Significance
Many patients with B cell lymphoma carry alterations in the gene coding for the transcription factor Foxp1. High Foxp1 expression has been linked to poor prognosis in those malignancies; however, the physiological functions of Foxp1 in mature B cells remain unknown. By employing genetic mouse models, we show that Foxp1 deletion results in reduced B cell numbers and impaired antibody production upon T cell-independent immunization. Foxp1-deficient mature B cells are impaired in survival and exhibit an increased proliferation capacity, and transcriptional analysis identified defective expression of the prosurvival Bcl-xl gene. Our results provide insight into the regulation of mature B cell survival by Foxp1 and have implications for understanding the role of Foxp1 in the development of B cell malignancies.
Keywords: immunology, transcriptional regulation, B cell survival, B cell quiescence
Abstract
The transcription factor Foxp1 is critical for early B cell development. Despite frequent deregulation of Foxp1 in B cell lymphoma, the physiological functions of Foxp1 in mature B cells remain unknown. Here, we used conditional gene targeting in the B cell lineage and report that Foxp1 disruption in developing and mature B cells results in reduced numbers and frequencies of follicular and B-1 B cells and in impaired antibody production upon T cell-independent immunization in vivo. Moreover, Foxp1-deficient B cells are impaired in survival even though they exhibit an increased capacity to proliferate. Transcriptional analysis identified defective expression of the prosurvival Bcl-2 family gene Bcl2l1 encoding Bcl-xl in Foxp1-deficient B cells, and we identified Foxp1 binding in the regulatory region of Bcl2l1. Transgenic overexpression of Bcl2 rescued the survival defect in Foxp1-deficient mature B cells in vivo and restored peripheral B cell numbers. Thus, our results identify Foxp1 as a physiological regulator of mature B cell survival mediated in part via the control of Bcl-xl expression and imply that this pathway might contribute to the pathogenic function of aberrant Foxp1 expression in lymphoma.
Forkhead box (Fox) transcription factors constitute a large family of regulatory proteins that play critical roles in organ development, cell division, survival, and metabolism in multiple tissues and in particular in the immune system (1–3). Foxp1 is one of four subfamily members that can form homo- or heterodimers to interact with transcription factors such as NFAT and SMRT in larger transcription cofactor complexes (4, 5) to activate or inhibit gene transcription depending on the cellular context (6–10). Foxp1 is expressed throughout B cell development as well as in mature naïve B cells and B-1 B cells (6, 11). Interestingly, Foxp1 is deregulated via recurrent chromosomal translocations in mature B cell lymphoma, including diffuse large B cell lymphoma (DLBCL) or mucosa-associated lymphoid tissue lymphoma (12–15), and high-level expression of Foxp1 correlates with inferior clinical outcome (16–18). Nevertheless, the physiological function of Foxp1 in mature naive B cells is not well defined.
Gene targeting and germline inactivation of Foxp1 in mice results in embryonic lethality (19). Transplantation of Foxp1-deficient fetal liver cells into recombination activating gene (Rag) 2-deficient mice revealed that, during early B cell development, Foxp1 acts as a transcriptional activator of Rag1 and Rag2 genes, thereby influencing V(D)J Igh gene rearrangement and hence B cell lymphopoiesis (6). Consequently, Foxp1 deficiency in early lymphoid precursors results in a block at the transition of pro-B to pre-B cell stage and severely reduces the peripheral mature B cell compartment. Similarly, down-regulation of Foxp1 via siRNA or miR-34a overexpression in bone marrow (BM) resulted in a partial developmental B cell block and diminished mature B cell frequencies (20). Although these experiments clearly define the essential roles of Foxp1 in early B cells, the Foxp1-dependent differentiation block prevents a genetic loss-of-function analysis of Foxp1 mature B cells.
To overcome these problems, we generated a conditional Foxp1 allele. By using CD19-Cre–mediated deletion or acute deletion of Foxp1 via Mb1CreERT2, we report here that Foxp1 plays an essential role in the survival, maintenance, and quiescence of peripheral mature B cells and is crucial for the development of specific B cell subpopulations.
Results and Discussion
Generation of Fetal Liver Chimera and B Cell-Specific Foxp1 Conditional KO Mice.
To study cell type-specific functions of Foxp1 in vivo, we introduced loxP sites into the Foxp1 gene locus flanking the regions of exons 10–12 that encode the major part of the DNA-binding forkhead domain (Fig. S1A). Crossing these mice to a ubiquitous Cre-deleter mouse strain resulted in germline deletion of Foxp1 and confirmed lethality at approximately embryonic day (E) 15.5 (Fig. S1B) (19). Moreover, transfer of fetal liver cells from Foxp1del/del into Rag2−/− mice also confirmed the essential requirement of Foxp1 for the transition from pro-B (B220+CD43+) to pre-B cell stage (B220+CD43−) and the subsequent generation of immature B cells (B220loIgM+) in the BM and mature B cells in the periphery (Fig. S1 C–E) (6). Foxp1-depleted T cells were generated in these mice as previously published (Fig. S1E) (6). Next, we crossed mice carrying the Foxp1flox allele to CD19-Cre transgenic mice, in which the earliest deletion of loxP-flanked regions starts at the pro-B cell stage and continues throughout B cell development (21, 22). Southern blot analysis of B cells isolated from offspring mice (Foxp1flox/del;CD19) revealed an efficient deletion of the Foxp1 locus (Fig. S1F). Consistently, the WT Foxp1 protein is absent in peripheral B cells (Fig. S1G). A mutated Foxp1 protein (Foxp1-ΔE10-12) that is expressed at a low level in Foxp1flox/del;CD19 B cells does not exhibit DNA binding activity (Fig. S1H). The fact that heterozygous Foxp1wt/del or Foxp1wt/flox;CD19 mice do not exhibit any apparent developmental defects or abnormalities in the immune system further indicates that the remaining Foxp1-ΔE10-12 does not exert any dominant-negative or gain-of-function activities.
