Significance
Macrophages can be functionally reprogrammed by the tumor microenvironment to further tumor growth and malignancy. In this study, we have discovered that this pathological process is dependent on the ERK5 MAPK. Accordingly, we demonstrated that inactivation of ERK5 in macrophages blocked the phosphorylation of STAT3, a transcription factor crucial for determining macrophage polarity, and impaired the growth of melanoma and carcinoma grafts. These results raise the possibility that targeting protumor macrophages via anti-ERK5 therapy constitutes a very attractive strategy for cancer treatment. This is important given that the detection of large numbers of macrophages in human tumors often correlates with poor prognosis, but also with a poor response of the tumor to anticancer agents.
Keywords: ERK5, MAPK, STAT3, macrophages, tumors
Abstract
Owing to the prevalence of tumor-associated macrophages (TAMs) in cancer and their unique influence upon disease progression and malignancy, macrophage-targeted interventions have attracted notable attention in cancer immunotherapy. However, tractable targets to reduce TAM activities remain very few and far between because the signaling mechanisms underpinning protumor macrophage phenotypes are largely unknown. Here, we have investigated the role of the extracellular-regulated protein kinase 5 (ERK5) as a determinant of macrophage polarity. We report that the growth of carcinoma grafts was halted in myeloid ERK5-deficient mice. Coincidentally, targeting ERK5 in macrophages induced a transcriptional switch in favor of proinflammatory mediators. Further molecular analyses demonstrated that activation of the signal transducer and activator of transcription 3 (STAT3) via Tyr705 phosphorylation was impaired in erk5-deleted TAMs. Our study thus suggests that blocking ERK5 constitutes a treatment strategy to reprogram macrophages toward an antitumor state by inhibiting STAT3-induced gene expression.
The inflammatory microenvironment is arguably one of the most influential components of most, if not all, tumors (1). Central to cancer-related inflammation is a population of macrophages—namely, tumor-associated macrophages (TAMs)—which primarily originate from blood monocytes that are continuously recruited to the tumor site. In early tumors, TAMs display an inflammatory and tumoricidal “M1-like” phenotype. However, as tumors progress, TAMs are functionally reprogrammed by tumor-derived signals to exhibit a trophic, angiogenic, and immune-inhibitory “M2-like” phenotype that contributes to further tumor growth and malignancy (2). Accordingly, the detection of large numbers of TAMs in cancer correlates with poor prognosis, but also with a poor response of the tumor to anticancer agents (3). Based on these observations, macrophage-targeted interventions have been tested in preclinical models with encouraging results (4). However, these strategies are limited because very few molecular targets can be exploited therapeutically to discriminate between different macrophage populations and specifically ablate the presence of TAMs in tumors. In particular, whereas a number of transcription factors have been implicated in the rearrangement of the transcriptional profile that sustains protumor TAM activities, the signaling mechanisms that govern macrophage reprogramming to aid disease progression are poorly understood.
Recently, the extracellular-regulated protein kinase 5 (ERK5) has emerged as a critical component of prooncogenic signaling (5). ERK5 belongs to the family of mitogen-activated protein kinases (MAPKs) that play key roles in transducing extracellular cues into a wide variety of cellular responses, including changes in gene expression (6). The catalytic core of ERK5 situated in the N-terminal half of the protein is most similar to that of the classical MAPKs ERK1/2. Despite this homology, ERK5 displays several distinct structural and functional properties that set it apart from ERK1/2 and the other members of the MAPK family. In particular, its unique extended C terminus comprises two proline-rich sequences, a bipartite nuclear localization sequence, and a transcriptional activation domain (6). These features clearly indicate unique mechanisms of downstream regulation of targets. The importance of ERK5 in signal transduction was further supported by genetic evidence that the pathway exerts nonredundant functions in vivo (7). Notably, consistent with its requirement for the maintenance of vascular integrity during development, ERK5 was implicated in the neovascularization of melanoma and carcinoma grafts by supporting endothelial cell survival (8, 9). Moreover, we have shown that suppressing inflammation by inducing erk5 deletion in neoplastic keratinocytes reduced tumor burden, and this effect was potentiated by the concurrent administration of subtherapeutic doses of the chemotherapeutic agent doxorubicin (10). This study is a demonstration that ERK5 in epithelial cells can remodel the inflammatory microenvironment to support cancer development.
In parallel, other groups have shown that ERK5 can contribute to immune cell fate. For example, ERK5 signaling mediates the mitogenic response of macrophages to colony-stimulating factor 1 (CSF1) stimulation (11). Interestingly, CSF1-induced malignant progression of murine mammary cancer was associated with enhanced infiltration of macrophages into primary mammary tumors (12). Moreover, ERK5 has been implicated in the production of proinflammatory mediators in endothelial cells and monocytes (13). The differentiation of monocytes to functioning macrophages was found to be dependent on ERK5, but not ERK1/2, signaling (14). The recent analysis of the LysMCre;erk5F/F mouse model further showed that erk5-deleted bone marrow-derived macrophages (BMDMs) displayed low-level expression of M2-related genes, but increased expression of M1-related genes at steady state, indicative of macrophage ERK5 favoring polarization toward an M2 phenotype (15). Based on these findings, we investigated the possibility that ERK5 might also fuel cancer progression by enabling TAMs to acquire tumorigenic properties.
Results
Inactivation of ERK5 in the Myeloid Lineage Prevents Tumor Growth.
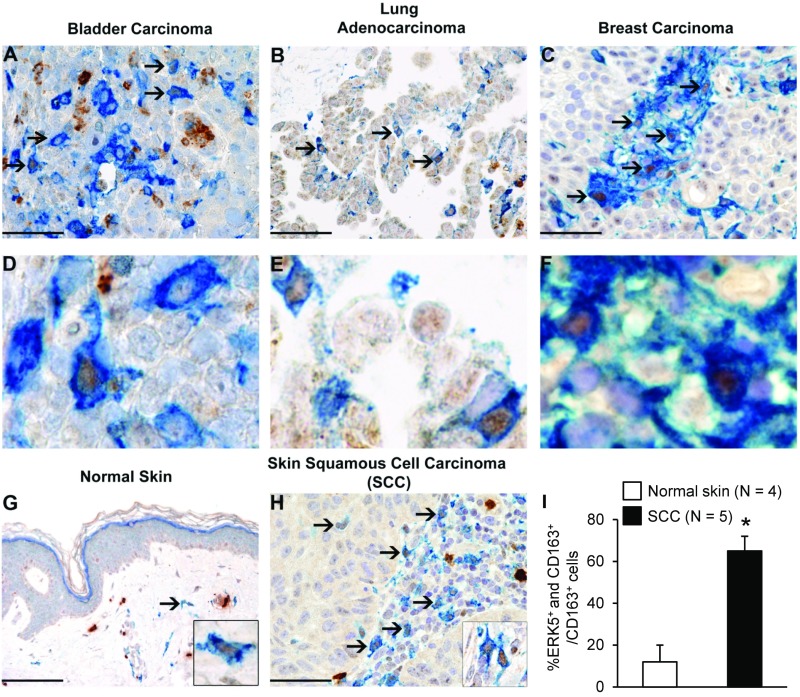
We used CD163 as a selective biomarker to detect macrophages exhibiting a tumor-promoting and immunosuppressive phenotype (16) in formalin-fixed paraffin-embedded human tissue blocks retrieved from the tissue bank of the Department of Pathology (Azienda Socio Sanitaria Territoriale degli Spedali Civili di Brescia, Brescia, Italy). These included transitional bladder cancer (1 case), lung adenocarcinoma (2 cases), breast carcinoma (4 cases), skin squamous cell carcinoma (SCC; 12 cases), and normal skin (4 cases). We found a significant association between CD163 (blue) and ERK5 (brown) expression in human cancer (Fig. 1 A–F and H). Quantification of the immunostaining confirmed an increased number of CD163+ macrophages exhibiting nuclear ERK5 in SCC compared with normal skin tissue (Fig. 1 G–I). To investigate the functional relevance of macrophage ERK5 in tumor development, we developed a murine model in which the erk5 gene could be selectively ablated in the myeloid lineage. This was achieved by crossing mice harboring LoxP sites in the erk5 allele [referred to as the flox (F) allele; ref. 17] with animals carrying a Cre transgene inserted into the ATG start site of the M lysozyme (LysM) gene (18) to generate LysMCre+/−;erk5F/F animals. The recent analysis of the LysMCre+/−;erk5F/F mouse model showed that macrophage ERK5 deficiency tipped the balance in favor of an M1 phenotype at steady state (15). In contrast, no notable difference was reported in total blood cell counts between LysMCre+/−;erk5F/F mice and nontransgenic littermate controls. Moreover, the two genotypes displayed similar numbers of monocytes (CD115+/Gr1+) in bone marrow and a similar level of peritoneal fluid F4/80+ macrophages (15). Immunoblot analysis confirmed the loss of ERK5 expression in LysMCre+/−;erk5F/F BMDMs at day 7 in culture (Fig. 2A).
