Significance
Our physical senses are separated not only into distinct experiences but also into specialized regions within the cerebral cortex. Synesthesia is a neurological phenomenon that causes unusual links between sensory experiences, and its molecular basis is completely unknown. We demonstrate that three families who experience color when listening to sounds are connected by rare genetic variants affecting genes that contribute to axonogenesis, a process essential for neuronal connections within and across brain regions. Multiple genes with similar activity patterns during neural development fall within parts of the genome previously linked to the condition. Our results connect synesthetes’ altered structural and functional connectivity to genes that support the development of those connections.
Keywords: synaesthesia, synesthesia, perception, axonogenesis, whole-exome sequencing
Abstract
Synesthesia is a rare nonpathological phenomenon where stimulation of one sense automatically provokes a secondary perception in another. Hypothesized to result from differences in cortical wiring during development, synesthetes show atypical structural and functional neural connectivity, but the underlying molecular mechanisms are unknown. The trait also appears to be more common among people with autism spectrum disorder and savant abilities. Previous linkage studies searching for shared loci of large effect size across multiple families have had limited success. To address the critical lack of candidate genes, we applied whole-exome sequencing to three families with sound–color (auditory–visual) synesthesia affecting multiple relatives across three or more generations. We identified rare genetic variants that fully cosegregate with synesthesia in each family, uncovering 37 genes of interest. Consistent with reports indicating genetic heterogeneity, no variants were shared across families. Gene ontology analyses highlighted six genes—COL4A1, ITGA2, MYO10, ROBO3, SLC9A6, and SLIT2—associated with axonogenesis and expressed during early childhood when synesthetic associations are formed. These results are consistent with neuroimaging-based hypotheses about the role of hyperconnectivity in the etiology of synesthesia and offer a potential entry point into the neurobiology that organizes our sensory experiences.
There are natural individual differences in the tuning of our senses. Apart from changes related to aging or disease, people vary in their sensory experience of the world. This variation is, in part, mediated by genetic factors, as demonstrated by the heritability of pain sensitivity and the well-replicated influence of polymorphisms in taste receptor genes on perception of bitter flavors (1–3). Although we have learned much from model organisms about the molecules and signaling pathways orchestrating the organization of the neural cortex, their human counterparts and the consequences of genetic variation are less well understood (4).
The history of neuroscience is punctuated by case studies of rare phenomena that advanced our knowledge of neural systems. The neurological phenomenon of synesthesia provides such a window into the separation of our senses. In synesthesia, stimulation of one sense simultaneously yields an automatic and consistent perception in another sense (5, 6). While it appears that any two bits of sensory data can be linked, synesthesia often involves color associations (e.g., “Wednesday is yellow”) (7, 8). Indeed, prevalence studies show that color is the most common secondary experience, induced by sequences such as letters of the alphabet or numbers in about one percent of the population (7). Thus, synesthesia represents a compelling model for studying variation in sensory perception occurring outside neurological conditions such as schizophrenia or autism, which have a more complex set of features.
Investigations of the neural correlates of synesthetic experiences revealed increased structural and functional connectivity in people with synesthesia compared with nonsynesthetes and differences in functional connectivity when exposed to triggering stimuli (9–12). These results contributed to a major hypothesis in synesthesia research: That such stable, cross-modal sensory experiences arise from alterations to the neural connections between brain regions that process the entwined sensory signals (13, 14). Longitudinal studies support a developmental basis for synesthesia, as the number and strength of these sensory links grows during early childhood (15). Importantly, synesthesia mainly occurs in individuals who are otherwise neurotypical. It has been argued that synesthesia may result from a breakdown in the neural “modularity” that each sense typically develops (16). An alternative explanation emphasizes how reduced inhibition of specialized cortical regions by control areas could also permit sensory spillover (17).