CD19 Cre-Mediated Deletion of Foxp1 Alters B Cell Development.
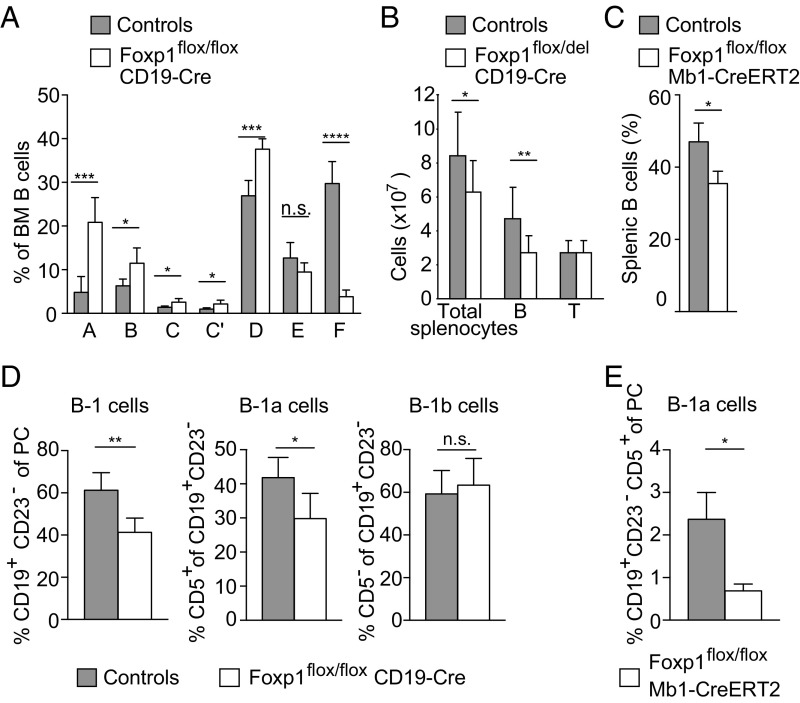
To investigate the function of Foxp1 in B cell development beyond the pro-B cell stage, we characterized BM B cell populations in Foxp1flox/del;CD19 mice by flow cytometry. The percentages of total BM B cells of Foxp1flox/flox;CD19 mice were only slightly reduced compared with controls (Fig. S2A). Further analysis of the B cell fractions according to Hardy et al. (23) revealed that the B cell development in Foxp1flox/flox;CD19 mice is disturbed, with a relative increase from fractions A–D and a significant decrease for fraction E (Fig. 1A and Fig. S2 B and C). Next, we measured the frequencies and numbers of B cell subsets in peripheral lymphoid organs by flow cytometry. We detected reduced B cell frequencies and a decrease in total B cell numbers of ∼40% in spleens of Foxp1flox/del;CD19 mice compared with control mice, whereas the total numbers of splenic T cells did not differ between the two groups (Fig. 1B and Fig. S2D). To bypass consequences of the Foxp1 deletion during early B cell differentiation, we also crossed the Foxp1flox/flox strain to the B cell-specific tamoxifen-inducible Cre line Mb1CreERT2 (24) and administered tamoxifen for five consecutive days to the offspring (Foxp1flox/flox;Mb1CreERT2). After this acute deletion of Foxp1 in B cells, we again detected reduced numbers of mature B cells in the spleen and BM of Foxp1flox/flox;Mb1CreERT2 mice (Fig. 1C and Fig. S2E), although the analysis of the Hardy fractions did not yet reveal a reduction in B cell compartments later in development (fractions E and F; Fig. S2F).
Fig. 1.
Reduction of B-1 and B-2 cell subsets in Foxp1flox/del;CD19 mice. Flow cytometric analysis of BM (A and B), spleen (C and D), or peritoneal lavage (E and F) from control and Foxp1flox/flox;CD19Cre mice. (A) Quantifications of B cell populations during development according to Hardy et al. (23) of control (n = 5) and Foxp1flox/flox;CD19 (n = 6) mice. (B) Absolute numbers of total splenocytes and B (B220+) and T cells (CD3+) from control Foxp1wt/wt;CD19 mice and Foxp1flox/del;CD19 mice (n ≥ 17 mice per group). (C) Inducible deletion using Foxp1flox/flox;Mb1CreERT2 and control mice treated for five consecutive days with tamoxifen. Percentages of splenic B220+ cells are shown (n = 3, respectively). (D) B-1 B cells were identified by the expression of CD19+ and CD23− and further discriminated based on expression of CD5. Quantitative analysis is shown in the bar graph for control (n = 5) and Foxp1flox/flox;CD19 (n = 6) mice. (E) Inducible deletion using Foxp1flox/flox;Mb1CreERT2 and control mice treated for five consecutive days with tamoxifen. Percentages of B-1a cells are shown (n = 3, respectively). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. n.s., not significant.
The decrease in splenic B cell numbers was mainly caused by a reduction in follicular B cells (B220+CD21loCD23hi), whereas the number of marginal zone (MZ) B cells (B220+CD21hiCD23lo) was unaltered in Foxp1flox/del;CD19 compared with controls (Fig. S2G). Surface expression of IgM and IgD on splenic B cells, as well as the transitional type 1 and type 2 B cell populations, did not differ between control and Foxp1flox/del;CD19 mice (Fig. S2 H and I). Analysis of the innate-like B-1 cell population in the peritoneal cavity revealed reduced frequencies of CD19+CD23− B-1 B cells in Foxp1flox/del;CD19 mice, with a prominent reduction in the CD5+ B-1a compartment (Fig. 1D and Fig. S2J; confirmed by an alternative gating strategy adapted from ref. 25; Fig. S2K). Consistent with these observations, the B-1a B cell population was similarly reduced after acute Foxp1 deletion in Foxp1flox/flox;Mb1CreERT2 mice (Fig. 1E). Thus, these results indicate a cell-intrinsic role of Foxp1 in mature and B-1a B cells.
