Significance
We developed a method enabling the noninvasive study of fine cellular responses that we applied to macrophage activation. The technique is based on a multimodal label-free microscopy system that simultaneously retrieves both morphological and molecular information based on quantitative phase imaging and Raman spectroscopy, respectively. The parameters obtained from these measurements are processed through a machine learning algorithm that makes it possible to reliably assess the macrophage activation state at single-cell level. We found that while each parameter set (morphology and Raman) can detect the activation state, they provide complementary information. Morphology is symptomatic of downstream phenotypes that make the detection dose-dependent, while Raman is indicative of upstream molecular changes that enable the detection of selective inhibition of activation pathways.
Keywords: live single cell, label-free imaging, immune response, machine learning, cellular heterogeneity
Abstract
We present a method enabling the noninvasive study of minute cellular changes in response to stimuli, based on the acquisition of multiple parameters through label-free microscopy. The retrieved parameters are related to different attributes of the cell. Morphological variables are extracted from quantitative phase microscopy and autofluorescence images, while molecular indicators are retrieved via Raman spectroscopy. We show that these independent parameters can be used to build a multivariate statistical model based on logistic regression, which we apply to the detection at the single-cell level of macrophage activation induced by lipopolysaccharide (LPS) exposure and compare their respective performance in assessing the individual cellular state. The models generated from either morphology or Raman can reliably and independently detect the activation state of macrophage cells, which is validated by comparison with their cytokine secretion and intracellular expression of molecules related to the immune response. The independent models agree on the degree of activation, showing that the features provide insight into the cellular response heterogeneity. We found that morphological indicators are linked to the phenotype, which is mostly related to downstream effects, making the results obtained with these variables dose-dependent. On the other hand, Raman indicators are representative of upstream intracellular molecular changes related to specific activation pathways. By partially inhibiting the LPS-induced activation using progesterone, we could identify several subpopulations, showing the ability of our approach to identify the effect of LPS activation, specific inhibition of LPS, and also the effect of progesterone alone on macrophage cells.
Recent research in cell biology has demonstrated the need to be able to analyze large populations at single-cell level, as individual variations have to be taken into account to accurately assess complex biological behaviors such as response to drugs (1), cell fate (2), or concurrent mechanisms of cellular death (3). This cellular heterogeneity has been studied through various techniques, providing insight at different levels. The most prominent method in past years has been fluorescence-activated cell sorting (FACS), which enables the identification of cell subpopulations with high throughput (4). More recently, new techniques have emerged, such as single-cell sequencing enabling detailed studies of RNA expression (5), as well as multiplexing analytic approaches for the parallel detection of secreted molecules (6, 7), that provide very specific insight, but at the cost of being destructive.
Microscopy also provides a natural way of retrieving single-cell information and assessing heterogeneity of the population, although its throughput is usually lower than commonly used methods such as FACS. The specificity of imaging approaches is typically provided by the use of chemically specific labels, such as to study the heterogeneity of protein expression under stimulation (8). Instead, we use here a multimodal label-free imaging approach that combines quantitative phase microscopy (QPM), Raman spectroscopy, and autofluorescence (AF) imaging, hence simultaneously providing quantitative information on both cellular morphology and molecular content (9). We then use that information to generate models based on training measurements that can be used to noninvasively study cellular states and responses to stimuli at single-cell level.
These methods have been used independently for various studies in cell biology. In particular, QPM has been used, for example, to derive indicators based on the dry mass to study cell cycle (10, 11), extract temporal signals as early indicators of cell death processes (12), or as an overall indicator of bacterial infection in macrophages (13) and inflammation in histological preparations (14). More recently, parameters derived from QPM have also been used with machine learning methods for applications such as cell viability (15), red blood cell classification (16), identification of bacterial spores (17), or algae assessment in flow cytometry (18, 19). Machine learning and statistical analysis based on feature extraction from QPM images can also be used for microorganism (20) and stem cell (21) identification and classification.
Raman spectroscopy has also been extensively used to discriminate cell types based on differences in their molecular content, such as in the case of cancer cells (22) or tumor detection (23, 24). It has also been used for temporal studies, for instance, to detect the initiation of cell death processes (25, 26). Raman spectroscopy has often relied on multivariate analysis and chemometrics for extracting information from the highly multidimensional spectral data (27), as well as advanced machine learning methods (28). It has been shown recently that the combination of QPM and Raman could also be used to discriminate various types of immune cells (29).
We here combine these two approaches (imaging and spectroscopy) and compare their respective performances, characteristics, and stability on measurements performed over several months to study macrophage activation, which is a complex process involving numerous pathways as well as down-regulating mechanisms, where it has also been shown that different activation pathways entail different cellular functions (30). In particular, we study the response of macrophage-like cells to lipopolysaccharide (LPS), which is known to act mainly through the Toll-like receptor 4 (TLR4) (31), although several other receptors are also involved (32).
The label-free methods used here embody a global detection approach, where measurements are indicative of the overall cellular phenotype and the presence of endogenous molecules. Indicators that can be derived from these data through empirical statistical analysis show highly useful information, which can complement more classical techniques that rely on labeling a priori known effector molecules. Especially for biological phenomena where no single definitive marker is available for identification (33), or for applications requiring dynamic studies (34), our approach can elucidate behaviors at the single-cell level and with relatively high throughput.
Results
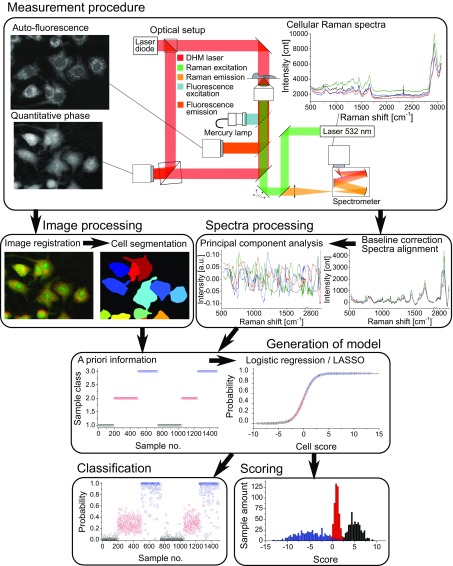
We developed a multimodal system that can measure several label-free signals that are then used for assessing cellular state or response, such as in the case of macrophage activation. The overall procedure, including measurement protocols and generation of statistical models, is described in detail in Materials and Methods, and an overview of the whole process is given in Fig. 1. Briefly, the system allows simultaneous acquisition of QPM images through an interferometric microscope and Raman cellular spectra with a scanning microscope (9, 35). A set of flipping mirrors also enable the recording of AF images through a wide-field epifluorescence system.
Fig. 1.
Measurement and data treatment principle, where morphological and spectral parameters are used to generate a statistical model that allows for analysis of new cellular data taken in later experiments for classification and scoring.
Images were then processed to get matched fields of view, and cells were segmented through an automated software (CellProfiler), which also extracted morphological features based, for instance, on size or intensity values by using the quantitative values from QPM and AF images. The Raman spectra were baseline compensated and decomposed into orthogonal signals through principal component analysis (PCA). The measured values retrieved from spectroscopy were often correlated, as in the neighborhood of a spectral band; generating the model based on the uncorrelated PCA coefficients ensured a better-conditioned problem.
The morphological parameters and PC scores were initially used along with the a priori information about stimulation conditions from training datasets to generate a statistical model fitted with penalized logistic regression (see SI Text for details). This model could then be used to assess new samples, without a priori information, including data measured on later days or with different conditions. The model outputs a probability of activation, suitable for binary classification. To analyze population distributions, we also linearized the probability values distribution along a logistic function for scoring the degree of individual cell response in various situations. Unless specified otherwise, all our displayed data are predictions on unknown samples measured on a given day, but based on a model generated from data measured on different days, typically days or weeks before.
Label-Free Indicators Can Detect Macrophage Activation.