Synesthesia clusters in families. Indeed, detailed accounts of families with multiple relatives affected with synesthesia date to the late 1800s (18). In the past decade, linkage mapping of such multiplex families indicated that synesthesia is genetically heterogeneous and unlikely to be explained by a single genetic locus (19–21). These initial studies highlighted chromosomal regions of potential interest, but the supporting evidence was largely suggestive, and the regions span several hundred kilobases, containing hundreds of genes. Targeted sequencing of hypothesis-driven candidate genes from these regions failed to detect rare variants segregating with the phenotype (19). Our understanding of the developmental processes that define the boundaries between our senses is impeded by our lack of knowledge about the genetic underpinnings.
We performed a genome-wide sequence-level analysis of synesthesia to begin addressing the major neurobiological hypotheses in this field from a genetic perspective. We focused on three unrelated families, ascertained on the basis of at least five members affected with sound–color synesthesia across three or more generations, validated through an established test battery (22). Using whole-exome sequencing (WES), we identified rare coding variants that perfectly segregate with the phenomenon within each family. Variants preferentially fell within genes tied to the processes of axonogenesis and cell migration, forming a common theme across families, one that aligns with contemporary theories about the neurodevelopmental origins of synesthesia. These results suggest that molecular approaches can help increase understanding of the neurobiology of our sensory experiences, beyond pathology.
Results
We used WES to identify genetic variants within coding parts of the genome for three multigenerational families with sound–color synesthesia (Fig. 1). For each family, we obtained exome sequences from four or five verified synesthetes across three generations plus at least one nonsynesthetic family member. All cases in this study had sound–color synesthesia as confirmed using the established Test of Genuineness (SI Materials and Methods), although it is possible that some also experience additional, more common types of synesthesia (e.g., colored weekdays). The families were identified from the Cambridge Synesthesia Research Group database and were originally included in a 2009 study using microsatellite markers to look for evidence of genetic linkage across 43 families of varying sizes with forms of auditory–visual synesthesia (20). The families in the present study were chosen as the most feasible for studying with WES based on size, structure, and availability of suitable DNA samples.
Fig. 1.
Sound–color synesthesia in three multiplex families from the Cambridge Synaesthesia Research Group. (A) Pedigrees of the families. Circles indicate females, squares refer to males, and gray shading indicates synesthesia. Blue outlines show which members underwent WES. (B) An illustration of sound–color matching over three trials (colored boxes) for three hypothetical individuals presented with two auditory stimuli. A synesthete (boxes on the left) would show high consistency across trials, while a nonsynesthete (boxes on the right) would be inconsistent in their color choices.
Using nonparametric linkage analysis and assuming genetic heterogeneity, the authors of the 2009 study found three regions with suggestive linkage (5q33, 6p12, and 12p12) (23). Chromosome 2q24 showed significant linkage after gene-drop simulations (heterogeneity logarithm of the odds score, HLOD 3.02, P value 0.047) (20). An independent investigation of five other families with sequence–color synesthesia (color associations for sequences such as numbers or months) reported suggestive evidence for a locus on chromosome 16q, but this was supported by only two of the five families (19). These studies suggest that synesthesia involves considerable genetic heterogeneity, with different genetic factors contributing in different families.
Following the Genome Analysis Toolkit’s best practices guidelines for calling DNA variants, we identified 11,597 variants across our three sound–color families after removing low-quality variants and those with low sequencing depth (family 2: 8,195; family 11: 9,202; and family 16: 8,074) (24). To elevate potentially causative variants, we applied filtration criteria based on our limited knowledge of synesthesia’s genetic architecture and the prevalence of the sound–color variety (familial or sporadic). A 2006 study established that up to 4.4% of the UK population may experience at least one form of synesthesia, but the prevalence of sound–color synesthesia is not well studied (7, 23, 25). Estimates range from 1 in 500 unselected individuals in the United Kingdom (from the same prevalence study) to 41% of self-referred Dutch and German synesthetes (7, 25). Given the uncertainty in these estimates, we chose a relatively inclusive maximum minor allele frequency (MAF) of 0.01 for highlighting variants of potential interest. In total, there were 3,864 variants across the families that were rare enough to be considered further (family 2: 1,812; family 11: 2,727; family 16: 1,862; note that, prior to the further filtering described below, these included some partially overlapping variants across families).