Foxp1 Deficiency in B Cells Results in Impaired T Cell-Independent Antibody Production.
Considering the known essential role of Foxp1 in Rag gene expression (6), and hence a potential impaired V(D)J recombination that might influence immune responses, we next analyzed the B cell receptor (BCR) repertoire from splenic B cells isolated from Foxp1flox/flox;CD19 or control mice. However, we did not observe considerable changes in the BCR repertoires from Foxp1flox/del;CD19 compared with control mice as determined by the analysis of pairwise shared clones (Fig. S3A, Left), the clone’s relative abundance (Fig. S3A, Right), the CD3R length distributions (Fig. S3B), or V, D, and J gene usage of IgH or IgL (IgH and IgL V usage exemplarily shown in Fig. S3 C and D). These results exclude the possibility that the observed effects in B cell numbers are simply secondary to a skewed B cell receptor repertoire.
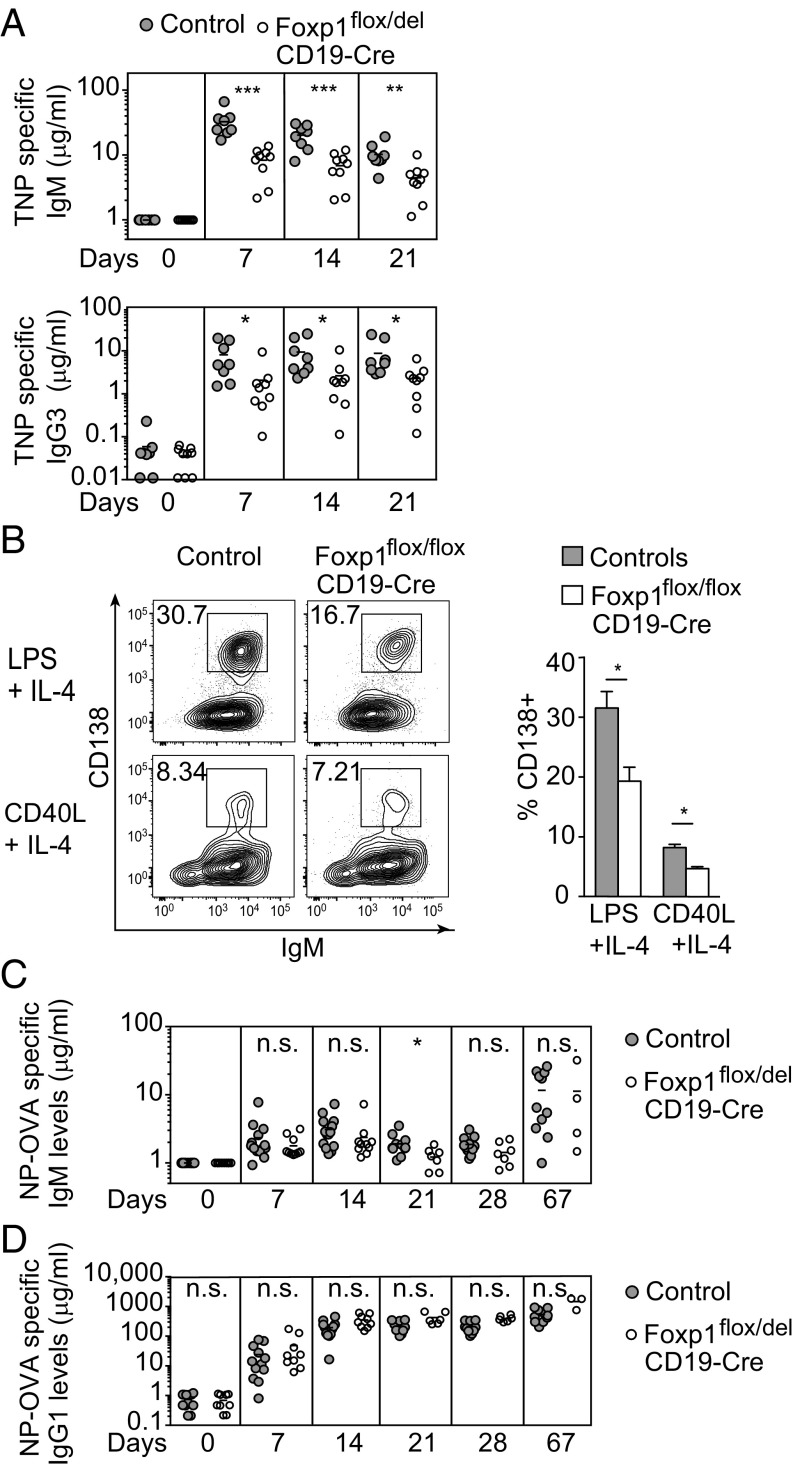
To study the functional consequences of Foxp1 deficiency in B cells, we next measured Ig concentrations in sera of unchallenged Foxp1flox/del;CD19 mice and controls. Whereas IgM, IgG2, and IgG3 concentrations were largely comparable to control levels, IgG1 antibody titers were slightly elevated and IgA titers were reduced in Foxp1flox/del;CD19 mice (Fig. S4A). Next, we immunized Foxp1flox/del;CD19 mice with the T cell-independent antigen 2,4,6-trinitrophenyl (TNP)-Ficoll and analyzed the subsequent antigen-specific immune responses in the sera of immunized mice by ELISA. As TNP-specific IgM was reduced in sera of Foxp1flox/del;CD19 mice even before immunization, we normalized levels to baseline titers at day 0 to determine the increase in IgM levels upon challenge. After T-independent immunization, Foxp1flox/del;CD19 mice principally retained the capacity to produce TNP-specific antibodies, presumably via their functional MZ B cell compartment (26, 27). However, TNP-specific IgM and IgG3 antibody titers were reduced in sera of Foxp1flox/del;CD19 mice compared with controls after immunization (Fig. 2A). We next isolated peritoneal B cells and performed in vitro differentiation assays by cultivating these cells with LPS or CD40L in the presence of IL-4. We found that the frequency of CD138+ plasma blasts derived from Foxp1-deficient B-1 cells was reduced compared with control cells (Fig. 2B), indicating an intrinsic differentiation defect of Foxp1-deficient B-1 cells into antibody secreting cells. Thus, the observed reduction of TNP-specific antibody titers might be explained by the diminished peritoneal B-1 B cell numbers in vivo (28) in addition to reduced differentiation capacity of Foxp1-deficient B-1 B cells to plasma blasts in vitro.