Fig. 1.
ERK5 is highly expressed in human TAMs. (A–H) Biopsies obtained from human carcinomas and from normal skin were coimmunostained for ERK5 (brown) and the macrophage marker CD163 (blue). Sections were counterstained with hematoxylin. Arrows point to CD163+ macrophages exhibiting strong expression of ERK5 in the nucleus. Compared with cutaneous SCC, only a minor fraction of CD163+ macrophages in normal skin express nuclear ERK5 (G–I); strong cytoplasmic ERK5 is observed in dermal mast cells. [Original magnification: 200× (G; Scale bar, 100 μm), 400× (A–C and H; scale bar, 50 μm; D–F are digital magnification from the corresponding microphotograph above), and 600× (G and H, Insets).] (I) For quantitative analysis, double positive (ERK5+ and CD163+) cells were counted in 4 normal skin (G) and 5 SCC cases (H) within a total area of 1 mm2, corresponding to 5 high-power fields (HPF). The mean per HPF ± SD was calculated. *P = 0.002 compares the percentage of CD163+ cells expressing ERK5 in SCC and in normal skin.
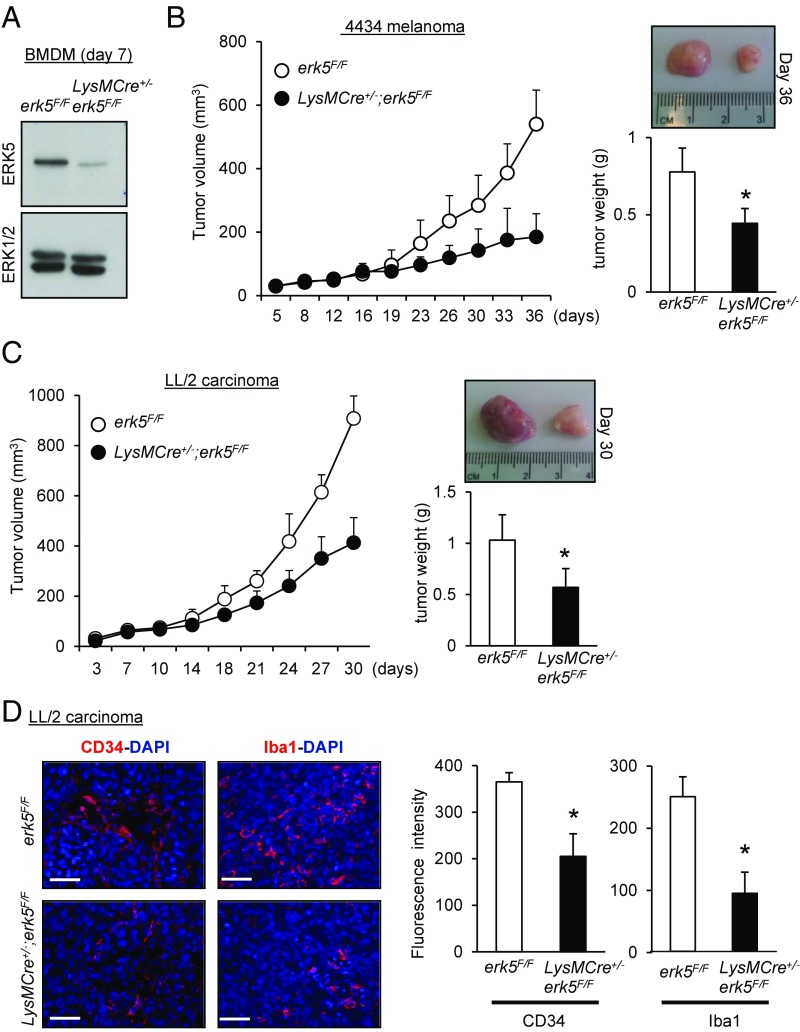
Fig. 2.
ERK5 expression in myeloid cells supports tumor growth in vivo. (A) Immunoblot analysis comparing the level of ERK5 expressed in macrophages obtained from the bone marrow of erk5F/F and LysMCre+/−;erk5F/F animals. Total ERK1/2 expression was used as a loading control. (B and C) The 4434 melanoma (B) or LL/2 carcinoma (C) cells were s.c. implanted into the back of erk5F/F mice that did or did not carry the LysMCre transgene. The growth of tumor grafts was monitored over time after initial cell injection. The data correspond to the mean of tumor volume ± SD (n = 5 mice in each group). Representative pictures of tumor grafts and the mean of tumor weight ± SD excised from erk5F/F and LysMCre+/−;erk5F/F mice killed at the end of the experiment are shown. (D) Sections of carcinoma grafts were analyzed by immunofluorescence using specific antibodies to CD34 (Left) and the pan macrophage marker Iba1 (Right). The immune complexes were detected with a secondary antibody conjugated to Cy3 (red). DNA was stained with DAPI (blue). (Scale bars, 50 μm.) The immunofluorescent signal corresponding to CD34 or Iba1 was quantitated with ImageJ. The data correspond to the mean ± SD (n = 3 tumors). *P < 0.05 (compares tumor grafts from erk5F/F and LysMCre+/−;erk5F/F mice).
Melanoma 4434 cells isolated from a transgenic mouse line carrying the BrafV600E mutation (a gift from Professor Richard Marais, Cancer Research UK Manchester Institute, Manchester; ref. 19) or LL/2 cells derived from a murine Lewis lung carcinoma (20) were injected s.c. in the flanks of syngeneic erk5F/F or LysMCre+/−;erk5F/F C57BL/6 animals. We observed that tumor growth was markedly impeded in the absence of macrophage ERK5, as demonstrated by a significant reduction in tumor volume in LysMCre+/−;erk5F/F mice compared with control erk5F/F animals (Fig. 2 B and C). Postmortem studies confirmed that macrophage ERK5 deficiency caused a significant decrease in tumor weight (Fig. 2 B and C). To further characterize the tumor growth defect associated with myeloid erk5 deletion, excised carcinoma grafts were processed by immunofluorescence staining (Fig. 2D). Antibodies to the endothelial cell-specific marker CD34 and to Iba1, a commonly used cell surface marker to label total TAMs (21, 22), were utilized to visualize angiogenesis and tumor macrophage infiltration. The results showed that tumor vasculature and TAM density were both reduced by ∼50% in tumors derived from LysMCre+/−;erk5F/F mice compared with controls (Fig. 2D). Collectively, these observations strongly suggested a link between ERK5 and macrophages to support tumor growth.
Protumor Macrophage Polarization Is Dependent on ERK5.