Based on pedigree structures of the three families, we next retained variants that followed dominant inheritance with full penetrance (Fig. 1A). Given the findings of prior genetic screens, we hypothesized that while familial sound–color synesthesia may be due to single variants within a particular family, such variants were unlikely to be shared across different families (20). To test this, we looked in our retained variants for rare, heterozygous mutations that perfectly segregated with synesthesia within each separate family. This yielded a total of 37 variants across 37 genes (Table 1). No variants were shared across all synesthetes in all families, consistent with the hypothesis of genetic heterogeneity. A missense variant in RGS21 was detected in all synesthetes from family 16 but was found in only one synesthete from family 2. Further supporting genetic heterogeneity in synesthesia, no single gene contained a perfectly segregating variant in all three families. None of the 37 highlighted variants fell within the suggestive linkage peaks reported in prior studies (19, 20).
Table 1.
Rare variants segregating with sound–color synesthesia within each family
| Family | Position | Ref/Alt | Gene | ExAC MAF | Impact | CADD | Eigen score | PolyPhen | SIFT |
| 11 | 1:1231208 | C/T | ACAP3 | 0.002 | Missense | 18.65 | −0.59 | Benign | Deleterious |
| 2 | 1:146696486 | C/G | FMO5 | 0 | Splice donor | 27.50 | 1.18 | NA | NA |
| 2 | 1:151665566 | T/C | SNX27 | 0.005 | Regulatory | 1.22 | 0.42 | NA | NA |
| 2 | 1:154574699 | T/G | ADAR | 0 | Missense | 24.80 | 0.14 | Possibly damaging | Deleterious |
| 2 | 1:156899248 | G/A | LRRC71 | 0.007 | Intron | 5.29 | 0.29 | NA | NA |
| 2 | 1:162760613 | C/G | HSD17B7 | 0 | Missense | 29.00 | 0.80 | Probably damaging | Deleterious |
| 2 | 1:162825523 | AGAG/A | C1orf110 | 0 | Regulatory | 14.18 | NA | NA | NA |
| 2,16 | 1:192321228 | A/G | RGS21 | 0 | Missense | 25.30 | 0.98 | Probably damaging | Deleterious |
| 16 | 1:205899176 | C/A | SLC26A9 | 0 | Synonymous | 11.48 | 1.43 | NA | NA |
| 16 | 3:10280460 | A/G | IRAK2 | 0.002 | Missense | 17.57 | −0.21 | Benign | Tolerated |
| 11 | 4:20619178 | C/T | SLIT2* | 0.005 | Missense | 22.50 | −0.56 | Benign | Tolerated |
| 11 | 4:25667731 | G/A | SLC34A2 | NA | Intron | 1.73 | −0.037 | NA | NA |
| 2 | 4:72620719 | G/A | GC | NA | Synonymous | 0.39 | −0.058 | NA | NA |
| 16 | 4:155511967 | G/C | FGA | NA | Regulatory | 0.94 | 0.65 | NA | NA |
| 16 | 5:1335280 | G/A | CLPTM1L | 0.003 | Missense | 28.60 | 0.23 | Possibly damaging | Deleterious |
| 16 | 5:16671591 | G/A | MYO10* | 0.006 | Synonymous | 13.99 | 1.56 | NA | NA |
| 2 | 5:44811207 | T/G | MRPS30 | 0.003 | Missense | 28.10 | 0.73 | Probably damaging | Deleterious |
| 2 | 5:52347346 | G/A | ITGA2* | NA | Missense | 24.60 | 0.46 | Probably damaging | Tolerated |
| 2 | 5:64492917 | A/G | ADAMTS6 | 0 | Synonymous | 3.33 | 1.50 | NA | NA |
| 2 | 7:140386681 | G/A | ADCK2 | NA | Upstream | 0.78 | −0.33 | NA | NA |
| 11 | 9:139342231 | A/G | SEC16A | NA | Intron | 1.72 | −0.054 | NA | NA |
| 16 | 10:49937707 | C/A | WDFY4 | NA | Intron | 10.42 | 0.16 | NA | NA |