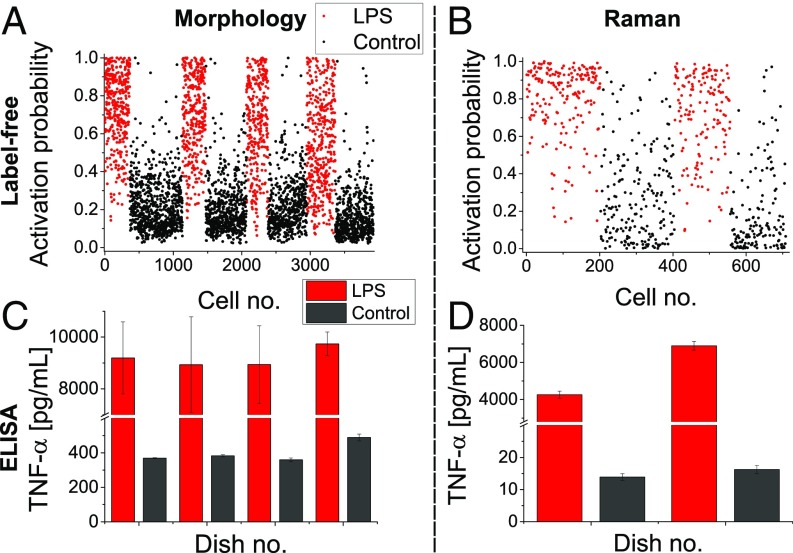
We applied this approach to macrophage cells stimulated with LPS (1,000 ng/mL) during 24 h and generated models based on the known state of stimulation (control or exposed to LPS) either from morphological or Raman measurements. We then used these models to retrieve an activation probability for individual cells measured on a different day, as shown in Fig. 2 A and B, respectively (data from several days including training and test sets are shown in Figs. S1 and S2). The activation status at the population level for each culture dish was confirmed by considering the levels of tumor necrosis factor (TNF-) in the culture medium (Fig. 2 C and D). The results showed that the overall activation probabilities were in agreement with cytokine production.
Fig. 2.
Single-cell activation probability measured with morphological (A and C) and spectral (B and D) indicators. For each dataset, the activation levels are validated by measuring the expression of TNF- in the culture medium of the same dishes used to extract the label-free indicators (C and D). All results are representative of at least three experiments; error bars indicate SD between triplicates.
Models were derived from data measured on given days (morphology: 1 d, 8 dishes, n = 2,235; Raman: 3 d, 12 dishes, n = 1,824) and used for binary classification assessing exposure to LPS, leading to an overall accuracy of 84–87%, as shown in Table 1, where confusion matrices are shown for both models (morphology and Raman) and for training and testing conditions. It is possible to see that both results are consistent in all subclasses. Despite their comparable overall performance, the two indicators have different features. In particular, the morphological one seems biased toward resting cells, with a high accuracy for control cells, but with a significant portion of LPS-exposed cells being identified as resting. In contrast, the Raman indicator shows more balanced ratios between false positives and false negatives. This suggests that a subpopulation of cells exposed to LPS has a morphology similar to resting cells, while still expressing molecules related to activation.
Table 1.
Confusion matrices of activation classification for both morphological and Raman measurements, given for both training and test data
| Predicted, % (training/test) | ||
| Actual | Resting | Stimulated |
| Morpho (84.34/87.79) | ||
| Control | 92.94/96.38 | 7.06/3.62 |
| LPS | 30.13/27.05 | 69.87/72.95 |
| Raman (86.4/85.59) | ||
| Control | 85.20/84.46 | 14.80/15.54 |
| LPS | 12.39/13.28 | 87.61/86.72 |
One should note that the assessment of statistical models is often performed through cross-validation or by keeping a subset of the data out of the training set for further tests. These validations can, however, provide overfitted models for data measured in rather similar conditions, which are then unstable when used on data taken at a later time or under different conditions. Validation was therefore performed with data obtained on different days to test the model’s performance in cases where system and specimen conditions can vary. We ensured that our training set size was sufficiently large to generate accurate models by computing the cross-entropy of our computed activation probabilities (see SI Text for details) based on models generated from an increasingly large amount of samples and for different values of the penalty parameter (Figs. S1 and S2). These tests, based on ensuring reproducibility between training and test datasets, indicated that the chosen (0.033 and 0.015 for morphology and Raman, respectively) are suitable. Higher values led to larger cross-entropy, indicating less accurate models, while lower values increased the difference between training and test data, which is symptomatic of overfitted models. Based on these results, the required sample size for generating stable statistical models was 500 samples in the case of morphological data and 750 for spectral data. This larger requirement in the Raman case is consistent with our observation that spectral data were more sensitive to day-to-day variations, so that the model had to be generated from several days of experiments to reach an accurate representation. Furthermore, the clearer separation obtained with Raman indicators in the results above was also visible through the cross-entropy, which reached 0.51 at full sample size for Raman, while morphological indicators attained 0.59.
These results also demonstrate the stability of the models, which can provide reproducible results over months, as shown in Fig. S1, where the cross-entropy is shown for data measured at different times after the training set, ranging from 1 d to 4 mo. For datasets taken at later times, it is possible to identify a small reduction in accuracy when increasing the amount of variables. This shows the effect of overfitting in the model, where accuracy can be increased, but only for data taken in rather similar conditions. It is also possible to identify that the accuracy obtained for the test morphological data is better than for the training set (Table 1). This can be explained by the fact that some cells in the training sets are not in the expected state, such as exposed but not activated by LPS, or spontaneously activated cells. This is also an indication of the stability of the models with strong penalty values, where no significant loss of accuracy can be identified in our results.
The required sample size derived above is significantly higher than some reported requirements for Raman spectroscopy (36), which can be understood by considering that the biological differences observed here (i.e., reversible changes within identical cells) should be finer in nature than cell-to-cell differences in previous studies that performed only classification between different cell types. On the other hand, the sample sizes required here are comparable with those reported in the case of deep learning methods based on label-free imaging for cell-type classification (18).
Models Allow for Biological Interpretation.
A significant advantage of linear models, as used here, is that the results can be easily interpreted, since the obtained coefficients are linearly related to the importance of the underlying parameter in terms of ability to separate the two classes. Furthermore, the lasso approach constrains the parameters so that variables with less separating power are shrunk to zero, and the amount of variables to interpret is then significantly reduced in the final model.
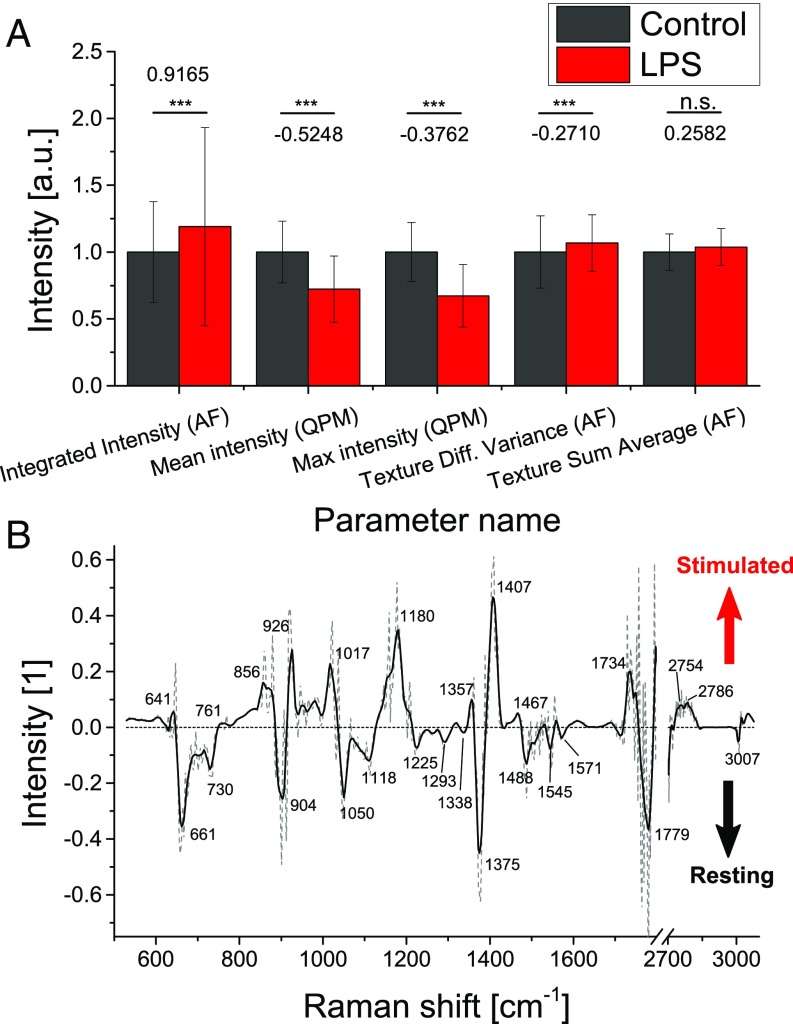
In the case of the morphological model, 15 of the 301 variables were retained (all coefficients in Fig. S3A and Table S1). It is interesting to note that some variables of all types (shape, intensity, texture, etc.) were selected in the automatically generated model, meaning that activation is related to a variety of features rather than just one type, which highlights the usefulness of using a multivariate approach. The values of the five largest coefficients for control and stimulated cells in the training dataset are shown in Fig. 3A, illustrating the degree of difference induced by individual variables. It is also important to note that, although statistical significance is valuable information, it is not always directly correlated with the capability of a variable to discriminate between two classes, as illustrated by the fifth best variable, which does not appear as significant.
