Significance
Some plant cells, such as root hairs, pollen tubes, and moss protonemata, expand in a highly polarized manner known as tip growth. That calcium and actin play essential roles in tip growth has been well established, but how they interact with one another in vivo has mostly been modeled from in vitro studies. Here, we use a wavelet analysis approach to dissect the complex calcium homeostasis found in the moss Physcomitrella patens. By perturbing the actin cytoskeleton with drugs and targeted genome editing we find that the shortest calcium oscillation is directly coupled to actin dynamics. This approach to analyzing calcium oscillation profiles sets the stage for subtle dissection of the calcium–actin interaction mechanism.
Keywords: calcium, actin, tip growth
Abstract
Tip-growing cells elongate in a highly polarized manner via focused secretion of flexible cell-wall material. Calcium has been implicated as a vital factor in regulating the deposition of cell-wall material. However, deciphering the molecular and mechanistic calcium targets in vivo has remained challenging. Here, we investigated intracellular calcium dynamics in the moss Physcomitrella patens, which provides a system with an abundant source of genetically identical tip-growing cells, excellent cytology, and a large molecular genetic tool kit. To visualize calcium we used a genetically encoded cytosolic FRET probe, revealing a fluctuating tipward gradient with a complex oscillatory profile. Wavelet analysis coupled with a signal-sifting algorithm enabled the quantitative comparison of the calcium behavior in cells where growth was inhibited mechanically, pharmacologically, or genetically. We found that cells with suppressed growth have calcium oscillatory profiles with longer frequencies, suggesting that there is a feedback between the calcium gradient and growth. To investigate the mechanistic basis for this feedback we simultaneously imaged cytosolic calcium and actin, which has been shown to be essential for tip growth. We found that high cytosolic calcium promotes disassembly of a tip-focused actin spot, while low calcium promotes assembly. In support of this, abolishing the calcium gradient resulted in dramatic actin accumulation at the tip. Together these data demonstrate that tipward calcium is quantitatively linked to actin accumulation in vivo and that the moss P. patens provides a powerful system to uncover mechanistic links between calcium, actin, and growth.
A subset of cell types that play essential roles in plant development, such as pollen tubes, root hairs, moss protonemata, and algal rhizoids, expand by constraining cell-wall expansion to the apex of the cell. This form of growth is commonly referred to as tip growth. Many tip-growing cells display an oscillating tipward calcium gradient in which periodic changes in the cytoplasmic calcium concentration correlate with periodic variations in growth rate (1). Interestingly, in both pollen tubes and root hairs the maximum calcium concentration peaks shortly after a maximum in the growth rate, suggesting that calcium dynamics are dependent on growth dynamics (2). When ionophores are used to experimentally increase the local cytosolic calcium concentration, growth is arrested (3). Additionally, calcium-channel inhibitors (GdCl3 or LaCl3) abolish the calcium gradient, typically resulting in arrested growth (4) and, in the case of root hairs, rupture of the tip (2). Additionally, growth orientation of pollen tubes and root hairs depends at least in part on calcium gradients (3, 5). Thus, calcium dynamics have an impact on growth dynamics, implicating calcium as a key component of the signal transduction machinery regulating apical cell wall expansion and thus tip growth.
One favored hypothesis is that cytosolic calcium levels control the actin cytoskeleton, which in turn regulates secretion and thus cell wall expansion (for review see ref. 6). In support of this hypothesis, actin-depolymerizing drugs such as latrunculin B (LatB) abolish the dynamic tipward calcium gradient and growth in pollen tubes and root hairs (7–10). Cytosolic calcium likely decreases because growth stops. Stretch-activated calcium channels in the plasma membrane have been proposed to link growth status to calcium by responding to cell-wall or membrane curvature, thereby modulating cytosolic calcium uptake (11, 12). However, how calcium regulates the actin cytoskeleton in tip-growing cells remains unclear.
In fact, there are myriad possible mechanistic connections between calcium and actin filament dynamics and organization. At the biochemical level, the activity of many actin-binding proteins is calcium-dependent and can thus modulate F-actin architecture directly (13). Specifically, profilin binding activity has been shown to be calcium-dependent in vitro (14), and profilin deficient tip-growing cells are impaired (15, 16). Villins, gelsolin-like proteins, both bind and sever actin in a calcium-dependent manner (17) and are necessary for pollen tube growth (18). Additionally, cytosolic calcium levels directly regulate calcium-dependent protein kinases (CDPKs), which do not require an interaction with calmodulin for activation. In support of this, CDPK-like null plants have aberrant actin structures within pollen tubes, suggesting CDPKs could regulate actin by phosphorylating specific actin-modulating proteins (19, 20), with ACTIN DEPOLYMERIZING FACTOR(ADF)/Cofilin being a well-studied example (21–23). Similarly, calcium levels are implicated in controlling small Rho/Rac of Plants GTPase activity (24)—proteins critical for tip growth. Thus, calcium impacts many actin-dependent cellular processes, making it challenging to uncover the mechanistic links between calcium and actin.
Establishing a mechanistic understanding of the interaction between calcium and growth via the regulation of the actin cytoskeleton in vivo has remained challenging due to the many cellular processes affected by calcium (25). An additional challenge has been identifying an in vivo tip-growing plant system amenable to rapid molecular genetic manipulation with excellent cytology. Here, we present evidence that the moss Physcomitrella patens provides the ideal in vivo system to dissect the interaction between calcium, growth, and actin. The juvenile tissue of mosses is composed of tip-growing cells, called protonemata, that can be easily propagated asexually by either moderate tissue homogenization or regenerating whole plants from single protoplasts, resulting in thousands of genetically identical tip-growing cells (26, 27). Furthermore, P. patens has excellent genetic resources: a sequenced and annotated genome (28), an expression atlas (29), well-established transformation protocols (30), high frequency of successful homologous recombination allowing for precise gene knock in/knock out (31), robust methods for RNAi (32–34), and CRISPR/Cas9 genome editing (35). However, calcium signaling in P. patens remains understudied. Early work using reconstituted aquaporins and coelenterazine demonstrated a wavelength-specific calcium response in protonemal tissue (36). A decade later, Koselski et al. (37) identified light-responsive calcium channel-dependent action potentials. Similarly, Tucker et al. (38) used Fura-2-dextran to report UV-A–induced calcium waves in apical protonemal cells. However, none of these studies investigated mechanistic connections between calcium and tip growth.
To simultaneously monitor actin and calcium dynamics we generated a P. patens line that stably expresses both Yellow-Cameleon 65, a FRET-based calcium probe (39, 40), and Lifeact-mRuby, a validated live-cell actin binding probe (41–43). Employing calcium-channel inhibitors and RNAi to silence specific calcium pumps we showed that calcium homeostasis is essential for protonemal growth. Using microfluidic chambers amenable to long-term live-cell imaging (44), we discovered that protonemata have a highly dynamic calcium gradient that has complex periodic behavior at the tip of growing cells. By perturbing growth pharmacologically, genetically, and mechanically we found that intracellular calcium dynamics depend on growth. By simultaneously imaging calcium and actin we demonstrated that tip-focused actin filaments also exhibited complex oscillatory behavior that is anticorrelated with cytosolic calcium. Our study represents a direct, quantitative dissection of the interaction between calcium and actin in live, tip-growing plant cells.
Results
Calcium Homeostasis Is Essential for Long-Term Protonemal Growth.
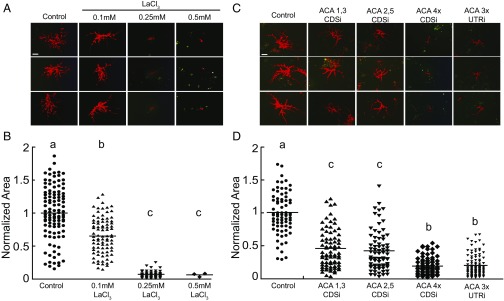
To determine whether calcium is important for protonemal tip growth we used lanthanum chloride (LaCl3) to inhibit calcium-channel proteins. Because LaCl3 forms a precipitate in normal growth medium, we grew plants regenerated from protoplasts on plates containing only water and agar. We found that after exposure to 0.1 mM LaCl3 for 3 d plants were 65% the size of the control (Fig. 1). Higher concentrations of LaCl3 further inhibited growth, and plants grown on 0.25 mM LaCl3 only were 7% the size of control (Fig. 1). The dramatic reduction in area was coupled with reduced viability (n = 3 plants, 0.5 mM LaCl3; n = 48, 0.25 mM LaCl3; n = 85, 0.1 mM LaCl3; n = 108, control). Together these results indicate that inhibiting calcium transport negatively affects growth and viability.