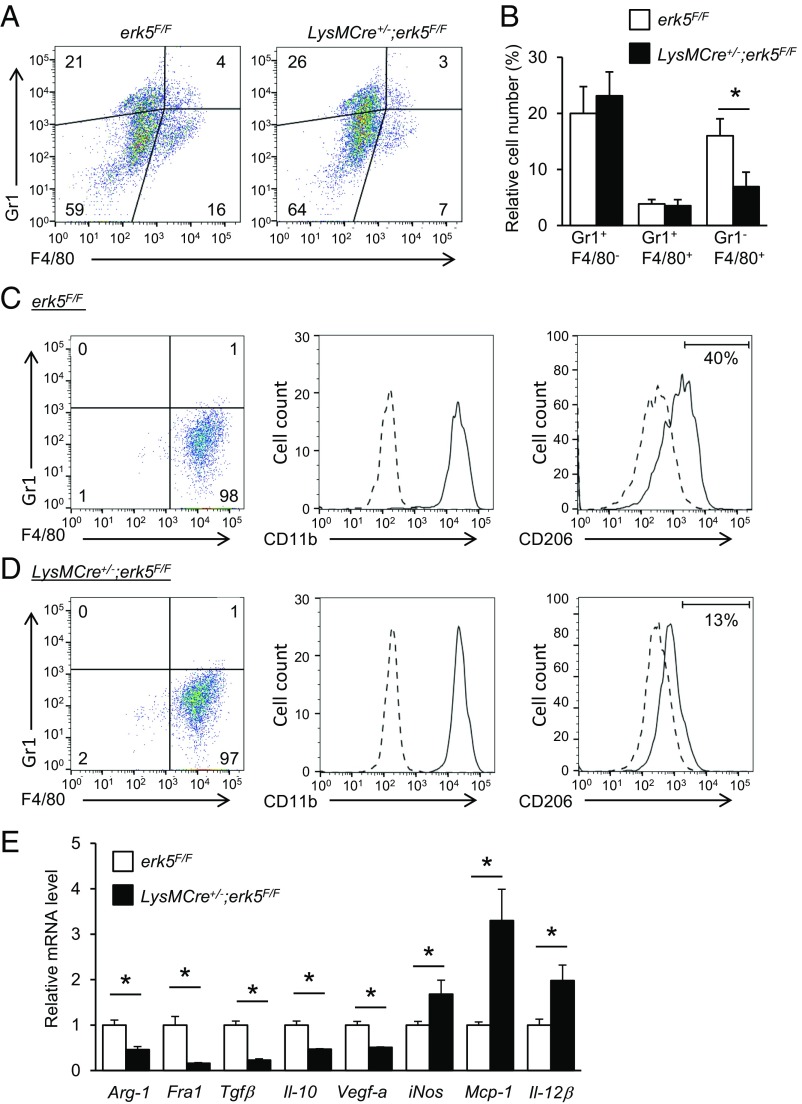
Next, the impact of myeloid ERK5 deficiency on the accumulation of immune cells in tumors was analyzed by using isolated cells from tumor grafts that were gated for live cell analysis (Fig. S1A). Gr1 and F4/80 were used as markers for identification of granulocytes (Gr1+/F4/80−), monocytes (Gr1+/F4/80+), and macrophages (Gr1−/F4/80+) (23, 24). Consistent with Iba1 staining (Fig. 2D), carcinoma and melanoma grafts extracted from LysMCre+/−;erk5F/F mice exhibited a significantly lower number of macrophages compared with tumors from control animals (Fig. 3 A and B and Fig. S1D). In contrast, no marked difference was detected in the number of granulocytes and monocytes between the two genotypes (Fig. 3B and Fig. S1D). Further analysis of the sorted F4/80+ macrophage population demonstrated that the absence of ERK5 did not affect the proportion of CD11b+ TAMs in carcinoma (Fig. 3 C and D). We subsequently utilized CD206 to distinguish tumor-supportive M2-like macrophages (25). The data showed that 40% of Gr1−/F4/80+ macrophages in carcinoma derived from erk5F/F mice were positive for CD206 staining (Fig. 3C), while only 13% were identified as TAMs on the M2 spectrum in tumor grafts from LysMCre+/−;erk5F/F mice (Fig. 3D). Tumor-promoting macrophages contribute to cancer development and progression by producing growth factors and immunosuppressive mediators which stimulate tumor cell proliferation and angiogenesis, while dampening the immune response to the tumor (2, 3). Therefore, to determine whether the polarization defect caused by ERK5 deficiency correlated with changes in transcription, we measured the mRNA expression level of various M2 markers in F4/80+ cells purified by magnetic-assisted cell sorting. We found that macrophages isolated from carcinoma grafts derived from LysMCre+/−;erk5F/F mice exhibited a notable down-regulation of arginase-1 (Arg-1), Fos-related antigen 1 (Fra1), transforming growth factor β (Tgfβ), Il-10, and vascular endothelial growth factor-a (Vegf-a) mRNAs (Fig. 3E). Interestingly, this correlated with a significant up-regulation of certain M1 markers—e.g., inducible nitric oxide synthase (iNos), monocyte chemoattractant protein-1 (Mcp-1), and Il-12β (Fig. 3E)—indicating that ERK5 not only controlled TAM density, but also maintained the M2 subtype of TAMs in tumors.
Fig. 3.
ERK5 suppression specifically reduces the relative fraction of M2 tumor-supportive macrophages in vivo. (A) Representative flow-cytometry analysis and quantification of live (DAPI−) myeloid cell populations in carcinomas 24 d after s.c. inoculation of LL/2 cells in erk5F/F and LysMCre+/−;erk5F/F mice. (B) Graphical analysis of A showing a reduced TAM fraction in carcinoma grafts excised from LysMCre+/−;erk5F/F mice. The data correspond to the mean ± SD (n = 3 tumors). (C and D) Flow-cytometry analysis of tumor-derived macrophages from erk5F/F (C) and LysMCre+/−;erk5F/F (D) mice after sorting with magnetic microbeads ultrapure conjugated to antibodies against F4/80. The expression of CD11b and CD206 (continuous line) in sorted Gr1−/F4/80+ cells from B and C was defined by increased staining over the isotype control (dotted line). (E) Quantitative PCR (qPCR) analysis of multiple M1 and M2 markers in Gr1−/F4/80+ sorted macrophages from tumor grafts. The data correspond to the mean ± SD (n = 3 tumors). *P < 0.05 (compares tumor grafts from erk5F/F and LysMCre+/−;erk5F/F mice).
To provide direct evidence of the protumoral role of macrophage ERK5, we switched to an in vitro system that allowed characterization of cell-autonomous effects. Bone marrow cells were isolated from femurs of CMV-CreER;erk5F/F mice. Ablation of ERK5 was induced by incubating the cells with 4-hydroxytamoxifen (4-HT) (Fig. S2A). This did not prevent the maturation of bone marrow cells in vitro (Fig. S2B). We used IL-6 and leukemia inhibitory factor (LIF), or tumor cell-conditioned medium, to polarize F4/80+/CD115+ macrophages toward a trophic, angiogenic, and immune-inhibitory M2d-like or TAM phenotype, respectively (26, 27). Inflammatory and tumoricidal M1 macrophages were obtained by activation with IFN-γ in combination with lipopolysaccharide (LPS) (23). Nonpolarized macrophages growing in complete medium were used as controls. The phenotypes were confirmed by flow-cytometry staining of surface (F4/80 and CD86) and intracellular (CD206) markers (Fig. S3). Interestingly, the intensity of F4/80 staining was slightly higher in activated erk5Δ/Δ compared with erk5F/F macrophages. This was consistent with our previous observation suggesting that the loss of erk5 notably advanced macrophage differentiation in vitro (Fig. S2B). Additionally, ERK5 deficiency caused a reduction in the number of CD206+ macrophages (Fig. S3), confirming a role of ERK5 in protumor macrophage activation.
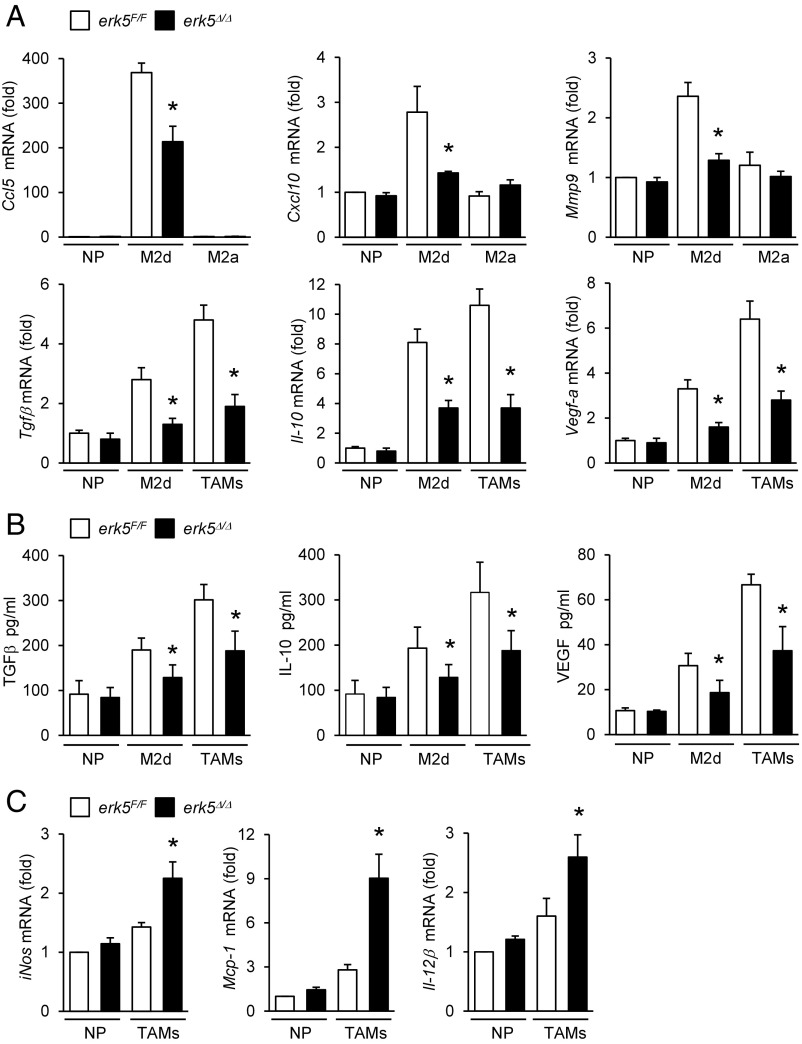
To distinguish specific features of macrophages activated by IL-6 and LIF, and of TAMs generated upon exposure to melanoma-conditioned medium, we analyzed the level of expression of M2 markers by quantitative real-time PCR. We confirmed that IL-6 and LIF-induced macrophages exhibited M2d-like characteristics, as demonstrated by elevated expression of CC chemokine ligand 5 (Ccl5), chemokine (C-X-C motif) ligand 10 (Cxcl10), matrix metalloproteinase 9 (Mmp9), and indoleamine 2,3-dioxygenase (Ido-1) mRNAs compared with M2a macrophages induced by IL-4 (Fig. 4A and Fig. S4). No significant induction of Cxcl16 in M2d polarized macrophages was observed (Fig. S4). Typical of M2 polarization, M2d macrophages and TAMs expressed high levels of Fizz-1and Ym-2 (Fig. S4). Compared with M2d macrophages, TAMs expressed more immunosuppressive (e.g., Tgfβ, Il-10, and Arg-1) and trophic (e.g., Vegf-a and Mmp9) factors (Fig. 4A and Fig. S4), but similar amounts of Ido-1 (Fig. S4). Likewise, TAMs generated upon exposure to carcinoma-conditioned medium exhibited high levels of Arg-1, Tgfβ, Il-10, and Vegf-a (Fig. S5A). Accordingly, melanoma-induced TAMs produced more TGFβ, IL-10, and VEGF than M2d macrophages (Fig. 4B). Consistent with our in vivo data (Fig. 3E), erk5Δ/Δ M2d and erk5Δ/Δ TAMs failed to fully up-regulate these hallmark markers of protumor macrophage activities (Fig. 4A and Figs. S4 and S5A). Notably, the secretion of TGFβ, IL-10, and VEGF by polarized macrophages was significantly impaired in the absence of ERK5 (Fig. 4B). In contrast, erk5Δ/Δ TAMs exhibited increased expression of proinflammatory M1 factors (iNos, Mcp1, and Il-12β) (Fig. 4C and Fig. S5B). These data supported the idea that ERK5 acted as a switch to turn on protumor macrophage properties, while shutting down antitumor immunity.
