Significance
Increased added sugar is contributing to a rise in aging-related diseases. Here, we use the nematode, Caenorhabditis elegans, which store sugar as both glycogen and trehalose. We demonstrate that by modifying sugar storage, we can prevent the harmful effects of a high-sugar diet. Our data show that a metabolic shift increasing the glucose disaccharide trehalose, and decreasing the glucose polysaccharide glycogen, extends lifespan and promotes healthy aging. The positive effects of trehalose require the DAF-16 transcription factor and the process of autophagy. Our data reveal the benefits of trehalose for prolonged health in the face of our high-sugar environment.
Keywords: glycogen, gsy-1, trehalose, daf-16, lifespan
Abstract
As Western diets continue to include an ever-increasing amount of sugar, there has been a rise in obesity and type 2 diabetes. To avoid metabolic diseases, the body must maintain proper metabolism, even on a high-sugar diet. In both humans and Caenorhabditis elegans, excess sugar (glucose) is stored as glycogen. Here, we find that animals increased stored glycogen as they aged, whereas even young adult animals had increased stored glycogen on a high-sugar diet. Decreasing the amount of glycogen storage by modulating the C. elegans glycogen synthase, gsy-1, a key enzyme in glycogen synthesis, can extend lifespan, prolong healthspan, and limit the detrimental effects of a high-sugar diet. Importantly, limiting glycogen storage leads to a metabolic shift whereby glucose is now stored as trehalose. Two additional means to increase trehalose show similar longevity extension. Increased trehalose is entirely dependent on a functional FOXO transcription factor DAF-16 and autophagy to promote lifespan and healthspan extension. Our results reveal that when glucose is stored as glycogen, it is detrimental, whereas, when stored as trehalose, animals live a longer, healthier life if DAF-16 is functional. Taken together, these results demonstrate that trehalose modulation may be an avenue for combatting high-sugar-diet pathology.
Changes in the human diet, primarily the increasing amount of added sugar, are contributing to an alarming rise in both obesity and type 2 diabetes (1–3). According to the US Department of Agriculture, the average individual consumed almost 100 pounds of sugar in 2015, more than double the amount consumed in 1900 (4). The effects of a high-sugar diet can be modeled in Caenorhabditis elegans, as the addition of glucose to the standard Escherichia coli diet has been shown to cause glucose toxicity, which results in a decrease in lifespan (5–8), an increase in triglycerides (7, 8), neuronal defects (9, 10), and a decrease in locomotion (11, 12). Intracellularly, excess glucose can also lead to a greater incidence of advanced glycation end products (AGEs) in both humans and C. elegans (13). AGEs are formed from the nonenzymatic reaction between reducing sugars and either proteins, nucleic acids, or lipids. Formed naturally over time, AGEs alter the structure and function of their substrates and have been implicated as a detrimental complication of diabetes (14, 15).
C. elegans provide a unique opportunity to dissect the metabolic and phenotypic effects of added dietary sugars. In most animals, including both humans and C. elegans, glycogen, a branched polymer of glucose, is the main form of stored glucose and a primary source for energy reserves (16). The enzyme, glycogen synthase, is a key enzyme in the synthesis of glycogen (17). In humans, glycogen is stored primarily in skeletal muscle and liver, and each of these tissues has a different isoform of glycogen synthase (16). In contrast, C. elegans have a single glycogen synthase encoded by the gene gsy-1 (18–20). Modulation of glycogen storage is critical, as there are at least 13 known human glycogen storage diseases, most notably von Gierke’s disease (21, 22) and Pompe’s disease (23, 24). These diseases are caused by mutations in enzymes that result in abnormal levels of stored glycogen (22, 24, 25).
In addition to the glucose polymer glycogen, C. elegans can store sugar in the form of trehalose, a glucose disaccharide (26). Addition of trehalose to the standard E. coli diet promotes longevity (27) in contrast to the toxicity caused by the addition of glucose (5–8). Furthermore, mutants have been identified that are long lived and store increased trehalose (27). Therefore, an association between longevity and trehalose has been established. Mammals do not store sugar as trehalose but do possess the enzyme trehalase to breakdown ingested trehalose (28). Further, oral supplementation of trehalose has been shown to improve glucose tolerance in individuals at high risk for developing type 2 diabetes (29) and can improve recovery from traumatic brain injury in mice (30).
In C. elegans, the well-conserved insulin/IGF1 signaling (IIS) pathway is central for lifespan regulation and responds to a high-glucose diet (6, 31). Downstream of the DAF-2/IIS receptor is a PI 3-kinase signaling cascade that functions to negatively regulate the FOXO transcription factor DAF-16 (32). The activity of the pathway modulates the ability of DAF-16 to enter the nucleus to bind and activate/represses numerous target genes involved in lifespan regulation, stress response, dauer formation, and metabolism (33–39).
Here, we examine how added dietary glucose modulates sugar storage, lifespan, and healthspan. We find that either the addition of dietary glucose to or normal aging on a standard diet results in increased stored glycogen. Moreover, lowering the levels of stored glycogen extends lifespan and promotes healthspan, even in the presence of a high-sugar diet. Decreasing glycogen storage leads to a metabolic shift, resulting in increased internal trehalose. In addition, two other methods that increase internal trehalose (a diet high in trehalose as well as mutation in the enzyme trehalase) also resulted in lifespan extension. The benefits of the increased internal trehalose are entirely dependent on a functional DAF-16 FOXO transcription factor and autophagy which lead to a longer, healthier life.
Results
High-Glucose Diet Decreases Lifespan and Healthspan.
The continually increasing amount of added sugar in the human diet is contributing to an alarming rise in both obesity and type 2 diabetes (2, 3). To better understand the metabolic and phenotypic consequences of this change, we modeled a high-glucose diet in C. elegans. Addition of glucose to the media decreased lifespan similar to previous studies (5–8, 12), and higher concentrations of supplemental glucose resulted in correspondingly shorter lifespans (Fig. 1A and SI Appendix, Table S1). Importantly, comparable effects were seen with either live OP50 E. coli (SI Appendix, Fig. S1A and Table S1) or UV-killed OP50 E. coli (Fig. 1A and SI Appendix, Table S1), demonstrating that the effects of glucose on C. elegans lifespan are not due to glucose-induced changes in E. coli metabolism.
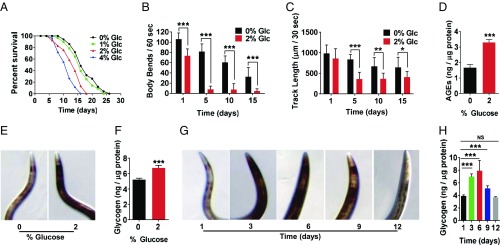
Fig. 1.
A high-glucose diet decreases lifespan and healthspan and increases glycogen storage. (A) Lifespan of wild type grown on NGM with 0%, 1%, 2%, or 4% glucose, 200 µM FUdR, and UV-killed OP50 E. coli. (B) Locomotion in liquid measured by number of body bends per minute, wild type grown for 1, 5, 10, and 15 d on NGM with 0% or 2% glucose, 200 µM FUdR, and OP50 E. coli. (C) Locomotion on solid media measured by track length traveled per 30 s, wild type grown for 1, 5, 10, and 15 d on NGM with 0% or 2% glucose, 200 µM FUdR, and OP50 E. coli. (D) AGEs of wild type grown on NGM with 0% or 2% glucose for 3 d, OP50 E. coli. AGEs standardized to protein content. (E and F) Glycogen stores of wild type grown on NGM with 0% or 2% glucose for 1 d, OP50 E. coli. (E) Iodine staining. (F) Total glycogen assay-glycogen content standardized to protein concentration. (G and H) Glycogen stores of aging wild type on NGM at 1, 3, 6, 9, and 12 d of adulthood, OP50 E. coli. (G) Iodine staining. (H) Total glycogen assay-glycogen content standardized to protein concentration (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, NS, not significant).
In addition to a reduction in lifespan, a high-glucose diet affects multiple measures of healthspan (5–8, 12). We analyzed healthspan in wild type grown on either control (0%) or a high-glucose (2%) diet at days 1, 5, 10, and 15 of adulthood. One healthspan metric is locomotion, which can be tested by both movement in liquid (measured by body bends) also known as thrashing/swimming and movement on solid media (measured by track length over a given time) (40). As shown in Fig. 1 B and C, locomotion both in liquid and on solid media declined with age on both control and high-glucose diets. By 5 d of age, animals grown on 2% added glucose declined approximately threefold faster than control for locomotion both in liquid and on solid media. Interestingly, previous studies demonstrated that a high-glucose diet decreases locomotion on solid media when animals are middle aged (day 10 or 14 of adulthood) (11, 12). Therefore, these data show that a high-glucose diet decreases mobility even in young animals.