Fig. 2.
Foxp1 deficiency in B cells affects humoral immune responses. (A) TNP-specific IgM and IgG3 concentrations measured at indicated days after immunization with TNP-Ficoll of Foxp1wt/wt;CD19 and Foxp1flox/del;CD19 mice. Means are indicated by horizontal lines. IgM values were normalized to day 0 for each mouse. (B) Peritoneal B-1 cells were stimulated with LPS or CD40L and IL-4 and analyzed for their capacity to differentiate into plasma blasts as determined by CD138 and IgM expression on day 3 by FACS analysis. The percentage of CD138 expressing B cells is indicated (Right; n = 4, respectively). (C and D) NP-specific IgM (C) and IgG1 (D) concentrations measured at indicated time points after immunization of Foxp1wt/wt;CD19 and Foxp1flox/del;CD19 mice with NP-OVA. Horizontal lines indicate means. (C) Relative increase after normalization to day 0 (i.e., before immunization). *P < 0.05, **P < 0.01, ***P < 0.001. n.s., not significant.
Next, we immunized mice with the antigen 4-hydroxy-3-nitrophenylacetyl hapten conjugated to ovalbumin (NP-OVA) to investigate T cell-dependent immune responses. The relative increase of NP-OVA–specific IgM, as well as the total NP-OVA–specific IgG1 antibody titers in the sera of Foxp1flox/del;CD19 mice, were comparable to antibody titers in sera of control mice after immunization (Fig. 2 C and D). The formation of germinal center (GC) B cells and plasma blasts and the frequency of NP-BSA–binding B cells in the spleen of immunized Foxp1flox/del;CD19 mice were similar to those in control mice (Fig. S4 B–D). When cultured on feeder cells expressing BAFF and CD40L in the presence of IgM and IL-4 in vitro (29), Foxp1-deficient splenic B cells differentiated into GC-like B cells expressing GL-7 and IgG1, similar to control cells (Fig. S4E). These results are consistent with the observed down-regulation of Foxp1 during GC B cell function (30), arguing against a critical function of Foxp1 during the GC reaction. Stimulated with LPS or CD40L in the presence of IL-4, splenic Foxp1-deficient and control B cells secreted comparable amounts of IgM (Fig. S4F). Taken together, our results demonstrated that the transcription factor Foxp1 is required for the generation of functional peritoneal B-1 B cells and is therefore essential for optimal T cell-independent immune responses, but is dispensable for T cell-dependent humoral immune responses.
Foxp1 Regulates the Survival of Mature B Cells.
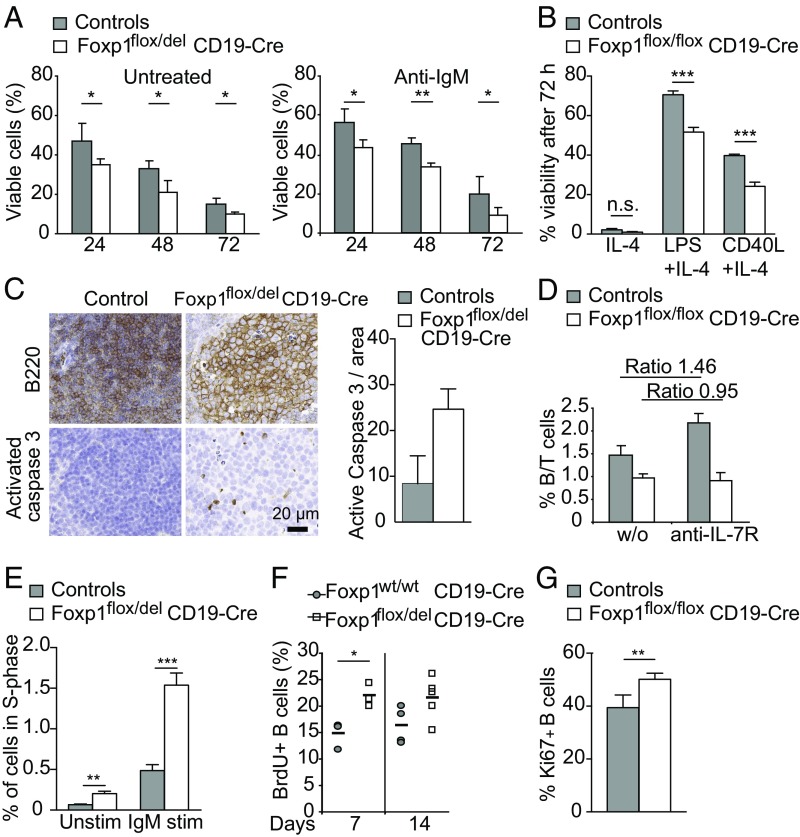
Acute deletion of Foxp1 led to reduced follicular and B-1a B cell numbers, pointing at a role of Foxp1 in B cell survival. We thus next isolated mature B cells from Foxp1flox/del;CD19 and control mice and analyzed their viability in vitro by flow cytometry. Untreated and anti-IgM–stimulated Foxp1-deficient B cells exhibited reduced survival compared with WT cells (Fig. 3A and Fig. S5A). When stimulated with signals supporting survival of B cells like IL-4 alone or in combination with LPS or CD40L, Foxp1-deficient B cells again exhibited reduced survival in vitro (Fig. 3B). Next, we used staining for B220 and activated Caspase 3 in immunohistochemistry and confirmed increased B cell apoptosis in the absence of Foxp1 in vivo (Fig. 3C). To further analyze survival of peripheral Foxp1-deficient B cells in the absence of de novo lymphopoiesis (31), we blocked the influx of B cells from the BM by injecting mice with anti–IL-7 receptor (IL-7R) antibodies, which resulted in an increased B/T cell ratio in the spleen of WT mice (Fig. 3D) (32). This effect, however, was not observed in mice with Foxp1-deficient B cells, indicating reduced B cell survival (ratio of 1.46 in controls vs. 0.95 in Foxp1flox/del;CD19Cre mice; Fig. 3D). Therefore, peripheral Foxp1-deficient B cells also exhibited reduced survival in vivo.