Fig. 1.
Calcium-channel inhibition inhibits protonemal growth. Representative fluorescence images of 7-d-old plants (A) and the quantification of plant size (B) grown on the indicated concentration of LaCl3. Representative 7-d-old plants transformed with the indicated constructs (C) and the quantification of plant size (D). Plants were visualized by imaging chloroplast autofluorescence with a fluorescence stereomicroscope. Plant area was determined by measuring the area of the chlorophyll autofluorescence, normalized to control conditions. Letters indicate statistical groups with α < 0.05 from an ANOVA analysis. (Scale bars, 100 μm.)
To investigate proteins potentially involved in calcium homeostasis during tip growth we knocked down the expression of key calcium-channel pumps via RNAi. In Arabidopsis thaliana, plants lacking the Autoinhibitory Calcium-ATPase 9 (AtACA9) have pollen tube growth defects, and fusions of this protein with GFP localize to the plasma membrane of pollen tubes (45). In P. patens, the gene product of transiently overexpressed PpACA1-GFP in protoplasts localized to the membrane of small vacuole-like structures (46). Interestingly, knockouts of PpACA1 resulted in plants with a decrease in both salt tolerance in gametophores and the ability to sequester cytosolic calcium in protonemata (46). In fact, there are five ACA genes in P. patens (28). Based on the published expression atlas (29), four of the ACA genes are expressed in protonemata: ACA1 (Pp3c8_9970V3), ACA2 (Pp3c19_22060V3), ACA3 (Pp3c22_18760), and ACA5 (Pp3c13_24540V3). ACA4 (Pp3c14_19110V3) is only significantly expressed in archegonia (29), the female organs, and thus was not targeted in the RNAi assays. For ACA1, -2, -3, and -5, a 500-bp fragment was cloned from the 5′ end of each coding sequence (CDS). After ligating these fragments we then generated RNAi constructs (CDSi) that simultaneously targeted either two or four PpACA genes in a manner described previously (33). ACA1,3 and ACA2,5 RNAi constructs resulted in a 55% and 57% reduction in plant size, respectively (Fig. 1). When ACA1, -2, -3, and -5 were targeted simultaneously the resulting plants were 80% smaller than the control.
To ensure that the coding sequence RNAi construct is specific for the expressed ACA genes we designed a second RNAi construct (UTRi) that targeted the UTRs of the ACAs. In trying to isolate the 5′UTR of ACA3, which did not have a predicted 5′ UTR (28), we discovered that under our culturing conditions ACA3 is not expressed in 7-d-old protonemata (Fig. S1). Thus, the UTRi RNAi construct only contains the UTRs of ACA1, -2, and -5. Nevertheless, regenerated plants expressing this plasmid phenocopy the coding-sequence-targeting constructs, as plants were 79% smaller than the control (Fig. 1 C and D). These data demonstrate that ACAs are required for protonemal growth and suggest that their regulation of calcium homeostasis likely underlies their role in growth.
Yellow-Cameleon Nano 65 Reveals a Calcium Gradient in Growing Protonemata.
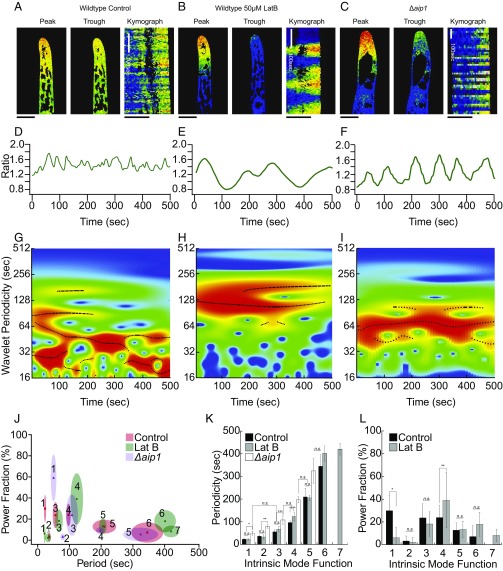
To visualize cytosolic calcium levels, we generated stable lines where expression of the FRET-based calcium probe Yellow-Cameleon Nano 65 (YCN) was driven by a constitutive promoter (39, 40). For confocal imaging, we regenerated YCN protonemal tissue in polydimethylsiloxane (PDMS) microfluidics devices, enabling imaging over extended time scales (44). By dividing the FRET-acceptor (YFP) signal by the FRET-donor (CFP) signal we generated ratiometric representations of cytosolic calcium. Protonemal filaments have two cell types: slower-growing chloronemata and faster-growing caulonemata. Here, we analyzed caulonemal cells because due to their growth rates it is easier to identify actively growing cells. We observed a manifest tipward calcium gradient (Fig. 2A) in growing caulonemal cells, similar to that observed in other tip-growing plant cells (47–51). We found that calcium fluctuated on the order of seconds and did so persistently when imaged for tens of minutes (Fig. 2 and Movie S1).
Fig. 2.
Visualization of tipward calcium gradient in protonemata. Representative images from a time-lapse acquisition showing the peak and trough of the calcium fluctuations in WT (A), 50 μM LatB-treated WT (B), and Δaip1 (C) cells (Movie S1). (Scale bars, 10 μm.) Kymographs are made from a 20-μm line drawn through the middle of the cell, parallel to the access of growth. Line traces of mean intensity in an apical region ROI for WT (D), LatB-treated WT (E), and Δaip1 (F) cells. Wavelet analysis output represented as a heat map, from WT (G), LatB-treated WT (H), and Δaip1 (I) cells. Black lines are ridges of statistically significant oscillations. (J) Period–power fraction plot of IMFs in each treatment. Size of ellipse is SD on each axis. (K) Period plot of each IMF. Stars denote statistical difference (Fisher’s LSD, P < 0.05). (L) Power fraction plot for each IMF in control and LatB-treated cells. Power of the IMFs shifts to favor longer periods, specifically IMF4 (Fisher’s LSD, P < 0.05).
Interestingly, these episodic changes do not form stationary oscillations but rather display a complex time course, with brief oscillations of different frequencies appearing and disappearing throughout the time course. We found that methods typically used to analyze oscillatory signals, such as autocorrelation and Fourier transformations (52), were not able to capture the temporal structure of the calcium changes. Instead, we used wavelet analysis, which scans the time series, identifying frequencies of brief oscillations (wavelets) occurring across any oscillatory dataset (53). Summing together the significant wavelet frequencies recapitulates the observed complex oscillation profile. In every experiment wavelet analysis revealed several statistically significant components shown in the heat map as red and as black lines across the frequency-versus-time profile (Fig. 2 G–I). To extract component frequencies we employed a signal-sifting algorithm that decomposes the complex oscillations into separate simple ones: empirical mode decomposition (EMD) (54). This process generates a series of intrinsic mode functions (IMFs) that correspond to discrete periods found in the complex signal. Additionally, this method allows us to quantify the degree to which each oscillation contributes to the net signal, referred to here as the power fraction. We extracted the component frequencies identified as significant in each experiment and averaged these values across experiments. For control cells there are six oscillatory components, summarized in Table 1 and shown in Fig. 2 J and K. Additionally, component frequencies had unequal contributions to the overall pattern, as seen in their power fraction (Fig. 2 J and L and Table 1). In control cells IMFs 1, 3, and 4 contribute to the bulk of the signal (Fig. 2L and Table 1). Both the number of component frequencies and their varied power fraction contribute to the complex oscillation pattern of the apical calcium gradient.
Table 1.