| 11 | 11:124747839 | G/T | ROBO3* | 0.003 | Missense | 27.90 | 0.23 | Probably damaging | Deleterious |
| 11 | 12:30834682 | G/A | IPO8 | NA | Synonymous | 15.41 | 0.72 | NA | NA |
| 11 | 12:49952781 | G/A | MCRS1 | 0.01 | Downstream | 4.74 | 0.15 | NA | NA |
| 2 | 13:47263384 | A/G | LRCH1 | NA | Intron | 3.22 | −0.12 | NA | NA |
| 2 | 13:49710555 | G/A | FNDC3A | 0 | Missense | 33.00 | 0.72 | Probably damaging | Deleterious |
| 11 | 13:110866346 | G/A | COL4A1* | 0.004 | Missense | 26.50 | 0.72 | NA | Deleterious |
| 16 | 14:23848194 | C/T | CMTM5 | 0 | Downstream | 7.20 | 0.43 | NA | NA |
| 2 | 16:71101385 | G/A | HYDIN | NA | Intron | 8.53 | 0.28 | NA | NA |
| 2 | 17:71354180 | T/C | SDK2 | 0 | Intron | 1.60 | −0.16 | NA | NA |
| 2 | 17:73832045 | G/A | UNC13D | 0 | Intron | 0.28 | −0.079 | NA | NA |
| 11 | 17:80708522 | A/C | FN3K | 0.001 | Missense | 15.58 | −0.49 | Benign | Tolerated |
| 11 | 19:44513250 | C/T | ZNF230 | 0.007 | Missense | 0.73 | −1.16 | Benign | Tolerated |
| 11 | 20:62867995 | C/T | MYT1 | 0 | Synonymous | 11.20 | 1.14 | NA | NA |
| 2 | 22:17671193 | G/A | CECR1 | NA | Upstream | 0.34 | −0.39 | NA | NA |
| 11 | X:135126891 | A/T | SLC9A6* | 0.003 | 3′ UTR | 6.21 | NA | NA | NA |
ExAC, Exome Aggregation Consortium; PolyPhen, polymorphism phenotyping; Ref/Alt, reference/alternative; SIFT, sorting intolerant from tolerant.
Genes associated with axonogenesis (GO:0007409) and cell migration (GO:0016477), biological categories with relevance to the hyperconnectivity account of synesthesia.
To further define shared mechanisms across the three families, we assessed whether there were enriched gene ontology terms within our set of putative candidate genes from WES. We excluded inferred electronic annotations, as these are considered less informative than manually curated entries. The ontology analysis identified significant enrichment for a narrow set of terms primarily related to neural development (Table 2). Notably, six genes were associated with axonogenesis (GO:0007409) and cell migration (GO:0016477), biological categories with clear relevance to the hyperconnectivity account of synesthesia. These genes, SLIT2, MYO10, ROBO3, ITGA2, COL4A1, and SLC9A6 (marked by an asterisk in Table 1), span the three families and may point to particular processes that can be investigated at higher levels (e.g., hyperconnectivity).
Table 2.
Gene ontology terms enriched in the combined set of synesthesia-associated variants
| Gene ontology term | Corrected P value | Terms | Overlap | Intersecting genes |
| BP: axonogenesis | 0.0459 | 605 | 6 | SLIT2, MYO10, ROBO3, ITGA2, COL4A1, SLC9A6 |
| BP: substrate-dependent cell migration | 0.0252 | 18 | 2 | SLIT2, ITGA2 |
| BP: cell morphogenesis involved in differentiation | 0.0345 | 799 | 7 | SLIT2, MYO10, ROBO3, ITGA2, FGA, COL4A1, SLC9A6 |
| MF: proteoglycan binding | 0.0225 | 17 | 2 | CECR1, SLIT2 |
| MF: hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines | 0.0452 | 24 | 2 | CECR1, SLIT2 |
| MF: deaminase activity | 0.0490 | 25 | 2 | CECR1, SLIT2 |
BP, biological process; MF, molecular function.
