Significance
Regulation of iron homeostasis is perturbed in numerous pathologic states. Thus, identifications of mechanisms responsible for iron metabolism have broad implications for disease modification. Here, we link the sulfur assimilation pathway to iron-deficiency anemia. Deletion of bisphosphate 3′-nucleotidase (Bpnt1), a key component of the sulfur assimilation pathway, leads to accumulation of phosphoadenosine phosphate (PAP), causing iron deficiency anemia in part due to inhibition of hypoxia-inducible factor 2-α. Reduction of PAP through introduction of a hypomorphic mutation in 3′-phosphoadenosine 5-phosphosulfate synthase 2 gene (Papss2, the enzyme responsible for PAP production) rescues the iron deficiency phenotype.
Keywords: Bpnt1, phosphoadenosine phosphate, nucleotidase, lithium, iron metabolism
Abstract
Sulfur assimilation is an evolutionarily conserved pathway that plays an essential role in cellular and metabolic processes, including sulfation, amino acid biosynthesis, and organismal development. We report that loss of a key enzymatic component of the pathway, bisphosphate 3′-nucleotidase (Bpnt1), in mice, both whole animal and intestine-specific, leads to iron-deficiency anemia. Analysis of mutant enterocytes demonstrates that modulation of their substrate 3′-phosphoadenosine 5′-phosphate (PAP) influences levels of key iron homeostasis factors involved in dietary iron reduction, import and transport, that in part mimic those reported for the loss of hypoxic-induced transcription factor, HIF-2α. Our studies define a genetic basis for iron-deficiency anemia, a molecular approach for rescuing loss of nucleotidase function, and an unanticipated link between nucleotide hydrolysis in the sulfur assimilation pathway and iron homeostasis.
Iron is a critical dietary micronutrient that serves as a cofactor for numerous metabolic reactions and is necessary for the production of red blood cells (RBCs) that transport oxygen throughout the body (1–3). Disease states result from an imbalance in systemic iron homeostasis including hereditary hemochromatosis, neurodegenerative disease, and anemia (1, 2). Hereditary hemochromatosis, the most common genetic cause of iron dysregulation in humans, results in tissue iron overload and eventual organ failure (4). In contrast, anemia due to chronic iron deficiency is a pervasive world-wide problem, affecting as many as two billion people (5). Discovery of mechanisms affecting regulation of iron are paramount to understanding these pathophysiologies.
Sulfur assimilation is the process of incorporating inorganic sulfate from the environment into sulfur-containing amino acids and sulfate-containing metabolites and is a feature shared among bacteria, yeast, plants, and mammals (6–8). Key regulators of metabolic flow in the sulfur assimilation pathway are the enzymes phosphoadenosine phosphosulfate synthetase (Papss2) and bisphosphate 3′-nucleotidase (Bpnt1) (Fig. 1A, Top); the latter is a member of a structurally related family of phosphatases (9). Mice and humans encode two such 3′-nucleotidases: Golgi-resident 3′-phosphoadenosine 5′-phosphate phosphatase (gPAPP) and the cytoplasmic Bpnt1. Loss of gPAPP activity in mice results in impaired chondroitin sulfation that underlies chondrodysplasia, pulmonary insufficiency, and bone joint defects (10), which are mimicked in human patients carrying recessive mutations (11). In contrast, animals deficient for Bpnt1 develop anasarca, hepatic insufficiency, and impaired ribosomal biogenesis (12), consistent with compartment-specific roles for sulfur assimilation and nucleotide hydrolysis in mammalian physiology. Here, we uncover a tissue-specific role for cytoplasmic nucleotidase Bpnt1 in iron homeostasis, define a genetic basis for iron deficiency anemia, and provide a molecular strategy that rescues loss of the nucleotidase.