Period and power fraction of IMFs
| Period, s | Power fraction, % | |||||
| IMF | Control | LatB | Δaip1 | Control | LatB | Δaip1 |
| IMF1 | 23.2 ± 3.39 | 22.1 ± 2.7 | 50.1 ± 8.7 | 30.0 ± 13 | 6.2 ± 9.1 | 58.9 ± 22.2 |
| IMF2 | 36.7 ± 3.8 | 32.9 ± 9.3 | 79.0 ± 7.5 | 3.3 ± 4.4 | 2.2 ± 4.1 | 3.1 ± 3.1 |
| IMF3 | 56.0 ± 8.4 | 67.1 ± 12.0 | 108.0 ± 19.0 | 23.7 ± 12.4 | 18.2 ± 10.8 | 23.6 ± 19.5 |
| IMF4 | 97.1 ± 6.88 | 123.1 ± 19.7 | 197.23 ± 13.5 | 24.0 ± 10.7 | 39.0 ± 24.3 | 9.0 ± 3.2 |
| IMF5 | 210.1 ± 37.1 | 205.2 ± 11.7 | 324.3 ± 54.88 | 12.8 ± 6.5 | 13.3 ± 7.4 | 5.4 ± 6.4 |
| IMF6 | 345.2 ± 49.9 | 401.4 ± 33.8 | — | 7.24 ± 9.6 | 18.0 ± 10.5 | — |
| IMF7 | — | 420.2 ± 25.9 | — | — | 8.2 ± 5.5 | — |
To probe the signaling significance of the calcium fluctuations observed in growing cells we investigated the change in calcium dynamics after inhibiting growth using LatB. LatB is a potent actin-depolymerizing drug, and its effects on protonemal growth have been well documented (55–58). Cells treated with 50 μM LatB, which is sufficient to depolymerize actin filaments in microfluidic chambers (44), continued to exhibit calcium oscillations (Fig. 2 B and E and Movie S1). LatB-treated cells stopped growing but continued to swell at an average rate of 2.64 ± 1.5 μm/h, a 4.7-fold reduction from the WT expansion rate. Through EMD, we found that the more rapid IMFs (1–3) were unchanged by the LatB treatment, but both IMFs 4 and 6 slowed down, taking on longer periods (Fig. 2 J and K and Table 1). While the period of IMF 1 did not change significantly, the power fraction of IMF 1 dropped substantially to 6.2 ± 9.1%, making it a weak contributor to the net signal (Fig. 2L and Table 1, n = 6). Simultaneously, the power fraction of IMF 4 increased significantly to 39.0 ± 24.3%, suggesting that LatB treatment results in oscillations composed of longer periods (Fig. 2L). In support of this, we detected an additional IMF with a period of 420.2 ± 25.9 s in LatB-treated cells (Fig. 2 J–L and Table 1). These results show that cells lacking actin had slower calcium oscillations.
LatB depolymerized all actin filaments, leading to immediate growth inhibition. To more subtly perturb growth we used a genetic approach to modulate actin dynamics. Null mutants in ACTIN INTERACTING PROTEIN1 (AIP1), which works synergistically with ADF to sever actin filaments (59, 60), exhibit slow-growing protonemata, dramatically reduced actin filament dynamics, and accumulation of cytosolic actin bundles (61). To analyze calcium behavior in this mutant we took advantage of P. patens’ efficient rate of homologous recombination to knock out AIP1 in the YCN/Lifeact-mRuby line (Fig. S2). The YCN/Lifeact-mRuby Δaip1 line had a morphology similar to Δaip1 mutants generated previously and grew at 4.67 ± 1.9 μm/h, 2.7-fold slower than the control YCN/Lifeact-mRuby line. Similar to LatB treatment, growing Δaip1 cells also exhibited a tipward calcium gradient (Movie S1). Similar to LatB-treated cells, both the time trace (Fig. 2F) and wavelet heat map (Fig. 2I) show a shift to favor longer periods in Δaip1. However, the periods of the component frequencies of the calcium oscillations were different from those of LatB-treated cells (Fig. 2). In Δaip1 cells, the first IMF that the EMD analysis identified was 50.1 ± 8.7 s—statistically dissimilar from IMF1 of both control and LatB-treated cells but statistically indistinguishable from control IMF 3 [Fisher’s least significant difference (LSD), P < 0.05; Fig. 2 J and K]. Likewise, Δaip1 IMF3 and IMF4 are most similar to control and LatB IMF4 and IMF5, respectively, suggesting that the component frequencies comprising the calcium oscillation in Δaip1 cells have shifted to longer periods (Fisher’s LSD, P < 0.05; Fig. 2 J and K). Thus, a reduction in actin dynamics results in a dramatic difference in calcium behavior.
Physical Barriers to Growth Alter the Tipward Cytosolic Calcium Behavior.
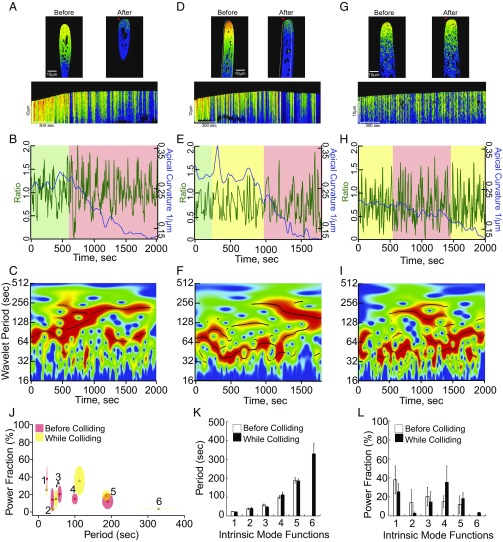
LatB treatment and deletion of AIP1 significantly alter growth by shifting the pool of monomeric and filamentous actin to either extreme, and the oscillation profile of the apical calcium gradient is likewise aberrant. To perturb growth in a less-invasive manner we sought to restrain growth mechanically, thereby not altering cell physiology, and then monitoring intracellular calcium behavior. We used microfluidic chambers engineered with a series of barriers around which growing protonemata must navigate. We imaged YCN caulonemal cells growing toward, interacting with, and finally pushing against barriers (Fig. 3 and Movie S2). Due to differences in the angle of interaction, and microtopography of the barriers, there was considerable variability in the responses from cells growing into barriers.
Fig. 3.
The tipward calcium oscillations were altered by inhibiting growth mechanically. (A, D, and G) Representative images and kymographs from a time-lapse acquisition of cells interacting with barriers. Deformed tips denoted by red arrows. White outline marks the cell’s initial position before it translocated along the barrier. (B, E, and H) YCN ratio trace (green line) and apical curvature (blue line); note that apical curvature decreases as a function of tip deformation. Green denotes time period before collision, yellow denotes time period when cell exhibits intermediate expansion rates, and red denotes time period when cell exhibits minimal expansion rates. (C, F, and I) Wavelet analysis showing a shift to favor long periods after maximal decrease in expansion rate (Fig. S3). (J) Period–power fraction plot of IMFs obtained from the green/yellow regions of traces before the maximal collision state (red regions). Size of ellipse denotes SD on each axis. (K) Period of each IMF before and during impact, showing that period length is unaffected. (L) Power fraction of each IMF shows a trend to favor IMF4 upon impact.
Fig. 3 shows three examples of the types of interactions observed. Growth inhibition can be readily observed for two of the cells in their respective kymographs. However, a decrease in the apical curvature (Fig. 3 B, E, and H, blue line) and in the expansion rates (Fig. S3) more accurately describes the collision behavior. The cell in Fig. 3 A–C exhibits a period of normal growth (Fig. 3B, green region) before colliding with the barrier. Upon collision, there is a drop in the apical calcium (Fig. 3B, green line). Afterward, normal growth is inhibited, as evidenced by a change in the apical curvature (Fig. 3B, blue line) and expansion still continues at 54.21 μm2/h, a 5.1-fold decrease from precollision expansion rates (Fig. S3A). Once normal growth is inhibited, the wavelet analysis (Fig. 3C) shows a shift in the oscillation profile to favor longer periods. The cell in Fig. 3 D–F slid along the barrier (Fig. 3E, yellow region), during which there was a slight decline in the apical curvature (Fig. 3E, blue line) and a decrease in the expansion rate (Fig. S3B). However, the cell was observed to fully collide once the apical curvature dropped below 0.25/μm (Fig. 3E, blue line), and the expansion rate was 7.0-fold lower than precollision expansion rates (Fig. S3B). The full collision was concurrent with a shift in the calcium oscillation profile. The structures due to the shorter frequencies are overshadowed by the intense, longer-frequency IMFs (Fig. 3E, red region, Fig. 3F). Finally, the cell in Fig. 3 G–I never stopped growing, as evidenced by the 1.6-fold decrease in expansion rate (Fig. S3C). However, at the minimal expansion rate (77.8 ± 5.1 μm2/h), concurrent with the apical curvature dropping below 0.17/μm, the calcium oscillation profile has a predominant longer period wavelet (Fig. 3I).