Fig. 3.
(A) Average values of parameters from training data having the largest coefficients (value indicated in graph) in the morphological model for control and stimulated cells, respectively (values are normalized in regards to control ones, error bars indicate SD between coefficients). Each morphological parameter is extracted from either AF or QPM. Diff., difference. ***P < 0.001; n.s., P 0.05. (B) Difference Raman vector between stimulated and control cells obtained from model coefficients (dashed, original values; line, LOESS-smoothed data).
Furthermore, several variables can then be interpreted in terms of biological meaning. For example, the most important parameter, AF integrated intensity, is of direct biological significance, as it is known that activated macrophages have higher levels of intracellular reactive oxygen species such as nitric oxide (NO) (37) that can be observed by AF (38). Other parameters indicate changes of the cell shape and intracellular density distribution that are consistent with previous studies showing that the activation state influences cell morphology, although these variations strongly depend on the cell type (33).
Similarly, the coefficients for Raman indicators could also be extracted (Fig. S3B), where each coefficient is related to a PCA loading vector. It is possible to see that all 36 retained coefficients were within the first 66 scores, which accounted for 89.22% of the variance in the data (Fig. S3C), with coefficient values decreasing for higher coefficients. The linear combination of the coefficients with their corresponding loading vector leads to a continuous function in the spectral domain, as shown in Fig. 3B. This represents the spectral features that characterize either resting or stimulated cells and should be related to the compositional differences between the states. As measured cells were genetically identical clones, observed spectral differences should be related to transcription/translation in the cell resulting from stimulation. A combination of proteins, transcription factors, mRNA, and other molecules would then be expected to compose this spectral separation vector.
The main feature to note in the separation vector is actually the absence of most of the common molecules typically observed in the Raman spectrum of a cell cytosol, such as lipids, proteins, and amino acids. In particular, the CH (2,873; 2,931 cm−1) as well as CH2 (2,888; 2,926 cm−1) stretching, indicative of lipids, was mostly negligible. Furthermore, several peaks indicative of protein secondary structure (39), such as H-bonded CO stretch (1,650; 1,670–1,680 cm−1) were not present. This suggests that the differences between control and stimulated cells were more subtle than just a change in overall molecular concentrations. Additionally, this absence of significant lipid contribution shows that the results were not directly affected by the presence of LPS. Several features in the separation vector toward control can be related to nucleotides, with adenine (730; 1,488 cm−1) and guanine (661; 1,118 cm−1) being particularly indicative of nonstimulated cells, along with other features (tyrosine, 1,180 cm−1; ribose, 1,017 cm−1) (40, 41). Spectral features associated with stimulation are also indicative of ribose (926; 1,017 cm−1) and phenylalanine (1,180 cm−1).
Cellular Heterogeneity Influences Models.
As shown previously, the classification performance for determining activation on a single-cell level is 85%. While this level of specificity may not appear extremely high, it should be noted that this assessment relies on the hypothesis that all measured cells in a control dish are resting, while all cells exposed to LPS have a 100% probability of being activated, which can be expected to be overly simplistic. It is known, for example, that cells can experience LPS tolerance after an initial stimulation (42).
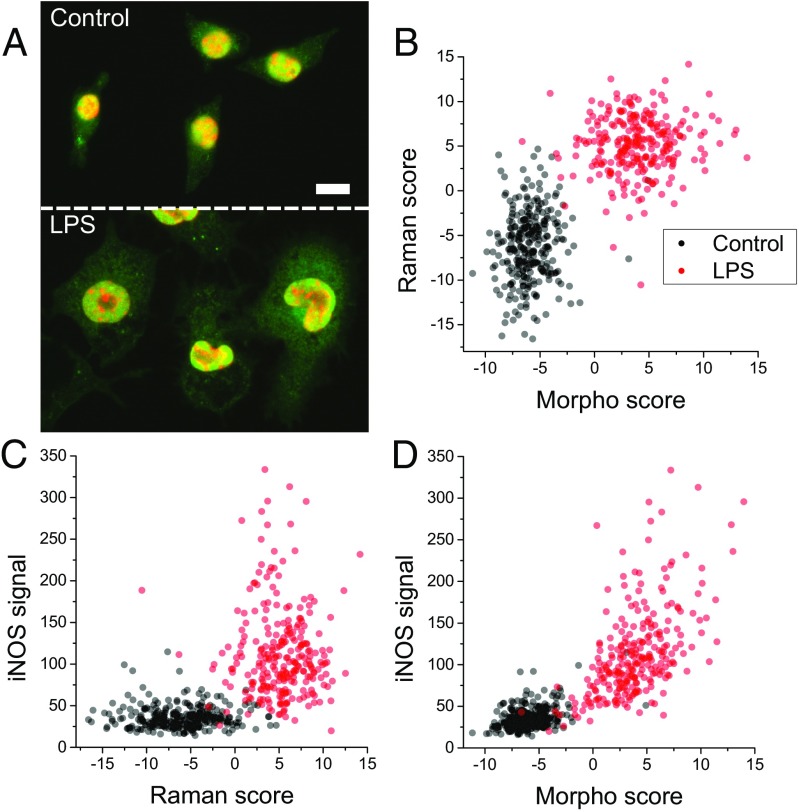
To further validate our approach, we compared our label-free classification with the intracellular levels of inducible NO synthase (iNOS), an enzyme known to be involved in the immune response by promoting the production of NO (43). This enzyme was selected as an activation indicator, as it displays a gradual increase over several hours upon LPS stimulation (44, 45). Following label-free measurements, cells were immediately fixed for subsequent immunostaining, enabling the measurement of the same cells measured with our approach, as shown in Fig. 4A. To derive a metric of activation level based on its expression, we segmented the cytoplasm of individual cells and used the overall iNOS signal as an indicator of activation.
Fig. 4.
(A) Typical immunofluorescence images of iNOS (green) with nuclear counterstaining (red) on control and LPS-stimulated Raw264 cells. (Scale bar: 10 m.) (B–D) Paired measurements of label-free activation scores (both morphological and spectral) compared with cytosolic levels of iNOS (n = 541; 2 d), showing good agreement in the identification of activated cells.
The comparison of the iNOS levels with both spectral (Fig. 4C) and morphological (Fig. 4D) predictions showed good agreement, where activation scores (obtained by linearizing the probability values; SI Text) were displayed. This result also held for outlier cells that appeared activated despite the absence of stimulation or cells appearing as resting, even under LPS exposure. This indicated that most cells which received an indeterminate activation score in one method were consistently shown as having an unclear activation state in the two other independent methods, suggesting that the binary hypothesis where all cells exposed to LPS become stimulated was not correct, leading to an underestimation of the performance of the model. This implied that we would be able to further probe the performance of our method and likely also improve the models at the training stage, if a fail-safe external indication of the activation at the single-cell level was available, and by only using strongly activated cells during training, or by taking into account the activation degree directly into the model.
Interestingly, the two label-free indicators actually appeared to provide better separation between resting and activated cells (Fig. 4B) than the iNOS levels. This may have been due to various factors, such as the difficulty of generating accurate indicators based on fluorescence, where photobleaching or staining efficiency may affect the measured intensity. It has also been shown that the expression of specific molecules in individual activated cells can vary significantly (6), so that the monitoring of a single molecule may be less accurate than global indicators such as those measured in our approach.