Having identified variants in a core set of six candidate genes with potential functional relevance for neurobiological accounts of synesthesia etiology, we went on to study their expression in neural tissues. We used two complementary datasets, the Genotype-Tissue Expression project (GTEx) based on RNA-sequencing (RNAseq) and the microarray-based Allen Human Brain Atlas (ABA), to determine expression patterns of the candidates (26, 27). GTEx includes data from >100 postmortem human brains with RNA sampled from 13 sites, including the frontal cortex (26). Each of the six genes had detectable expression in these samples, with SLC9A6 and ITGA2 having the highest and lowest expression, respectively (Table S1).
As protein levels are not always well correlated with RNA expression (28), we combined these findings with immunohistochemistry data from the Human Protein Atlas, which includes manually quantified protein expression from human cerebral cortical tissue of three adult donor brains (age range, 37–70 y) (29). The Atlas included results for each of the relevant proteins, except ROBO3. All five remaining proteins were observed in neuronal cells, albeit to varying degrees (Table S1 shows ranges when multiple antibodies produced different results). Beyond neurons, multiple candidates showed staining in neuropil and endothelial cells, while only SLIT2 was observed at high levels in glia. These results support the GTEx RNAseq data, indicating that the six genes are active in adult brain tissue.
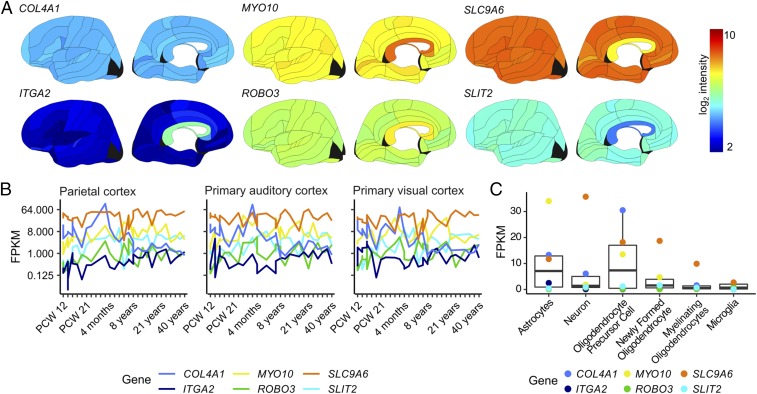
The GTEx and Human Protein Atlas resources lack data from several cortical regions with relevance for synesthesia studies, and the tissues were primarily collected from middle-aged adults. Thus, we next sought to determine if the highlighted genes are active in the auditory, visual, and parietal cortices and to examine their expression patterns during development. Reports vary on the neuroanatomical regions and activity patterns that may mediate synesthetic experiences; some support a role for the visual cortex, and others emphasize sensory integration in the parietal lobe (11, 14, 30). We used microarray data from six postmortem brains, sampled at ∼500 locations, with the data mapped to structural MRI scans (ABA), to visualize gene expression in a more fine-grained fashion (27). Despite the differing methodologies, expression values from the GTEx cortical samples were well correlated with frontal cortical data from the ABA (Pearson’s, r = 0.95, P = 0.004). In the ABA, we found that each of the six genes was widely expressed across the brain (Fig. 2A), including the auditory, visual, and parietal cortices.
Fig. 2.