We also examined the accumulation of AGEs, which have been suggested to be an indication of the physiological age. AGEs represent glucose toxicity caused by methylglyoxal, a precursor of AGEs, and are increased as a result of a high-glucose diet (13, 41, 42). As shown in Fig. 1D, the high-glucose diet significantly increased AGEs compared with control diet by day 3 of adulthood.
Alterations of Glycogen Stores by a High-Glucose Diet and Aging.
To understand the metabolic mechanisms that underlie the effects of a high-glucose diet, we examined whether the high-glucose diet caused a change in the polysaccharide glycogen, the main form of stored sugar (25, 43). We measured glycogen storage in whole animals using two different methods: iodine staining (18, 19) and a total glycogen assay (Biovision). Animals grown on a high-glucose diet showed an increase in glycogen storage in both assays: Fig. 1E (iodine staining) and Fig. 1F (total glycogen assay ∼30% increase in glycogen). Therefore, similar to Gusarov et al. (44), these data reveal that excess dietary glucose is processed and stored via the glycogen synthesis pathway.
We also examined glycogen storage as the animals aged. We found that on a standard diet, as animals aged, glycogen storage increased. As shown in Fig. 1G (iodine staining) and Fig. 1H (total glycogen assay), aging adult animals (1, 3, 6, 9, and 12 d old) had a ∼30–70% increase in stored glycogen by day 6. After day 6, glycogen levels started to decrease as animals continued to age. Therefore, together these data demonstrate that glycogen storage in C. elegans is modulated in a similar manner in response to both a high-sugar diet and aging.
Disruption of Glycogen Synthesis Extends Lifespan and Promotes Healthspan.
Next, we tested whether reducing glycogen storage could affect the aging process. Glycogen synthase is a critical enzyme in the synthesis of glycogen (17). In C. elegans, glycogen synthase is encoded by the gene gsy-1. We used two methods to test the effects of reducing levels of gsy-1: wild-type animals grown on gsy-1 RNAi and gsy-1(tm6196) mutants. We confirmed the effectiveness of gsy-1 RNAi by qPCR and found that RNAi reduced gsy-1 mRNA expression by 65% (SI Appendix, Fig. S1B). Iodine staining (Fig. 2A and SI Appendix, Fig. S1C) showed that both gsy-1 mutants and animals grown on gsy-1 RNAi had a dramatic decrease in stored glycogen similar to Gusarov et al. (44). These results were confirmed using a total glycogen assay, which revealed that gsy-1 RNAi lowered glycogen stores by ∼60% (SI Appendix, Fig. S1D).
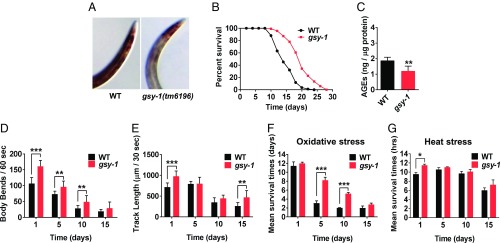
Fig. 2.
Disruption of glycogen synthesis extends lifespan and promotes healthspan. (A) Iodine staining of wild type and gsy-1(tm6196) mutants, day 3 adults. (B) Lifespan of wild type and gsy-1 mutants. (C) AGEs of wild type and gsy-1 mutants, day 3 adults, standardized to protein content. (D) Locomotion measured by number of body bends per minute in liquid, wild type and gsy-1 mutants grown for 1, 5, 10, and 15 d. (E) Locomotion measured by track length traveled per 30 s on solid media, wild type and gsy-1 mutants grown for 1, 5, 10, and 15 d. (F) Resistance to oxidative stress, wild type and gsy-1 mutants grown for 1, 5, 10, and 15 d. (G) Resistance to heat stress, wild type and gsy-1 mutants grown for 1, 5, 10, and 15 d (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). All animals were grown on NGM with 200 µM FUdR, OP50 E. coli.
Next, we tested animals grown on gsy-1 RNAi and gsy-1 mutants for lifespan. Both animals grown on gsy-1 RNAi (44) and gsy-1 mutants showed an ∼20% increase in mean lifespan (Fig. 2B and SI Appendix, Fig. S1E and Table S1) and similar lifespan results were observed without 5-fluoro-2′-deoxyuridine (FUdR) in the media (SI Appendix, Fig. S1F and Table S1). We also measured AGEs and found gsy-1 mutants had lower levels of AGEs compared with wild type (Fig. 2C). Thus, these data demonstrate that decreasing glycogen storage increases mean lifespan and decreases age-associated AGEs for animals grown on a standard E. coli diet.
Previous work from the H.A.T. laboratory shows that healthspan, in addition to lifespan, is a key measure for longevity interventions: animals should not simply live longer, but also display prolonged activity that resembles younger animals (40). Moreover, our work in C. elegans (40) and studies in mice (45, 46) have suggested that lifespan and healthspan can be separated since long-lived animals do not necessarily show prolonged health. Therefore, lifespan and healthspan need to be examined separately.
Using previously established healthspan assays (40), including locomotion in liquid and on solid media, resistance to heat stress, and resistance to oxidative stress, we tested whether the long-lived animals with decreased glycogen stores displayed prolonged healthspan. For locomotion in liquid media, both gsy-1 mutants and animals grown on gsy-1 RNAi exhibited increased body bends at every age tested (day 1, 5, 10, and 15) compared with wild type or wild type grown on vector control, respectively (Fig. 2D and SI Appendix, Fig. S3A). For locomotion on solid media, gsy-1 mutants and animals grown on gsy-1 RNAi increased the distance traveled at 1, 5, 10, and 15 d compared with wild type or wild type grown on vector control, respectively (Fig. 2E and SI Appendix, Fig. S3B). Although the rates of decline are similar between gsy-1 mutants and wild type at every age tested, gsy-1 mutants display greater levels of locomotion. Therefore, day 5 adult gsy-1 mutants look similar to day 1 wild-type adults, indicating prolonged healthspan.
To test healthspan by examining resistance to oxidative stress, we aged wild type and gsy-1 mutants for 1, 5, 10, and 15 d on a standard diet and then measured their ability to survive following a 2-h exposure to 250 mM paraquat. Fig. 2F shows that mean survival time of gsy-1 mutants was significantly increased compared with wild type at days 5 and 10. Therefore, in middle age, gsy-1 mutants show increased health in this parameter. To test healthspan by examining resistance to heat stress, we exposed aged animals to 37 °C and measured their survival every 2 h. gsy-1 mutants displayed strong resistance to heat stress compared with wild type at day 1, with a significant increase in survival time (20%) (Fig. 2G). Taken together, reduction of function in gsy-1 extends lifespan and prolongs multiple aspects of healthspan.
Importantly, our data reveal that gsy-1 mutants and animals grown on gsy-1 RNAi exhibit similar phenotypes for lifespan, multiple measures of healthspan, and glycogen storage. Therefore, for further analysis, we used gsy-1 mutants or animals grown on gsy-1 RNAi interchangeably.
Decreased gsy-1 Attenuates the Negative Effects of High-Glucose Diet on Longevity.
Since decreased glycogen storage leads to increased lifespan and healthspan on a standard diet (Fig. 2), we examined whether decreased glycogen storage would affect animals grown on a high-glucose diet. gsy-1 mutants grown on a high-glucose diet had a lifespan similar to wild type without added glucose (SI Appendix, Fig. S3D and Table S1). Similar results were also seen with gsy-1 RNAi in accordance with ref. 44 (Fig. 3A and SI Appendix, Table S1). Therefore, modulating gsy-1 and glycogen storage can attenuate glucose toxicity.
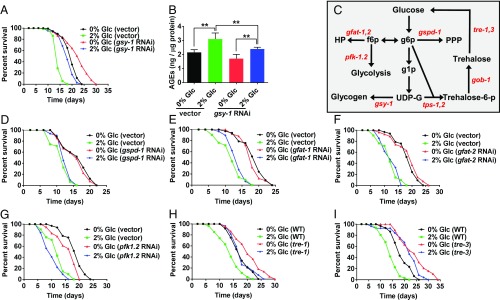
Fig. 3.
