the BSR-K1 protein to OsPAL1 mRNA in vitro.
Fig. 4.
BSR-K1 binds to OsPAL1 mRNA and regulates its turnover. (A) qRT-PCR verification of OsPAL1 mRNA enrichment in two independent RIP experiments (Dataset S3C). The enrichment was calculated as the fold change of RNA abundance comparing GST-BSR-K1 pulldown with GST pulldown. The Os11g0671000 gene was used as a negative control. (B) RNA pull-down assays with biotinylated OsPAL1 RNA. BSR-K11–1,196aa, full-length BSR-K1; BSR-K11–105aa, the truncated BSR-K1 protein in the Bsr-k1KO plant; BSR-K11–288aa, the truncated BSR-K1 protein in the bsr-k1 mutant. The red triangles in input indicate bands of target proteins. This experiment was repeated twice with similar results. (C) Enrichment of OsPAL1 RNA in 3×HA-Bsr-k1 plants as detected by qRT-PCR. The enrichment was calculated as the fold change in the abundance of RNA immunoprecipitated in 3×HA-Bsr-k1 plants compared with Kitaake. Os11g0671000 was used as a negative control (Ctrl). (D) OsPAL1 mRNA levels in bsr-k1 and Kitaake. (E) OsPAL1 mRNA turnover rates (mean ± SEM, n = 3) in bsr-k1 and Kitaake. This experiment was repeated twice with similar results. (F) qRT-PCR detection of OsPAL1 mRNA (mean ± SEM, n = 3) in two overexpression lines. (G) Punch inoculation of wild-type TP309 and two independent OsPAL1 overexpression lines. Two leaves each of TP309, TP309/35S:OsPAL1-#6, and TP309/35S:OsPAL1-#7 are shown. (H) Lesion lengths (mean ± SD, n > 10) of TP309, TP309/35S:OsPAL1-#6, and TP309/35S:OsPAL1-#7 inoculated with the blast isolate Zhong1. (I) Punch inoculation of OsPAL1KO/bsr-k1 plants. Two leaves each of Kitaake, bsr-k1, OsPAL1KO/bsr-k1-#1, and OsPAL1KO/bsr-k1-#2 are shown. (J) Lesion lengths (mean ± SD, n > 10) of Kitaake, bsr-k1, OsPAL1KO/bsr-k1-#1, and OsPAL1KO/bsr-k1-#2 inoculated with the blast isolate Zhong1. The same letter above bars indicates the same statistical group (P > 0.05, Tukey’s multiple-comparison test). (K) Expression levels (mean ± SEM, n = 3) of OsPAL genes and other defense-related genes detected by RIP-seq (Dataset S3C) in bsr-k1 mutant and Kitaake. This experiment was repeated twice with similar results. **P ≤ 0.01, t test.
We then generated transgenic plants carrying the native promoter-3×HA-Bsr-k1, which encodes HA-tagged BSR-K1 protein and complements the bsr-k1 mutant (Fig. S5D). We detected and immunoprecipitated the 3×HA-BSR-K1 fusion protein using anti-HA antibody (Fig. S5 E and F). We then performed an in vivo RIP assay using transgenic and Kitaake plants. qRT-PCR analysis showed that the OsPAL1 mRNA was pulled down by the 3×HA-BSR-K1 protein (Fig. 4C), confirming the binding of the BSR-K1 protein to OsPAL1 mRNA in vivo.
BSR-K1 Regulates the OsPAL1 mRNA Level Through Modulating the OsPAL1 mRNA Turnover.
We next tested whether the OsPAL1 mRNA level is altered in bsr-k1. The results showed that OsPAL1 mRNA accumulated to a higher level (∼threefold) in bsr-k1 than in Kitaake plants (Fig. 4D), indicating that BSR-K1 suppresses OsPAL1 mRNA accumulation in Kitaake plants.