Fig. 4.
ERK5 deficiency in macrophages impairs the production of protumoral factors. erk5F/F and erk5Δ/Δ macrophages were polarized with IL-6 + LIF (M2d), IL-4 (M2a), or 4434 cell-conditioned medium (TAMs). Nonpolarized (NP) macrophages were used as controls. (A and C) The expression of M2 (A) and M1 (C) markers was analyzed by qPCR. The data correspond to the mean ± SD of three (A) or two (C) independent experiments performed in triplicate. (B) The secretion of TGFβ, IL-10, and VEGF by NP and M2d macrophages or by TAMs was measured by ELISA. The data represent the mean ± SD of three independent experiments. *P < 0.05 (compares polarized erk5F/F vs. erk5Δ/Δ macrophages).
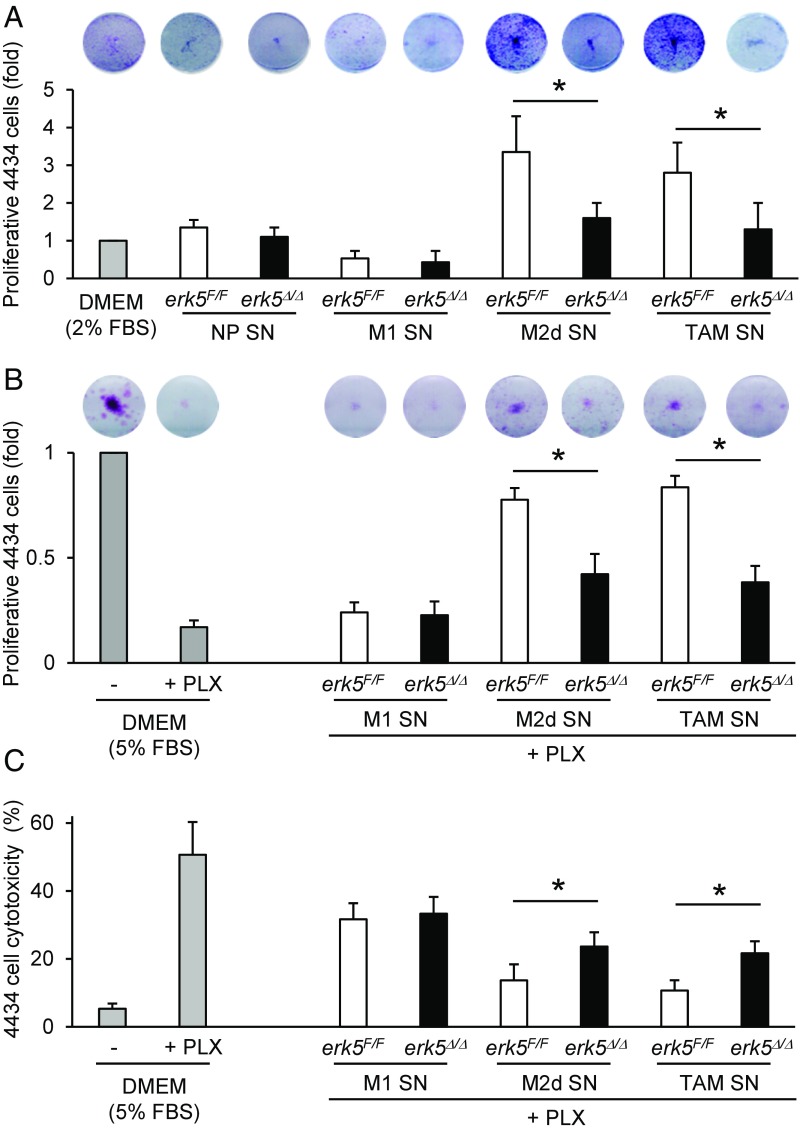
Activated macrophage supernatants were subsequently collected and added onto melanoma or carcinoma cells plated at very low density (Fig. S2C). As expected, supernatants from M2d-like macrophages or TAMs stimulated the proliferation of cancer cells (Fig. 5A and Fig. S5C). This was significantly impaired in response to supernatants from similarly polarized erk5Δ/Δ macrophages (Fig. 5A and Fig. S5C). Moreover, erk5Δ/Δ macrophages exposed to IL-6 and LIF, or to melanoma cell-conditioned medium, exhibited a reduced ability to protect 4434 cells against vemurafenib (PLX4032) treatment compared with erk5F/F macrophages activated under the same conditions (Fig. 5 B and C). Likewise, pharmacological inhibition of ERK5 using the novel JWG-045 compound (28) impaired the ability of human TAMs to support melanoma cell proliferation and to protect melanoma cells against PLX4032 toxicity (Fig. S6), thereby demonstrating the therapeutic implication of our findings. In contrast, supernatants from nonpolarized or M1 macrophages displayed no trophic activity toward murine or human tumor cells (Fig. 5A and Figs. S5C and S6A) and provided no protection to murine 4434 melanoma cells against PLX4032 (Fig. 5 B and C). We observed a slight protective effect of nonpolarized macrophage supernatant in human A375 cells incubated with PLX4032, but this was independent of ERK5 (Fig. S6 B and C). Collectively, these data demonstrated the requirement of ERK5 for macrophages to acquire tumor-promoting activities in response to tumor-derived signals.
Fig. 5.
ERK5 is required for macrophage-induced tumor cell proliferation and drug resistance. (A and B) Colony formation assay of murine 4434 melanoma cells incubated for 7 d with supernatants (SN) from nonpolarized (NP) macrophages or from macrophages activated with IFNγ + LPS (M1), IL-6 + LIF (M2d), or 4434 cell-conditioned medium (TAMs). The 4434 cells growing in low-serum medium were used as controls. Where indicated, PLX4032 (PLX; 1 μM) was added to the medium for the duration of the assay. (C) The 4434 cell cytotoxicity was determined by using CellTox green and analyzed with Gen5. The data correspond to the mean ± SD of two independent experiments performed in triplicate. *P < 0.05 (compares tumor cell proliferation induced by SN from polarized erk5F/F vs. erk5Δ/Δ macrophages).
STAT3 Activation Is Impaired in ERK5-Deficient Macrophages.