Fig. 3.
Foxp1 affects B cell survival, proliferation, and quiescence. (A and B) Viability of B cells isolated from control mice (n = 3) and Foxp1flox/del;CD19 mice (n = 3) cultured (A) in the absence (Left) or presence of 10 μg/mL anti-IgM (Right) for indicated time points or (B) with IL-4 alone or in combination with LPS or CD40L for 72 h defined as Annexin V− and propidium iodide− by flow cytometry. (C) Representative immunohistochemistry analysis of spleen sections for the expression of B220 or activated Caspase 3 harvested from control or Foxp1flox/del;CD19 mice. Bar graph depicts quantification of activated Caspase 3-positive cells normalized to the area of the analyzed histology sections. Data are shown for two biological replicates for each genotype. (D) Foxp1wt/wt;CD19 control mice and Foxp1flox/del;CD19 mice were injected with PBS solution without (w/o) or with anti–IL-7R antibodies for 14 d, and frequencies of splenic B and T cells were assessed by flow cytometry. Data show ratio means ± SD (n = 4) to take into account that IL-7R blocking affects T cells. (E) Proliferation of splenic B cells purified from Foxp1flox/wt mice and Foxp1flox/del;CD19 mice after stimulation with anti-IgM (10 μg/mL) for 16 h as measured by EdU incorporation. Data show ratio means ± SD (n = 3). (F) BrdU incorporation in control and Foxp1flox/del;CD19 B cells at days 7 and 14 after BrdU administration. (G) Splenic B cells from control (n = 5) and Foxp1flox/flox;CD19 (n = 6) mice were fixed and stained ex vivo for the expression of B220 and Ki-67. Percentages of B220+ and Ki-67+ cells are shown. *P < 0.05, **P < 0.01, ***P < 0.001. n.s., not significant.
Next, we investigated B cell proliferation in the presence and absence of Foxp1. First, we monitored cell division of unstimulated splenic CD19+ B cells after EdU incorporation by flow cytometry and observed significantly more Foxp1-deficient B cells in S-phase compared with control B cells. IgM stimulation revealed an even more pronounced proliferation of Foxp1-deficient B cells (Fig. 3E and Fig. S5B). This was further confirmed by using carboxyfluorescein succinimidyl diester (CFSE) dilution assay after IgM stimulation at 24 h (Fig. S5C). However, optimal in vitro B cell stimulation using CD40L or LPS in combination with IL-4 induced similar levels of proliferation in Foxp1-deficient compared with WT B cells (Fig. S5D). To investigate the proliferation of Foxp1-deficient B cells in vivo, we fed BrdU to Foxp1flox/del;CD19 or control mice for 7 or 14 d. Subsequent FACS analysis demonstrated a higher percentage of BrdU-positive splenic B cells in Foxp1flox/del;CD19 compared with control mice (Fig. 3F). Similarly, ex vivo analysis of Ki-67 expression confirmed higher proliferation levels of Foxp1-deficient mature B cells (Fig. 3G). Analysis of the expression of activation markers in splenic Foxp1-deficient B cells revealed an increase in cell size and up-regulation of CD86, MHCII, and CD43, as well as down-regulation of CD62L, compared with WT cells (Fig. S5E). These findings indicate an activated B cell phenotype and a potential transition of Foxp1-deficient B cells from a quiescent to an activated state.
Foxp1 Regulates Apoptosis-Related Genes for the Maintenance of B Cell Survival.