We then compared the turnover rate of OsPAL1 mRNA in bsr-k1 and Kitaake. We designed primers for qPCR to detect the 5′ end and 3′ end of OsPAL1 mRNA, respectively (Fig. S6A), because mRNA degradation generally starts from the 3′ end, and thus the 5′-end/3′-end ratio represents its turnover rate (28). The reverse primers for each fragment were used to prime separate cDNA synthesis reactions (28). We found that Kitaake has a 5′-end/3′-end ratio approximately twice as high as bsr-k1 (Fig. 4E), suggesting that the turnover rate of OsPAL1 mRNA is compromised in bsr-k1. Together, these results indicate that BSR-K1 binds OsPAL1 mRNA and promotes its turnover in Kitaake.
To determine whether the elevated OsPAL1 mRNA level contributes to resistance to rice blast, we overexpressed the OsPAL1 gene in a susceptible rice variety, TP309 (Fig. 4F), and performed blast inoculation. We found that the OsPAL1 overexpression lines showed enhanced resistance to blast with lesion lengths significantly (three- to sevenfold) shorter than those observed for TP309 (Fig. 4 G and H), supporting the role of OsPAL1 in bsr-k1–mediated resistance. However, KO of OsPAL1 in bsr-k1 or Kitaake plants did not compromise resistance to blast (Fig. 4 I and J and Fig. S6 B–E), suggesting that other OsPAL members may function redundantly and compensate for the loss of OsPAL1. In support, we also find that bsr-k1 accumulates higher levels of OsPAL1–OsPAL7 (especially OsPAL1–OsPAL4) mRNAs than Kitaake (Fig. 4K) and that BSR-K1 binds OsPAL1–OsPAL7 mRNAs and facilitates their turnover in vivo (Fig. S6 F–H). For comparison, we also analyzed other defense-related genes, Os11g0703900, Os05g0343400 (WRKY53), and Os05g0293500, detected in RIP sequencing (RIP-seq) (Dataset S3C). BSR-K1 showed very weak or no binding to these genes (Fig. S6G). Consistently, none of their mRNA turnover was remarkably repressed (Fig. S6H); their mRNAs did not significantly accumulate compared with OsPAL1-7 in the bsr-k1 mutant (Fig. 4K). Together, these results indicate that BSR-K1 preferentially promotes OsPAL mRNA turnover in rice plants and that loss of function of BSR-K1 (bsr-k1) results in mRNA accumulation of multiple OsPAL genes, leading to enhanced resistance in bsr-k1.
Plants Carrying Homozygous bsr-k1 Alleles Exhibit Enhanced Resistance with No Major Yield Penalty.
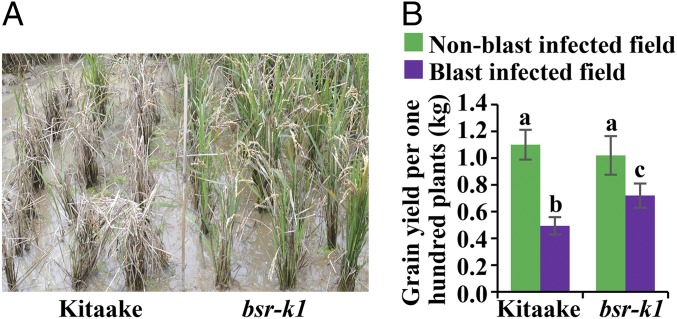
To assess the usefulness of the bsr-k1 allele for rice blast control, we performed field trials in two rice-growing regions. Field one (Pujiang, Sichuan, China) was a field with high blast infection (29); as a control, field two (Lingshui, Hainan, China) had little or no M. oryzae present. We found that the bsr-k1 plants displayed high levels of leaf and panicle resistance to M. oryzae in field one, whereas the Kitaake control plants succumbed to infection (Fig. 5A and Fig. S7).
Fig. 5.