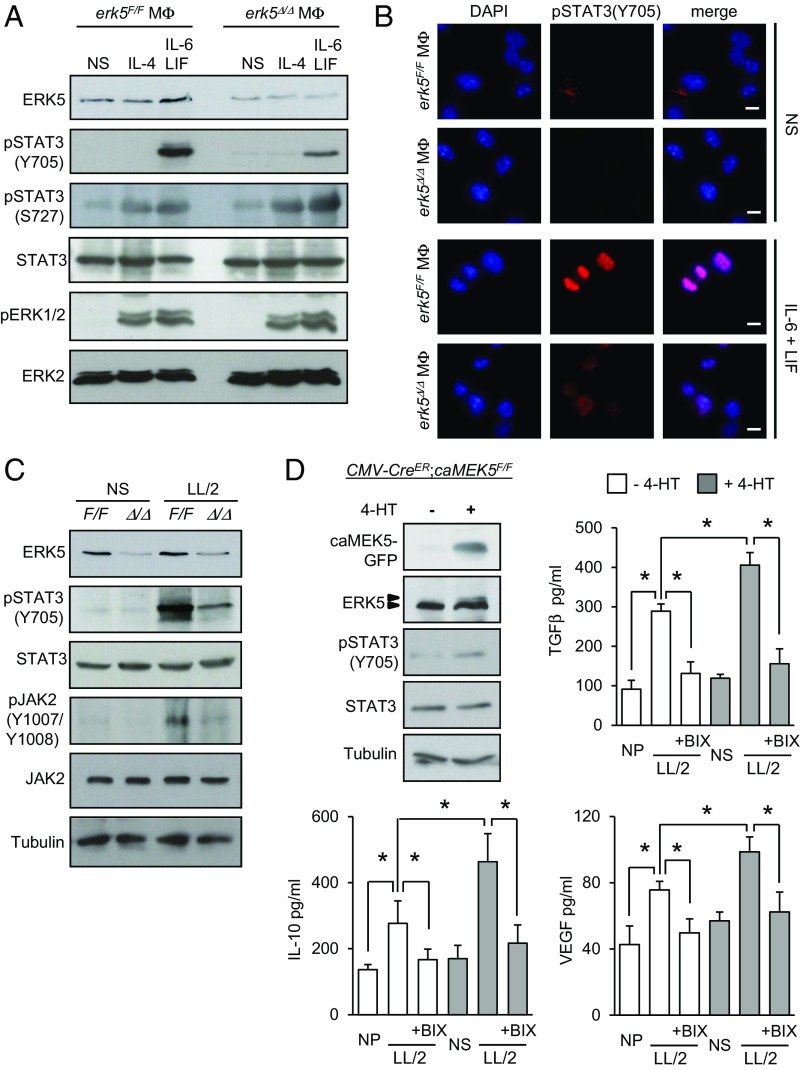
Central to macrophage activation and immune receptor signaling pathways are the transcription factors of the signal transducer and activator of transcription (STAT) family. In particular, STAT3 is activated upon phosphorylation at Tyr705 by protein kinases of the JAK family, enabling nuclear translocation and induction of genes associated with protumor macrophage phenotypes, while suppressing proinflammatory markers (29, 30). Consistently, STAT3 was tyrosine-phosphorylated and accumulated in the nuclei of macrophages stimulated with IL-6 and LIF (Fig. 6 A and B). Remarkably, this was prevented in the absence of ERK5 (Fig. 6 A and B and Fig. S7 A and B). In contrast, erk5F/F and erk5Δ/Δ macrophages displayed similar levels of STAT3 phosphorylation at Ser727 and ERK1/2 phosphorylation at the Thr-Glu-Tyr (TEY) motif, suggesting that the loss of ERK5 expression did not cause a general defect in IL-6 signaling (Fig. 6A). We confirmed by flow cytometry that erk5F/F and erk5Δ/Δ macrophages exhibited a comparable level of expression of the common gp130 subunit, a transmembrane protein that interacts with the IL-6 receptor for signal transduction (Fig. S7C). As expected, we found no evidence that IL-4 activated STAT3 in macrophages (Fig. 6A). Nonetheless, like IL-6 and LIF, IL-4 induced STAT3 phosphorylation at Ser727 and phosphorylation of ERK1/2 independently of ERK5 (Fig. 6A).
Fig. 6.
STAT3 phosphorylation at Tyr705 in macrophages is dependent on ERK5 signaling. (A–C) After starvation in 1% FBS medium overnight, erk5F/F and erk5Δ/Δ macrophages were stimulated with IL-6 and LIF, or IL-4, or exposed to LL/2-conditioned medium (LL/2) for 30 min, as indicated. Nonstimulated (NS) macrophages were used as controls. (A and C) Protein lysates were analyzed by immunoblot with indicated antibodies. Similar results were obtained in two independent experiments. (B) Immunofluorescence was performed with a specific antibody to pSTAT3(Y705). The immune complexes were detected with a secondary antibody conjugated to Cy3 (red). DNA was stained with DAPI (blue). (Scale bars, 10 μm.) (D) CMV-CreER;caMEK5F/F macrophages were mock-treated (−) or incubated with 4-HT (0.1 μM; +) at day 5 in culture for 48 h. Protein lysates were analyzed by immunoblot with antibodies against GFP, ERK5, pSTAT3(Y705), STAT3, and tubulin. Similar results were obtained in two independent experiments. Alternatively, the cells were incubated with BIX02189 (1 µM; BIX), as indicated, for 2 h before being exposed to LL/2-conditioned medium (LL/2). Nonpolarized (NP) macrophages were used as controls. After 36 h, macrophages were rinsed with PBS and cultured in DMEM supplemented with 5% FBS for a further 24 h. The amount of TGFβ, IL-10, and VEGF secreted in supernatants was quantified by ELISA. The data represent the mean ± SD of two independent experiments. *P < 0.01 (compares differences between treatments).
Increased STAT3 phosphorylation in macrophages exposed to LL/2-conditioned medium was also impaired by genetic inactivation of the erk5 allele (Fig. 6C) or pharmacological inhibition of ERK5 activity (Fig. S8A). Additionally, ERK5 deficiency prevented JAK2 phosphorylation at Tyr1007/1008 (Fig. 6C). Therefore, we tested the possibility that ERK5 controlled STAT3 phosphorylation by blocking the expression of negative regulators of the JAK/STAT pathway. These include suppressors of cytokine signaling (SOCS), a family of cytokine-inducible proteins which have been implicated in shaping M1/M2 macrophage polarization via negative-feedback loops (31). For example, SOCS3 suppressed IL6R-mediated STAT3 activation (31). Moreover, reduced expression of SOCS3 in macrophages correlated with the induction of M2-type phenotypes (32, 33). Here, we observed a slight increase in SOSC3 protein expression in macrophages stimulated with IL-6 and LIF (Fig. S8B). This was clearly enhanced upon pharmacological inhibition of ERK5 (Fig. S8B). Consistently, LL/2-stimulated erk5Δ/Δ macrophages exhibited an elevated level of Socs3 transcript (Fig. S8C). In parallel, we examined the expression of the Krüppel-like factor 4 (KLF4), a known downstream target of ERK5 (34) which cooperates with STAT6 to up-regulate M2 genes, in particular Arg-1 and Fizz-1, while inhibiting M1 genes such as iNos, in response to IL-4 stimulation (35). We found no evidence that klf4 was involved in mediating the response of macrophages to tumor-derived signals (Fig. S8C).
The role of ERK5 in controlling STAT3-mediated transcription was further demonstrated by luciferase reporter assay in HeLa cells exhibiting inducible expression of full-length (FL) ERK5 (Fig. S8D). The catalytic activity of ERK5 was required to mediate this effect, given that enhanced transcription by STAT3 in EGF-stimulated cells was blocked by pretreatment with XMD8-92 at a concentration reported to inhibit ERK5 with limited off target effect on BRD4 (Fig. S8D and ref. 28). Interestingly, ectopic expression of a C-terminal truncated mutant of ERK5 (ERK5-ΔC) failed to enhance STAT3 activity to the same level as observed with ERK5-FL (Fig. S8D), indicative of the involvement of the C-terminal region of ERK5 in mediating STAT3-increased transcription.
ERK5-Dependent Regulation of STAT3 Underpins Protumor Macrophage Activation.