Modulating glycogen storage, but not other glucose-processing pathways, attenuates the effects of a high-glucose diet on longevity. (A) Lifespan of wild type on 0% or 2% glucose, 200 µM FUdR, empty vector or gsy-1 RNAi. (B) AGEs of day 3 adults on 0% or 2% glucose, on empty vector or gsy-1 RNAi. AGEs results standardized to protein content. (C) Model of glucose metabolic pathways in C. elegans, f6p, fructose-6-phosphate; g1p, glucose-1-phosphate; g6p, glucose-6-phosphate; HP, hexosamine pathway; PPP, pentose phosphate pathway; UDP-G, uridine diphosphate glucose. (D–G) Lifespan of wild type on 0% or 2% glucose, 200 µM FUdR, with either empty vector, gspd-1, gfat-1, gfat-2, or pfk-1.2 RNAi. (H and I) Lifespan of wild type, tre-1(ok327), and tre-3(ok394) mutants on 0% or 2% glucose, 200 µM FUdR, fed UV-killed OP50 E. coli (**P ≤ 0.01).
Similar to Fig. 1D, a high-glucose diet increased the levels of AGEs in wild type, and on a 0% glucose diet, animals grown on gsy-1 RNAi showed lower levels of AGEs compared with wild type (Fig. 3B). Interestingly, animals grown on gsy-1 RNAi on a high-glucose diet also increased AGEs; however, the levels were significantly lower than wild type on a high-glucose diet (Fig. 3B). Therefore, a high-glucose diet increases the level of AGEs dependent on the expression of gsy-1 or the level of glycogen storage. Taken together, these data suggest that animals could live a wild-type lifespan, even when grown on a high-sugar diet, if the excess sugar is not stored as glycogen.
Only Modification of gsy-1 Attenuates Glucose Toxicity.
Next, we examined the possible metabolic alternatives for the excess glucose, if it cannot be stored as glycogen. As shown in Fig. 3C, glucose is metabolized by several different pathways, including glycogen synthesis, pentose phosphate, hexosamine, and phoshofructokinase. We examined whether decreased metabolic flux through any of these pathways would protect animals from the harmful effects of a high-glucose diet similar to the protection seen when glycogen storage is blocked by loss of function of gsy-1.
We tested the initial enzymes in the pentose phosphate (gspd-1), hexosamine (gfat-1, gfat-2), and phoshofructokinase (pfk-1.2) pathways on a standard and high-glucose diet. RNAi of gspd-1, gfat-1, gfat-2, or pfk-1.2 resulted in lifespans similar to wild type on the standard diet (Fig. 3 D–G and SI Appendix, Table S1). On a high-glucose diet, RNAi of gspd-1, gfat-1, gfat-2, or pfk-1.2 resulted in lifespan similar to wild-type (Fig. 3 D–G and SI Appendix, Table S1). Therefore, blocking these metabolic pathways failed to phenocopy blocking glycogen storage (Fig. 3A and SI Appendix, Table S1). This suggests that modulating glycogen storage, but not other glucose processing pathways, can attenuate glucose toxicity.
Metabolic Shift from Glycogen to Trehalose.
Fig. 3C shows that UDP-glucose is an intermediary for both glycogen synthesis as well as trehalose synthesis. In addition, C. elegans can store dietary glucose as both glycogen (a glucose polymer) and trehalose (a glucose–glucose disaccharide) (26). Previous studies have found that supplementing the C. elegans diet with 5 mM trehalose extends lifespan and enhances thermotolerance (27). Trehalose is also necessary for desiccation tolerance (47). Opposing the trehalose synthesis pathway, trehalose is broken down via the enzyme trehalase, to produce two glucose molecules and C. elegans have five trehalase genes (tre-1, tre-2, tre-3, tre-4, and tre-5) (48).
Consequently, we tested the possibility that mutation in C. elegans trehalase could mimic gsy-1 mutants’ attenuation of glucose toxicity and lifespan extension. Of the five C. elegans trehalase genes, only tre-1 and tre-3 mutants showed changes in lifespan and both tre-1 and tre-3 mutants had increased internal trehalose (SI Appendix, Fig. S3 G and H and Table S1). On a high-glucose diet, similar to gsy-1 mutants, tre-1 and tre-3 mutants exhibited attenuation of glucose toxicity (Fig. 3 H and I and SI Appendix, Table S1). Therefore, either modulation of glycogen storage or modulation of trehalose storage result in lifespan extension and protection from a high-glucose diet.
To begin to examine the relationship between glycogen storage, trehalose storage, lifespan, and a high-glucose diet, we measured the levels of internal trehalose in the pathways outlined in Fig. 3C, including gspd-1, gfat-1, gfat-2, and pfk-1.2 RNAi, and gsy-1, tre-1, and tre-3 mutants. As shown in Fig. 4A, wild type on a high-glucose diet had slightly lower levels of trehalose than wild type on a standard diet. Animals grown on RNAi of either gspd-1, gfat-1, gfat-2, or pfk-1.2 had similar levels of trehalose on both diets. However, gsy-1, tre-1, and tre-3 mutants had higher levels of internal trehalose on the high-glucose diet (Fig. 4A). Although all of the genetic modulations show a decrease in internal trehalose in response to the high-glucose diet, the animals with the highest level of internal trehalose were most protected from the glucose toxicity (Figs. 3 A, H, and I and 4A). Therefore, the two metabolic modulations that led to lifespan extension and protection from a high-glucose diet both showed increased levels of internal trehalose.
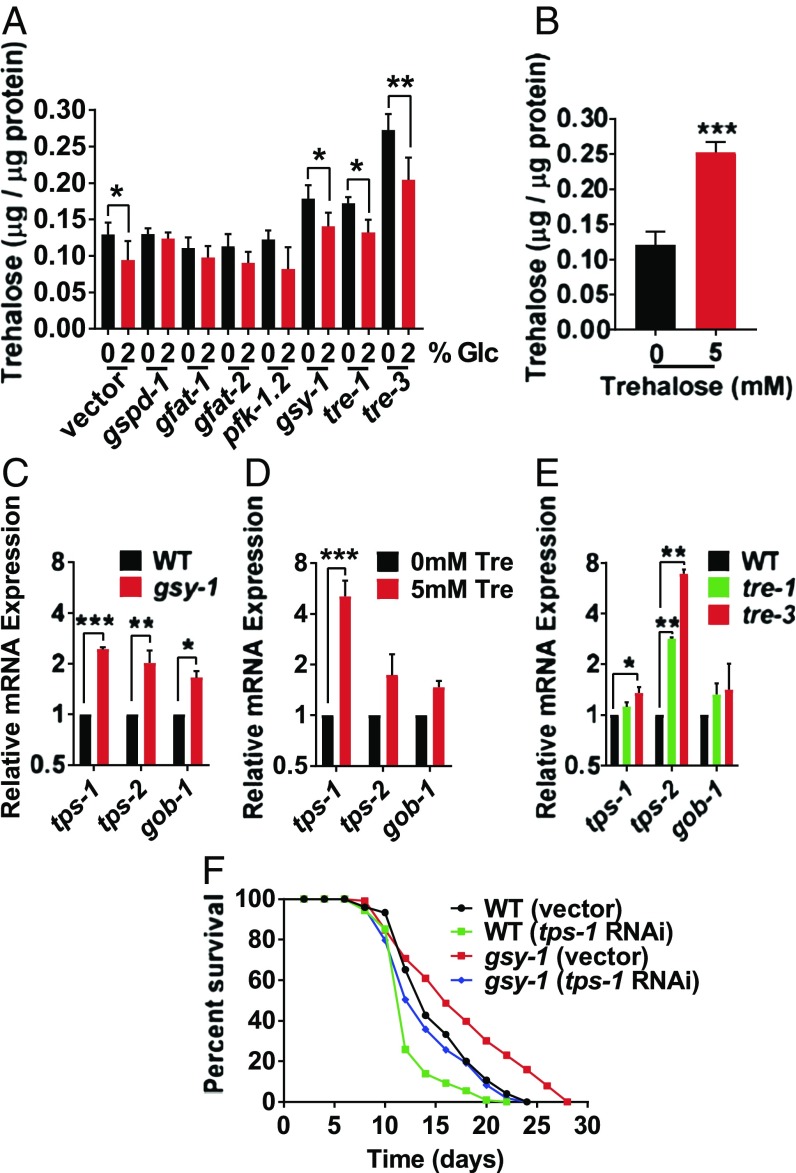
Fig. 4.