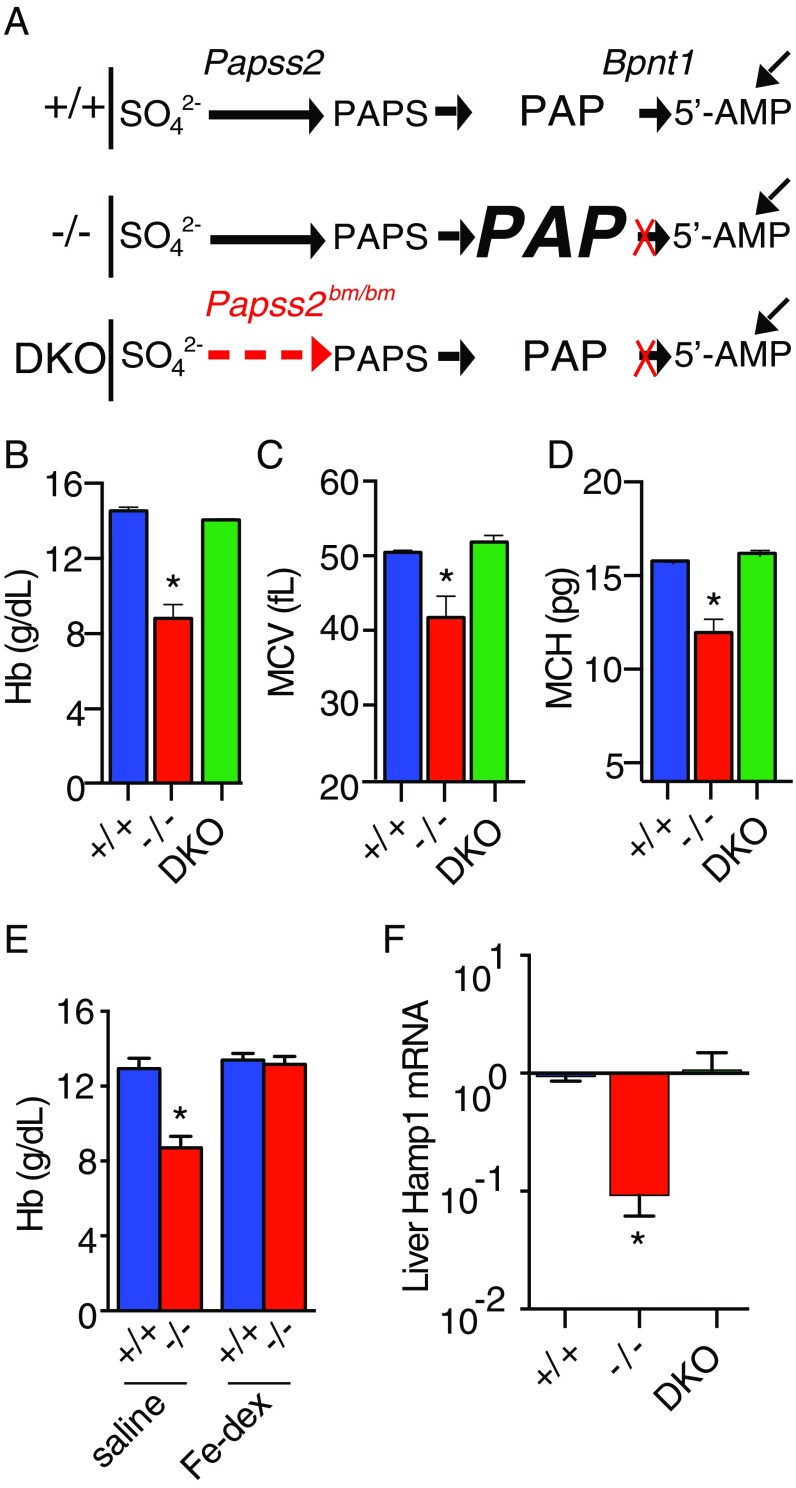
Fig. 1.
Bpnt1 global knockout mice develop PAP-dependent iron deficiency anemia. (A) Top schematic (+/+) shows abbreviated molecular components of the sulfur assimilation pathway and PAP metabolism by 3′-phosphoadenosine 5-phosphosulfate synthase 2 (Papss2) and Bpnt1 in normal cells. Horizontal arrows represent cellular reactions, while diagonal arrows represent contribution from the predominant sources of 5′-AMP independent of the sulfur assimilation pathway. Middle schematic (−/−) represents the pathway in Bpnt1-deficient cells (red “X”) with PAP, enlarged font and bolded, symbolizing up to 40-fold elevation of cellular concentrations, yet normal levels of other metabolites as reported in normal and mutant tissue (12). Bottom schematic (DKO) represents the double mutant in which tissue possess both a partial loss of PAPSS2 activity (dashed red arrow) and Bpnt1 deletion (red X) illustrating the metabolic reduction of PAPS synthesis and PAP levels, even in the context of 3′-nucleotidase loss. (B–D) Hematological parameters of 8-wk-old wild-type (+/+; blue), Bpnt1−/− knockouts (−/−; red), and Bpnt1−/−Papss2bm/bm double mutants (DKO; green) fed standard grain-based chow. (E) Hb levels of 1-y-old wild-type and knockout mice that received s.c. saline or Fe–dextran once a week for 3 wk before sacrifice at the end of the fourth week. (F) qRT-PCR of Hamp1 transcript relative to beta actin from total liver RNA of 8-wk-old mice. *P < 0.05.
Results
Our phenotypic analysis of Bpnt1-deficient mice revealed an age-dependent hair loss (alopecia), a clinical manifestation associated with iron deficiency anemia. Thus, we analyzed complete blood counts (CBCs) from wild-type and Bpnt1−/− mice. Bpnt1−/− animals had significantly lower hemoglobin (Hb), smaller mean corpuscular volumes (MCVs), and reduced average cellular hemoglobin (MCH), indicative of microcytic hypochromic anemia (Fig. 1 B–D). Microcytic anemia is frequently due to systemic iron deficiency. To probe whether the anemia observed in Bpnt1−/− animals was the result of inadequate iron stores or a primary defect in the hematopoietic system, we supplemented wild-type and Bpnt1−/− mice with s.c. Fe–dextran or saline once a week for 3 wk and assessed CBCs at the end of the fourth week. Bpnt1−/− mice that received Fe–dextran displayed normalized Hb levels (Fig. 1E), confirming that the anemia was due to a defect in iron homeostasis. Thus, global loss of Bpnt1 impairs the maintenance of sufficient bodily iron stores for normal hematopoiesis.