Our previous findings convincingly demonstrated a genetic link between ERK5 and STAT3 signaling in macrophages. To test whether ERK5 activation promoted M2 subtype polarization via STAT3, we generated macrophages carrying a transgene encoding a constitutive active (ca) mutant MEK5 fused with green fluorescent protein (GFP) (36). Importantly, a PGK-Neo-STOP cassette flanked with LoxP sites was placed in front of the caMEK5-GFP CDNA to enable caMEK5 expression in a temporally controlled manner. Induced caMEK5 expression in CMV-CreER;caMEK5F/F macrophages incubated with 4-HT caused ERK5 hyperphosphorylation and increased STAT3 phosphorylation at Tyr705 (Fig. 6D). This primed macrophage polarization toward the M2 subtype, as evidenced by elevated secretion of M2 markers following exposure to LL/2-conditioned medium (Fig. 6D). This effect was abolished following pharmacological inhibition of MEK5 by BIX02189 (Fig. 6D). A similar observation was made in ERK5-deficient macrophages coinfected with adenoviruses encoding Flag-tagged wild-type (WT) ERK5 and HA-tagged caMEK5 (Fig. S9). In contrast, HA-caMEK5 failed to phosphorylate STAT3 and to increase the level of M2 transcripts in macrophages expressing a dominant negative mutant form of ERK5 (F-ERK5-AEF) (Fig. S9 B and C).
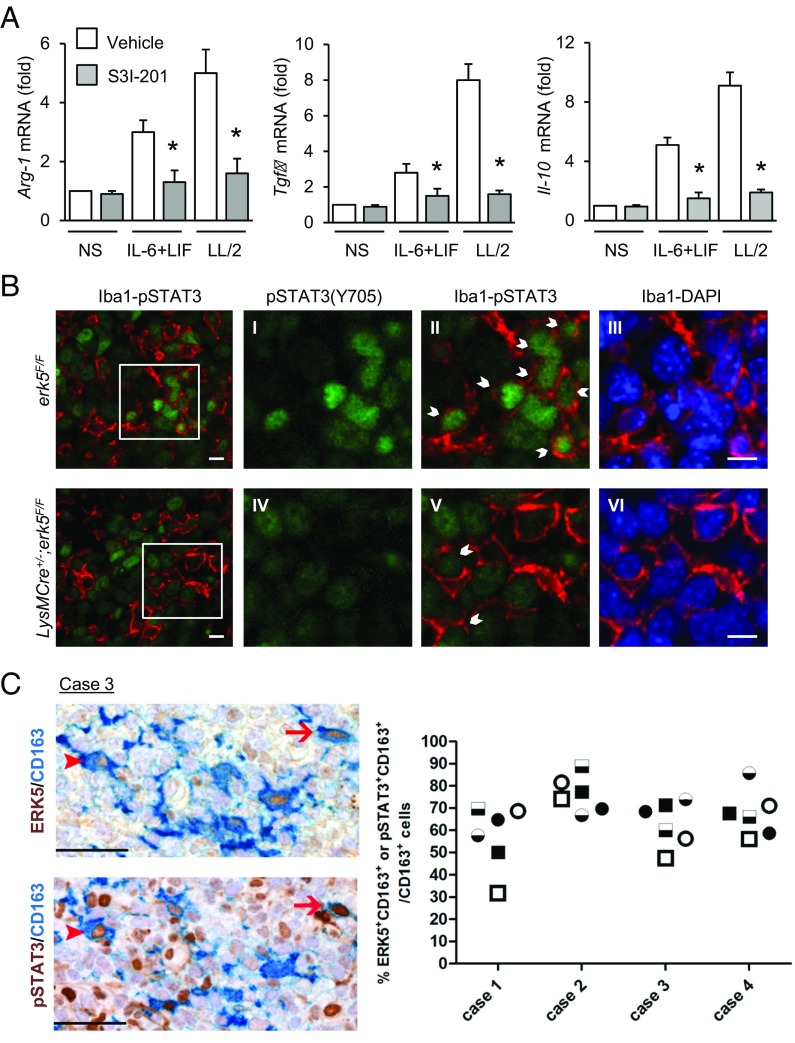
To confirm the importance of STAT3 to activate M2d macrophage and TAM polarization, we tested the requirement of STAT3 in controlling M2 marker expression. STAT3 activity was inhibited by preincubating the cells with S3I-201 (Fig. S10A). We found that S3I-201 significantly impaired increased expression of Arg-1, Tgfβ, and Il-10 transcripts in response to IL-6 and LIF stimulation, or following exposure of macrophages to LL/2-conditioned medium (Fig. 7A). We also analyzed STAT3 activation in tumor-infiltrated macrophages. We used Iba1 to detect TAMs in sections of murine carcinoma grafts. The data showed that the percentage of TAMs (red) exhibiting phospho-Tyr705 STAT3 (green) significantly decreased in tumors excised from LysMCre+/−;erk5F/F animals (Fig. 7B and Fig. S10B). To evaluate whether this finding was clinically relevant, we analyzed biopsies of human SCC (Fig. 1H). Consistent with the idea that STAT3 activation is dependent on ERK5, we found a high proportion of CD163+ macrophages coexpressing ERK5 and phospho-Tyr705 STAT3 in human skin tumors (Fig. 7C).
Fig. 7.
STAT3 is phosphorylated at Tyr705 in TAMs. (A) erk5F/F macrophages were preincubated with S3I-201 (50 μM; Selleck) for 2 h before being stimulated with IL-6 and LIF or with LL/2-conditioned medium (LL/2) for 1 h. Nonstimulated (NS) macrophages were used as controls. Expression of M2 markers was analyzed by qPCR. The data correspond to the mean ± SD of three independent experiments performed in triplicate. *P < 0.05 [compares cells mock-treated (vehicle) vs. treated with S3I-201]. (B) Carcinoma grafts excised from erk5F/F and LysMCre+/−;erk5F/F mice were processed by immunofluorescence with specific antibodies to pSTAT3(Y705) or to the pan macrophage marker Iba1. The immune complexes were detected with Fluorescein Avidin D (green) or Texas Red Avidin D (red). DNA was stained with DAPI (blue). (Scale bars, 10 μm.) B, I–VI are digital magnifications from the corresponding microphotograph. (Scale bars, 15 μm.) Arrows indicate pSTAT3(Y705) positive macrophages. (C) Serial sections obtained from biopsies of human SCC (four cases) were immunostained for ERK5 (brown; Upper) or pSTAT3(Y705) (brown; Lower), coupled with the macrophage marker CD163 (blue). Two HPFs are shown, and the arrows/arrowheads identify two ERK5+pSTAT3+CD163+ triple-positive cells. (Original magnification: 400×; scale bars, 50 μm.) The frequency of double-positive macrophages for ERK5 (circle) or pSTAT3 (square) in three distinct corresponding tumor areas (indicated by black, black/white, and white) is reported in the graph.
Discussion
This investigation into the signaling mechanisms by which myeloid cells contribute to tumorigenesis has led us to uncover a role for ERK5 signaling in regulating the functionality of protumor macrophages in cancer. This is substantiated by our demonstration that macrophages rely on ERK5 to produce trophic and immunosuppressive factors. Moreover, ERK5 activation caused by ectopic expression of caMEK5 in macrophages enhanced the secretion of M2 markers after stimulation with IL-6 and LIF or exposure to LL/2-conditioned medium. ERK5-mediated Arg-1 and Vegf-a expression may support tumor growth by providing the substrates for cancer cells and by inducing neovascularization. Accordingly, decreased tumor growth observed in LysMCre;erk5F/F mice as a consequence of impaired protumor macrophage activation correlated with the lessened ability of Δerk5 macrophages to support tumor cell proliferation and tumor angiogenesis. Additionally, an ERK5-dependent increase in Tgfβ and Il-10 expression may be critical to establish an immune-suppressive microenvironment that dampens effective antitumor T cell responses. For this reason, interesting future directions may include investigating the effect of selective blockade of ERK5 in myeloid lineage cells on tumor infiltration of T cells exhibiting cytotoxic activities. Further analyses of the antitumor immune response in combinatory studies looking at the synergetic effect of targeted inhibition of ERK5 in macrophages with new immune checkpoint inhibitors, such as anti-PDL1 and -CTL4 antibody therapies, would have obvious therapeutic implications (37). In addition to contributing to tumorigenesis, TAMs play a critical role in tumor-drug resistance and disease relapse after therapy (3, 4). In this study, we have found that TAMs lacking ERK5 exhibited a reduced ability to protect melanoma cells against vemurafenib. Consequently, it may also be possible to enhance the responsiveness of tumors to first-line therapy by inhibiting ERK5 in macrophages. In light of our findings, we believe that these additional studies will establish ERK5 as an important drug target in combination with immune-based therapies and conventional chemotherapy and radiotherapy to achieve long-term control of malignancy and durable remission for cancer patients.