Animals with increased internal trehalose exhibit lifespan extension. (A) Internal trehalose levels of wild type on 0% or 2% glucose, with either empty vector, gspd-1, gfat-1, gfat-2, or pfk-1.2 RNAi; and gsy-1, tre-1, and tre-3 mutants on 0% or 2% glucose, UV-killed OP50 E. coli. (B) Internal trehalose levels of day 1 adults, on 0 mM or 5 mM trehalose, UV-killed OP50 E. coli, internal trehalose levels standardized to protein content. (C) RT-qPCR of trehalose synthesis genes in wild-type day 1 adults and gsy-1 mutants day 1 adults, OP50 E. coli. (D) RT-qPCR of trehalose synthesis genes in wild-type day 1 adults on 0 mM or 5 mM trehalose, UV-killed OP50 E. coli. (E) RT-qPCR of trehalose synthesis genes in wild-type, tre-1, and tre-3 mutants day 1 adults, OP50 E. coli. (F) Lifespan of wild type and gsy-1 mutants, 200 µM FUdR, with either empty vector or tps-1 RNAi (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Given our results showing an association between the increased internal levels of trehalose and positive life benefits, we examined another method which may increase internal trehalose: supplementing the diet with trehalose. As shown in Fig. 4B and SI Appendix, Fig. S3F, animals fed a diet supplemented with 5 mM trehalose had increased internal trehalose and showed lifespan extension. Therefore, across three methods: reducing the levels of gsy-1, reducing the levels of tre-1 or tre-3, and supplementing the diet with trehalose, internal trehalose was increased and lifespan was extended.
In agreement with our finding that gsy-1 mutants store more trehalose, gsy-1 mutants also had significantly elevated mRNA expression of genes critical for trehalose synthesis: tps-1, tps-2 (trehalose-6-phosphate synthase 1 and 2), and gob-1 (trehalose-6-phosphatase) (Fig. 4C). In addition, RT-qPCR of animals fed 5 mM trehalose as well as tre-1 and tre-3 mutants showed a similar up-regulation of the trehalose synthesis genes (Fig. 4 D and E).
Thus far, it was unclear whether the lifespan extension in the gsy-1 mutants (or gsy-1 RNAi) was due to either the decrease in glycogen or the increase in trehalose. Therefore, we next examined whether blocking trehalose synthesis would eliminate the lifespan extension observed upon reduction of function of gsy-1 using the tps-1/2 genes which have been shown to regulate the levels of internal trehalose (49). Similar to previous studies (27), RNAi of tps-1 decreased mean lifespan ∼15% (Fig. 4F and SI Appendix, Table S1). In the gsy-1 mutant background, RNAi of tps-1 resulted in a lifespan similar to wild type (Fig. 4F and SI Appendix, Table S1), suggesting that the increased trehalose stores are responsible for the lifespan extension when glycogen stores were decreased. This is unlike daf-2 mutants, which still show extended lifespan when trehalose is reduced (27, 49).
Through our three methods: gsy-1 mutation, exogenous trehalose feeding, and trehalase mutants, we observed both an increase in internal trehalose as well as an up-regulation of the trehalose synthesis genes. These data demonstrate that a metabolic shift toward increased internal trehalose, either by blocking glycogen synthesis, dietary intervention, or inhibited trehalose breakdown, extends lifespan.
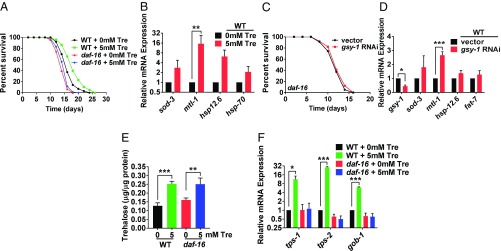
Trehalose Benefits Are Dependent on DAF-16.
DAF-16 is the downstream target of the IIS pathway and has been shown to respond to a high-glucose diet (6, 31). Animals fed a 5 mM trehalose diet exhibited lifespan extension dependent on DAF-16 similar to Honda et al. (27) (Fig. 5A and SI Appendix, Table S1) and increased expression of DAF-16 target genes (such as sod-3, mtl-1, hsp12.6, and fat-7) (33, 39, 50) (Fig. 5B). Therefore, next, we examined whether animals grown on gsy-1 RNAi or gsy-1 mutants with increased internal trehalose exhibited similar phenotypes and gene expression. As shown in Fig. 5C and SI Appendix, Fig. S7A and Table S1, lifespan extension of either animals grown on gsy-1 RNAi similar to Gusarov et al. (44) or gsy-1 mutants was suppressed by daf-16 (RNAi or mutation). Consistently, DAF-16 target genes were also up-regulated in wild-type animals grown on gsy-1 RNAi (Fig. 5D).
Fig. 5.
Positive benefits of increased internal trehalose requires functional DAF-16. (A) Lifespan of wild type and daf-16(mgDf50) on 0 mM or 5 mM trehalose, 200 µM FUdR, UV-killed OP50 E. coli. (B) RT-qPCR of DAF-16 targets of day 1 adults on 0 mM or 5 mM trehalose, UV-killed OP50 E. coli. (C) Lifespan of daf-16 mutant with either vector or gsy-1 RNAi, 200 µM FUdR. (D) RT-qPCR of DAF-16 targets in wild-type day 1 adults with either vector or gsy-1 RNAi. (E) Internal trehalose concentration of wild type and daf-16 mutants on 0 mM and 5 mM trehalose, UV-killed OP50 E. coli, day 3 adults, internal trehalose concentration standardized to protein content. (F) RT-qPCR of trehalose synthesis genes in wild type and daf-16 mutants on 0 mM and 5 mM trehalose, UV-killed OP50 E. coli, day 3 adults (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
In response to a diet of added trehalose, wild type and daf-16 mutants had similar increases in levels of trehalose (Fig. 5E). Importantly, despite the increase in internal trehalose storage, daf-16 mutants did not up-regulate trehalose synthesis genes (Fig. 5F). Therefore, even with the elevated levels of internal trehalose, lifespan extension benefits require functional daf-16.
DAF-16 has three isoforms (a, b, and d/f), where daf-16a and daf-16d/f are the isoforms important for longevity (50–54). Using transgenic strains which bear a complete loss of daf-16 and then a single isoform expressed from a transgene, we revealed that gsy-1 RNAi resulted in lifespan extension in the daf-16d/f background but not daf-16a background (SI Appendix, Fig. S7B and Table S1). Consistent with this, we found that genes transcriptionally regulated by DAF-16d/f but not DAF-16a (such as sod-3, mtl-1, hsp12.6, and fat-7) had increased expression in both wild-type and daf-16d/f transgenic animals grown on gsy-1 RNAi (Fig. 5D and SI Appendix, Fig. S7 C and D). These data suggest that the DAF-16d/f, but not the DAF-16a isoform may be preferential for lifespan extension in response to the changes in glycogen and trehalose storage mediated by gsy-1.
High Levels of Trehalose Promote Lifespan Through Up-Regulation of Autophagy.
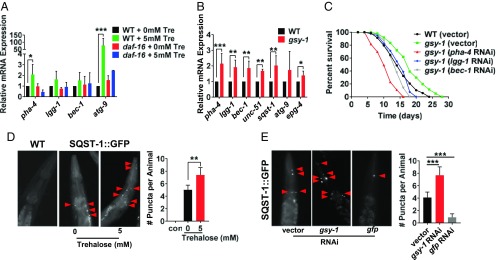
Previously, trehalose has been shown in mammalian cell culture to enhance the removal of protein aggregates through the process of autophagy (55). In addition, increased autophagy can result in C. elegans lifespan extension (56–61). To test the connection between trehalose, daf-16, and autophagy, first we examined the animals with increased internal trehalose, similar to previous C. elegans autophagy studies (60, 62, 63). Wild-type animals fed 5 mM trehalose had increased mRNA expression of autophagy-related genes (Fig. 6A). However, dietary trehalose had little effect on expression of these same genes in a daf-16 mutant background (Fig. 6A). In addition, gsy-1 mutants on a standard diet displayed similar up-regulation of the same autophagy-related genes as well as others (unc-51, sqst-1, and epg-4) (Fig. 6B). Together, these data revealed that increased internal trehalose leads to up-regulation of genes involved in regulation of autophagy but only if animals have a functional DAF-16.