Although each cell imaged was unique, commonalities are apparent. IMF period values and power fractions were measured comparing regions of the time course before the collisions (green/yellow) versus during the collisions (red). As each cell interacts with a barrier there is a shift in the power of IMFs to favor the longer frequencies (Fig. 3 J and L). Although shorter frequencies were diminished in power they were still detectable and their period seemed unchanged. Interestingly, the oscillation profile for a cell that experienced an abrupt change in expansion rate (Fig. 3 A–C) upon contact with a barrier resembled what we observed after growth was inhibited with LatB (Fig. 2). Since mechanical, pharmacological, and genetic inhibition of growth similarly alter the calcium oscillation behavior these data provide a quantitative framework for analyzing the link between calcium and growth.
Cytosolic Calcium and Apical Actin Are Anticorrelated During Tip Growth.
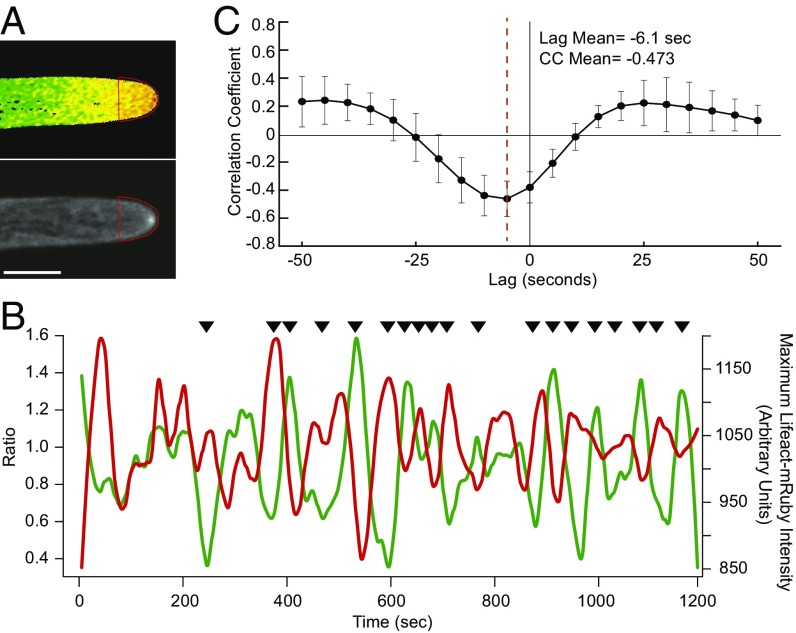
Since dynamic actin is required for tip growth we investigated the relationship between actin and calcium during growth. We quantified the tipward calcium gradient fluctuations and the intensity levels of a focal point of tip-localized actin filaments (the actin spot, Fig. 4). While persistent over long time periods, this actin spot is constantly polymerizing and depolymerizing on a time scale of seconds, leading to changes in actin-spot position and intensity during growth (Movie S3). We reasoned that if calcium modulates actin turnover in the apical actin spot then the calcium and actin signals would be correlated. To ensure that decreases in intensity were not due to the actin spot moving out of the imaging plane we increased the size of the pinhole capturing light from above and below the imaging plane. For calcium, we measured the mean intensity of a region of interest (ROI) at the cell tip, starting ∼6 μm back from the apex. For actin we measured the maximum intensity in the same ROI to track changes in the actin spot (Fig. 4A). When these intensities were plotted simultaneously we observed that high levels of calcium qualitatively correlated with low levels of actin (Fig. 4B; black arrows point to anticorrelated peaks). To quantitatively test the observed correlation we used cross-correlation analysis (Fig. 4C). Cross-correlation analysis quantifies the similarity of two datasets by comparing one to the other along all of the points in the series. By shifting the actin data along the calcium data the cross-correlation analysis revealed that calcium and actin signals were significantly anticorrelated (−0.473, n = 9). Additionally, this analysis revealed a −6.1-s lag associated with the calcium signal and the corresponding actin response. We obtained similar values when we performed cross-correlation analysis of the apical calcium and the area of the actin spot (Fig. S4). Cross-correlation analysis takes into account the shape of each intensity change. Therefore, the temporally local decrease in Lifeact-mRuby signal is proportional to the previous rise in YCN ratio, demonstrating that the intensity of the actin at the apical spot is predicted by the preceding calcium activity.
Fig. 4.
Actin and calcium are anticorrelated at the tip of growing cells. (A) Images of YCN and Lifeact-mRuby acquired simultaneously. Red trace depicts the ROI used for analysis (Movie S3). (Scale bar, 10 μm.) (B) Intensity plot of mean YCN intensity within the ROI (green) and maximum Lifeact-mRuby intensity within the ROI (red). Black arrows indicate qualitative regions of anticorrelation. (C) Cross-correlation average trace (n = 9 cells) revealing a maximum correlation coefficient and lag between calcium and actin.
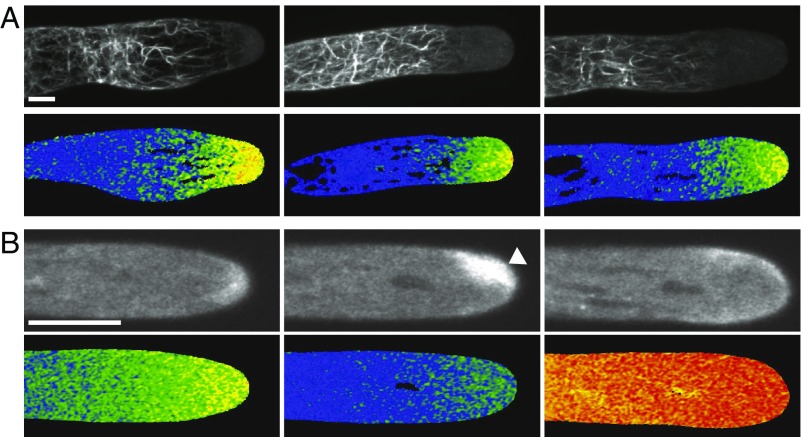
To further investigate the relationship between calcium and actin we took advantage of the Δaip1 mutant since it has altered actin and calcium dynamics. We performed simultaneous acquisition of both YCN and Lifeact-mRuby in YCN/LAmRuby/Δaip1 protonemata. Similar to previous observations (61) and distinct from what is present in WT cells, we observed dramatic actin filament bundles that undergo retrograde movement (Fig. 5A and Movie S4). Interestingly, in growing cells, actin bundles originated in a subapical region precisely correlating with the boundary between the apical calcium gradient and background cytosolic calcium levels (Fig. 5A and Movie S4). This observation is consistent with the formation of actin filaments in the low-calcium state. However, actin filaments, particularly the prominent apical actin spot, were nearly absent from the apical cytoplasm even in the presence of an oscillating calcium gradient (Fig. 2). It is possible that actin monomers are sequestered in the subapical actin bundles and thus are unavailable to polymerize at the cell apex because actin filaments turn over slowly in Δaip1 plants (61). Thus, in a cell where the equilibrium between monomeric and filamentous actin has been shifted toward the latter, actin network formation was still anticorrelated with calcium activity.
Fig. 5.
Aberrant actin and calcium. (A) Maximum intensity projections of Z-stacks (Top) of Lifeact-mRuby and medial plane selections of YCN (Bottom) from three representative YCN/LA/Δaip1 apical cells. Note that the basal end of the calcium gradient coincides with the apical limit of the actin bundles (Movie S4). (B) Single-focal-plane images of Lifeact-mRuby (Top) and YCN (Bottom) from a cell treated with 0.1 mM LaCl3. The first pair is pretreatment, the second pair shows the accumulated apical actin the absence of cytosolic calcium (white arrow), and the third pair shows the abolishing of accumulated apical actin after rupture (Movie S5). (Scale bars, 10 µm.)