Our discovery that ERK5 signaling was required for STAT3 phosphorylation at Tyr705 further underscored its crucial role in tumor macrophage activation. Indeed, stat3 gene deletion in bone marrow cells caused antitumor immune response and significant growth inhibition of melanoma grafts (38). Moreover, STAT3 activation in the immune microenvironment was previously associated with TAMs on the M2 spectrum (29, 39). Accordingly, we found that STAT3 inhibition suppressed increased expression of M2 markers in macrophages stimulated with IL-6 and LIF or with LL/2-conditioned medium. We predict that, in addition to requiring ERK5 catalytic activity, the transactivation domain located in the C-terminal region of ERK5 may be important for the acquisition of M2-like markers downstream of STAT3 (40). Moreover, we observed that JAK2 activation by phosphorylation was impaired in Δerk5 macrophages. Conversely, SOCS3 expression was increased after genetic inactivation of erk5 or inhibition of ERK5 activity. Interestingly, up-regulation of SOCS3 has been associated with increased expression of the M1 marker, iNos (32). A key role of SOCS3 in M1 polarization was further supported by evidence that reduced expression of SOCS3 in macrophages correlated with the induction of M2-type phenotypes (33). Therefore, ERK5-mediated suppression of SOCS3 expression is in line with the priming of TAMs to elicit protumorigenic activities via JAK/STAT3 signaling. This hypothesis is consistent with the genetic demonstration that hyperactivation of STAT3 in macrophages caused by SOCS3 deficiency produced less inflammation (41). However, in contrast to expectation, this study showed that myeloid suppression of SOCS3 prevented cancer metastasis through promoting antitumor immunity (41). This finding highlights the great difficulty in predicting the impact of immune receptor signaling in tumor biology. Hence, further experiments will be required to rigorously establish the importance of SOCS3 in ERK5-induced macrophage activation associated with malignancy.
Overall, our in vitro approach has modeled the effect of STAT3-activating cytokines (i.e., IL-6 and LIF) produced by tumor cells on macrophages. However, many regulated genes induced by STAT3 activate the same cytokine–STAT3 pathway in autocrine and paracrine feed-forward loops going between tumor cells and tumor-interacting immune cells (30, 38). For example, tumor epithelial expression of STAT3 elicits an antitumor immune response involved in mammary tumor growth and breast cancer metastasis (42). We have previously demonstrated that ERK5 was required in neoplastic epithelial cells to propagate tumor-associated inflammation through the production of two major neutrophil chemoattractants, namely, CXCL1 and CXCL2, and increased expression of component of the IL-1/COX-2 pathway (10). Furthermore, our current analysis of human cancer biopsies showed a number of tumor cells exhibiting high ERK5 expression and strong phosphorylation of STAT3 at Tyr705. These findings directly impact the question of whether the cross-talk between ERK5 and STAT3-mediated signal transduction also forms a critical axis in cancer cells to control and shape the local tumor microenvironment. Therefore, the role of ERK5-dependent epithelial STAT3 activation as a mechanism to reduce tumor immunogenicity and evade immune surveillance will deserve further exploration.
Materials and Methods
Tumor Grafts and Isolation of Single Cells from Mouse Tumors.
Mice were inoculated in the flanks with 5 × 105 LL/2 carcinoma cells or 5 × 106 4434 melanoma cells. All animal procedures were performed under license in accordance with the UK Home Office Animals (Scientific Procedures) Act (1986) and approved by the Animal Welfare and Ethical Review Body of the University of Manchester, Manchester, United Kingdom. Tumor size was determined by caliper measurements of tumor length, width, and depth, and volume was calculated as follows: volume = 0.5236 × length × width × depth (millimeters). To characterize tumor-infiltrating immune cells, mice were euthanized 24 d after tumor cell injection. Tumors (0.5 g) were isolated, minced in a Petri dish on ice, and enzymatically dissociated in HBSS, as described (43). The duration of enzymatic treatment was optimized for greatest yield of live F4/80+ cells per tumor type. After neutralization with 1% BSA containing HBSS, cell suspensions were filtered through a 70-μm cell strainer. Red blood cells were solubilized with red cell lysis buffer (Pharm Lyse; BD Biosciences), and the resulting suspension was filtered through a cell strainer to produce a single-cell suspension. Cells were washed once with PBS before use in flow-cytometry analysis or magnetic bead purification.
Magnetic Bead Purification of TAMs.
Single-cell preparations from tumors were incubated with FcR-blocking reagent (mouse seroblock FcR; Bio-Rad) for 15 min and then with 20 μL of magnetic microbeads ultrapure conjugated to an antibody against F4/80 (catalog no. 130-110-443; Miltenyi Biotec MACS Microbeads) per 107 cells for 30 min at 4 °C. After washing, cells bound to magnetic beads were purified according to the manufacturer’s instructions.
Flow Cytometry.
Mononuclear single-cell suspensions were analyzed by fluorescence-activated cell sorting (FACS). In brief, cells were pelleted, washed twice, and resuspended in MACS solution (PBS containing 10% FBS and 1 mM EDTA). Cell viability was assessed by Trypan blue exclusion or DAPI (catalog no. D1306; Molecular Probes) to discriminate dead from live cells. For surface staining, cells were incubated with an anti-mouse FcR antibody (mouse seroblock FcR; Bio-Rad) for 20 min at 4 °C, before being stained with the following antibodies conjugated to phycoerythrin (PE): CD115-PE (catalog no. MCA1898PET; Bio-Rad), CD86-PE (catalog no. A16385; Molecular Probes), CD130-PE (catalog no. 12-1302-80; eBioscience), CD11b-PE (catalog no. 12-0112-81; eBioscience), Gr1-PE (anti-mouse Ly-6G and Ly-6C; catalog no. 553128; BD-Pharmingen), and F4/80-Alexa Fluor 488 (catalog no. MF48020; Life Technologies). For intracellular staining, cells were blocked with the anti-mouse FcR antibody in permeabilization buffer (eBioscience) for 45 min at 4 °C, before incubation with anti–CD206-PE (catalog no. MCA2235PET; Bio-Rad). Flow-cytometry analysis was carried out with a BD FACScan instrument and analyzed by using the FlowJo software.
Colony Formation and Cell Cytotoxicity Assays.
Tumor cells were plated in 12-well plates (colony formation assay) or in clear-bottom, black, 96-well plates (cell cytotoxicity assay) and allowed to adhere overnight in DMEM complete medium. The next day, the medium was replaced with control medium containing 2% or 5% FBS or with medium derived from nonpolarized, M1 or M2d polarized macrophages, or from TAMs (1/2 diluted). Where indicated, vemurafenib (PLX4032; 1 μM) was added to the medium. For the colony formation assay, cells were fixed with 4% PFA after 7 d in culture and stained with 0.1% crystal violet. Dried cells were incubated with 10% acetic acid to solubilize the crystal violet. Absorbance was read at 590 nm. For the cell cytotoxicity assay, cells were incubated with CellTox green (catalog no. G8741; Promega) for 15 min in the dark, after 5 d in culture. The number of dead cells was measured by using the fluorimeter Synergy HT (BioTek). Immediately after this first reading, a second reading was taken after incubating the cells for 30 min at 37 °C in Triton X-100 to measure total cell number.
Immunoblot Analysis.
Proteins were extracted in radioimmunoprecipitation assay buffer containing inhibitors of proteases and protein phosphatases. Extracts (30 μg) were resolved by SDS/PAGE and electrophoretically transferred to an Immobilon-P membrane (Millipore, Inc.). The membranes were saturated in 3% nonfat dry milk or 3% BSA (when using phosphospecific antibodies) and probed overnight at 4 °C with antibodies from Cell Signaling Technology (1:1,000 dilution, unless indicated otherwise) to pERK1/2 (catalog no. 4370), ERK1/2 (catalog no. 4695), ERK5 (catalog no. 3372), STAT3 (catalog no. 12640), pSTAT3-Y705 (1:2,000; catalog no. 9145), pSTAT3-S727 (catalog no. 9134), pJAK2-Y1007/1008 (catalog no. 3771), JAK2 (catalog no. 3230), GFP (catalog no. 2956), SOCS3 (catalog no. 2923), or tubulin (catalog no. 2125) or with an antibody from Santa Cruz to ERK2 (1:500; catalog no. C-14). Immunocomplexes were detected by enhanced chemiluminescence with IgG coupled to horseradish peroxidase as the secondary antibody (GE Healthcare).
Quantitative Real-Time PCR.
Total RNA was isolated from cells by using the RNeasy kit (Qiagen), and cDNA synthesis was carried out, as described (17). Quantitative real-time PCRs were performed by using the SYBR Green I Core Kit (Eurogentec). Sequences of the forward and reverse primers are indicated in Table S1. PCR products were detected in the ABI-PRISM 7700 sequence detection systems (Applied Biosystems). Results were analyzed by using the 2−ΔΔG method. The level of expression of mRNA was normalized to Gapdh and Pgk1 mRNA.