Field trial of the bsr-k1 rice line for blast resistance and yield property. (A) Field trial of bsr-k1 and Kitaake plants on resistance to blast. Photographs were taken at the mature stage. (B) Evaluation of grain yield of bsr-k1 and Kitaake plants in Lingshui and Pujiang. Grain yield was calculated as per 100 plants (mean ± SD, n > 3). Different letters above bars indicate different statistical groups (P < 0.05, Tukey’s test).
We then evaluated the grain yield of bsr-k1 plants and Kitaake control plants in the two fields. In field two, where no obvious blast infection occurred, the bsr-k1 plants yielded a similar amount of grain as Kitaake plants (1.02 vs. 1.10 kg) (Fig. 5B). In contrast, in field one, where heavy blast infection occurred, the yield of bsr-k1 plants was significantly higher than that of Kitaake plants (0.72 vs. 0.49 kg) (Fig. 5B). Furthermore, in three field tests at two locations, elite rice cultivars carrying bsr-k1, including Shuihui527bsr-k1/bsr-k1, Minghui63bsr-k1/bsr-k1, and Mianhui725bsr-k1/bsr-k1 (Fig. S8), showed agronomic traits and yields similar to those of their parents, except for slightly lower yields of Shuhui527bsr-k1/bsr-k1 and Minghui63bsr-k1/bsr-k1 in one test (in Pujiang, 2016) (Dataset S4).
Discussion
We have demonstrated that loss of function of the Bsr-k1 gene encoding a TPR-domain protein leads to enhanced resistance to all tested isolates of M. oryzae and Xoo. We conclude that the bsr-k1 mutant clearly confers broad-spectrum disease resistance. None of the previously reported genes (7–15) conferring broad-spectrum resistance encodes proteins with RNA-binding activity. Thus, BSR-K1 represents a class of protein regulating a pathway leading to broad-spectrum resistance. Loss of functional BSR-K1 (bsr-k1) results in mRNA accumulation of multiple OsPAL (OsPAL1–OsPAL7) genes (Fig. 4K). We hypothesize that BSR-K1 promotes mRNA degradation of these OsPAL genes and that loss of function of BSR-K1 results in lower turnover of their mRNAs, leading to mRNA accumulation.
Previous studies have shown that the expression of PAL1 genes is required for the biosynthesis of secondary metabolites, including lignin and SA, a phytohormone associated with defense against biotrophic pathogens (27). Our results show that bsr-k1 accumulates more lignin, but not total SA (Fig. S9 A and B), despite the elevation of defense-related genes such as OsPR1a in bsr-k1 (Fig. 1E). Consistently, the lignin content also accumulates in OsPAL1 overexpression plants (Fig. S9C) that exhibit enhanced resistance to rice blast (Fig. 4 F–H), but the SA content remains unchanged (Fig. S9D). These results indicate that higher expression levels of OsPAL genes increase the biosynthesis of lignin, instead of SA, and resistance to pathogens in the bsr-k1 mutant. However, how the elevation of lignin content is associated with the activation of defense genes remains unknown.
TPR-containing proteins are involved in the regulation of RNA metabolism in various organisms (23–25). Our results demonstrate that the BSR-K1 protein regulates mRNA turnover of multiple OsPAL genes (Fig. 4E and Fig. S6H). Thus, BSR-K1 regulates plant immunity through a mechanism different from those of previously identified RNA-binding proteins (30–32). For example, Arabidopsis MOS2 is required for proper splicing of SNC1, and loss of MOS2 function results in suppressing the autoimmune responses in snc1 (30). Bean PRP-BP participates in the elicitor-induced destabilization of PvPRP1 mRNA, which encodes a cell wall-strengthening protein, implicating PRP-BP in plant defense (31). Human HuR regulates the splicing of Dlst mRNA, encoding a subunit of the 2-oxoglutarate dehydrogenase (a-KGDH) complex; absence of HuR results in large amounts of reactive oxygen species and B cell death (32).