Fig. 6.
Positive benefits of increased internal trehalose requires autophagy. (A) RT-qPCR of autophagy-related genes in wild type and daf-16 mutants on 0 mM and 5 mM trehalose, UV-killed OP50 E. coli, day 3 adults. (B) RT-qPCR of autophagy-related genes in wild type and gsy-1 mutants, OP50 E. coli, day 1 adults. (C) Lifespan of wild type and gsy-1 mutants, 200 µM FUdR, with either empty vector, pha-4, lgg-1, or bec-1 RNAi. (D) Photographs and quantification of wild-type control and sqst-1::GFP on 0 mM or 5 mM trehalose, UV-killed OP50 E. coli, day 3 adult pharynx. Red arrows denote GFP positive puncta, which were quantified. (E) Photographs and quantification of sqst-1::GFP on either empty vector, gsy-1, or gfp(control) RNAi, day 3 adult pharynx. Red arrows denote GFP positive puncta, which were quantified (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Next, we investigated whether interfering with autophagy could abrogate the lifespan extension of gsy-1 mutants. Similar to previous studies (64), RNAi of the autophagy-related gene, pha-4 reduced wild-type lifespan (SI Appendix, Fig. S8 and Table S1). In contrast, in a gsy-1 mutant background, RNAi of the three autophagy-related genes tested, eliminated the lifespan extension (Fig. 6C and SI Appendix, Table S1), suggesting that increasing internal trehalose up-regulates autophagy and promotes lifespan extension.
To monitor autophagy in a live animal, similar to previous C. elegans autophagy studies (62, 63), we used sqst-1::GFP transgenics where puncta represent autophagic vesicles. Animals grown on 5 mM trehalose had a modest 1.4-fold increase in the number of puncta in the pharynx and upper intestine (Fig. 6D). Furthermore, gsy-1 RNAi knockdown led to a significant 1.8-fold increase in the number of puncta (Fig. 6E), supporting the idea that increased internal trehalose promotes lifespan extension through autophagy.
In summary, our findings suggest that increasing trehalose storage, whether through dietary intervention or through a metabolic shift that limits either glycogen storage (reduction of function glycogen synthase) or trehalose breakdown (reduction of function trehalase), extends lifespan and promotes healthspan in a DAF-16 and autophagy-dependent manner.
Discussion
The changing human diet, in particular the large amount of added dietary sugar, has contributed to the increase in diabetes, obesity, and cardiovascular disease (1–3). Recent studies in mice found that a high-glucose diet significantly impaired healing following a tendon injury (65, 66), demonstrating that high-sugar diets have far-reaching effects. Similar to previous studies (5–8, 10–13) and data in Fig. 1, we found that addition of glucose to the C. elegans diet leads to shortened lifespan, diminished healthy aging or healthspan, and increased AGEs.
Interestingly, not all high-sugar diets are detrimental. A recent study reports that addition of 1–2% fructose prolongs C. elegans lifespan (67). In mammals, fructose is processed differently than glucose. While glucose is used immediately for energy, or processed to be stored as glycogen, excess fructose is mainly processed by the liver and stored as lipids (68). Therefore, the detrimental effects of excessive sugar diets may be highly dependent on how the sugar is processed. Interestingly, Lee et al. (69) used a FA/retinol-binding protein 3 (far-3) promoter (far-3p::GFP) as an in vivo glucose reporter in a genome-wide RNAi screen to identify genes involved in the shortening of lifespan by glucose. They found that glucose toxicity arises due to the accumulation of saturated fat. Up-regulation of C. elegans SREBP homolog SBP-1 and the transcriptional coregulator mediator-15 (MDT-15) could prevent the accumulation of saturated fat and thus results in alleviation of glucose toxicity (69). Given that we have identified the trehalose storage as a beneficial glucose-processing pathway, it will be interesting to test whether and how this pathway intersects with the regulation of saturated fat, including SBP-1/MDT-15.
In this study, we found that when glycogen stores were decreased, either by RNAi or mutation in the enzyme glycogen synthase encoded by the gene gsy-1, animals exhibited lifespan extension and prolonged healthspan (Fig. 2 and SI Appendix, Fig. S2). Moreover, we demonstrate that reduction of function of gsy-1 protects animals from the toxicity of a high-glucose diet, as shown by lifespan [similar to Gusarov et al. (44)] and AGEs (Fig. 3). Based on C. elegans physiology and metabolism (70), we suggest that reduction of gsy-1 leads to high levels of UDP-glucose, which is a substrate for both gsy-1 (for glycogen) and tps-1 (for trehalose). Accordingly, reduction of function of gsy-1 leads to a metabolic shift, where expression of the trehalose synthesis genes tps-1, tps-2, and gob-1 are up-regulated and levels of stored trehalose increase (Fig. 4).
In contrast to addition of dietary glucose, addition of dietary trehalose, a disaccharide of two glucose molecules, prolongs lifespan (27) and promotes healthspan in C. elegans. Trehalose is used as a glucose storage molecule in prokaryotes and metazoans, including worms and flies, where its primary role has been associated with desiccation, heat shock response, cold response, and oxidation (47, 48, 71). Chemically, trehalose is a stable nonreducing carbohydrate which does not participate in spontaneous nonenzymatic reactions with proteins or lipids, which may also reduce its potential to generate AGEs. Accordingly, our data show that the animals with increased internal trehalose, have decreased AGEs (Fig. 4).
Using three different methods: dietary intervention, a metabolic shift that limited glycogen synthesis (glycogen synthase modification), or reduced trehalose breakdown (trehalase modification), we have increased C. elegans internal trehalose levels. In all three interventions, the increased internal trehalose resulted in an increase in expression of trehalose synthesis genes, tps-1, tps-2, and gob-1, as well as lifespan extension (Fig. 4).
A recent publication confirms much of our findings with gsy-1 (44), although this study finds that gsy-1 modification is associated with increased oxidative stress and altered NADPH. This is distinct from our results which suggest that the increased internal trehalose is responsible for the lifespan and healthspan benefits of blocking glycogen synthase. Interestingly, Penkov et al. (72) found that increased trehalose synthesis diverts glucose-6-phosphate from the pentose phosphate pathway, reducing NADPH and increasing dauer formation. Consistently, we find that reduction of function of gsy-1 leads to an increase in dauer formation and trehalose synthesis (SI Appendix, Fig. S3 C and E, respectively). Therefore, it is possible that findings of Gusarov et al. (44) could be accounted for by increased trehalose.
Our data reveal the requirement of daf-16 in the metabolic shift by gsy-1. Importantly, our data show that simply increasing internal trehalose is not beneficial unless DAF-16 is functional (Fig. 5). Interestingly, very recently, studying the distinct process of survival during starvation, Hibshman et al. (49) also found a connection between daf-16 function and trehalose synthesis.
daf-16 has three isoforms (daf-16d/f, daf-16b, and daf-16a), and daf-16d/f and daf-16a are the isoforms that are involved in longevity (50, 54, 73). Using multiple assays, including lifespan and target gene expression, our data suggest the metabolic shift to increased trehalose via gsy-1 is dependent on daf-16d/f. RNAi knockdown of gsy-1 up-regulated several antistress genes, such as sod-3, mtl-1, hsp12.6, and fat-7, which have been shown to be specific targets of daf-16d/f, but not daf-16a (SI Appendix, Fig. S7 C and D). Further, this change in gene expression was confirmed with animals grown on the 5 mM trehalose diet.
In addition to functional DAF-16, autophagy is required for increased trehalose storage to lead to lifespan extension. Previously, studies in mammalian cell culture have linked trehalose to autophagy (55, 74), but this has not been shown in a whole organism. We observed that increased internal trehalose up-regulates pha-4, a transcription factor important for autophagy, as well as genes for phagophore synthesis (lgg-1, bec-1, sqst-1, and unc-51) and autophagosome maturation (epg-4 and atg-9). Moreover, interfering with autophagy by RNAi knockdown of either pha-4, lgg-1, or bec-1 eliminated the beneficial effect of the increased internal trehalose in gsy-1 mutants (Fig. 6). These results together indicate that the high level of trehalose built up as a result of limiting glycogen synthesis improves healthy aging through daf-16 and autophagy.