To investigate actin’s response we sought to specifically alter calcium. The anticorrelation between calcium and actin predicts that under sustained periods of low calcium the apical actin spot should undergo unregulated actin polymerization. Similarly, under sustained periods of high calcium the apical actin spot would depolymerize. We tested this prediction by monitoring the response of both the actin spot and cytosolic calcium levels when growing cells were exposed to 0.1 mM LaCl3, which inhibits calcium entry. Within seconds of the addition of LaCl3, growth stopped and the strong tipward gradient was abolished (Fig. 5B and Movie S5). Concomitant with the loss of the calcium gradient the actin spot dramatically increased in size (Fig. 5B, white arrow head), taking up a larger region of the cell tip. Interestingly, the spot remained quite mobile during this transition. The remaining cytosolic calcium dropped over the course of minutes. Strikingly, each cell treated with 0.1 mM LaCl3 ultimately exploded after several minutes (Movie S5). Before explosion, there was a dramatic increase in cytosolic calcium that appeared to emanate from the tip, suggesting that a small rupture of the plasma membrane occurred at the tip. This dramatic increase was accompanied by an immediate decrease in the accumulated actin spot. Taken together, these data suggest that elevated calcium inhibits apical actin filament accumulation in vivo.
Discussion
Here, we have shown that P. patens tip-growing cells exhibit a complex and dynamic calcium oscillation profile. To gain insight into the significance of the component frequencies identified in the calcium oscillations we manipulated growth pharmacologically, genetically, and mechanically. We found all treatments that result in a reduction in growth alter the oscillation profile. The most consistent change is that upon growth retardation longer frequencies predominate in the oscillation profile. These data indicate that growth is tightly coupled to calcium homeostasis. Furthermore, we probed the relationship between intracellular calcium and the actin cytoskeleton, which is essential for polarized growth in P. patens. By simultaneously imaging calcium and actin in cells we demonstrated calcium activity predicts the size and intensity of the apical actin spot.
The complex calcium behavior at the apex of growing P. patens cells was not readily analyzed with commonly employed approaches, such as Fourier and spectral analyses (52). In fact, these methods failed to pull out robust frequencies, presumably because the calcium signal is a summation of multiple frequencies resulting from distinct biological processes. However, using wavelet analysis and EMD sifting we were able to reliably extract frequencies represented with statistical significance from the calcium data. Wavelet analysis has been increasingly used to dissect a variety of complex oscillatory data, from hydrology (62) to brain wave activity (63) to financial markets (64). P. patens apical calcium oscillation profile has six predominant frequencies which modulate the temporally local calcium signal in a variable combinatorial manner. Calcium oscillations have been detected in several other systems with vastly different growth rates, including pollen tubes (11, 65, 66) and the tip-growing hyphae of Aspergillus nidulans (67), both of which exhibit calcium oscillations with a single frequency. Recently, a two-frequency oscillation profile was reported in Arabidopsis root hairs (68). It is unclear why some tip-growing cells, such as pollen tubes, have a single oscillation frequency and others, such as root hairs and moss caulonemata, have more complex oscillations. One possibility considers the more permanent nature of moss protonemata. Pollen tubes are highly specialized cells with the singular purpose of growing toward the ovule. Root hairs must integrate information about soil composition into their growth patterns but are ultimately ephemeral structures. Protonemata, however, represent the entire body of the juvenile plant and therefore may have calcium signals that are observed in the more permanent structures of seed plants (25).
Additionally, the longer-frequency oscillations may represent differential uptake/export from distinct intracellular or extracellular calcium stores. If, for example, in pollen tubes the power of the longer-frequency calcium oscillations is small in comparison with the power of the changes in the short-frequency oscillation then they may not be detected. In turn, the power of each frequency may reflect differences in how much calcium is buffered in the cell wall or intracellular compartments. As seen by the EMD, the power fraction for each IMF is varied, and it may be that other tip-growing systems have a greater difference in component frequencies such that only one is favored. By taking advantage of the excellent cytology and genetic manipulability of P. patens future computational modeling of calcium homeostasis, coupled to genetic perturbations of molecules involved in calcium regulation, will help to identify the molecular basis of the longer-frequency calcium oscillations.
Notably, altering actin dynamics in P. patens shifted the predominant frequencies to longer time scales. For example, in the absence of actin filaments we found that the contribution of the short-frequency calcium oscillation is diminished and a new longer-frequency oscillation (IMF 7) is observed. Additionally, IMF4 increases in power upon LatB treatment and trends toward the same during collisions with barriers. It could be that the trend to favor longer oscillations is indicative of a stress response, of which calcium has been known to play a part (69–71). When actin dynamics were drastically inhibited in Δaip1 cells we observed a shift of the more rapid IMFs in periodicity to slower times, but significantly faster than in the absence of actin filaments. Indeed, it appears that what was identified as IMF3 in control cells completely disappears in Δaip1 cells, and IMF 1 and 2 shift to compensate (Fig. 2J). These data suggest that dynamic actin influences the calcium oscillatory behavior. However, it is difficult to uncouple growth and actin dynamics, since cells lacking actin do not grow and cells with drastically impaired actin dynamics such as the Δaip1 cells grow very slowly. To address this we imaged cells with normal actin dynamics that are physically restrained by encountering a barrier. Barrier navigation is analogous to how protonemata might negotiate and colonize uneven soil. Interestingly, we also observed shifts to predominantly longer frequencies when the cells grew against the barriers. Together these data suggest that when growth is affected the calcium oscillatory frequencies slow down, indicating a change in growth status.
Since the calcium behavior is oscillatory, it suggests that there is active feedback between calcium and growth. Intriguingly, and in contrast to pollen tubes, moss caulonemata do not exhibit obvious growth oscillations (72), which could simply be an issue of resolution. If a feedback mechanism is present, then we would expect that in the absence of calcium oscillations growth would be inhibited. We were able to abolish all calcium oscillations in growing caulonemata by adding LaCl3 to the growth medium. As predicted, caulonemal cells exposed to 0.1 mM LaCl3 immediately stopped growing and exploded within a few minutes. Because LaCl3 abolishes the calcium gradient by inhibiting calcium import, it suggests that calcium oscillatory behavior requires entry of extracellular calcium. Furthermore, calcium oscillations during tip growth are required for cellular integrity. Interestingly, prolonged exposure to 0.1 mM LaCl3, as in our growth assays (Fig. 1), was not lethal. This suggests that plants are able to compensate for the deleterious effects of LaCl3, perhaps by changing the composition of the cell wall, or by sequestering LaCl3. When imaging for our growth assay, plants are predominately composed of chloronemata, as caulonemata occur later in development. Together, these observations could suggest that chloronemal cells are more resilient than caulonemata to the effects of LaCl3.
Having identified that entry of extracellular calcium is essential we reasoned that calcium sequestration would likely play an important role in growth. Calcium sequestration could be achieved by rapidly transporting calcium into intracellular compartments, or pumped out of the cell entirely, by Ca2+-ATPases. In P. patens the autoinhibitory Ca2+-ATPases (ACAs) have been shown to be important for calcium homeostasis. Specifically, Δaca1 protonemata are unable to restore cytosolic calcium levels to baseline after salt-induced calcium uptake. In that study, the authors also showed that 35S::PpACA1-GFP gene product localizes to small vacuoles (46). Together these data suggest that PpACA1 plays a role in the rapid sequestration of calcium into membranous vacuolar compartments. Furthermore, plant ACA activity requires calcium-dependent calmodulin binding to the N-terminal autoinhibitory domain (45, 46, 73). Thus, calcium influxes across the plasma membrane, such as those observed in caulonemata, could activate the machinery required for immediate calcium transport by ACAs localized to the plasma membrane, endoplasmic reticulum, vacuoles, chloroplasts, or mitochondria. Here, we silenced the three ACA genes expressed in protonemata (Fig. S1) and found that plants were severely impaired in growth, which suggests that this family of calcium ATPases is critical for regulating calcium homeostasis in moss. Future studies of ACA localization and the impact of ACA loss-of-function mutations on intracellular calcium dynamics will begin to connect the regulatory mechanisms that link calcium oscillatory behavior to growth.