Up to this point, the two measurements (morphology and Raman) have been used independently to analyze their performance separately. It is also possible to join all parameters, potentially yielding more accurate models by accumulating uncorrelated variables for prediction, as suggested by the separation displayed in Fig. 4B. However, tests at this point performed on larger datasets spread on several weeks only showed a small increase in performance to 88% (unbiased). As discussed above, this could be attributed to the fact that the perceived lack of performance (<90% accuracy) was in part resulting from real population heterogeneity at the single cell level, and it would not be expected that combining the discriminatory power of the two independent modes should change this significantly.
Morphological Indicators Are Dose-Dependent, but Spectral Ones Are Not.
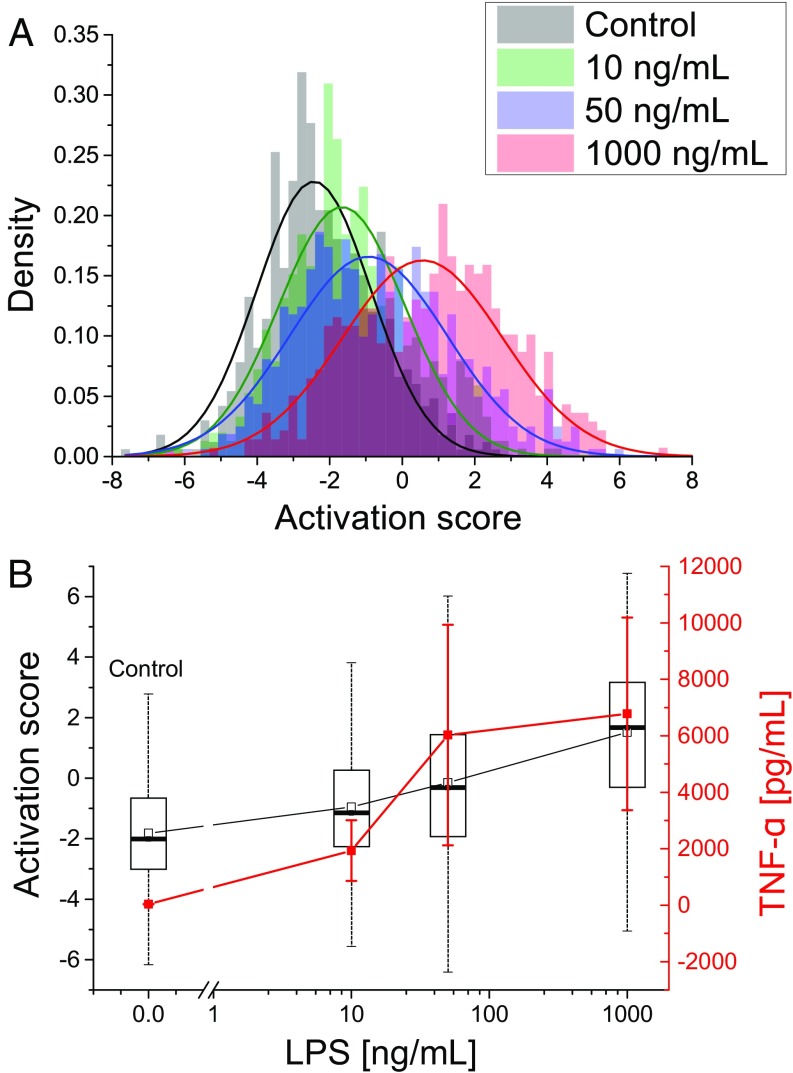
To assess the detection sensitivity of the label-free indicators, we performed measurements with various concentrations of LPS. We first performed dosage experiments for TNF- production in well plates, and from the results (Fig. S4), we used a range where the cytokine production underwent the most significant rise, as well as a 1,000 ng/mL stimulation, where the response already appeared saturated.
As described previously, a model was trained based on an experiment comparing control and stimulated cells with 1,000 ng/mL LPS. The model was then used to derive the activation scores of cells exposed to concentrations of 0, 10, 50, and 1,000 ng/mL. As shown in Fig. 5A, the average morphological score of each cell population gradually increased with dosage. Furthermore, the distribution spread broadened compared with control responses, suggesting more heterogeneous features from stimulated cells. The single cell responses were compared with TNF- production (Fig. 5B), where the activation scores showed a more linear dependence on LPS dose than cytokine production, which was concentrated in the 10- to 50-ng/mL region, along with saturation at higher dosage, in accordance with standard tests (Fig. S4). While the cytokine levels across the whole dish showed much lower spread than the single cell responses, due to the thousands of cells globally assessed in the TNF- assay, there were still significant differences in cytokine concentration, which can be attributed, for example, to the varying number of cells per dish.
Fig. 5.
(A) Activation score distributions based on morphological indicators for stimulation from different LPS concentrations, showing gradual increase with concentration (n = 2,284). Results are representative of at least three experiments. (B) Comparison of scores with TNF- production. Box plots illustrate the spread within cells populations (box, quartiles; whiskers, minimum/maximum values); error bars indicate the SD within nine dishes.
For these dose–response experiments, the Raman indicators here exhibited a different behavior compared with morphology (Fig. S5). While shifts in average scores could be identified for stimulation concentrations at 10 and 50 ng/mL, no gradual increase was observed, and these populations had scores much closer to control than to cells stimulated with 1,000 ng/mL, suggesting that the molecular changes measured by Raman were not changing linearly with the stimulation concentration. This would be consistent with the complexity of the cellular immune response that involves multiple up- and down-regulating pathways.
Label-Free Indicators Can Detect Selective Stimulation Inhibition.
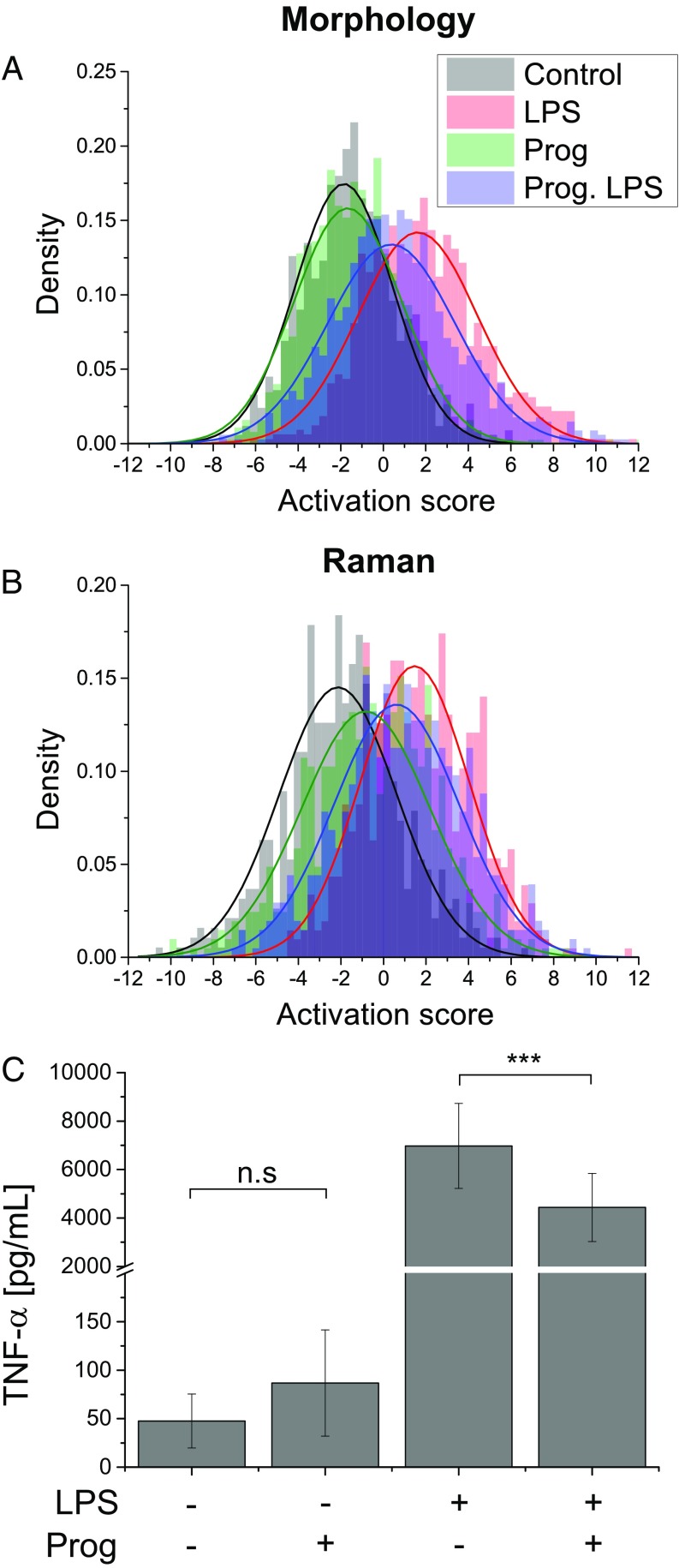
One potential advantage of label-free indicators is that by measuring the cell state as a whole, rather than one particular indicator of activation, it may be possible to discern more subtle changes in individual cells, rather than simply activated vs. nonactivated classes. To assess this, we also tested our approach in combination with a selective inhibitor of LPS activation. Progesterone (Prog) is known to inhibit the release of TNF- (46) as well as NO (47) in macrophages. It selectively inhibits the transcription of NF-B (48) and also promotes the expression of SOCS1, a cytokine known to negatively regulate the TLR4 cascade (49).