Although vertebrates do not store glucose as trehalose and the trehalose synthesis enzymes have been lost from their genomes, they can consume trehalose and do express a trehalase enzyme highly homologous to C. elegans trehalase (48). In addition, a recent study showed that the mammalian GLUT8 receptor transports trehalose into hepatocytes (75–77). Humans express trehalase and GLUT8 on the intestinal villae and kidney brush border, presumably to help digest foods high in trehalose such as shrimp, crab, and mushrooms (78, 79). As shown in Figs. 3I and 4A, tre-3 mutants showed attenuation to the life-shortening effects of glucose. This may reveal the possibility that despite our inability to synthesize trehalose, dietary interventions or trehalase modulation in humans may provide resistance to a high-glucose diet.
Taken together, these results demonstrate that a metabolic shift from glycogen to trehalose is beneficial for healthy aging as well as increased lifespan. These data provide an insight into the possibility to manipulate carbohydrate metabolism in response to aging, stress, and a high-sugar diet. Our findings reveal the possible benefit of trehalose for prolonged health in the face of our high-sugar environment.
Materials and Methods
Strains.
All strains were maintained at 15 °C using standard C. elegans techniques except where indicated (80). A standard diet refers to NGM plates seeded with OP50 E. coli. Strains used in this study: N2, tre-1(ok327) I, daf-16(mgDf50) I, gsy-1(tm6196) II, daf-2(e1370) III, tre-3(ok394) V, and HZ589 [him-5(e1490); bpIs151[sqst-1p::sqst-1::GFP + unc-76(+)]]III. Transgenic strains HT1883 [daf-16(mgDf50); daf-2(e1370); daf-16d/f::GFP] and HT1881 [daf-16(mgDf50); daf-2(e1370); daf-16a::GFP] were generated by the H.A.T. laboratory (50). gsy-1(tm6196) was provided by the T. Mitani laboratory through the National BioResource Project of the MEXT, Japan. gsy-1(tm6196) was backcrossed to wild type five times before use. Wherever possible, mutants were used for analysis.
Lifespan Assay.
All lifespan assays were performed at 20 °C. Strains were semisynchronized by allowing gravid adults to lay embryos on standard NGM or RNAi plates overnight, and animals developed for several days until they reached L4. Then, ∼100 L4s were transferred to NGM or RNAi plates containing 200 µM FUdR (Pharma Waldhof GmbH), with/without added sugar and kept at 20 °C (∼33 animals per plate). Animals were scored by gently tapping with a platinum wire every 2–3 d. Those that did not respond were scored as dead. Animals that crawled off the plate, bagged, or died from vulva bursting were censored from the analysis. Each figure shows one trial, with additional trials shown in SI Appendix, Table S1. The mean lifespan, SD, and P values were calculated via two-tailed unpaired t tests. Survival graphs and statistical analyses were produced using GraphPad Prism 7.00 (GraphPad Software).
Glucose and Trehalose Diet.
For addition of glucose and trehalose to the diet, filter-sterilized water (control), d-(+)-glucose, or d-(+)-trehalose dihydrate (Sigma) was added to cooled molten NGM to the desired concentration (2% for glucose, 5 mM for trehalose) with 0.1 mg/mL ampicillin (Fisher Scientific) before pouring plates. Plates were seeded with OP50 E. coli grown overnight in LB. After 6 h of drying, plates were exposed to UV for 4.5 min at 4,000 µW/cm2 (power setting: 9,999) using the Stratagene Stratalinker 2400 (Agilent Genomics). OP50 was then streaked onto an LB agar plate and cultured at 37 °C overnight to confirm lack of growth. Plates were stored at 4 °C until use.
RNA Interference.
All RNAi clones were obtained from the Ahringer RNAi library (81). RNAi plates were made by adding filter-sterilized reagents to cooled molten NGM to a final concentration of 0.1 mg/mL ampicillin (Fisher Scientific), 1 mM isopropyl-β-d-thiogalactoside (US Biological), and for lifespan assays 200 μM FUdR. The plates were seeded with HT115 E. coli, first grown overnight from a single colony at 37 °C in 0.1 mg/mL ampicillin and 12.5 μg/mL tetracycline (Genlantis), then diluted 1/100 in LB, and cultured again in 0.1 mg/mL ampicillin for 6 h at 37 °C. Plates were dried at room temperature for 24 h and then stored at 4 °C until use. Before use, plates were placed at room temperature for at least 1 h. Animals were grown on RNAi for two generations before being used for experiments. When epistasis was performed only with RNAi, a caveat should be noted as this may not be a complete loss of function.
Iodine Staining.
L4s grown on NGM at 20 °C were picked onto NGM plates with 0% or 2% added glucose at 20 °C for 24 h before staining. Aging animals were picked to NGM plates supplemented with 200 μM FUdR and grown at 20 °C until they reached the desired stage before staining. For RNAi experiments, L4s grown on vector or gsy-1 RNAi were transferred to new plates for 24 h before staining. Then, 20 animals were picked onto unseeded NGM plates and inverted and placed on top of a bottle of iodine flakes (Fisher Chemical) for 1 min. Animals were then photographed using a 5.0 MP Microscope Digital Camera (AmScope) outfitted onto a dissecting microscope. Methods were adapted from the M. B. Roth laboratory (19).
AGEs Assay.
Approximately 500 L4s grown on NGM at 20 °C were transferred to plates containing 200 µM FUdR and incubated at 20 °C for 3 d. Animals were then washed off plates with M9 buffer and subsequently rinsed twice with M9 buffer. Then 200 µL NET Lysis Solution [150 mM NaCl, 1 mM EDTA (pH 8.0), 50 mM Tris (pH 7.5), 0.5% CHAPS (adapted from ref. 82)] was added to each sample. Samples were put on ice, and then sonicated using a Microson XL2000 Qsonica P-3 at 4 W (Fisher Scientific) for a total of 30 s, with 3 × 10 s bursts. AGEs were determined using the OxiSelect Advanced Glycation End Product Competitive ELISA Kit (Cell Biolabs) according to the manufacturer’s instructions. The absorbance was measured at 450 nm using the SpectraMax M5 Microplate Reader (Molecular Devices).
Glycogen Storage.
Approximately 500 L4s grown on NGM at 20 °C were transferred to 0% or 2% glucose plates for 24 h before the assay. Aging animals were picked to NGM plates supplemented with 200 μM FUdR and grown at 20 °C until they reached the desired stage before lysis. Then, aged animals were washed off plates using ddH2O and subsequently rinsed twice with ddH2O and put on ice. Samples were then sonicated using a Microson XL2000 Qsonica P-3 at 4 W (Fisher Scientific) for 30 s with 3 × 10 s bursts. The lysates were then boiled for 10 min and then centrifuged at 15,800 × g for 10 min at 4 °C. Supernatants were then collected and total glycogen was measured using the Glycogen Colorimetric/Fluorometric Assay Kit (BioVision) and a SpectraMax M5 Microplate Spectrophotometer (Molecular Devices). Glycogen levels were normalized to protein content measured with Pierce Coomassie Plus (Bradford) Assay Kit (Thermo Scientific).
Trehalose Storage.
Approximately 500 L4s grown on NGM at 20 °C were transferred to NGM, 2% glucose, 5 mM trehalose, or RNAi plates for 24 h before assaying. Animals were then washed off plates using M9 and subsequently rinsed twice with M9 to 100 µL. Next, 200 µL of NET Lysis Solution [150 mM NaCl, 1 mM EDTA (pH 8.0), 50 mM Tris (pH 7.5)], and 0.5% CHAPS was added to each strain. The samples were then put on ice and sonicated using a Microson XL2000 Qsonica P-3 at 4 W (Fisher Scientific) for a total of 30 s, with 3 × 10 s bursts. The lysates were then centrifuged at 15,800 × g for 13 min at 4 °C. Supernatants were collected and trehalose was measured using the Trehalose Assay Kit (Megazyme) and a SpectraMax M5 Microplate Spectrophotometer (Molecular Devices). Trehalose levels were normalized to protein content measured with Pierce Coomassie Plus (Bradford) Assay Kit (Thermo Scientific). Methods were adapted from Miersch et al. (82).
RNA Isolation and Real-Time PCR.