Probing the mechanistic links between calcium, actin, and growth has been challenging, because calcium affects a multitude of cellular processes (25) and therefore has a complex intracellular regulatory infrastructure. Consequently, the focus of the tip-growth field has been on calcium modulation of the actin cytoskeleton ultimately impacting secretion (6). The majority of these studies have analyzed correlations between intracellular activities (calcium/actin/secretion) and growth, not between the intracellular activities themselves (reviewed in ref. 74). Recently, however, Takeshita et al. (67) approached this question in the hyphae of the fungus A. nidulans. By knocking out known calcium channels they dramatically reduced the correlated calcium and F-actin oscillations seen in WT. Here, we modulated the actin dynamics in a plant system and measured the consequences for the calcium oscillation profile. Additionally, by hindering calcium uptake with LaCl3 we documented a robust reciprocal interaction whereby actin polymerization prevails when the apical region is in a low-calcium state.
With simultaneous imaging of calcium and actin probes we saw that calcium and the intensity and size of an apical actin structure are anticorrelated. We found that in WT cells with normal actin dynamics high calcium levels lead to depolymerization of the apical actin spot. Low levels promoted formation of the actin spot. In the subapical region of the cell that has constitutively low levels of calcium actin bundles were observed. However, in cells with reduced actin dynamics, such as in Δaip1 cells, we observed a shift toward actin bundling due to the longer lifetime of individual filaments (61). As a result, in the subapical regions, which directly correlated with low calcium levels, we observed massive bundling, which likely deprived the apical domain of sufficient monomers to generate the apical actin spot, even though Δaip1 cells exhibited sustained periods of low calcium at the cell tip.
In the absence of the calcium gradient, which was achieved in cells treated with LaCl3, we observed the actin spot grow in intensity, corroborating our observations that calcium inhibits actin accumulation in WT cells. Importantly, we observed that the enlarged actin spot lost focus and occupied a larger portion of the tip, suggesting that calcium not only controls the dynamics of the apical actin spot but also spatially restricts the actin spot in the apical dome. Treatment with LaCl3 led to cell rupture, suggesting that an enlarged actin spot leads to unregulated cell expansion. Thus, our data suggest that oscillatory changes in calcium precisely modulate the apical actin spot, leading to regulation of cell expansion at the cell tip. Moving forward, coupling modeling of the rich and complex calcium oscillation profile in P. patens caulonemata with the superb cytology and extensive genetic resources available in P. patens provides an opportunity to uncover the molecular linkages between growth, calcium, and actin.
Materials and Methods
Plant Culture and Transformation.
All lines were aseptically propagated weekly by moderate homogenization in water and pipetting onto a permeable cellophane-covered solid media, as described previously (58). Petri dishes of tissue were grown at room temperature in Percival growth chambers, under 85 μmolphotons/m2⋅s light with long-day conditions. Stable lines were generated with a PEG-mediated transformation protocol, described previously (30). Yellow-Cameleon 65 in the pTH-Ubi gate expression vector (33) was a gift from Simon Gilroy, University of Wisconsin–Madison, Madison, WI. In this expression vector YCN65 expression is driven by the maize ubiquitin promoter. Linearized plasmid is transformed into protoplasts from 7-d-old WT tissue. Selection for transgenic plants was done as described previously (30), and hygromycin-resistant plants were screened visually for expression of the probe. Similarly, Lifeact was fused to mRuby via site-specific recombination-based cloning into pTZ-Ubi gate (58) and stably transformed into the YCN line. The Δaip1 line was generated as described (61), with the exception that the neomycin resistance cassette was incorporated into the knock-out construct and G418 was used for selection.
Growth Assays.
For LaCl3 treatments, LaCl3 was added to water agar after autoclaving to achieve the final concentration described in the text. Four-day-old regenerated protoplasts were transferred to the media containing LaCl3 and were imaged on the seventh day. Plants were visually screened by chlorophyll autofluorescence. For RNAi of ACA genes we used a method described previously (32). In short, a 500-bp fragment from each gene was ligated together via restriction enzyme sites introduced into the PCR primers (see Table S1). This product was then directionally cloned into the pENTR/D-TOPO vector (Invitrogen) and sequenced. LR enzyme-mediated site-specific recombination with the entry clone and an RNAi destination vector (33) produced the final RNAi expression vector. A moss line stably expressing a fusion protein of a nuclear localization signal, GFP, and β-glucuronidase (GUS) was transformed and allowed to regenerate. On the fourth day, the plants were transferred to hygromycin selection. On the seventh day, plants were visually screened both by chlorophyll autofluorescence and for the lack of nuclear GFP-GUS, an indicator of active silencing (33). Area data were calculated by thresholding the chlorophyll autofluorescence via ImageJ and measuring the area. Each condition was then normalized to the RNAi control (GUS silencing). For both growth assays all images were taken with a Leica MZ 16 FA microscope equipped with a PlanAPO 1.0× objective and a Leica DFC 300FX camera. Image analysis was perfomed with ImageJ.
Growth in Microfluidic Chambers.
Microfabricated PDMS microfluidic chambers were designed and built in collaboration with John Oakey at the University of Wyoming and described in detail previously (44). Seven-day-old protonemal tissue was gently homogenized by pipetting up and down and then loaded into the chamber, which was flooded with liquid Hoagland’s media [4 mM KNO3/2 mM KH2PO4/1 mM Ca(NO3)2/89 μM Fe citrate/300 μM MgSO4/9.93 μM H3BO3/220 nM CuSO4/1.966 μM MnCl2/231 nM CoCl2/191 nM ZnSO4/169 nM KI/103 nM Na2MoO4]. For LaCl3 treatment the nutritional media was replaced with water a day before imaging and was refreshed three times to dilute anything that might precipitate out with the LaCl3.
Confocal Imaging and Image Analysis.
YCN and Lifeact-mRuby images were acquired at the Nikon Center of Excellence at the University of Massachusetts Amherst. Specifically, a Nikon A1R confocal with a resonance scanner was utilized for all imaging. YCN was excited via 445-nm laser and mRuby was excited via a 561-nm laser. The A1R is equipped with a triple dichroic mirror, allowing for the simultaneous acquisition of YCN and mRuby emission. With this dichroic we collected three ranges of emitted light: 465–505 nm (CFP), 525–550 nm (YFP), and 570+ nm (mRuby). The RatioPlus plugin for ImageJ was used to generate ratiometric images, with the “Blue Green Red” lookup table provided by the NucMed plugin used to color the images, described in full detail in ref. 75. For WT and LatB experiments we acquired images every 5 s to balance between temporal resolution and photodamage to the fluorophores. Δaip1 lines required 10 s between each acquisition. Each single focal plane time lapse was registered via StackReg plugin with the Rigid Body transformation so that a fixed ROI could be drawn over the apical region.
Wavelet and EMD.
For the wavelet analysis the calcium ratio signal in a 4-µm-wide slice across the cell between 4 and 8 µm distal to the cell tip was averaged at each time point. After lightly smoothing to remove very-high-frequency noise (single sample noise, 5- to 10-s period) time courses were analyzed using the WaveletComp package in R, with default settings. For the EMD we used the R package Rlibeemd, utilizing the ceemdan and coredata functions.
Cross-Correlation Analysis.
To quantify calcium–actin correlation through time we conducted a cross-correlation analysis. A ROI was drawn over the apex of a registered cell. For each frame of the ratiometric data we measured the mean intensity of this ROI. To quantify the apical actin spot we took two approaches. For the maximum intensity we used the same ROI used to measure the mean intensity of the ratiometric data and found the maximum intensity of Lifeact-mRuby within that ROI. For area a cropped region of apex was thresholded such that each frame had a single shape to measure. Both ratio and actin spot data were imported into R, lightly smoothed, and subjected to cross-correlation analysis (CCF function), with default settings.
Curvature Determination.