In rice, it has been reported that TPR proteins mainly act as interaction scaffolds in the formation of multiprotein complexes involved in the immune response, such as in the defensome of chitin-induced immunity (33). This defensome complex includes two TPR proteins, OsSGT1 and OsSti1a, which associate with the HSP90 immune complex to regulate chitin-triggered immunity and resistance against rice blast fungus (33). In contrast, BSR-K1 does not directly interact with the rice defensome complex (Fig. S10A). Thus, the action of the BSR-K1 TPR protein represents a mechanism leading to immunity regulated through interaction with mRNA. Because the TPR motifs are highly conserved in diverse organisms (34), we hypothesize that TPR proteins, especially those sharing high homologies to BSR-K1, including Arabidopsis AtSKI3 (Fig. S10B), may also be involved in regulating immune responses via a similar mechanism.
In practice, most breeding programs rely on dominant resistance (R) genes (35), which often confer race-specific resistance and are overcome by the evolution of new pathogen races within a few years (36). Although a number of genes or QTLs for broad-spectrum and durable resistance have been identified (7–15), the tight linkage to undesirable agricultural traits has often hindered breeding. The bsr-k1 mutant confers broad-spectrum resistance with no major penalty on main agronomic traits (Fig. 1F and Fig. S1B). The moderately elevated activation of defense-related genes in bsr-k1 (Fig. 1E) is sufficient to suppress pathogen attack, similar to the putative characteristics of pi21-mediated resistance (10). Indeed, the bsr-k1 allele strikingly improves rice resistance in Kitaake in a naturally blast-diseased field compared with the wild-type Bsr-k1 allele (Fig. 5A and Fig. S7). Three field tests of elite rice cultivars suggest that the bsr-k1 allele does not carry a major penalty affecting main agronomic traits or yield (Dataset S4), making it a good candidate for rice breeding.
Materials and Methods
Rice blast and Xoo infections and disease assessments were performed as described (21, 22, 29). Purified recombinant proteins from Escherichia coli were used for in vitro RIP assays and RNA pulldown assays. Immunoprecipitated recombinant proteins from transgenic rice were used for in vivo RIP assays. In vitro RIP assays and in vivo RIP assays were performed as described (37). The RNA pulldown assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit following the manufacturer’s instructions (Thermo Scientific). Determination of mRNA turnover was performed as described (28). The blast isolates used to identify Kitaake-compatible blast isolates are listed in Dataset S5. The primers used for qRT-PCR are listed in Dataset S6. All other materials and details of experimental methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
X.C. was supported by National Science Foundation of China (NSFC) Grants 31171622 and 31571994, National Key Research and Development Program of China Grant 2016YFD0100600, Transgenic Projects from the Chinese Ministry of Agriculture (TPCMA) Grant 2014ZX0800903B, Program for New Century Excellent Talents in University from the Ministry of Education in China Grant NECT-13-0920, and “Hundred Talents Plan” Foundation Youth Foundation Grant 13QNJJ0076. J.Y. was supported by NSFC Grant 31601290. M.C. and P.C.R. were supported by National Science Foundation Grant PGRP IOS 1237975, NIH Grant GM59962, and US Department of Agriculture National Institute of Food and Agriculture Grant 2017-67013-26590. Jing Wang was supported by NSFC Grant 31401351. W.L. was supported by NSFC Grant 31501627, and B.M. was supported by TPCMA Grant 2016ZX08001002.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 2859.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705927115/-/DCSupplemental.