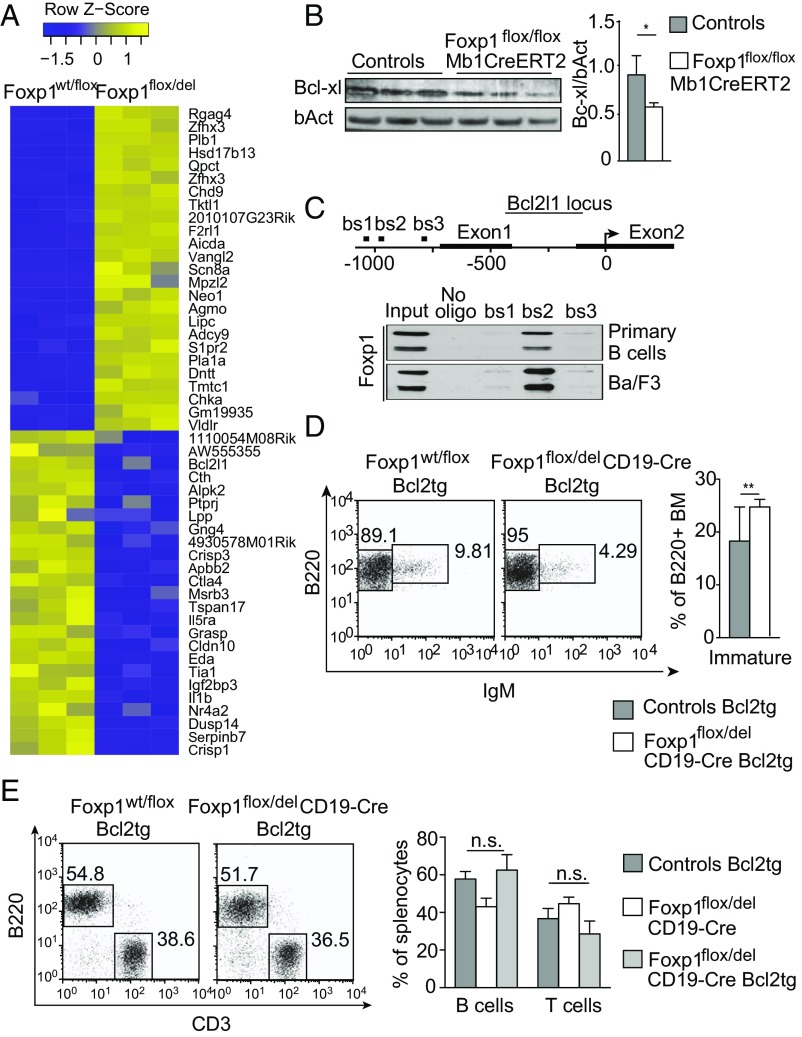
To gain mechanistic insights into how the transcription factor Foxp1 regulates B cell survival and homeostasis, we compared the transcriptome of splenic B cells from Foxp1flox/del;CD19 mice and control mice by using microarrays (Fig. 4A). One of the most significantly down-regulated transcripts was a key regulator of cell survival, Bcl2l1, encoding Bcl-xl. This down-regulation and that of other apoptotic genes was verified by using quantitative RT-PCR (Fig. S6A). In line with these results, acute deletion of Foxp1 in IL-7–expanded pro-B cells from Foxp1flox/flox mice in vitro demonstrated a significant down-regulation of Foxp1 (Fig. S6B). Furthermore, acute in vivo deletion of Foxp1 in Foxp1flox/flox;Mb1CreERT2 mice led to a reduced expression of Bcl-xl in mature B cells as shown by immunoblot (Fig. 4B). Together these data demonstrate the requirement for Foxp1 in Bcl-xl expression. To investigate whether Foxp1 directly controls Bcl2l1 transcription, we performed a sequence analysis of the Bcl2l1 promoter region. We identified putative Foxp1 binding sites within the first 1.5 kb in the 5′ region of the Bcl2l1 start codon (Fig. 4C, Top) in different orientations (6). To test direct Foxp1 binding to these regions, we performed pull-down experiments with oligonucleotides containing the individual putative Foxp1 binding sites, derived from the murine Bcl2l1 promoter. We detected highly specific binding of Foxp1 from primary B cells or Ba/F3 cells to one of three regions (Fig. 4C), indicating that Foxp1 binds to the Bcl2l1 promoter region and may directly regulate the transcription of Bcl2l1.
Fig. 4.
Foxp1 is a regulator of Bcl-xl transcription, and Bcl-2 transgene expression rescues the survival defect of Foxp1-deficient B cells. (A) Heat map of top regulated genes in purified splenic B cells from Foxp1flox/del;CD19 mice or Foxp1wt/flox control mice. DNA microarray data from three independent experiments are shown. (B) Bcl-xl protein levels were determined in splenic B cells of Foxp1flox/flox;Mb1CreERT2 and control mice treated with tamoxifen. Densitometric quantification is shown in the bar graph. (C) Schematic representation of the murine Bcl-xl promoter region (Top). Oligonucleotide pull-down assays with lysates of primary C57BL/6 B cells and Ba/F3 cells. Biotinylated oligonucleotides containing putative Foxp1 binding sites were coupled to Strep-Tactin resins and incubated with cell lysates, and nucleotide-bound proteins were analyzed by immunoblotting for Foxp1 (Bottom). (D) Flow cytometry of BM of Foxp1wt/flox;Bcl2tg control and Foxp1flox/del;CD19;Bcl2tg mice stained for pro-B/pre-B cells (B220loIgM−) and immature B cells (B220loIgM+). Numbers represent percentages of viable splenic cells and B220+IgD− BM cells, respectively. Analysis of four mice per group is shown in the bar graph. (E) Flow cytometry of spleens from Foxp1wt/flox;Bcl2tg control and Foxp1flox/del;CD19;Bcl2tg mice stained for B220 and CD3. Bar graph summarizes results of six individual mice per group. Controls are pooled results of Bcl2tg control mice and WT mice, which did not differ in B and T cell numbers. *P < 0.05 and **P < 0.01. n.s., not significant.