Six synesthesia candidate genes are widely expressed across neural development. (A) Expression data from the ABA, mapped to a unified anatomic framework (27). (B) Gene expression in human parietal, primary auditory, and primary visual cortices from 12 wk post conception to age 40 y, from the BrainSpan atlas (one to three independent measurements per gene, region, and time point); the y axis uses a log2 scale to visualize change over time for each gene. (C) Neural gene expression in specific cell types isolated from adult mice. Boxes represent the 25th to 75th percentiles; whiskers extend to 1.5 times the interquartile range. Data used in C from the Barres laboratory are available at https://web.stanford.edu/group/barres_lab/brain_rnaseq.html. FPKM, fragments per kilobase of transcript per million mapped reads.
Longitudinal studies show that synesthetic associations often form during primary school years, and axonogenesis primarily occurs from early fetal development through the first years of life, prompting us to examine expression of our candidate genes across human brain development (15). To do this, we used data from the Allen Institute’s BrainSpan project, which includes human brain tissue from age 8 wk post conception to 40 y, sampling an average of 13 regions (range, 1–17) from one to three brains per time point, measured using RNAseq (31). Each of the six genes of interest has detectable expression in auditory, visual, and parietal cortices during fetal development and early childhood (Fig. 2B). Finally, to determine whether the six candidates were preferentially expressed in any specific neural cell types, we used an RNAseq dataset of six cell types isolated from the mouse cortex with a series of antibody-based purification steps to ensure high cell-type specificity (32). The candidate genes were broadly expressed across the neural and glial transcriptomes, with relatively lower expression levels in microglia (Fig. 2C).
Previous synesthesia genetics studies not only faced limitations of small family sizes but also were restricted by the use of lower-precision methods than those adopted here. We thus sought to determine whether the results of the current study might be relevant for those earlier reports. Coexpression patterns can reveal genes involved in similar neurodevelopmental processes, and so we tested whether genes coexpressed with the six candidates fell within the putative linkage peaks of prior studies, while acknowledging that most evidence from previous reports was at the suggestive level (33). The peaks reported by Asher et al. (20) were unlikely to be a result of linkage within the three families used in the present study, as we found no segregating variants within those regions (Table 1). Thus, the peaks from 5q33, 6p12, 12p12, 2q24 (sound–color synesthesia) and 16q12.2–23.1 (sequence–color synesthesia) represent signals from other families. Using the six putative candidates as seed genes, we found 109 genes coexpressed at r > 0.7 during neural development (31). The complete set of coexpressed genes contained six located within a previously reported synesthesia linkage peak. However, a formal test of enrichment did not achieve statistical significance (Fisher’s exact test, odds ratio = 2.069, P = 0.08). Mapping known protein–protein interactions from Reactome within this extended set yielded a network that was relatively sparse but included ITGA2 and COL4A1 as hubs within a subnetwork (Fig. S3).
Finally, recent findings suggest that people with autism spectrum conditions (henceforth, “autism”) and savant abilities are more likely to experience synesthesia, while synesthetes as a group show atypical sensory sensitivities similar to those seen in autism (34, 35). We thus hypothesized that genetic variants uncovered in synesthetic individuals without autism may show links to genetic pathways implicated in autism. The presence of comorbid neurological conditions (including autism) was an exclusion criterion when recruiting the families for this work (20). Three of the 37 variants that tracked with synesthesia status in our families are within genes found in the Simons Foundation AutDB catalog of autism-associated genes: FGA (family 16), HYDIN (family 2), and SLC9A6 (family 11) (36). However, the specific genetic variants seen in the synesthesia families (Table 1) are not shared with the autism cases currently contained in AutDB. Also, the three genes did not represent a significant enrichment (Fisher’s exact test, odds ratio = 2.01, P value = 0.20); thus the presence of these variants is not unusual, given the large number of genes associated with autism. Four of the 109 coexpressed genes were previously associated with autism (Fig. S3), although, again, this did not represent significant enrichment (Fisher’s exact test, odds ratio = 2.084, P = 0.13).