References
- 1.Azizi P, et al. Toward understanding of rice innate immunity against Magnaporthe oryzae. Crit Rev Biotechnol. 2016;36:165–174. doi: 10.3109/07388551.2014.946883. [DOI] [PubMed] [Google Scholar]
- 2.Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 3.Hu KM, Qiu DY, Shen XL, Li XH, Wang SP. Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant. 2008;1:786–793. doi: 10.1093/mp/ssn039. [DOI] [PubMed] [Google Scholar]
- 4.Miah G, et al. Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol Biol Rep. 2013;40:2369–2388. doi: 10.1007/s11033-012-2318-0. [DOI] [PubMed] [Google Scholar]
- 5.Kou Y, Wang S. Toward an understanding of the molecular basis of quantitative disease resistance in rice. J Biotechnol. 2012;159:283–290. doi: 10.1016/j.jbiotec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Kou Y, Wang S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr Opin Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Xiao S, et al. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- 8.Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, et al. STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat Commun. 2014;5:4768. doi: 10.1038/ncomms5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuoka S, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 11.Deng Y, et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–965. doi: 10.1126/science.aai8898. [DOI] [PubMed] [Google Scholar]
- 12.Li W, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126.e15. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Krattinger SG, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 14.Fu D, et al. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Büschges R, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Wen Y, Berkey R, Xiao S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell. 2009;21:2898–2913. doi: 10.1105/tpc.109.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Ronald PC. Innate immunity in rice. Trends Plant Sci. 2011;16:451–459. doi: 10.1016/j.tplants.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gou JY, et al. Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell. 2015;27:1755–1770. doi: 10.1105/tpc.114.134296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Iqbal M, Yang RC, Spaner D. Effect of Lr34/Yr18 on agronomic and quality traits in a spring wheat mapping population and implications for breeding. Mol Breed. 2016;36:53. [Google Scholar]
- 20.Piffanelli P, et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CH, et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X, et al. Quantitative trait Loci mapping for bacterial blight resistance in rice using bulked segregant analysis. Int J Mol Sci. 2014;15:11847–11861. doi: 10.3390/ijms150711847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goebl M, Yanagida M. The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 24.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 25.Izumiyama T, Minoshima S, Yoshida T, Shimizu N. A novel big protein TPRBK possessing 25 units of TPR motif is essential for the progress of mitosis and cytokinesis. Gene. 2012;511:202–217. doi: 10.1016/j.gene.2012.09.061. [DOI] [PubMed] [Google Scholar]
- 26.Kim DS, Hwang BK. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot. 2014;65:2295–2306. doi: 10.1093/jxb/eru109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan L, Liu H, Li X, Xiao J, Wang S. Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant. 2014;152:486–500. doi: 10.1111/ppl.12192. [DOI] [PubMed] [Google Scholar]
- 28.Dorcey E, et al. Context-dependent dual role of SKI8 homologs in mRNA synthesis and turnover. PLoS Genet. 2012;8:e1002652. doi: 10.1371/journal.pgen.1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye HL, et al. Study on control effects of rice mixture on rice blast in different ecological zones. Xi Nan Nong Ye Xue Bao. 2010;23:1510–1514. [Google Scholar]
- 30.Copeland C, Xu S, Qi Y, Li X. MOS2 has redundant function with its homolog MOS2H and is required for proper splicing of SNC1. Plant Signal Behav. 2013;8:e25372. doi: 10.4161/psb.25372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Mehdy MC. Binding of a 50-kD protein to a U-rich sequence in an mRNA encoding a proline-rich protein that is destabilized by fungal elicitor. Plant Cell. 1994;6:135–145. doi: 10.1105/tpc.6.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Muñoz MD, et al. The RNA-binding protein HuR is essential for the B cell antibody response. Nat Immunol. 2015;16:415–425. doi: 10.1038/ni.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, et al. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe. 2010;7:185–196. doi: 10.1016/j.chom.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 35.Pavan S, Jacobsen E, Visser RGF, Bai Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed. 2010;25:1–12. doi: 10.1007/s11032-009-9323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panstruga R, Dodds PN. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing D, Wang Y, Hamilton M, Ben-Hur A, Reddy AS. Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell. 2015;27:3294–3308. doi: 10.1105/tpc.15.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.