Our previous studies of 3′-nucleotidase mutant mice revealed two distinct mechanisms that contributed to observed phenotypes: The first involved the toxic accumulation of PAP substrate as depicted in the −/−-labeled pathway (Fig. 1A) (12), whereas the second was consistent with impaired sulfation through defective import of Golgi PAPS and/or failure to produce 5′AMP (10). To further probe if either mechanism explained the observed microcytic anemia, we generated a double-mutant mouse (Bpnt1−/− Papss2bm/bm, DKO) deficient for Bpnt1 and harboring a hypomorphic mutation in the phosphoadenosine phosphosulfate synthase, Papss2. We surmised according to the first model that a double mutant would rescue iron deficiency anemia because of concomitant down-regulation of PAP synthesis depicted by the DKO pathway (Fig. 1A), whereas if the second model were true, an exacerbation of the anemia would be expected. The double-mutant mice exhibited dramatically reduced PAP accumulation relative to Bpnt1 deficiency alone and indeed displayed no detectable hematological abnormalities (Fig. 1 B–D). We conclude Bpnt1 is a key regulator of PAP and aberrant accumulation of PAP results in toxic modulation of pathways relevant to the onset of microcytic anemia. Importantly, we also define a potential strategy to overcome iron deficiency anemia caused by loss of 3′-nucleotidase through concomitant inhibition of PAPS synthase.
Iron homeostasis is exquisitely maintained by a series of complex physiological pathways including hepatic production of the small peptide hormone Hepcidin (Hamp1). Hamp1 executes its function at the duodenum through binding the basolateral iron transporter ferroportin (Fpn), thereby regulating its trafficking and stability (13). Since impaired iron absorption can result from the pathological overproduction of Hamp1, as in anemia of inflammation (14), and because Bpnt1−/− mice display significant defects in hepatic function (12), we postulated that aberrant overproduction of Hamp1 might be responsible for the iron deficiency observed. Surprisingly, we found that relative to wild type, Bpnt1−/− livers had ∼10-fold lower levels of Hamp1 mRNA (Fig. 1F). These data are consistent with, but do not confirm, that the hepatic Hamp1 signaling axis responds normally to the iron-deficient state yet is insufficient to overcome the disease pathology. In addition, the Bpnt1−/− animals displayed heavier, pale, dilated, and abnormal nucleolar morphology in the intestine, suggesting a marked effect of PAP accumulation in the intestine (Fig. S1 A–D). Taken alongside the s.c. rescue by Fe–dextran (Fig. 1E), our data suggest the anemia may be a result of impaired iron absorption in the intestine.
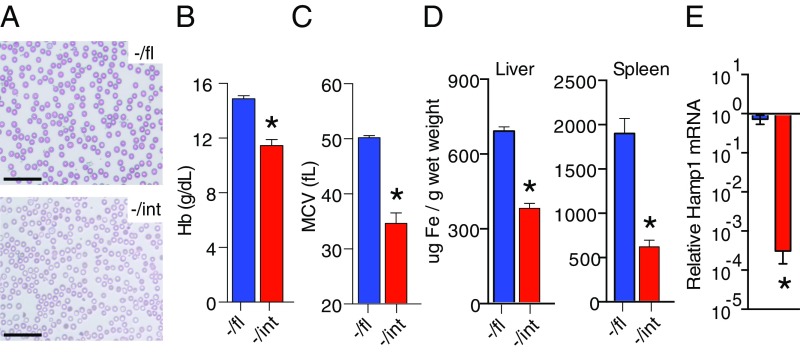
The aberrant intestinal morphology coupled with the hypothesis that Bpnt1 may specifically regulate iron absorption in enterocytes prompted the generation of tissue-specific Bpnt1 deficiency by crossing mice harboring a floxed allele of Bpnt1 with animals expressing Cre recombinase driven by the intestine-specific villin promoter (Fig. S2A). Intestine-specific loss was confirmed by immunoblotting for Bpnt1 and analyzing PAP levels in the small intestine (Fig. S2 B and C). Bpnt1−/int mice appeared normal at birth and display no detectable differences in weight gain following weaning (Fig. S3A). By 8 wk postnatal, Bpnt1−/int mice developed moderate alopecia similar to global knockouts (Fig. S3B). CBCs and blood smears of Bpnt1−/int animals revealed lower Hb levels and MCV as well as small hypochromic RBCs (Fig. 2 A–C). Bpnt1−/int mice also displayed aberrant intestinal morphology and nucleolar condensation similar to Bpnt1−/− mice (Fig. S3C). We also analyzed bodily iron stores and found that Bpnt1−/int mice have roughly 50% and 30% lower hepatic and splenic iron content, respectively (Fig. 2D). Consistent with the results from the global Bpnt1 mutant, we observed a dramatic down-regulation of Hamp1 transcript in livers derived from Bpnt1−/int animals (Fig. 2E). While the intestine-specific loss of Bpnt1 recapitulates the iron-deficiency anemia observed in the global Bpnt1 knockouts, we note that other phenotypes reported for the global Bpnt1 knockout, such as anasarca, have not been observed in the intestine-specific mutant at any age. Collectively, our data confirm that proper metabolic flow through the sulfur assimilation pathway in the small intestine is required for normal iron absorption and mice with impaired Bpnt1 function are predisposed to developing anemia despite maintaining an iron-sufficient diet, the latter of which has potential significance in identifying unmapped inherited forms of anemia.