Discussion
Synesthesia represents the outer edges of natural variation in sensory perception. In this study, we identified rare genetic variants perfectly cosegregating with the trait in three unrelated multigenerational families with at least five people affected with sound–color synesthesia. A core set of six genes was related to axonogenesis, the process by which immature neurons send out their primary process to connect with other brain regions by following secreted guidance cues. Among other cortical sites, these genes were expressed throughout development in auditory and visual cortex, consistent with a potential role in developmental processes spanning both brain regions. Using a network approach, we uncovered six additional genes with similar neural expression patterns that fall within suggestive linkage regions originally identified in screens of larger numbers of families, which could be further investigated in follow-up studies (20).
Potentially supporting emerging data that synesthesia shares some phenotypic and mechanistic features with autism, the overrepresented ontologies fit with data from the autism neural transcriptome. A 2011 study of 17 autism cases and 19 controls generated a weighted coexpression network from microarray data from three brain regions (37). The set of genes most closely correlated with autism status was enriched for down-regulated genes related to neuronal projection. On the other hand, we did not see significant convergence between our synesthesia findings and published data on autism when examining specific genes or the rare mutations themselves. Thus, etiological overlaps might be most apparent at the level of biological processes rather than at the level of individual genetic loci. With further investigation in model systems, these candidate genes for synesthesia may provide molecular support for hyperconnectivity between brain regions as an underlying mechanism.
The segregating mutations in these three families did not fall within regions suggested in the 2009 linkage-mapping study of 43 families of varying size; it is very likely that the current families did not contribute to the suggestive signals highlighted in that prior investigation (20). Note that we did not filter our list of rare variants to select exceptionally damaging mutations, e.g., by using high CADD (Combined Annotation-Dependent Depletion) or haploinsufficiency scores (38). While such approaches are appropriate for neurodevelopmental disorders, synesthesia is not a disorder and is unlikely to reduce biological fitness, so we do not expect causative variants to be subject to negative selection. The six candidates also did not fall within linkage regions suggested by Tomson and colleagues; however, that previous study focused on a different form of synesthesia (sequence–color synesthesia), and the reported linkage evidence did not meet strict thresholds for significance (19). It is too early to tell whether different forms of synesthesia share underlying mechanisms (most recent studies have focused on grapheme–color synesthesia), although that is probable given that many synesthetes experience multiple varieties (7).
The top candidates we identified include the well-studied SLIT/ROBO chemoattractant system plus other genes with roles in shaping neuronal processes. In family 11, we identified rare segregating variants in SLIT2 and ROBO3, which regulate axon guidance through separate signaling pathways (39). Family 16 contained a variant in MYO10, which encodes an actin-based motor protein that acts in neurite growth cones (40). Although the MYO10 variant is synonymous, it results in a rare cysteine codon with no corresponding transfer RNA genes in humans (41). Such nonoptimal codon usage can reduce mRNA stability and slow ribosomal progression, disrupting translational regulation (41, 42). We observed a variant in ITGA2, encoding an integrin receptor subunit, which segregated with sound–color synesthesia in family 2. While studies of the functions of ITGA2 in neurodevelopment are sparse, it acts as a heterodimer with integrin β1 in embryonic retinal cells and has been observed in adult rat hippocampal mossy fiber tracks (43, 44). Integrin receptors on axon growth cones are a key mechanism by which developing neurons sense and respond to chemoattractants in the extracellular matrix (44). Furthermore, COL4A1 (family 11) encodes an alpha subunit of type IV collagen, which binds αβ integrin heterodimers to promote axonal growth and branching (45).
Synesthesia is a nonpathological anomaly of sensory integration, and our results suggest that rare genetic variants may shape the outcome of this developmental process within a normal range of experience. There are multiple ways that slight alterations in axonogenesis could lead to hyperconnectivity, such as incorrect length or localization, unusual branching, or subtle changes in morphology. The idiosyncratic sensory associations experienced by synesthetes expand and become more consistent during childhood (15). Thus, genetic variants increasing the likelihood of synesthesia may act in concert with early life experiences to form synesthetes’ unique secondary mappings (such as C♯ appearing orange, or B♭ as a deep brown) (13). Are the mechanisms that create a synesthetic brain developmentally restricted or do they involve processes also relevant in adulthood? That the top candidates from this study are expressed throughout development and are not neuron-specific but are also seen in glia and epithelial cells leaves this question open. The broad expression of these genes in the developing and adult brain fits with the wider synesthesia phenotype, which includes enhanced memory performance as well as altered sensory sensitivity (34, 46).