When applying a model of activation based on cells stimulated with the same LPS concentration (50 ng/mL), pretreatment with Prog (10 M) resulted in cells that still indicated activation, but showed significantly reduced scores both in morphology and Raman indicators, as shown in Fig. 6 A and B, respectively. This result was consistent with the cytokine levels, where pretreatment by Prog significantly reduced TNF- production in stimulated cells (Fig. 6C). On the other hand, unstimulated cells pretreated with the inhibitor displayed similar scores in morphology as control, but showed significant differences in their Raman scores. These results suggested that, while Prog did not significantly affect the morphology when used in isolation, it caused by itself changes in intracellular content that can be discerned by Raman measurements.
Fig. 6.
(A and B) Activation score distributions with 50 ng/mL LPS and pretreatment with progesterone (Prog) for both morphological (A; n = 3,760) and spectral (B; n = 2,487; 5 d) indicators, showing a decrease of the scores for inhibition of LPS activation by Prog, compared with positive control. Results are representative of at least three experiments. (C) TNF- production for LPS stimulation and Prog pretreatment. Results are the average of nine dishes per experiment; error bars represent SD. ***P < 0.001; n.s., P 0.05.
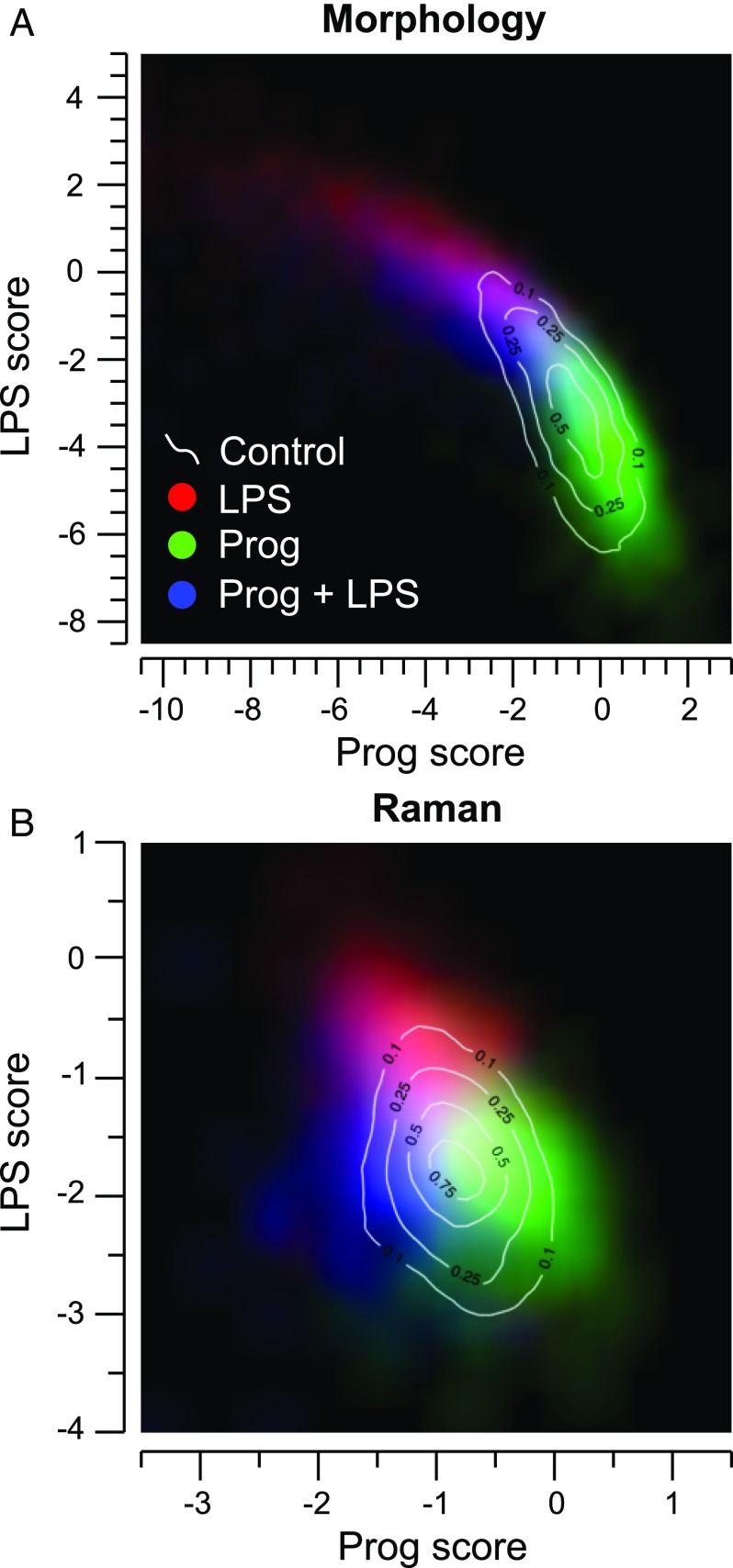
Since the above results indicate that the cells treated with LPS and/or Prog exhibit a more complex behavior than pure activation, we also used a multinomial classification scheme, which generates independent models to separate each class from each other (Fig. S6), yielding probabilities of belonging to each class. This approach allows for the separation of effects induced by LPS and Prog and for studying behaviors induced by these two molecules which may not be purely additive. We used the scores of having been exposed to LPS or Prog to assess the links between the two treatments, as shown in Fig. 7 A and B for morphology and Raman indicators, respectively.
Fig. 7.
Density maps for cell populations having been exposed to LPS and/or Prog, obtained from multinomial classification, for both morphological (A; n = 10,094) and Raman (B; n = 1,741) indicators.
In cell morphology, it is possible to see a strong anticorrelation between the effect of LPS and Prog, where stimulated cells have an increased LPS score, along with a lower Prog score. Conversely, pretreated cells are close to control ones, but show a wider spread, with a tendency toward lower LPS scores. Concurrently exposed cells follow this trend, by being located mostly between control and LPS ones. This suggests a “continuum” in morphology features, along which cells are located, providing mostly the same information as the binomial analysis given in Fig. 6A.
This anticorrelation can also be identified in the Raman indicators for LPS and Prog cells, but the Prog cells are more clearly separated from the control ones. Furthermore, LPS+Prog cells are also more separated, with comparable Prog scores as exposed to LPS, but with a much lower LPS score. This suggests that it is possible to detect underlying molecular changes, both from exposure to Prog and also from the inhibition of LPS activation by this molecule. Interestingly, it is also possible to identify several similarities between the four Raman separation vectors in a four-class multinomial model and the activation vector obtained previously in the binomial case (Fig. S6 A2–D2 and Fig. 3B, respectively). While this is expected between the two LPS vectors, one should still note that the derivation procedure is different in the two cases, as all classes, including Prog and Prog+LPS, are separated from LPS in the multinomial case. Nevertheless, several features, including the peak shift at 1,400 cm−1 and the negative region at 1,100 cm−1, as well as the main peak at 904 cm−1 are present in both vectors. Furthermore, it is also possible to see that some features present in the Prog+LPS vector—such as the large positive peak 1,180 cm−1—are also visible in the main LPS vector, but not the multinomial one, which may indicate that some spectral features are pathway-dependent.