Fig. 2.
Inactivation of Bpnt1 specifically in the small intestine results in iron-deficiency anemia. (A) Representative photograph of Wright-stained whole blood smear from Bpnt1−/fl and Bpnt1−/int showing microcytic hypochromic RBCs. (Scale bar: 25 μm.) (B and C) Hematological parameters of Bpnt1−/fl and Bpnt1−/int 8-wk-old mice demonstrating anemia (n = 6–8 per group). (D) Quantification of tissue iron stores in livers and spleens of 8-wk-old male Bpnt1−/fl and Bpnt1−/int mice. (E) qRT-PCR of Hamp1 mRNA relative to beta actin from total liver RNA in Bpnt1−/fl and Bpnt1−/int mice (n = 4). *P < 0.05.
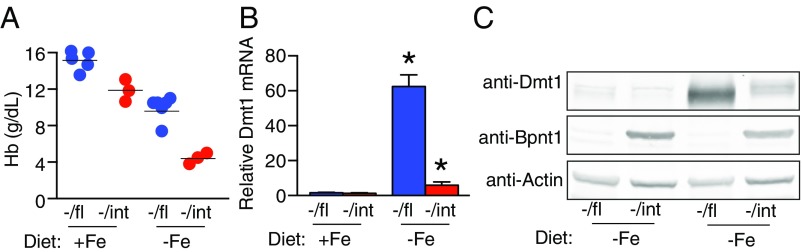
Because Bpnt1−/int mice appear unable to adequately modulate iron stores, we wondered whether Bpnt1-deficient enterocytes might also be susceptible to diet-induced iron starvation. To test this, mice were fed iron-deficient (2–6 ppm) or control (200 ppm) diets ad libitum for 5 wk from weaning, after which we measured CBCs. Bpnt1−/int animals displayed average Hb concentrations of 4.1 g/dL relative to 9.5 g/dL for Bpnt1−/fl controls (Fig. 3A). To understand why Bpnt1−/int mice were unable to respond to the low iron stress, we looked for defects in basolateral and apical iron transport. When basolateral transport is impaired, either through up-regulated Hamp1 or Fpn deficiency, intracellular iron accumulates in the duodenal enterocytes and is visible as ferritin granules by histological iron staining (15). Bpnt1−/int animals displayed no detectable increase in enterocyte iron stores, suggesting that defective basolateral transport was not primarily responsible for the iron deficiency (Fig. S3D). We next examined the functionality of iron import across the apical membrane by Dmt1. To test this, we analyzed the transcript and protein levels of Dmt1 from isolated duodenal enterocytes. Normally, when an organism is iron-starved, there is a profound compensatory response to induce Dmt1 expression, subsequently increasing iron absorption. Strikingly, while iron-depleted Bpnt1−/fl animals significantly up-regulated Dmt1, Bpnt1−/int enterocytes failed to increase Dmt1 transcript or protein levels (Fig. 3 B and C). Our data suggests that insufficient iron transport across the apical membrane by Dmt1 gives rise to the iron deficiency anemia in Bpnt1−/int mice.
Fig. 3.
Bpnt1−/int enterocytes have defects in apical iron transport. (A) Hb levels of mice on control or iron-deficient diets. Mice were fed a low iron (2 ppm) or control (200 ppm) diet for 5 wk after weaning (n = 3–5 per group). (B) qRT-PCR of Dmt1+IRE mRNA relative to actin in the duodena of control or iron starved mice (n = 3–5 per group). (C) Western blot analysis of Dmt1, Bpnt1, and actin loading control from isolated enterocytes of the same mice as in B. *P < 0.05.
Recently, a number of studies have implicated transcription factor hypoxia-inducible factor 2 alpha (HIF-2α) as a key mediator of the intestine’s response to iron deficiency (16–18). During iron starvation, degradation of HIF-2α is inhibited, leading to its accumulation and subsequent transcription of genes containing HIF response elements (HREs). Mice lacking intestinal HIF-2α display blunted responses to iron deficiency, including an inability to up-regulate Dmt1, and thus are unable to maintain sufficient serum iron and Hb levels. Because Dmt1 expression in Bpnt1−/int mice is not stimulated following iron deficiency, we surmised that HIF-2α or other genes under the control of HIF-2α might also be perturbed.