To measure the curvature of the cell tip we first used ImageJ to crop and rotate the fluorescent movies such that the cells were seen to grow horizontally left to right. We then used ImageJ to segment all individual fluorescent images using the Thresholding tool. The interior of each cell became black on a white background and we manually filled all “holes” in the cell image and then used the Find Edges tool (Sobel) followed by the Binary Skeletonize tool to create an outline of single black pixels. We used the Analyze Tools Save XY Coordinates tool to create a text file listing all edge xy coordinates for each frame of the video. Using a custom script in R the edge coordinates for each frame were first sorted in order from beginning (lower left corner) to end (top left corner) and the distance from point to point was tallied, creating a linear coordinate S along the edge. We sampled the edge in a moving window of 13 points, fitting a smooth spline and calculating the derivatives at each point. Curvature at each point was then calculated as
Analysis of Curvature Pattern.
Curvature along the edge of a spheroidal cell tip creates a characteristic pattern as the shape changes from prolate, typical for a normal growing protonemata, to oblate as the end of the cell encounters an obstacle. A prolate cell shows a single curvature maximum approximately along the central growth axis. As the tip flattens a “valley” of reduced curvature appears along the central axis and two curvature maxima appear further back from the tip, growing in size and in distance from one another as the tip flattens. Notably, as moss protonemata encounter a barrier one “shoulder” of the cell eventually becomes more curved than the other, perhaps indicating a reorientation of growth to a new direction, around the barrier. For this analysis we show the curvature of the tip as the curvature of the “valley.”
Expansion Rate Determination and Analysis.
For control, LatB-treated, and Δaip1 cells we measured the distance the tip traveled during the acquisition. This was done parallel to the growth axis of the cell or, in the case of LatB-treated cells, the growth axis of the cell before treatment. Growth rate was calculated by dividing the length of this line in microns by time.
For collision cells, expansion does not readily occur in the direction of the growth axis, but rather swelling occurs along a different axis. Because cells are roughly radially symmetrical, we assumed that measuring the area of a single optical slice would be an appropriate proxy for the volume of the cell. As such, a tight cropping of the apical region was taken, as this was the only part of the cell that changed shape. Time-lapse YCN movies were thresholded to exclude the background, and the ImageJ Analyze Particles tool was used to measure the thresholded area in each frame with the option to “Include Holes.” In this way, the whole apical region was considered one particle, and the organelles not containing the calcium probe (e.g., vacuole and plastids) were included. To analyze these data they were imported into R and smoothed via the smooth.spline function. The first derivative of the smoothed time series was calculated via the predict function.
Supplementary Material
Acknowledgments
We thank Profs. Madelaine Bartlett, Dong Wang, Caleb Rounds, and Peter Hepler for their close reading of the manuscript prior to submission; John Oakey for providing the microfluidics chambers necessary for this work; and Prof. Simon Gilroy for the pTHUBI::YCN 65 expression construct. This work was supported by National Science Foundation Grant MCB-1330171 (to M.B.) and the David and Lucile Packard Foundation. C.S.B. received support from the Plant Biology Graduate Program at the University of Massachusetts Amherst.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711037115/-/DCSupplemental.
References
- 1.Hepler PK. Calcium: A central regulator of plant growth and development. Plant Cell. 2005;17:2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monshausen GB, Messerli MA, Gilroy S. Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 2008;147:1690–1698. doi: 10.1104/pp.108.123638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova TN, Zhigilei A, Gilroy S. Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta. 1997;203:495–505. doi: 10.1007/s004250050219. [DOI] [PubMed] [Google Scholar]
- 4.Wang YF, Fan LM, Zhang WZ, Zhang W, Wu WH. Ca2+-permeable channels in the plasma membrane of Arabidopsis pollen are regulated by actin microfilaments. Plant Physiol. 2004;136:3892–3904. doi: 10.1104/pp.104.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malho R, Trewavas AJ. Localized apical increases of cytosolic free calcium control pollen tube orientation. Plant Cell. 1996;8:1935–1949. doi: 10.1105/tpc.8.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hepler PK, Winship LJ. The pollen tube clear zone: Clues to the mechanism of polarized growth. J Integr Plant Biol. 2015;57:79–92. doi: 10.1111/jipb.12315. [DOI] [PubMed] [Google Scholar]
- 7.Gibbon BC, Kovar DR, Staiger CJ. Latrunculin B has different effects on pollen germination and tube growth. Plant Cell. 1999;11:2349–2363. doi: 10.1105/tpc.11.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cárdenas L, Lovy-Wheeler A, Kunkel JG, Hepler PK. Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 2008;146:1611–1621. doi: 10.1104/pp.107.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketelaar T, de Ruijter NCA, Emons AMC. Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell. 2003;15:285–292. doi: 10.1105/tpc.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosero A, Žársky V, Cvrčková F. AtFH1 formin mutation affects actin filament and microtubule dynamics in Arabidopsis thaliana. J Exp Bot. 2013;64:585–597. doi: 10.1093/jxb/ers351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messerli MA, Créton R, Jaffe LF, Robinson KR. Periodic increases in elongation rate precede increases in cytosolic Ca2+ during pollen tube growth. Dev Biol. 2000;222:84–98. doi: 10.1006/dbio.2000.9709. [DOI] [PubMed] [Google Scholar]
- 12.Dutta R, Robinson KR. Identification and characterization of stretch-activated ion channels in pollen protoplasts. Plant Physiol. 2004;135:1398–1406. doi: 10.1104/pp.104.041483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hepler PK. The cytoskeleton and its regulation by calcium and protons. Plant Physiol. 2016;170:3–22. doi: 10.1104/pp.15.01506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovar DR, Drøbak BK, Staiger CJ. Maize profilin isoforms are functionally distinct. Plant Cell. 2000;12:583–598. doi: 10.1105/tpc.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidali L, Augustine RC, Kleinman KP, Bezanilla M. Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell. 2007;19:3705–3722. doi: 10.1105/tpc.107.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, et al. Profilin regulates apical actin polymerization to control polarized pollen tube growth. Mol Plant. 2015;8:1694–1709. doi: 10.1016/j.molp.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Huang S, Blanchoin L, Chaudhry F, Franklin-Tong VE, Staiger CJ. A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J Biol Chem. 2004;279:23364–23375. doi: 10.1074/jbc.M312973200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, et al. Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell. 2010;22:2749–2767. doi: 10.1105/tpc.110.076257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon GM, Dowd PE, Gilroy S, McCubbin AG. Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell. 2006;18:867–878. doi: 10.1105/tpc.105.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, et al. Arabidopsis thaliana calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca(2+) concentration. Plant Mol Biol. 2014;86:225–236. doi: 10.1007/s11103-014-0220-y. [DOI] [PubMed] [Google Scholar]
- 21.Allwood EG, Smertenko AP, Hussey PJ. Phosphorylation of plant actin-depolymerising factor by calmodulin-like domain protein kinase. FEBS Lett. 2001;499:97–100. doi: 10.1016/s0014-5793(01)02528-5. [DOI] [PubMed] [Google Scholar]
- 22.Smertenko AP, et al. Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. 1998;14:187–193. doi: 10.1046/j.1365-313x.1998.00107.x. [DOI] [PubMed] [Google Scholar]
- 23.Augustine RC, Vidali L, Kleinman KP, Bezanilla M. Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 2008;54:863–875. doi: 10.1111/j.1365-313X.2008.03451.x. [DOI] [PubMed] [Google Scholar]
- 24.Yan A, Xu G, Yang Z-B. Calcium participates in feedback regulation of the oscillating ROP1 Rho GTPase in pollen tubes. Proc Natl Acad Sci USA. 2009;106:22002–22007. doi: 10.1073/pnas.0910811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudla J, Batistic O, Hashimoto K. Calcium signals: The lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cove D. The moss Physcomitrella patens. Annu Rev Genet. 2005;39:339–358. doi: 10.1146/annurev.genet.39.073003.110214. [DOI] [PubMed] [Google Scholar]