We next crossed the Foxp1flox/del;CD19 mice to Bcl2 transgenic mice that constitutively express Bcl2 under the control of the vav promoter in all hematopoietic cells (33). Bcl-2 is a functional homolog of Bcl-xl, and, although the gene expression of these two factors is dependent on the differentiation state and context of lymphocyte subsets, Bcl2 can functionally compensate for Bcl-xl (34, 35). Transgenic expression of Bcl-2 was unable to overcome the defect in early B cell development as shown by the number of immature B cells (Fig. 4D and Fig. S6C). Nevertheless, splenic B cell frequencies in Foxp1flox/del;CD19;Bcl2tg mice were comparable to those in control mice, indicating a rescued survival of mature Foxp1-deficient B cells by Bcl-2 in vivo (Fig. 4E). Histology analysis confirmed reduction of active Caspase 3 upon Bcl2 overexpression (Fig. S6D). In vitro survival assays confirmed the survival benefit of Bcl-2 Foxp1-deficient B cells (Fig. S6E). However, Foxp1-deficient B cells expressing Bcl-2 still showed an activated phenotype (exemplarily shown by CD62L staining; Fig. S6F). Thus, our results demonstrate that increased Bcl-2 family-mediated apoptosis is a major contributor to the reduced B cell count in the periphery of Foxp1flox/del;CD19 mice.
In conclusion, in this study, we found genetic evidence that Foxp1 regulates the transcription of Bcl2l1, coding for Bcl-xl in mature primary B cells. Recent independent ChIP sequencing experiments in human B cell lymphoma cells additionally implicated direct Bcl-xl control by Foxp1 (36). In our microarrays, Foxp1 deletion also resulted in differential regulation of S1pr2, Lpp, and Aicda, which were previously identified as Foxp1 targets in DLBCL cell lines (36, 37). Down-regulated genes in Foxp1-deficient B cells included the dual-specific phosphatase Dusp14, which can negatively regulate antigen receptor signaling in T cells (38), and the negative regulator Ctla4, which can limit IgM-mediated early immune responses (39). Defective Dusp14 or Ctla4 expression are likely to contribute to the activated phenotype seen in Foxp1-deficient B cells, in line with the previously published function of Foxp1 in maintaining quiescence of T cells (40, 41). Interestingly, bioinformatic pathway analysis using the Kyoto Encyclopedia of Genes and Genomes and the Database for Annotation, Visualization, and Integrated Discovery did not reveal deregulations of quiescence controlling pathways such as mTOR or Foxo1 signaling as causes for the increased proliferation of Foxp1-deficient B cells. Thus, further studies are required to dissect the precise mechanisms of Foxp1-dependent quiescence control in B cells.
Overexpression of Foxp1 in human DLBCL lines and primary human B cells represses a number of proapoptotic genes, such as Bik, Eaf2, and Hrk, and cooperates with NF-κB activity to promote B cell survival (42). Together with the data presented here, we suggest that the high levels of Foxp1 expression in lymphoma cells are likely to prevent these cells from undergoing apoptosis. Furthermore, Foxp1 knockdown in human DLBCL cell lines induces increased MHCII expression (43, 44) similar to what we describe here after Foxp1 deletion. Thus, Foxp1 could possibly also affect tumor immunosurveillance during lymphomagenesis, which requires further investigations.
Materials and Methods
Mouse Strains and Breedings.
The complete targeting strategy for generation of the conditional Foxp1 allele is described in SI Materials and Methods. Expression of Bcl2 transgene in hematopoietic cells was achieved by crossing to vav-Bcl2 transgenic mice (33). Foxp1wt/flox littermates or Foxp1wt/wt;CD19 mice were used as controls for comparison with conditional functional KO Foxp1flox/del;CD19 mice. After crossing Foxp1flox/flox to Mb1CreERT2 mice, we treated Foxp1flox/flox;Mb1CreERT2 and control mice for five consecutive days via i.p. injections with 1 mg/d tamoxifen in resolved in 10% ethanol, 90% Miglyol. Organs were harvested on day 7 (2 d after the last tamoxifen injection) for further analysis. Mice were housed in a specific pathogen-free facility of the Technical University of Munich according to the Federation of European Laboratory Animal Science Associations recommendations (www.felasa.eu). All animal work was conducted in accordance with German Federal Animal Protection Laws and approved by the Institutional Animal Care and Use Committee at the Technical University of Munich.
DNA Microarray and PCR Array Analysis.
Splenic B220+ B cells of three biological replicates from Foxp1wt/flox (control) and Foxp1flox/del;CD19 mice were isolated by FACS, and RNA was extracted by the RNeasy Plus Mini Kit (Qiagen). Total RNA was processed according to Affymetrix standard protocols and hybridized to the murine expression array MOE430 2.0 (Affymetrix). Expression data were processed in R by using the Bioconductor packages “gcrma” and “limma.” Genes differentially expressed at least log 1.5-fold in control vs. Foxp1-deficient cells were determined with a P value cutoff of 0.05 and Benjamini–Hochberg false discovery rate correction. For annotation, the package “mouse4302.db” version 3.0.0 was used. Microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE85240.
Statistics.
P values were determined by Student’s two-tailed t test for independent samples on all experimental data sets with Microsoft Excel or Prism 6 (with exception of the microarray data analysis). All data are shown as means ± SD. In all figures, significant differences are indicated with a single asterisk for P < 0.05, double asterisks for P < 0.01, triple asterisks for P < 0.001, and quadruple asterisks for P < 0.0001. “n.s.” indicates not significant.
Supplementary Material
Acknowledgments
We thank Verena Laux, Tanja Ruff, Julian Hofmann, and Silvia Weidner for excellent technical assistance; Andreas Gewies for guidance and advice; Alison Banham for providing the JC12 antibody; Katja Steiger for the immunohistochemistry staining and evaluation; Marc Schmidt-Supprian for helpful discussion; and Reinhard Hoffmann, Julia Engelmann, and Claudio Lottaz for microarray analysis. This work was supported by research grants from the Helmholtz Alliance Preclinical Comprehensive Cancer Center; Deutsche Forschungsgemeinschaft (DFG) Grants SFB1054, SFB746, TRR130, EXC294, and RU 695/6-1; European Research Council FP7 Grant agreements 322865 and 322972 (to J.R. and M.R.); and German Cancer Aid (Deutsche Krebshilfe; M.B. and M.R.). I.F. is a member of the DFG-funded cluster of excellence ImmunoSensation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE85240).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711335115/-/DCSupplemental.