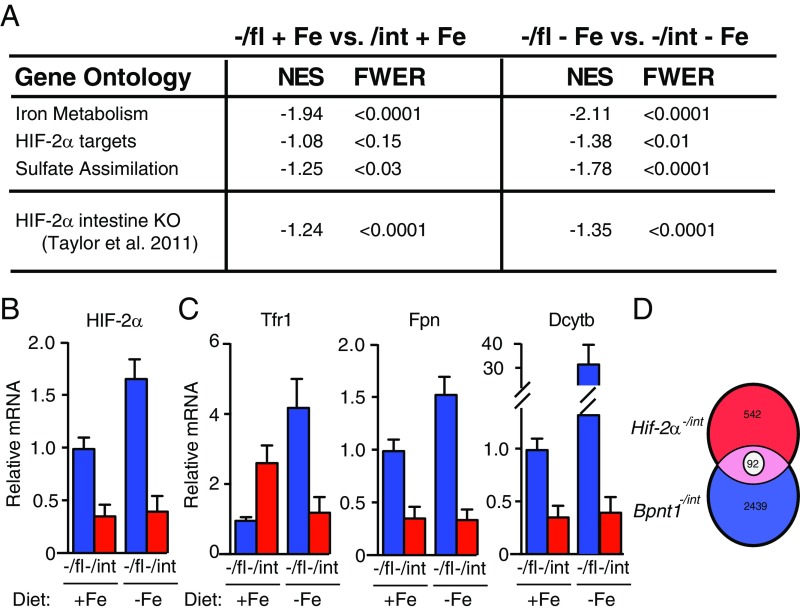
Thus, we performed RNA-sequencing (RNA-seq) on polyA-selected RNA isolated from enterocytes of Bpnt1−/fl and Bpnt1−/int mice fed either iron-replete or iron-deficient chow (Fig. S4A). Strikingly, the pathways and biological functions identified using this algorithm pointed to iron-related and hematologic parameters as functionally pertinent (Fig. S5B). Gene Set Enrichment Analysis (GSEA) was performed and revealed significant enrichment of iron metabolism-related genes, HIF-2α gene ontology signature, and sulfate assimilation gene ontology signatures (Fig. 4A). Of note, genes that are essential for proper iron homeostasis, import, and export including HIF-2α, transferrin receptor (Tfr1), Fpn, iron reductase (Dcytb), and Dmt1 (Figs. 3 B and C and 4A) were altered in Bpnt1−/int enterocytes, supporting the role of PAP accumulation in mediating control of iron metabolism at each step. Furthermore, only under conditions of iron deficiency does the HIF-2α–dependent transcriptional signature (19) become enriched (Fig. 4A and Fig. S4C). This suggests that an animal with a defect in the sulfur assimilation pathway may be genetically predisposed to develop iron deficiency anemia through aberrant HIF-2α signaling.
Fig. 4.
PAP accumulation induces transcriptional changes in iron metabolism-related genes, the HIF-2α pathway, and sulfur assimilation-related genes. (A) Enterocytes of Bpnt1−/fl vs. Bpnt1−/int mice fed either an iron-replete or iron-deficient diet for 5 wk (n = 3 per group) were isolated and subjected to polyA-selected RNA-seq. GSEA was performed on pertinent gene ontology signatures defined for targets of iron metabolism, the HIF-2α pathway, genes involved in sulfur assimilation, as well as sequencing data from HIF-2α−/int mice published by ref. 18. GSEA analysis comparing the Bpnt−/int and HIF-2α−/int transcriptional signatures was performed as follows. Genes that changed greater than threefold in HIF-2α−/int mice (542) and Bpnt1−/int mice (2,439) were compared using GSEA. NES, normalized enrichment score. Family wise error rate (FWER) < 0.05 was considered statistically significant. (B) qRT-PCR of HIF-2α mRNA relative to actin in the duodena of control or iron-starved mice (n = 3 per group). (C) RNA-seq reveals expression changes of key iron-regulatory genes. (D) Venn diagram showing unique and shared transcriptional signature of genes described in A. There were 92 genes whose expression was altered analogously in. HIF-2α−/int and Bpnt1−/int mice.
Both Bpnt1−/int and HIF-2α−/int animals are unable to stimulate Dmt1 production when fed iron-deficient chow (18). Thus, we surmised that PAP accumulation may recapitulate the molecular signature of a HIF-2α–deficient cell. We performed GSEA comparing the transcriptional signature of genes that changed more than threefold when Bpnt−/int and HIF-2α−/int animals were fed iron-deficient chow (Fig. 4A and Fig. S4C). We observed significant enrichment between the HIF-2α−/int genetic signature and Bpnt−/int mice that were fed either iron-rich or iron-deplete chow. We hypothesize this is likely due to repression of HIF-2α in Bpnt−/int enterocytes, independent of dietary iron content (Fig. 4B).
Of the total number of transcripts that were altered in HIF-2α−/int (542) and Bpnt−/int (2,439) animals, 92 genes were similarly altered between genotypes (Fig. 4C). Assessment of HIF-2α by immunohistochemistry in normal and mutant intestinal tissue confirmed that loss of Bpnt1 results in decreased HIF-2α protein levels (Fig. 5). We did not observe an appreciable difference in HIF-2α subcellular localization in Bpnt−/int intestine compared with wild type (Fig. 5A). As a control, we confirmed the specificity of the antibody signal observed by staining intestinal tissue from HIF-2α knockout tissue (Fig. 5). Furthermore, canonical targets of HIF-2α, but not other HIF family members, were largely repressed in Bpnt−/int enterocytes (Fig. S6). Intriguingly, Dmt1, Dcytb, and Fpn were all repressed in both HIF-2α−/int and Bpnt−/int animals. We suspect that PAP’s ability to impair Dmt1 transcription and translation (Fig. 3 B and C) is most important because there is no increase in iron stores in Bpnt−/int animals (Fig. S3D). However, modulation of transcriptional control of iron bioavailability at the level of reduction, import, nuclear transcriptional activity, and export represents exquisite control of iron metabolism at every key step. Collectively, these data are consistent with a role for Bpnt1 in the control of transcriptional responses to iron deficiency through modulation of HIF-2α–dependent and independent mechanisms.