Approximately 200 synchronous animals developed on experimental condition plates at 20 °C until they reached day 1 of adulthood before RNA extraction. Animals were washed off the plates with M9 buffer and then rinsed twice with DEPC-treated water. Total RNA was isolated using TRIzol Reagent with the Direct-zol RNA MiniPrep (Zymo Research). After RNA isolation, first-strand cDNA was synthesized from 1.0 μg of total RNA using dNTPs, Oligo(dT)12–18 and SuperScript II Reverse Transcriptase (Invitrogen). Quantitative PCR was done with an Applied Biosystems StepOne Plus Real-Time PCR system with Power SYBR Green PCR Master Mix (Applied Biosystems) per the manufacturer’s instructions, with triplicates done for each of three biological replicates, and actin-1 used as the endogenous control for data normalization. Sequences of primers can be found in SI Appendix, Table S2. Specificity of PCR amplification was determined by the melting curve for each reaction. The threshold cycle (CT) for each primer set was automatically determined by the StepOne Software 2.1 (Applied Biosystems). Relative fold changes of gene expression were calculated by using the 2−△△Ct method. The RQ values were then used to graph relative gene expression over at least three replicate experiments using GraphPad Prism 7.00 (GraphPad Software).
Healthspan: Track Length/Movement on Solid Media/Locomotion.
L4s grown on NGM at 20 °C were transferred to NGM plates containing 200 µM FUdR and incubated at 20 °C for 1, 5, 10, or 15 d. On the experimental day, 10 animals were picked onto unseeded NGM plates and each worm was recorded for 30 s using a mounted DMK 21AF04 Camera (The Imaging Source) outfitted onto a dissecting microscope. The average distance was calculated by measuring the tracks formed from each individual worm using WormLab software (MBF Bioscience) and then graphed using GraphPad Prism 7.00 (GraphPad Software).
Healthspan: Body Bends/Movement in Liquid Media.
L4s grown on NGM at 20 °C were transferred to plates containing 200 µM FUdR and incubated at 20 °C for 1, 5, 10, or 15 d. On the experimental day, individual animals were picked onto unseeded NGM plates, then 100 µL of M9 buffer was pipetted onto each animal. After 5 s, the number of body bends was recorded per 60 s using a mounted DMK 21AF04 camera (The Imaging Source) outfitted onto a dissecting microscope. At least 20 animals were recorded per strain or treatment and the average body bends per minute for each of the samples was calculated and graphed using GraphPad Prism 7.00 (GraphPad Software).
Healthspan: Resistance to Heat Stress.
Thirty L4s grown on NGM at 20 °C were transferred to four separate plates containing 200 µM FUdR and incubated at 20 °C for 1, 5, 10, or 15 d. On the experimental day, an individual plate was transferred to 37 °C and scored for survival every 2 h. Animals were touched with a platinum wire, and animals that did not respond to the stimulus were scored as dead. Two independent replicates of the experiment were performed and the percentage and mean survival were calculated using GraphPad Prism 7.00 (GraphPad Software).
Healthspan: Resistance to Oxidative Stress.
Thirty L4s grown on NGM at 20 °C were transferred to four separate plates containing 200 µM FUdR and incubated at 20 °C for 1, 5, 10, or 15 d. On the experimental day, animals were transferred to the paraquat plates at 20 °C and then scored daily for survival. Animals were touched with a platinum wire, and those that did not respond were scored as dead. Two independent replicates were performed and the percentage and mean survival were calculated using GraphPad Prism 7.00 (GraphPad Software). Paraquat plates were made by adding 1 mL of 250 mM paraquat solution to unseeded NGM plates. Then plates were put on a shaker for 1 h, followed by 1.5 h in a laminar flow hood to ensure plates were dry with the paraquat evenly distributed.
Fluorescence Quantification.
Approximately 30 HZ589 [sqst-1::GFP] animals were grown either on NGM plates supplemented with trehalose or RNAi plates at 20 °C from egg to day 3 adults. These animals were then picked onto glass slides with 2% agarose and immobilized with 100 mM NaN3. Fluorescence imaging of GFP was done using a Zeiss Axioskop 2 plus fitted with a Hamamatsu ORCA-ER camera. Quantification of the sqst-1::GFP positive puncta from the head of the animal to the start of the intestine was performed by three investigators independently.
Supplementary Material
Acknowledgments
We thank members of the H.A.T. laboratory; Drs. Scot Wolfe, Lucio Castilla, Nathan Lawson, Michael Brodsky, Cole Haynes, and David Greenstein for advice and suggestions; Nina Bhabhalia and Susan Lee for technical support; Drs. Mark Roth, Malene Hansen, Hong Zhang, and Eric Baehrecke for technical advice; and the reviewers for their time and effort as addressing their comments helped strengthen the manuscript. Some of the C. elegans strains were kindly provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). H.A.T. is a William Randolph Hearst Investigator. This project was funded in part by grants from the National Institute of Aging (AG025891) and the American Diabetes Association (1-17-IBS-176) (to H.A.T.) and an endowment from the William Randolph Hearst Foundation (to H.A.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714178115/-/DCSupplemental.
References
- 1.Bray GA, Popkin BM. Dietary sugar and body weight: Have we reached a crisis in the epidemic of obesity and diabetes? Health be damned! Pour on the sugar. Diabetes Care. 2014;37:950–956. doi: 10.2337/dc13-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, et al. Added sugar intake and cardiovascular diseases mortality among US adults. JAMA Intern Med. 2014;174:516–524. doi: 10.1001/jamainternmed.2013.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray GA. Energy and fructose from beverages sweetened with sugar or high-fructose corn syrup pose a health risk for some people. Adv Nutr. 2013;4:220–225. doi: 10.3945/an.112.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuker CS. Food for the brain. Cell. 2015;161:9–11. doi: 10.1016/j.cell.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Schlotterer A, et al. C. elegans as model for the study of high glucose- mediated life span reduction. Diabetes. 2009;58:2450–2456. doi: 10.2337/db09-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Garcia AM, et al. Glucose induces sensitivity to oxygen deprivation and modulates insulin/IGF-1 signaling and lipid biosynthesis in Caenorhabditis elegans. Genetics. 2015;200:167–184. doi: 10.1534/genetics.115.174631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salim C, Rajini PS. Glucose feeding during development aggravates the toxicity of the organophosphorus insecticide Monocrotophos in the nematode, Caenorhabditis elegans. Physiol Behav. 2014;131:142–148. doi: 10.1016/j.physbeh.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Salim C, Rajini PS. Glucose-rich diet aggravates monocrotophos-induced dopaminergic neuronal dysfunction in Caenorhabditis elegans. J Appl Toxicol. 2017;37:772–780. doi: 10.1002/jat.3426. [DOI] [PubMed] [Google Scholar]
- 11.Choi SS. High-glucose diets shorten lifespan of Caenorhabditis elegans via ectopic apoptosis induction. Nutr Res Pract. 2011;5:214–218. doi: 10.4162/nrp.2011.5.3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liggett MR, Hoy MJ, Mastroianni M, Mondoux MA. High-glucose diets have sex-specific effects on aging in C. elegans: Toxic to hermaphrodites but beneficial to males. Aging (Albany NY) 2015;7:383–388. doi: 10.18632/aging.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendler M, et al. daf-16/FOXO and glod-4/glyoxalase-1 are required for the life-prolonging effect of human insulin under high glucose conditions in Caenorhabditis elegans. Diabetologia. 2015;58:393–401. doi: 10.1007/s00125-014-3415-5. [DOI] [PubMed] [Google Scholar]
- 14.Aragno M, Mastrocola R. Dietary sugars and endogenous formation of advanced glycation endproducts: Emerging mechanisms of disease. Nutrients. 2017;9:E385. doi: 10.3390/nu9040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schalkwijk CG, Stehouwer CD, van Hinsbergh VW. Fructose-mediated non-enzymatic glycation: Sweet coupling or bad modification. Diabetes Metab Res Rev. 2004;20:369–382. doi: 10.1002/dmrr.488. [DOI] [PubMed] [Google Scholar]
- 16.Bollen M, Keppens S, Stalmans W. Specific features of glycogen metabolism in the liver. Biochem J. 1998;336:19–31. doi: 10.1042/bj3360019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer JC, et al. Control of glycogen deposition. FEBS Lett. 2003;546:127–132. doi: 10.1016/s0014-5793(03)00565-9. [DOI] [PubMed] [Google Scholar]
- 18.LaMacchia JC, Frazier HN, 3rd, Roth MB. Glycogen fuels survival during hyposmotic-anoxic stress in Caenorhabditis elegans. Genetics. 2015;201:65–74. doi: 10.1534/genetics.115.179416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazier HN, 3rd, Roth MB. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Curr Biol. 2009;19:859–863. doi: 10.1016/j.cub.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Possik E, et al. FLCN and AMPK confer resistance to hyperosmotic stress via remodeling of glycogen stores. PLoS Genet. 2015;11:e1005520. doi: 10.1371/journal.pgen.1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokal JE, Lowe CU, Sarcione EJ. Liver glycogen disease (von Gierke’s disease) Arch Intern Med. 1962;109:612–624. doi: 10.1001/archinte.1962.03620170110019. [DOI] [PubMed] [Google Scholar]