References
- 1.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 2.Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62:397–409. doi: 10.1007/s00018-004-4365-8. [DOI] [PubMed] [Google Scholar]
- 3.Wijchers PJ, Burbach JP, Smidt MP. In control of biology: Of mice, men and foxes. Biochem J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jepsen K, Gleiberman AS, Shi C, Simon DI, Rosenfeld MG. Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 2008;22:740–745. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 6.Hu H, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 7.Li B, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 8.Shi C, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112:4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi C, et al. Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1. J Clin Invest. 2004;114:408–418. doi: 10.1172/JCI21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- 11.Banham AH, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 12.Fenton JA, et al. t(3;14)(p14;q32) results in aberrant expression of FOXP1 in a case of diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2006;45:164–168. doi: 10.1002/gcc.20278. [DOI] [PubMed] [Google Scholar]
- 13.Haralambieva E, et al. Genetic rearrangement of FOXP1 is predominantly detected in a subset of diffuse large B-cell lymphomas with extranodal presentation. Leukemia. 2006;20:1300–1303. doi: 10.1038/sj.leu.2404244. [DOI] [PubMed] [Google Scholar]
- 14.Streubel B, et al. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- 15.Wlodarska I, et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia. 2005;19:1299–1305. doi: 10.1038/sj.leu.2403813. [DOI] [PubMed] [Google Scholar]
- 16.Banham AH, et al. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11:1065–1072. [PubMed] [Google Scholar]
- 17.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- 18.Sagaert X, et al. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:2490–2497. doi: 10.1200/JCO.2006.05.6150. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 20.Rao DS, et al. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 23.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hobeika E, et al. CD19 and BAFF-R can signal to promote B-cell survival in the absence of Syk. EMBO J. 2015;34:925–939. doi: 10.15252/embj.201489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tung JW, Parks DR, Moore WA, Herzenberg LA, Herzenberg LA. Identification of B-cell subsets: An exposition of 11-color (Hi-D) FACS methods. Methods Mol Biol. 2004;271:37–58. doi: 10.1385/1-59259-796-3:037. [DOI] [PubMed] [Google Scholar]
- 26.Fagarasan S, Honjo T. T-independent immune response: New aspects of B cell biology. Science. 2000;290:89–92. doi: 10.1126/science.290.5489.89. [DOI] [PubMed] [Google Scholar]
- 27.Li S, et al. Rapidly induced, T-cell independent xenoantibody production is mediated by marginal zone B cells and requires help from NK cells. Blood. 2007;110:3926–3935. doi: 10.1182/blood-2007-01-065482. [DOI] [PubMed] [Google Scholar]
- 28.Hardy RR. B-1 B cells: Development, selection, natural autoantibody and leukemia. Curr Opin Immunol. 2006;18:547–555. doi: 10.1016/j.coi.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Nojima T, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 30.Sagardoy A, et al. Downregulation of FOXP1 is required during germinal center B-cell function. Blood. 2013;121:4311–4320. doi: 10.1182/blood-2012-10-462846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudo T, et al. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grenningloh R, et al. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J Immunol. 2011;186:969–976. doi: 10.4049/jimmunol.1002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogilvy S, et al. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci USA. 1999;96:14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao DT, et al. Bcl-XL and Bcl-2 repress a common pathway of cell death. J Exp Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-García M, et al. bcl-XL is the major bcl-x mRNA form expressed during murine development and its product localizes to mitochondria. Development. 1994;120:3033–3042. doi: 10.1242/dev.120.10.3033. [DOI] [PubMed] [Google Scholar]
- 36.Dekker JD, et al. Subtype-specific addiction of the activated B-cell subset of diffuse large B-cell lymphoma to FOXP1. Proc Natl Acad Sci USA. 2016;113:E577–E586. doi: 10.1073/pnas.1524677113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flori M, et al. The hematopoietic oncoprotein FOXP1 promotes tumor cell survival in diffuse large B-cell lymphoma by repressing S1PR2 signaling. Blood. 2016;127:1438–1448. doi: 10.1182/blood-2015-08-662635. [DOI] [PubMed] [Google Scholar]
- 38.Yang CY, et al. Dual-specificity phosphatase 14 (DUSP14/MKP6) negatively regulates TCR signaling by inhibiting TAB1 activation. J Immunol. 2014;192:1547–1557. doi: 10.4049/jimmunol.1300989. [DOI] [PubMed] [Google Scholar]
- 39.Quandt D, Hoff H, Rudolph M, Fillatreau S, Brunner-Weinzierl MC. A new role of CTLA-4 on B cells in thymus-dependent immune responses in vivo. J Immunol. 2007;179:7316–7324. doi: 10.4049/jimmunol.179.11.7316. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115:510–518. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng X, et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Keimpema M, et al. FOXP1 directly represses transcription of proapoptotic genes and cooperates with NF-κB to promote survival of human B cells. Blood. 2014;124:3431–3440. doi: 10.1182/blood-2014-01-553412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown PJ, et al. FOXP1 suppresses immune response signatures and MHC class II expression in activated B-cell-like diffuse large B-cell lymphomas. Leukemia. 2016;30:605–616. doi: 10.1038/leu.2015.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown PJ, et al. N-terminally truncated FOXP1 protein expression and alternate internal FOXP1 promoter usage in normal and malignant B cells. Haematologica. 2016;101:861–871. doi: 10.3324/haematol.2016.142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.