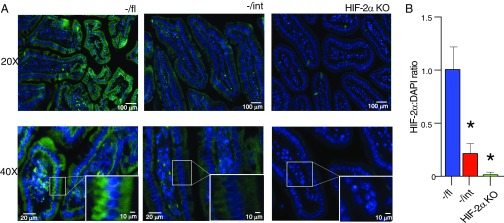
Fig. 5.
Bpnt1−/int enterocytes and small intestine display decreased HIF-2α and no difference in HIF-2α subcellular localization. (A) Representative images of immunofluorescence staining of proximal duodenum in −/fl WT control (Left), Bpnt1−/int (−/int, Center), and HIF-2α knockout (HIF-2α KO) for HIF-2α (green) and DAPI (blue) at 20× and 40× with magnified Inset. (B) Quantification of HIF-2α fluorescent signal relative to DAPI signal in WT, Bpnt1−/int, and HIF-2α KO (n = 3, 5 sections per animal) proximal duodenum sections as seen in B. *P < 0.05.
Discussion
This work uncovers an unanticipated link between an intestine-specific nucleotide hydrolysis in the sulfur assimilation pathway and iron homeostasis. Our data point to a mechanism by which inactivation of the 3′-nucleotidase Bpnt1 in intestinal tissue leads to a dramatic accumulation of its substrate PAP, thereby preventing the physiological response to low iron levels. Remarkably, by concomitantly reducing PAP synthesis in the context of nucleotidase deficiency, we are able to completely rescue the observed pathophysiology. This has profound implications in two areas: the first being a new complementation group for iron-deficiency anemia, and the second, albeit hypothetical at this point, is that if human patients are identified with loss-of-function mutations in Bpnt1, our work outlines a possible approach to overcome these specific genetic defects. Considering that shortly after publication of our work on a Golgi-localized nucleotidase gPAPP knockout mouse and associated chondrodysplasia (10) human patients were identified that had gPAPP mutations and similar joint defects and dwarfism observed in our mouse studies (11), it is reasonable to add Bpnt1 as a possible gene to sequence in patients with iron-deficiency anemia.
Mechanistically, we show that iron deficiency is likely due to a failure of Bpnt1-null mice to up-regulate Dmt1 at the apical enterocyte surface to promote iron uptake. In addition, we observed a striking parallel between phenotypic characterization of Bpnt1 and Hif-2α intestine knockout mice in that both animals are incapable of mounting a transcriptional response to iron starvation. Furthermore, Bpnt1 and Hif-2α intestine knockout mice display common transcriptional signatures—most notably decreased expression of Dmt1, Dcytb, Tfr1, and Fpn—thereby broadly regulating iron metabolism at all three essential steps in the metabolism of iron in the small intestine. However, the primary role of PAP appears to hinge on impairment of apical iron transport through Dmt1. These data suggest multiple potential functions of PAP in mediating iron metabolism.
Recent reports have demonstrated that the majority of the transcriptional response to iron starvation is mediated by Hif-2α (18). Our data link the sulfur assimilation pathway to iron homeostasis possibly through changes in Hif-2α levels. Loss of Bpnt1 in intestine tissue decreases both Hif-2α transcript and protein levels independent of iron levels in the diet. Of interest, the 70% reduction, rather than ablation, of Hif-2α protein levels in Bpnt1 mutant intestine tissue may explain why only a subset of Hif-2α transcriptional targets are altered—92 out of 542 based on RNA-seq data from published Hif-2α (Epas1−/int) intestine-specific knockout. That the transcripts for important iron transport machinery are among these 92 gene products elevates the significance of the link between nucleotidase activity and iron homeostasis through Hif-2α. Our data are consistent with a mechanism that toxic accumulation of a Bpnt1 substrate, such as PAP, directly or indirectly influences transcriptional (pre or post) and/or translational events (pre or post) that alter Hif-2α levels. Perhaps such alterations may induce an impairment in Hif-2α’s ability to bind promoter elements and activate downstream transcriptional programs in response to iron starvation. Alternatively, toxic PAP may alter Hif-2α function, for example through posttranslational modifications that alter conformation, translocation to the nucleus, stabilization, or binding partners. Given the broad differences observed in gene expression between WT and Bpnt−/int mice, this possible effect may reveal new genes involved in maintenance of iron homeostasis. However, further investigation is needed to address these possibilities.