- 22.Chou JY, Mansfield BC. Mutations in the glucose-6-phosphatase-alpha (G6PC) gene that cause type Ia glycogen storage disease. Hum Mutat. 2008;29:921–930. doi: 10.1002/humu.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cori GT, Cori CF. Glucose-6-phosphatase of the liver in glycogen storage disease. J Biol Chem. 1952;199:661–667. [PubMed] [Google Scholar]
- 24.Dasouki M, et al. Pompe disease: Literature review and case series. Neurol Clin. 2014;32:751–776,ix. doi: 10.1016/j.ncl.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: Some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanover JA, et al. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda Y, Tanaka M, Honda S. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell. 2010;9:558–569. doi: 10.1111/j.1474-9726.2010.00582.x. [DOI] [PubMed] [Google Scholar]
- 28.Richards AB, et al. Trehalose: A review of properties, history of use, human tolerance, and results of multiple safety studies. Food Chem Toxicol. 2002;40:871–898. doi: 10.1016/s0278-6915(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 29.Mizote A, et al. Daily intake of trehalose is effective in the prevention of lifestyle-related diseases in individuals with risk factors for metabolic syndrome. J Nutr Sci Vitaminol (Tokyo) 2016;62:380–387. doi: 10.3177/jnsv.62.380. [DOI] [PubMed] [Google Scholar]
- 30.Portbury SD, Hare DJ, Finkelstein DI, Adlard PA. Trehalose improves traumatic brain injury-induced cognitive impairment. PLoS One. 2017;12:e0183683. doi: 10.1371/journal.pone.0183683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mondoux MA, et al. O-linked-N-acetylglucosamine cycling and insulin signaling are required for the glucose stress response in Caenorhabditis elegans. Genetics. 2011;188:369–382. doi: 10.1534/genetics.111.126490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenyon C. The first long-lived mutants: Discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 34.Oh SW, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 35.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 36.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 37.Tullet JM. DAF-16 target identification in C. elegans: Past, present and future. Biogerontology. 2015;16:221–234. doi: 10.1007/s10522-014-9527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuster E, et al. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen VL, Gallo M, Riddle DL. Targets of DAF-16 involved in Caenorhabditis elegans adult longevity and dauer formation. Exp Gerontol. 2006;41:922–927. doi: 10.1016/j.exger.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 40.Bansal A, Zhu LJ, Yen K, Tissenbaum HA. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc Natl Acad Sci USA. 2015;112:E277–E286. doi: 10.1073/pnas.1412192112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinkas A, Aschner M. Advanced glycation end-products and their receptors: Related pathologies, recent therapeutic strategies, and a potential model for future neurodegeneration studies. Chem Res Toxicol. 2016;29:707–714. doi: 10.1021/acs.chemrestox.6b00034. [DOI] [PubMed] [Google Scholar]
- 42.Thornalley PJ. Glyoxalase I: Structure, function and a critical role in the enzymatic defence against glycation. Biochem Soc Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 43.Young FG. Claude Bernard and the discovery of glycogen; a century of retrospect. BMJ. 1957;1:1431–1437. doi: 10.1136/bmj.1.5033.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gusarov I, et al. Glycogen controls Caenorhabditis elegans lifespan and resistance to oxidative stress. Nat Commun. 2017;8:15868. doi: 10.1038/ncomms15868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Valles R, et al. Life-long spontaneous exercise does not prolong lifespan but improves health span in mice. Longev Healthspan. 2013;2:14. doi: 10.1186/2046-2395-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer KE, et al. A cross-sectional study of male and female C57BL/6Nia mice suggests lifespan and healthspan are not necessarily correlated. Aging (Albany NY) 2016;8:2370–2391. doi: 10.18632/aging.101059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erkut C, et al. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol. 2011;21:1331–1336. doi: 10.1016/j.cub.2011.06.064. [DOI] [PubMed] [Google Scholar]
- 48.Pellerone FI, et al. Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes. Int J Parasitol. 2003;33:1195–1206. doi: 10.1016/s0020-7519(03)00173-5. [DOI] [PubMed] [Google Scholar]
- 49.Hibshman JD, et al. daf-16/FoxO promotes gluconeogenesis and trehalose synthesis during starvation to support survival. Elife. 2017;6:e30057. doi: 10.7554/eLife.30057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 52.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 53.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 54.Chen AT, et al. Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics. 2015;201:613–629. doi: 10.1534/genetics.115.177998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 56.Hars ES, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 57.Lapierre LR, Hansen M. Lessons from C. elegans: Signaling pathways for longevity. Trends Endocrinol Metab. 2012;23:637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tóth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 59.Morselli E, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meléndez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 62.Lapierre LR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai LL, Gao JX, Zou CG, Ma YC, Zhang KQ. mir-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS Pathog. 2015;11:e1004606. doi: 10.1371/journal.ppat.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 65.Wu YF, et al. High glucose alters tendon homeostasis through downregulation of the AMPK/Egr1 pathway. Sci Rep. 2017;7:44199. doi: 10.1038/srep44199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korntner S, et al. A high-glucose diet affects Achilles tendon healing in rats. Sci Rep. 2017;7:780. doi: 10.1038/s41598-017-00700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng J, et al. Lower doses of fructose extend lifespan in Caenorhabditis elegans. J Diet Suppl. 2017;14:264–277. doi: 10.1080/19390211.2016.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farooqui AA. Metabolic Syndrome: An Important Risk Factor for Stroke, Alzheimer Disease, and Depression. Springer Science+Business Media; New York: 2013. Glucose- and fructose-induced toxicity in the liver and brain. [Google Scholar]
- 69.Lee D, et al. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev. 2015;29:2490–2503. doi: 10.1101/gad.266304.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braeckman BB, Houthoofd K, Vanfleteren JR. Intermediary metabolism. WormBook. 2009 doi: 10.1895/wormbook.1.146.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Behm CA. The role of trehalose in the physiology of nematodes. Int J Parasitol. 1997;27:215–229. doi: 10.1016/s0020-7519(96)00151-8. [DOI] [PubMed] [Google Scholar]
- 72.Penkov S, et al. Integration of carbohydrate metabolism and redox state controls dauer larva formation in Caenorhabditis elegans. Nat Commun. 2015;6:8060. doi: 10.1038/ncomms9060. [DOI] [PubMed] [Google Scholar]
- 73.Bansal A, et al. Transcriptional regulation of Caenorhabditis elegans FOXO/DAF-16 modulates lifespan. Longev Healthspan. 2014;3:5. doi: 10.1186/2046-2395-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X, et al. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Sci Rep. 2016;6:28423. doi: 10.1038/srep28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeBosch BJ, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016;9:ra21. doi: 10.1126/scisignal.aac5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mardones P, Rubinsztein DC, Hetz C. Mystery solved: Trehalose kickstarts autophagy by blocking glucose transport. Sci Signal. 2016;9:fs2. doi: 10.1126/scisignal.aaf1937. [DOI] [PubMed] [Google Scholar]
- 77.Mayer AL, et al. SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy. Sci Rep. 2016;6:38586. doi: 10.1038/srep38586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: A multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 79.Kalač P. A review of chemical composition and nutritional value of wild-growing and cultivated mushrooms. J Sci Food Agric. 2013;93:209–218. doi: 10.1002/jsfa.5960. [DOI] [PubMed] [Google Scholar]
- 80.Stiernagle T. Maintenance of C. elegans. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 82.Miersch C, Döring F. Paternal dietary restriction affects progeny fat content in Caenorhabditis elegans. IUBMB Life. 2012;64:644–648. doi: 10.1002/iub.1042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.