Immunohistochemical Analysis of Human Tissues.
Four-micrometer-thick human tissue sections were immunostained with an antibody from Santa Cruz to ERK5 (1:100 dilution; catalog no. sc-1284-R clone C-20). The reaction was revealed by using Novolink Polymer (Leica Microsystems) followed by DAB. Double immunohistochemistry was performed for detection of ERK5 (1:100 dilution; sc-1286) and p-STAT3(Y705) (1:120 dilution; catalog no. 9145 clone D3A7 from Cell Signaling Technology) in human TAMs (1:50 dilution; CD163 antibody clone 10D6 from Thermo Scientific). Briefly, after completing the first immune reaction, the second was visualized by using Mach 4 MR-AP (Biocare Medical), followed by Ferangi Blue (Biocare Medical) as chromogen and slightly counterstained with hematoxylin.
Immunofluorescence Analysis of Murine Tumor Grafts.
Freshly isolated murine tumor grafts were fixed and stained with a primary antibody to CD34 (RAM34; dilution 1:50; BD Biosciences) and detected by donkey anti-rat–Cy3 (1:10.000 dilution; Jackson Immuno). Double immunofluorescence was performed for detecting pSTAT3(Y705) in TAMs. Briefly, tumor sections were stained with the pSTAT3(Y705) antibody described above overnight and detected with a secondary biotinylated anti-rabbit (1:500 dilution; Vector Laboratories) and Fluorescein Avidin D (1:200 dilution; Vector Laboratories). To detect macrophages, tumor sections were stained with anti-Iba1 (1:200 dilution; Abcam catalog no. ab5076) for 1 h followed by a secondary biotinylated anti-goat (1:500 dilution; Vector Laboratories) and Texas Red Avidin D (1:200 dilution; Vector Laboratories). Sections were mounted with ProLong Gold antifade with DAPI (Molecular Probes). Fluorescence images were captured by using a Leica DM5000 B microscope. Image analysis was performed by using ImageJ software.
Data Availability.
All data supporting the findings of this study are available within the main text and Supporting Information or from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
We thank Peter March and Peter Walker (Bioimaging and Histology Facilities, University of Manchester) for very helpful advice and the staff at the University of Manchester Biological Safety Unit for looking after the mice. This work was supported by a Marie Curie Research Fellowship (to E.G.) and by a grant from Worldwide Cancer Research (to C.T.). S.L. is supported by Fondazione Beretta; A.P. is supported by a PhD studentship from Cancer Research UK; and W.V. is supported by Associazione Italiana per la Ricerca sul Cancro IG Grant 15378.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707929115/-/DCSupplemental.
References
- 1.Balkwill FR, Mantovani A. Cancer-related inflammation: Common themes and therapeutic opportunities. Semin Cancer Biol. 2012;22:33–40. doi: 10.1016/j.semcancer.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 3.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Allavena P. The interaction of anticancer therapies with tumor-associated macrophages. J Exp Med. 2015;212:435–445. doi: 10.1084/jem.20150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simões AE, Rodrigues CM, Borralho PM. The MEK5/ERK5 signalling pathway in cancer: A promising novel therapeutic target. Drug Discov Today. 2016;21:1654–1663. doi: 10.1016/j.drudis.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Nithianandarajah-Jones GN, Wilm B, Goldring CE, Müller J, Cross MJ. ERK5: Structure, regulation and function. Cell Signal. 2012;24:2187–2196. doi: 10.1016/j.cellsig.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi M, Lee JD. Role of the BMK1/ERK5 signaling pathway: Lessons from knockout mice. J Mol Med (Berl) 2004;82:800–808. doi: 10.1007/s00109-004-0602-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, et al. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18:258–267. doi: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi M, Fearns C, Eliceiri B, Yang Y, Lee J-D. Big mitogen-activated protein kinase 1/extracellular signal-regulated kinase 5 signaling pathway is essential for tumor-associated angiogenesis. Cancer Res. 2005;65:7699–7706. doi: 10.1158/0008-5472.CAN-04-4540. [DOI] [PubMed] [Google Scholar]
- 10.Finegan KG, et al. ERK5 is a critical mediator of inflammation-driven cancer. Cancer Res. 2015;75:742–753. doi: 10.1158/0008-5472.CAN-13-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rovida E, et al. ERK5/BMK1 is indispensable for optimal colony-stimulating factor 1 (CSF-1)-induced proliferation in macrophages in a Src-dependent fashion. J Immunol. 2008;180:4166–4172. doi: 10.4049/jimmunol.180.6.4166. [DOI] [PubMed] [Google Scholar]
- 12.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelmsen K, et al. Extracellular signal-regulated kinase 5 promotes acute cellular and systemic inflammation. Sci Signal. 2015;8:ra86. doi: 10.1126/scisignal.aaa3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. The MAPK ERK5, but not ERK1/2, inhibits the progression of monocytic phenotype to the functioning macrophage. Exp Cell Res. 2015;330:199–211. doi: 10.1016/j.yexcr.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo KS, et al. ERK5 activation in macrophages promotes efferocytosis and inhibits atherosclerosis. Circulation. 2014;130:180–191. doi: 10.1161/CIRCULATIONAHA.113.005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, et al. Activation of extracellular signal-regulated protein kinase 5 downregulates FasL upon osmotic stress. Cell Death Differ. 2006;13:2099–2108. doi: 10.1038/sj.cdd.4401969. [DOI] [PubMed] [Google Scholar]
- 18.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 19.Dhomen N, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Bertram JS, Janik P. Establishment of a cloned line of Lewis lung carcinoma cells adapted to cell culture. Cancer Lett. 1980;11:63–73. doi: 10.1016/0304-3835(80)90130-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa T, et al. Optimum immunohistochemical procedures for analysis of macrophages in human and mouse formalin fixed paraffin-embedded tissue samples. J Clin Exp Hematop. 2017;57:31–36. doi: 10.3960/jslrt.17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora M, et al. TLR4/MyD88-induced CD11b+Gr-1 int F4/80+ non-migratory myeloid cells suppress Th2 effector function in the lung. Mucosal Immunol. 2010;3:578–593. doi: 10.1038/mi.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: Function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4:376–389. [PMC free article] [PubMed] [Google Scholar]
- 26.Duluc D, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood. 2007;110:4319–4330. doi: 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, et al. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20:701–712. doi: 10.1038/cr.2010.52. [DOI] [PubMed] [Google Scholar]
- 28.Williams CA, et al. Erk5 is a key regulator of naive-primed transition and embryonic stem cell identity. Cell Rep. 2016;16:1820–1828. doi: 10.1016/j.celrep.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson HM. SOCS proteins in macrophage polarization and function. Front Immunol. 2014;5:357–374. doi: 10.3389/fimmu.2014.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold CE, et al. A critical role for suppressor of cytokine signalling 3 in promoting M1 macrophage activation and function in vitro and in vivo. Immunology. 2014;141:96–110. doi: 10.1111/imm.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, et al. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J Immunol. 2008;180:6270–6278. doi: 10.4049/jimmunol.180.9.6270. [DOI] [PubMed] [Google Scholar]
- 34.Ohnesorge N, et al. Erk5 activation elicits a vasoprotective endothelial phenotype via induction of Kruppel-like factor 4 (KLF4) J Biol Chem. 2010;285:26199–26210. doi: 10.1074/jbc.M110.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao X, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. Genetic activation of ERK5 MAP kinase enhances adult neurogenesis and extends hippocampus-dependent long-term memory. J Neurosci. 2014;34:2130–2147. doi: 10.1523/JNEUROSCI.3324-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 39.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 40.Yan C, Luo H, Lee JD, Abe J, Berk BC. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J Biol Chem. 2001;276:10870–10878. doi: 10.1074/jbc.M009286200. [DOI] [PubMed] [Google Scholar]
- 41.Hiwatashi K, et al. Suppression of SOCS3 in macrophages prevents cancer metastasis by modifying macrophage phase and MCP2/CCL8 induction. Cancer Lett. 2011;308:172–180. doi: 10.1016/j.canlet.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 42.Jones LM, et al. STAT3 establishes an immunosuppressive microenvironment during the early stages of breast carcinogenesis to promote tumor growth and metastasis. Cancer Res. 2016;76:1416–1428. doi: 10.1158/0008-5472.CAN-15-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassetta L, et al. Isolation of mouse and human tumor-associated macrophages. Adv Exp Med Biol. 2016;899:211–229. doi: 10.1007/978-3-319-26666-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the main text and Supporting Information or from the corresponding authors on reasonable request.