How do we know it is toxic accumulation of PAP in knockout tissue that is responsible for the phenotypes observed, or could it be the failure to produce a product or some other metabolite? Bpnt1 has been shown to hydrolyze a variety of substrates including 3′-nucleotides and inositol bisphosphate. Biochemical characterization of Bpnt1 substrates indicates PAP conversion to 5′ AMP is kinetically favored compared with other nucleotides and exhibits 1,000-fold diminished activity as an inositol bisphosphate phosphatase (20). Additionally, analysis of metabolites from Bpnt1−/− mouse embryonic fibroblasts demonstrates that by far the highest fold accumulation occurs in PAP (40-fold) compared with more modest changes in PAPS (fivefold) and normal levels of ATP, ADP, or AMP (12). Our forward genetics approach confirms that concomitantly decreasing PAP synthesis (Papss2bm/bm) in the context of a Bpnt1 knockout (the double mutant is referred to as DKO) is sufficient to rescue the phenotypes, thereby eliminating “moonlighting” inositol phosphatase activity and failure to produce 5′ AMP as plausible mechanisms. The DKO strategy does not eliminate the possibility that some of the phenotypes may be due to toxic accumulation PAPS, rather than PAP. For example, if PAPS accumulation hyperactivates sulfotransferase (SULT) reactions, then loss of Papss2 activity would be predicted to diminish this effect by restoring normal SULT activity. Two important considerations disfavor this hypothesis: (i) Both metabolic and kinetic data we have generated indicate that Bpnt1 prefers PAP over PAPS as a substrate (21, 22), and (ii) PAP has been shown to inhibit SULTs (20). Thus, if Bpnt1 loss results in 40-fold accumulation of an inhibitor (PAP) and a fivefold increase in an activator (PAPS), then the net effect on SULTs would likely be inhibitory, in which case the DKO strategy would be predicted to exacerbate the effect, which it does not. It is also noteworthy to consider that a number of studies have demonstrated that the 5′–3′ exoribonucleases 1 and 2 (Xrn1/2) are targets of PAP toxicity in a variety of cell types and organisms (12, 21–24). Interestingly, in contrast to our previous study (12), in which the production of highly abundant hepatic proteins is attenuated, Dmt1 protein is normally expressed at relatively low levels and is only robustly expressed upon iron starvation (25). Thus, while high levels of PAP directly inhibit nuclear Xrn2 (26) and lead to ribosomal RNA processing defects in hepatocytes, it is unclear how this defect would impact protein levels in enterocytes. Delineation of the direct targets of PAP requires more extensive mechanistic inquiry.
In summary, we define a genetic basis and complementation group for iron-deficiency anemia and demonstrate a molecular genetic approach for overcoming the specific nucleotidase deficiency-induced pathophysiology. This work uncovers an unanticipated link between an intestine-specific nucleotide hydrolysis in the sulfur assimilation pathway and iron homeostasis. Our data point to a mechanism by which loss of Bpnt1 leads to massive elevation in PAP, which prevents the physiological response to low iron levels by perturbing transcriptional up-regulation of iron homeostasis factors, possibly through modulation of Hif-2α activity. Our studies also provide a set of tools to study other roles for nucleotidases and sulfur assimilation. Given that Bpnt1 is potently inhibited by subtherapeutic doses of lithium used for the treatment of bipolar disorder (21, 22), it is possible that lithium may be mediating its toxic and/or therapeutic effects through accumulation of PAP.
Materials and Methods
Bpnt1 floxed mice were generated using a standard homologous recombination approach (Fig. S2A). Briefly, we recombined the fourth and fifth exons of the Bpnt1 locus in 129/SvEv ES cells with a construct containing flanking LoxP sites and a neomycin resistance cassette. Cells resistant to neomycin were confirmed by PCR and Southern blotting, injected into blastocysts, and implanted into pseudopregnant females by the University of North Carolina Animal Models Core. Chimeric founders were identified by Southern blotting and PCR and crossed with B6.Cg-Tg(ACTFLPe)9205Dym/J mice (The Jackson Laboratory) expressing FLP recombinase to remove the neomycin cassette. Bpnt1+/fl mice were then backcrossed four generations into the C57BL/6J background, intercrossed, and maintained as Bpnt1fl/fl animals. To obtain intestine-specific knockouts, we first crossed B6.SJL-Tg(Vil-cre)997Gum/J mice (The Jackson Laboratory) expressing Cre recombinase under the control of the villin promoter with Bpnt1+/− animals to obtain Bpnt1+/−Vil-Cre+ double heterozygotes. These animals were then crossed to Bpnt1fl/fl mice to generate Bpnt1+/fl, Bpnt1−/fl, Bpnt1+/int, and Bpnt1−/int mice. Wild-type and conventional knockout Bpnt1 alleles were genotyped by multiplex PCR using the following primers: (i) 5′-cctatagtcctagcacttgagagg-3′, (ii) 5′-accaaagaacggagccggttggcg-3′, and (iii) 5′-aggtcggaaccctgttctctagtc-3′. Floxed Bpnt1 alleles were genotyped by PCR using the following primers: (i) 5′-cttgtggtttgggttgaccccttag-3′ and (ii) 5′-ctctagcccagtcagacatgtcag-3′. Villin-Cre expression was determined by PCR using the following primers: (i) 5′-gcggtctggcagtaaaaactatc-3′ and (ii) 5′-gtgaaacagcattgctgtcactt-3′. All animals unless otherwise noted were maintained on Purina 5058 natural products chow. Animals receiving supplemental iron were injected into the scruff of the neck weekly for a total of 3 wk with 5 mg of sterile Fe–dextran (Sigma). After 3 wk, mice were killed and analyzed as described above. Animals challenged with iron-deficient diets were maintained on normal Purina 5058 chow until the time of weaning (P23), at which point they were given either iron-deficient AIN-93G–defined chow with 2–6 ppm total iron or an identical AIN-93G supplemented with 200 ppm iron(II) sulfate (Harlan-Teklad TD.120105 and TD.120106, respectively). Mice were killed after 5 wk of dietary treatment and analyzed for hematological parameters as described above. Animal care and experiments were performed in accordance with the National Institutes of Health guidelines and approved by the Duke University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank P. Matak and T. B. De Renshaw (Duke University) for technical assistance; P. Gros (McGill University) for the generous gift of Dmt1 antibody; and members of J.D.Y. laboratory and J. Hudson for helpful discussions, critical reading of the manuscript, and constructive comments. This work was supported by funds from the Vanderbilt Medical Scientist Training Program (5T32GM007347, to A.T.H.), Howard Hughes Medical Institute (J.D.Y. is an alumni Investigator), and Vanderbilt University (J.D.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715302115/-/DCSupplemental.