Discussion
The method we propose here, by generating statistical models based on label-free cellular morphology and molecular content parameters, can identify LPS-induced macrophage activation within a population of genetically identical cells. While the two indicators are independent, in terms of both acquisition procedure and of information they exploit, the results on the degree of activation assessed by paired measurements are in agreement, showing a correlation between morphological and molecular changes occurring during activation. The results obtained by the two indicators are also in good agreement with external validation performed by measuring iNOS levels, where the comparison showed that certain cells could be considered misclassified based purely on their initial conditions, which is not necessarily representative of the actual cellular activation state.
It is interesting that the two independent models give highly correlated predictors of individual cell states, since the two measurement approaches are significantly different. The morphological parameters are extracted from wide-field images that can provide a relatively high throughput due to the presence of numerous cells in each field of view. The throughput can be limited more by the computational power required to segment the cell image data and extract parameters (size, texture, etc.) rather than the actual measurement time. Raman spectroscopy, on the other hand, has inherently lower acquisition throughput due to the small vibrational cross-section of biomolecules. It is optimized here by taking one spectrum per cell while scanning the beam through the cell to obtain a representative spectrum (35). The point-based Raman detection method still does not reach the throughput of the wide-field imaging mode, although it could be improved by further automating the acquisition procedure. On the other hand, acquired spectra can then be used in a straightforward manner for the assessment of macrophage activation with much less data postprocessing compared with widefield imaging.
Our results also indicate that the two models are indicative of different biological processes and provide information that can be used in conjunction for a better understanding of the macrophage activation state. Morphological variables appear to be more symptomatic of downstream effects which affect the cellular phenotype as an end result, while Raman indicators can be related to upstream effects linked to intracellular molecular changes occurring upon LPS stimulation. These differences can be identified first by the fact that a significant subpopulation of LPS-exposed cells seem to show little change in morphology and appear similar to control cells, leading to a loss of specificity in detection. This is, however, not the case with molecular indicators, which have similar sensitivity/specificity rates (Table 1), showing that, despite comparable morphologies, these cells undergo molecular changes indicative of LPS-induced activation. In particular, the preferential display of features indicative to adenine, along with phosphate-related bands [904 cm−1 (41); Fig. 3B] toward control may indicate an ATP depletion in stimulated cells, a phenomenon that has been confirmed in alveolar macrophages (50).
The differences between the two indicators can also be identified in the subsequent results, where the morphological indicators respond in a dose-dependent manner, while Raman indicators at intermediate LPS doses (10 and 50 ng/mL) appear very close to control cells. This again suggests that morphological parameters are related to downstream effects that are dependent on the level of stimulation, while molecular changes occurring under different stimulation doses cannot be linearly related. The fact that the scores of both lower-dosage conditions are clearly closer to control than stimulation may also suggest a threshold behavior at detection level, where the spectral features used to identify strong LPS exposure (1,000 ng/mL) are not present at lower dosage.
Furthermore, the conjugate use of Prog, a known inhibitor of LPS-induced activation, shows that cell morphology indicators indeed exhibit the reduction of activation score by pretreatment with Prog, but no clear difference between control and pretreated cells was seen. On the other hand, molecular indicators clearly show distinct populations for the four possible combinations of LPS and Prog, indicative of upstream molecular changes for all conditions, despite similar phenotypes. A finer analysis, based on a model aimed at independently separating all classes, shows that Raman indicators can separate all experimental conditions, and provide clear subpopulations for control, LPS-induced activation, Prog exposure, and Prog-inhibited LPS stimulation. The capability of discriminating these conditions is in agreement with reported effects of Prog, which is known to act as a selective inhibitor of the TLR4 activation cascade (48, 49), but also to independently promote alternative activation of macrophages (51), which seemingly induces molecular changes without significant phenotype modifications in the time frames of our experiments. Furthermore, the separation vectors we retrieve in case of separating the different subclasses of activation with inhibition are very comparable with the one obtained with simple activation (Fig. 3B and Fig. S6 A2–D2). However, several features present in the original activation vector are displayed only in specific vectors of the multinomial classification, namely, activated with LPS but with or without progesterone inhibition. This suggests that some molecular changes observed here which indicate macrophage activation are also pathway-dependent.
The method we present is a global approach that takes overall parameters which are derived from endogenous contrast, making them less specific than labels bonding to particular molecules such as proteins or cytokines. The model derived from these variables is then made specific to the particular studied process through supervised learning. The results do, however, show that the label-free indicators can have better separation capabilities than specific immune response-related proteins, which could be explained by the overall approach used here, compared with the monitoring of a single molecule. The label-free models are, however, also influenced by other factors, as shown by the spread of control populations, even in the case of genetically identical clone cells, and processes such as the cell cycle can significantly affect both morphology and molecular content. Nevertheless, this approach can accurately identify a complex process such as macrophage activation. It is also interesting to note that stimulated populations have a larger spread than control ones, which indicates a higher degree of heterogeneity within the cell responses and within the activation state itself. This would be consistent with the effects of LPS-induced activation, which possesses numerous up- and down-regulating mechanisms that should affect single-cell response levels.
Overall, our technique provides a highly noninvasive approach to assess complex biological behaviors at the single-cell level. We showed that the models were providing stable assessment for data taken over several months, which is critical if this approach is to be used for reliable biological measurements, since it accounts for typical drifts in both the instrumentation and in cell conditions, which occur commonly but are challenging to quantify. The technique is sensitive enough to observe stimulation in a dose-dependent manner, as well as detecting the inhibition of particular activation pathways. While we applied it here to LPS-induced macrophage activation, the general approach can be used for the study of other phenomena, where specific models can be created based on the same measurement procedure in different conditions for the supervised learning process. It provides a means to discriminate subtle changes in cell response, either on the single-cell level or with greater statistical power on a population level. As the approach is noninvasive, it can be combined with other single-cell methodologies and existing wet lab techniques. Its nondestructiveness and the absence of any chemical alteration of the samples make this approach particularly suitable for dynamic measurements and temporal studies.
Materials and Methods
Cell Preparation and Stimulation.
Raw264 cells (Riken BioResource Center) were cultured in 10-cm Petri dishes and immersed in DMEM (Nacalai) supplemented with 10% FBS (Gibco) and penicillin/streptomycin (Sigma-Aldrich) with, respectively, 10,000 units and 10 mg/mL diluted at 10 mL/L and incubated at 37 °C in a humidified atmosphere containing 5% CO2. Cells were trypsinyzed with a solution containing 0.25% trypsin and 1 mM EDTA (Nacalai) for approximately 5 min at 37 °C to detach them from the dish. They were then plated on 3.5-cm quartz-bottom dishes (FPI) at a density of 20,000 cells per cm2 and then incubated again as described above. Quartz substrates were previously coated with poly-l-lysine (PLL) by immersing the surface in a 0.01% PLL solution (Sigma-Aldrich) during 30 min at room temperature (RT). The surface was then washed with deionized water and left to dry for 2–3 h at RT before plating the cells.
After ∼24 h, the culture medium was replaced by fresh medium containing the stimulating agent. For standard stimulation, LPS from Escherichia coli (Sigma-Aldrich) stock was prepared by diluting it in phosphate buffer solution (PBS; Nacalai) at a concentration of 1 mg/mL, which was further diluted in the culture medium in concentrations ranging from 10 ng/mL to 1 g/mL, depending on the experiment. In the case that Prog (Sigma-Aldrich) was used, stock was prepared by diluting it at a concentration of 10 mM in DMSO. Cells were pretreated during 4 h with Prog at a concentration of 10 M, after which the medium was replaced with a medium containing both Prog and LPS for stimulation. Medium changes for control dishes were always performed in a similar manner as stimulated ones.
After 24 h of stimulation, the medium was extracted for further analysis through sandwich enzyme-linked immunosorbent assay (ELISA), and the culture dishes were washed two or three times with PBS supplemented with glucose (5 mM) and MgCl2 (2 mM) before measurement.
Cell Measurements.
Cells were measured on a multimodal microscope extensively described in publications (9, 52). Quantitative phase images were recorded with an interferometric setup based on off-axis digital holography (53). A laser diode at 780 nm (Thorlabs) was split into object and reference beams. The object beam illuminated the sample, and the diffracted wave was collected with a Fluor 40 microscope objective (MO; Nikon; NA 0.75). The two beams were then recombined, and the interference pattern was recorded by a CCD camera (TXG20; Baumer) with an exposure time of typically 2 ms. Phase images were extracted from holograms through Fourier filtering and phase aberrations compensation (54, 55).