- 27.Prigge MJ, Bezanilla M. Evolutionary crossroads in developmental biology: Physcomitrella patens. Development. 2010;137:3535–3543. doi: 10.1242/dev.049023. [DOI] [PubMed] [Google Scholar]
- 28.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Ramírez C, et al. A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant. 2016;9:205–220. doi: 10.1016/j.molp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer D, Zrÿd JP, Knight CD, Cove DJ. Stable transformation of the moss Physcomitrella patens. Mol Gen Genet. 1991;226:418–424. doi: 10.1007/BF00260654. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer DG, Zrÿd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 32.Bezanilla M, Pan A, Quatrano RS. RNA interference in the moss Physcomitrella patens. Plant Physiol. 2003;133:470–474. doi: 10.1104/pp.103.024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol (Stuttg) 2005;7:251–257. doi: 10.1055/s-2005-837597. [DOI] [PubMed] [Google Scholar]
- 34.Nakaoka Y, et al. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell. 2012;24:1478–1493. doi: 10.1105/tpc.112.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Obando M, et al. Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 (Bethesda) 2016;6:3647–3653. doi: 10.1534/g3.116.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell AJ, Cove DJ, Trewavas AJ, Wang TL. Blue light but not red light induces a calcium transient in the moss Physcomitrella patens (Hedw.) B., S. & G. Planta. 1998;206:278–283. [Google Scholar]
- 37.Koselski M, Trebacz K, Dziubinska H, Krol E. Light- and dark-induced action potentials in Physcomitrella patens. Plant Signal Behav. 2008;3:13–18. doi: 10.4161/psb.3.1.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tucker EB, et al. UV-A induces two calcium waves in Physcomitrella patens. Plant Cell Physiol. 2005;46:1226–1236. doi: 10.1093/pcp/pci131. [DOI] [PubMed] [Google Scholar]
- 39.Horikawa K, et al. Spontaneous network activity visualized by ultrasensitive Ca(2+) indicators, yellow Cameleon-Nano. Nat Methods. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 40.Choi W-G, Toyota M, Kim S-H, Hilleary R, Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidali L, Rounds CM, Hepler PK, Bezanilla M. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One. 2009;4:e5744. doi: 10.1371/journal.pone.0005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, et al. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J Cell Biol. 2012;199:831–847. doi: 10.1083/jcb.201209044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bascom CS, Jr, Wu S-Z, Nelson K, Oakey J, Bezanilla M. Long-term growth of moss in microfluidic devices enables subcellular studies in development. Plant Physiol. 2016;172:28–37. doi: 10.1104/pp.16.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiøtt M, et al. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA. 2004;101:9502–9507. doi: 10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qudeimat E, et al. A PIIB-type Ca2+-ATPase is essential for stress adaptation in Physcomitrella patens. Proc Natl Acad Sci USA. 2008;105:19555–19560, and erratum (2011) 108:18566. doi: 10.1073/pnas.0800864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obermeyer G, Weisenseel MH. Calcium channel blocker and calmodulin antagonists affect the gradient of free calcium ions in lily pollen tubes. Eur J Cell Biol. 1991;56:319–327. [PubMed] [Google Scholar]
- 48.Clarkson DT, Brownlee C, Ayling SM. Cytoplasmic calcium measurements in intact higher plant cells: Results from fluorescence ratio imaging of fura-2. J Cell Sci. 1988;91:71–80. [Google Scholar]
- 49.Felle HH, Hepler PK. The cytosolic Ca2+ concentration gradient of Sinapis alba root hairs as revealed by Ca2+-selective microelectrode tests and fura-dextran ratio imaging. Plant Physiol. 1997;114:39–45. doi: 10.1104/pp.114.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller DD, Callaham DA, Gross DJ, Hepler PK. Free Ca2+ gradient in growing pollen tubes of Lilium. J Cell Sci. 1992;101:7–12. [Google Scholar]
- 51.Rathore KS, Cork RJ, Robinson KR. A cytoplasmic gradient of Ca2+ is correlated with the growth of lily pollen tubes. Dev Biol. 1991;148:612–619. doi: 10.1016/0012-1606(91)90278-b. [DOI] [PubMed] [Google Scholar]
- 52.Uhlén P. Spectral analysis of calcium oscillations. Sci STKE. 2004;2004:pl15. doi: 10.1126/stke.2582004pl15. [DOI] [PubMed] [Google Scholar]
- 53.Torrence C, Compo GP. A practical guide to wavelet analysis. Bull Am Meteorol Soc. 1998;79:61–78. [Google Scholar]
- 54.Kaleem M, Guergachi A, Krishnan S. Hierarchical decomposition based on a variation of empirical mode decomposition. Signal Image Video Process. 2017;11:793–800. [Google Scholar]
- 55.Harries PA, Pan A, Quatrano RS. Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell. 2005;17:2327–2339. doi: 10.1105/tpc.105.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Z, et al. The kinesin-like proteins, KAC1/2, regulate actin dynamics underlying chloroplast light-avoidance in Physcomitrella patens. J Integr Plant Biol. 2015;57:106–119. doi: 10.1111/jipb.12303. [DOI] [PubMed] [Google Scholar]
- 57.Burkart GM, Baskin TI, Bezanilla M. A family of ROP proteins that suppresses actin dynamics, and is essential for polarized growth and cell adhesion. J Cell Sci. 2015;128:2553–2564. doi: 10.1242/jcs.172445. [DOI] [PubMed] [Google Scholar]
- 58.Wu S-Z, Bezanilla M. Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. eLife. 2014;3:e03895. doi: 10.7554/eLife.03498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: New blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- 60.Ketelaar T, et al. The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol. 2004;14:145–149. doi: 10.1016/j.cub.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Augustine RC, Pattavina KA, Tüzel E, Vidali L, Bezanilla M. Actin interacting protein1 and actin depolymerizing factor drive rapid actin dynamics in Physcomitrella patens. Plant Cell. 2011;23:3696–3710. doi: 10.1105/tpc.111.090753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sang Y-F. A review on the applications of wavelet transform in hydrology time series analysis. Atmos Res. 2013;122:8–15. [Google Scholar]
- 63.Faragó P, et al. Interictal brain activity differs in migraine with and without aura: Resting state fMRI study. J Headache Pain. 2017;18:8. doi: 10.1186/s10194-016-0716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berger T. Forecasting based on decomposed financial return series: A wavelet analysis. J Forecast. 2015;35:419–433. [Google Scholar]
- 65.Pierson ES, et al. Tip-localized calcium entry fluctuates during pollen tube growth. Dev Biol. 1996;174:160–173. doi: 10.1006/dbio.1996.0060. [DOI] [PubMed] [Google Scholar]
- 66.Holdaway-Clarke TL, Feijo JA, Hackett GR, Kunkel JG, Hepler PK. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell. 1997;9:1999–2010. doi: 10.1105/tpc.9.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takeshita N, et al. Pulses of Ca2+ coordinate actin assembly and exocytosis for stepwise cell extension. Proc Natl Acad Sci USA. 2017;114:5701–5706. doi: 10.1073/pnas.1700204114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Candeo A, Doccula FG, Valentini G, Bassi A, Costa A. Light sheet fluorescence microscopy quantifies calcium oscillations in root hairs of Arabidopsis thaliana. Plant Cell Physiol. 2017;58:1161–1172. doi: 10.1093/pcp/pcx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Du L, Poovaiah BW. Calcium signaling and biotic defense responses in plants. Plant Signal Behav. 2014;9:e973818. doi: 10.4161/15592324.2014.973818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ranty B, et al. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front Plant Sci. 2016;7:327. doi: 10.3389/fpls.2016.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao X-Q, et al. Biotic and abiotic stresses activate different Ca2+ permeable channels in Arabidopsis. Front Plant Sci. 2017;8:83. doi: 10.3389/fpls.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furt F, Liu Y-C, Bibeau JP, Tüzel E, Vidali L. Apical myosin XI anticipates F-actin during polarized growth of Physcomitrella patens cells. Plant J. 2013;73:417–428. doi: 10.1111/tpj.12039. [DOI] [PubMed] [Google Scholar]
- 73.Pedersen CNS, Axelsen KB, Harper JF, Palmgren MG. Evolution of plant p-type ATPases. Front Plant Sci. 2012;3:31. doi: 10.3389/fpls.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michard E, Simon AA, Tavares B, Wudick MM, Feijó JA. Signaling with ions: The keystone for apical cell growth and morphogenesis in pollen tubes. Plant Physiol. 2017;173:91–111. doi: 10.1104/pp.16.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat Protoc. 2011;6:1835–1846. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.