References
- 1.Andrews NC. Forging a field: The golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: Regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Knutson MD. Iron-sensing proteins that regulate hepcidin and enteric iron absorption. Annu Rev Nutr. 2010;30:149–171. doi: 10.1146/annurev.nutr.012809.104801. [DOI] [PubMed] [Google Scholar]
- 4.Siddique A, Kowdley KV. Review article: The iron overload syndromes. Aliment Pharmacol Ther. 2012;35:876–893. doi: 10.1111/j.1365-2036.2012.05051.x. [DOI] [PubMed] [Google Scholar]
- 5.Cameron BM, Neufeld LM. Estimating the prevalence of iron deficiency in the first two years of life: Technical and measurement issues. Nutr Rev. 2011;69:S49–S56. doi: 10.1111/j.1753-4887.2011.00433.x. [DOI] [PubMed] [Google Scholar]
- 6.Hudson BH, York JD. Roles for nucleotide phosphatases in sulfate assimilation and skeletal disease. Adv Biol Regul. 2012;52:229–238. doi: 10.1016/j.advenzreg.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopriva S, et al. Editorial: Frontiers of sulfur metabolism in plant growth, development, and stress response. Front Plant Sci. 2016;6:1220. doi: 10.3389/fpls.2015.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 9.York JD, Ponder JW, Majerus PW. Definition of a metal-dependent/Li(+)-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc Natl Acad Sci USA. 1995;92:5149–5153. doi: 10.1073/pnas.92.11.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frederick JP, et al. A role for a lithium-inhibited Golgi nucleotidase in skeletal development and sulfation. Proc Natl Acad Sci USA. 2008;105:11605–11612. doi: 10.1073/pnas.0801182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vissers LE, et al. Chondrodysplasia and abnormal joint development associated with mutations in IMPAD1, encoding the Golgi-resident nucleotide phosphatase, gPAPP. Am J Hum Genet. 2011;88:608–615. doi: 10.1016/j.ajhg.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson BH, et al. Role for cytoplasmic nucleotide hydrolysis in hepatic function and protein synthesis. Proc Natl Acad Sci USA. 2013;110:5040–5045. doi: 10.1073/pnas.1205001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemeth E, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas G, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donovan A, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Anderson SA, et al. The IRP1-HIF-2α axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab. 2013;17:282–290. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mastrogiannaki M, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor M, et al. Hypoxia-inducible factor-2α mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology. 2011;140:2044–2055. doi: 10.1053/j.gastro.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer CF, et al. PID: The pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rens-Domiano SS, Roth JA. Inhibition of M and P phenol sulfotransferase by analogues of 3′-phosphoadenosine-5′-phosphosulfate. J Neurochem. 1987;48:1411–1415. doi: 10.1111/j.1471-4159.1987.tb05679.x. [DOI] [PubMed] [Google Scholar]
- 21.Spiegelberg BD, Dela Cruz J, Law TH, York JD. Alteration of lithium pharmacology through manipulation of phosphoadenosine phosphate metabolism. J Biol Chem. 2005;280:5400–5405. doi: 10.1074/jbc.M407890200. [DOI] [PubMed] [Google Scholar]
- 22.Spiegelberg BD, Xiong JP, Smith JJ, Gu RF, York JD. Cloning and characterization of a mammalian lithium-sensitive bisphosphate 3′-nucleotidase inhibited by inositol 1,4-bisphosphate. J Biol Chem. 1999;274:13619–13628. doi: 10.1074/jbc.274.19.13619. [DOI] [PubMed] [Google Scholar]
- 23.López-Coronado JM, Bellés JM, Lesage F, Serrano R, Rodríguez PL. A novel mammalian lithium-sensitive enzyme with a dual enzymatic activity, 3′-phosphoadenosine 5′-phosphate phosphatase and inositol-polyphosphate 1-phosphatase. J Biol Chem. 1999;274:16034–16039. doi: 10.1074/jbc.274.23.16034. [DOI] [PubMed] [Google Scholar]
- 24.Miki TS, Carl SH, Stadler MB, Großhans H. XRN2 autoregulation and control of polycistronic gene expresssion in Caenorhabditis elegans. PLoS Genet. 2016;12:e1006313. doi: 10.1371/journal.pgen.1006313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunshin H, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 26.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.