To acquire AF images, excitation was generated by a mercury lamp (Nikon) attenuated with a ND12 filter and spectrally filtered by a DAPI filter cube (Semrock); cells were illuminated in an epifluorescence configuration. The signal was acquired with the same MO as QPM and acquired with a scientific complementary metal oxide semiconductor (sCMOS) detector (Rolera Bolt; QImaging) with an exposure time of 100 ms.
The Raman excitation was performed with a continuous wave laser at 532 nm (Verdi V-6; Coherent), with a power density of 170 mW/m2. Point spectra were acquired while optically averaging the signal from a large region of the cell in a hybrid imaging configuration (35). The collected backscattered light was separated from the excitation laser with a long-pass dichroic mirror and sent into a 500-mm focal length Czerny–Turner spectrometer (Shamrock; Andor Technology). The light dispersed by the grating (300 lp/mm) was then measured with a low-noise sCMOS camera (Orca-Flash 4.0; Hamamatsu Photonics) with an exposure time of 3 s.
ELISA.
Collected medium was centrifuged at 1,500 g during 5 min to isolate cell debris. Supernatants were then collected and stored at −85 °C until TNF- level quantification was performed with ELISA in the following weeks. Thawed supernatants were adequately diluted and measured on immuno-plates (Thermo Scientific) according to the instructions of the ELISA kit manufacturer (Mouse TNF- MAX Standard; Biolegend), in the 7- to 500-pg/mL range. Absorbance at 450 nm was measured with a plate reader (SpectraFluor; Tecan), and quantitative levels were estimated with measurements of TNF- standards through a logistic fit.
Immunofluorescence.
Immediately after label-free measurements, cells were fixed with 2% paraformaldehyde for 10 min at RT and stored at 4 °C until further processing. The next day, cells were permeabilized with 0.1% Triton X-100 during 5 min at RT and incubated in Block Ace solution (DS Pharma Biomed) with 0.1% Tween 20 for 2 h at RT to block nonspecific binding. Cells were then incubated in anti-iNOS antibody conjugated with Alexa Fluor 488 (Santa Cruz Biotechnology) at 4 g/mL during 1 h at RT. Cells were then counterstained with Hoechst 33342 (Sigma) at 0.2 g/mL during 2 min at RT, washed three times with PBS, and stored at 4 °C until observation.
Fluorescent signals were observed on the same microscope as used for AF measurements, with a DAPI filter set (gain 1; 10-ms exposure) and an FITC filter set (gain 14; 300-ms exposure) for nuclear and iNOS signals, respectively.
Data Processing.
AF images were registered to provide the same field of view as QPM ones, by estimating the coefficients of the linear transformation by maximizing the autocorrelation on a subset of images which were then applied to the whole stack of images. Cell segmentation was then performed with the CellProfiler program (56), where pipelines specifically designed for these label-free images were used. Segmentation results were then manually checked for accuracy, and morphological indicators were extracted from segmented images with the same program, where parameters based on morphology (size and shape from the MeasureObjectSizeShape module) and intensity-based parameters (intensity statistics and texture within segmented cells, from the MeasureObjectIntensity, MeasureObjectRadialDistribution, and MeasureTexture modules) were extracted due to the quantitative nature of the measured images. A full list of the morphological parameters is provided in Table S2. Parameters retrieved from size, QPM, and AF measurements were then merged together (yielding 301 variables) for further processing.
Raman spectra were first processed to prevent potential spectral shifts when comparing data measured on different days, by interpolating spectra on a grid calibrated by extracting the main peaks of a spectrum of pure ethanol measured on the same day. Spectra were then baseline-corrected by extracting values having a low probability of being larger than the signal by taking the 0.2 quantile value in several windows within the spectrum. The baseline was then estimated on all data points with cubic spline interpolation (57). The silent region (1,800–2,700 cm−1) was then removed from spectra, which were then decomposed with PCA, and statistical analysis was performed on the PC scores.
To enhance spectral features on showed spectra, curves were smoothed with a locally weighted scatter-plot smoothing (LOESS) (58), which avoided reducing peak sharpness. The smoothing was performed directly with the loess function in R (59).
Statistical Models.
Before analysis, morphological parameters were first normalized by their individual variance. Analysis was performed in the statistical program R through penalized logistic regression (60) with the glmnet package. To ensure more stable predictions, parameters shrinkage was performed with the lasso approach, where a penalty term was introduced to minimize the amount of used parameters to the most significant ones for classification (SI Text). The selection of the penalty term was performed manually by considering the binomial deviance through 10-fold cross-validation, while ensuring reproducible performance between training and test datasets.
Statistical tests were performed as unpaired Student’s t tests (assuming unequal variance). In case of multiple groups, significance was first assessed with ANOVA, and individual differences were tested with post hoc Tukey’s range tests. All computations were performed with standard functions in R. Symbols used are: ***; **; *; and n.s. (not significant), .
Data Availability.
Representative raw data (DHM and AF images, Raman spectra, etc.) and all the parameters extracted from the various measurements shown in this article (including the values used as training data) are available at dx.doi.org/10.5281/zenodo.1135372.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science through the World Premier International Research Center Initiative Funding Program. Additional support has been provided by the Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering through a grant for research development and the Uehara Memorial Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data extracted from our images and Raman spectra are available at dx.doi.org/10.5281/zenodo.1135372.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711872115/-/DCSupplemental.
References
- 1.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiller DG, Wood CD, Rand DA, White MRH. Measurement of single-cell dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Yuan J, Lipinski MM. Live imaging and single-cell analysis reveal differential dynamics of autophagy and apoptosis. Autophagy. 2013;9:1418–1430. doi: 10.4161/auto.25080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Álvarez-Barrientos A, Arroyo J, Cantón R, Nombela C, Sánchez-Pérez M. Applications of flow cytometry to clinical microbiology. Clin Microbiol Rev. 2000;13:167–195. doi: 10.1128/cmr.13.2.167-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsson A, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci USA. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junkin M, et al. High-content quantification of single-cell immune dynamics. Cell Rep. 2016;15:411–422. doi: 10.1016/j.celrep.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay S, et al. Single-cell NF-B dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavillon N, Hobro AJ, Smith NI. Cell optical density and molecular composition revealed by simultaneous multimodal label-free imaging. Biophys J. 2013;105:1123–1132. doi: 10.1016/j.bpj.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rappaz B, et al. Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy. J Biomed Opt. 2009;14:034049. doi: 10.1117/1.3147385. [DOI] [PubMed] [Google Scholar]
- 11.Falck Miniotis M, Mukwaya A, Gjörloff Wingren A. Digital holographic microscopy for non-invasive monitoring of cell cycle arrest in L929 cells. PLoS One. 2014;9:e106546. doi: 10.1371/journal.pone.0106546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavillon N, et al. Early cell death detection with digital holographic microscopy. PLoS One. 2012;7:e30912. doi: 10.1371/journal.pone.0030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekpenyong AE, et al. Bacterial infection of macrophages induces decrease in refractive index. J Biophotonics. 2013;6:393–397. doi: 10.1002/jbio.201200113. [DOI] [PubMed] [Google Scholar]
- 14.Lenz P, et al. Digital holographic microscopy quantifies the degree of inflammation in experimental colitis. Integr Biol. 2013;5:624–630. doi: 10.1039/c2ib20227a. [DOI] [PubMed] [Google Scholar]
- 15.Kühn J, et al. Label-free cytotoxicity screening assay by digital holographic microscopy. Assay Drug Dev Techn. 2013;11:101–107. doi: 10.1089/adt.2012.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi F, Moon I, Javidi B. Cell morphology-based classification of red blood cells using holographic imaging informatics. Biomed Opt Express. 2016;7:2385–2399. doi: 10.1364/BOE.7.002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jo Y, et al. Holographic deep learning for rapid optical screening of anthrax spores. Sci Adv. 2017;3:e1700606. doi: 10.1126/sciadv.1700606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CL, et al. Deep learning in label-free cell classification. Sci Rep. 2016;6:21471. doi: 10.1038/srep21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo B, et al. High-throughput, label-free, single-cell, microalgal lipid screening by machine-learning-equipped optofluidic time-stretch quantitative phase microscopy. Cytometry. 2017;91:494–502. doi: 10.1002/cyto.a.23084. [DOI] [PubMed] [Google Scholar]
- 20.Javidi B, Moon I, Yeom S, Carapezza E. Three-dimensional imaging and recognition of microorganism using single-exposure on-line (SEOL) digital holography. Opt Express. 2005;13:4492–4506. doi: 10.1364/opex.13.004492. [DOI] [PubMed] [Google Scholar]
- 21.Moon I, Javidi B. Three-dimensional identification of stem cells by computational holographic imaging. J R Soc Interface. 2007;4:305–313. doi: 10.1098/rsif.2006.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crow P, et al. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br J Cancer. 2005;92:2166–2170. doi: 10.1038/sj.bjc.6602638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd GR, et al. Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis. Analyst. 2013;138:3900–3908. doi: 10.1039/c2an36579k. [DOI] [PubMed] [Google Scholar]
- 24.Kong K, et al. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc Natl Acad Sci USA. 2013;110:15189–15194. doi: 10.1073/pnas.1311289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verrier S, Notingher I, Polak JM, Hench LL. In situ monitoring of cell death using Raman microspectroscopy. Biopolymers. 2004;74:157–162. doi: 10.1002/bip.20063. [DOI] [PubMed] [Google Scholar]
- 26.Okada M, et al. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc Natl Acad Sci USA. 2012;109:28–32. doi: 10.1073/pnas.1107524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedegaard M, et al. Spectral unmixing and clustering algorithms for assessment of single cells by Raman microscopic imaging. Theor Chem Acc. 2011;130:1249–1260. [Google Scholar]
- 28.Goodacre R, et al. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology. 1998;144:1157–1170. doi: 10.1099/00221287-144-5-1157. [DOI] [PubMed] [Google Scholar]
- 29.McReynolds N, Cooke FGM, Chen M, Powis SJ, Dholakia K. Multimodal discrimination of immune cells using a combination of Raman spectroscopy and digital holographic microscopy. Sci Rep. 2017;7:43631. doi: 10.1038/srep43631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujihara M, et al. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: Roles of the receptor complex. Pharmacol Therapeut. 2003;100:171–194. doi: 10.1016/j.pharmthera.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 32.An H, et al. Involvement of ERK, p38 and NF–B signal transduction in regulation of TLR2, TLR4 and TLR9 gene expression induced by lipopolysaccharide in mouse dendritic cells. Immunology. 2002;106:38–45. doi: 10.1046/j.1365-2567.2002.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel DY, et al. Human macrophage polarization in vitro: Maturation and activation methods compared. Immunobiology. 2014;219:695–703. doi: 10.1016/j.imbio.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Gordonov S, et al. Time series modeling of live-cell shape dynamics for image-based phenotypic profiling. Integr Biol. 2016;8:73–90. doi: 10.1039/c5ib00283d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavillon N, Smith NI. Maximizing throughput in label-free microspectroscopy with hybrid Raman imaging. J Biomed Opt. 2015;20:016007. doi: 10.1117/1.JBO.20.1.016007. [DOI] [PubMed] [Google Scholar]
- 36.Beleites C, Neugebauer U, Bocklitz T, Krafft C, Popp J. Sample size planning for classification models. Anal Chim Acta. 2013;760:25–33. doi: 10.1016/j.aca.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 37.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 38.Kable EP, Kiemer AK. Non-invasive live-cell measurement of changes in macrophage NAD(P)H by two-photon microscopy. Immunol Lett. 2005;96:33–38. doi: 10.1016/j.imlet.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Ellis DI, Cowcher DP, Ashton L, O’Hagan S, Goodacre R. Illuminating disease and enlightening biomedicine: Raman spectroscopy as a diagnostic tool. Analyst. 2013;138:3871–3884. doi: 10.1039/c3an00698k. [DOI] [PubMed] [Google Scholar]
- 40.Takai Y, Masuko T, Takeuchi H. Lipid structure of cytotoxic granules in living human killer T lymphocytes studied by Raman microspectroscopy. Biochim Biophys Acta. 1997;1335:199–208. doi: 10.1016/s0304-4165(96)00138-9. [DOI] [PubMed] [Google Scholar]
- 41.Hobro AJ, Standley DM, Ahmad S, Smith NI. Deconstructing RNA: Optical measurement of composition and structure. Phys Chem Chem Phys. 2013;15:13199–13208. doi: 10.1039/c3cp52406j. [DOI] [PubMed] [Google Scholar]
- 42.Nomura F, et al. Cutting edge: Endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J Immunol. 2000;164:3476–3479. doi: 10.4049/jimmunol.164.7.3476. [DOI] [PubMed] [Google Scholar]
- 43.Anderson KV. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs AT, Ignarro LJ. LPS-induced expression of IFN- mediates the timing of iNOS induction in RAW 264.7 macrophages. J Biol Chem. 2001;276:47950–47957. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- 45.Xiao L, et al. Lipopolysaccharide-induced expression of microsomal prostaglandin E synthase-1 mediates late-phase PGE2 production in bone marrow derived macrophages. PLoS One. 2012;7:e50244. doi: 10.1371/journal.pone.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao TC, Van Alten PJ, Greager JA, Walter RJ. Steroid sex hormones regulate the release of tumor necrosis factor by macrophages. Cell Immunol. 1995;160:43–49. doi: 10.1016/0008-8749(95)80007-6. [DOI] [PubMed] [Google Scholar]
- 47.Miller L, Hunt JS. Sex steroid hormones and macrophage function. Life Sci. 1996;59:1–14. doi: 10.1016/0024-3205(96)00122-1. [DOI] [PubMed] [Google Scholar]
- 48.Miller L, Hunt JS. Regulation of TNF- production in activated mouse macrophages by progesterone. J Immunol. 1998;160:5098–5104. [PubMed] [Google Scholar]
- 49.Su L, et al. Progesterone inhibits Toll-like receptor 4-mediated innate immune response in macrophages by suppressing NF-B activation and enhancing SOCS1 expression. Immunol Lett. 2009;125:151–155. doi: 10.1016/j.imlet.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Dagvadorj J, et al. Lipopolysaccharide induces alveolar macrophage necrosis via CD14 and the P2X7 receptor leading to interleukin-1 release. Immunity. 2015;42:640–653. doi: 10.1016/j.immuni.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menzies FM, Henriquez FL, Alexander J, Roberts CW. Selective inhibition and augmentation of alternative macrophage activation by progesterone. Immunology. 2011;134:281–291. doi: 10.1111/j.1365-2567.2011.03488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pavillon N, Smith NI. Implementation of simultaneous quantitative phase with Raman imaging. EPJ Tech Instr. 2015;2:1–11. [Google Scholar]
- 53.Marquet P, et al. Digital holographic microscopy: A noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt Lett. 2005;30:468–470. doi: 10.1364/ol.30.000468. [DOI] [PubMed] [Google Scholar]
- 54.Cuche E, Marquet P, Depeursinge C. Simultaneous amplitude–contrast and quantitative phase–contrast microscopy by numerical reconstruction of Fresnel off–axis holograms. Appl Opt. 1999;38:6994–7001. doi: 10.1364/ao.38.006994. [DOI] [PubMed] [Google Scholar]
- 55.Colomb T, et al. Total aberrations compensation in digital holographic microscopy with a reference conjugated hologram. Opt Express. 2006;14:4300–4306. doi: 10.1364/oe.14.004300. [DOI] [PubMed] [Google Scholar]
- 56.Carpenter AE, et al. Cellprofiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pavillon N, Bando K, Fujita K, Smith NI. Feature-based recognition of surface-enhanced Raman spectra for biological targets. J Biophotonics. 2013;6:587–597. doi: 10.1002/jbio.201200181. [DOI] [PubMed] [Google Scholar]
- 58.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- 59.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2016. [Google Scholar]
- 60.Hastie T, Tibshirani R, Jerome F. 2008. The Elements of Statistical Learning, Springer Series in Statistics (Springer, Berlin), 2nd Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Representative raw data (DHM and AF images, Raman spectra, etc.) and all the parameters extracted from the various measurements shown in this article (including the values used as training data) are available at dx.doi.org/10.5281/zenodo.1135372.