Since studies of the genetic basis for synesthesia are in early stages, there were limitations to our approach and dataset. Only one prevalence study has asked about sound–color synesthesia, confirming its presence in 1 of 500 UK college students surveyed (7). This conflicts with a survey of Dutch and German synesthetes where a much higher percentage reported sound–color associations (25). We erred on the side of inclusivity when determining the MAF threshold to use here. Despite this, we note that most of the synesthesia-specific variants we report are rare, falling well below this cutoff. We further focused on variants that perfectly tracked with the phenotype in each family. The families in this study were the largest multiplex families for whom we could obtain sufficient DNA for WES and validation, but they are still relatively small for linkage-based approaches, and we were unable to narrow the segregating variants to a single site per family. Further family-based studies of sound–color synesthesia and other forms will help clarify the genetic architecture and the potential role of altered neuronal morphology.
Over 130 y after the first reports of familial synesthesia, these results provide a molecular starting point for studies addressing the origins of healthy variation in sensory integration. While cellular and animal models will likely prove illustrative, behavioral experiments to test cross-modal perception in animals may elude the field for some time. It remains to be seen if other forms of synesthesia will involve alterations in axon growth and guidance. This study focused on relatively rare genetic variation occurring in families with multiple generations of synesthetes; assessing potential roles of common variation represents an intriguing question for the future.
Materials and Methods
Three families with sound–color (auditory–visual) synesthesia, without a history of drug use or any neurological, ophthalmological, or psychiatric disorder, were identified from the Cambridge Synaesthesia Research Group database. Ethical approval was granted by the Human Biology Research Ethics Committee of the University of Cambridge (Ref: 2011.06). Informed consent was obtained from all participants. Variants in exome-sequencing data were called using the Genome Analysis Toolkit best practices pipeline (24). Gene ontology analyses were performed with gProfileR, and human gene-expression data were downloaded from GTEx, the Allen Human Brain Atlas, and the BrainSpan database, while mouse RNAseq data were accessed from Zhang et al. (26, 31, 32). The datasets generated during the current study are available upon request from The Language Archive (TLA: https://corpus1.mpi.nl/ds/asv/?0), a public data archive hosted by the Max Planck Institute for Psycholinguistics. An extended description of materials and methods appears in Supporting Information.
Supplementary Material
Acknowledgments
We thank the families from the Cambridge Synaesthesia Research Group Database for their contributions to this study; Allen Chan and Laura Murphy for assistance with resampling the families; Han Brunner, Joris Veltman, and Alex Hoischen for assistance with exome sequencing; and Anthony Monaco for his contribution to earlier phases of the genetic analysis for these families. Funding was provided by European Commission Grant H2020-MSCA-IF-2015 #704393 (to A.K.T.) and the Hertie Foundation through the Eric Kandel Young Neuroscientists Prize (to S.E.F.). K.S.K., A.V., A.K.T., and S.E.F. were supported by the Max Planck Society. The funders had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.M.H. is a guest editor invited by the Editorial Board.
Data deposition: The datasets generated during the current study are available upon request from The Language Archive (TLA: https://corpus1.mpi.nl/ds/asv/?0), a public data archive hosted by the Max Planck Institute for Psycholinguistics. The data are stored under the node IDs MPI1758324# and MPI1815362# and are accessible at https://hdl.handle.net/1839/00-0000-0000-001A-8756-4@view.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715492115/-/DCSupplemental.
References
- 1.Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–R794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 2.MacGregor AJ, Andrew T, Sambrook PN, Spector TD. Structural, psychological, and genetic influences on low back and neck pain: A study of adult female twins. Arthritis Rheum. 2004;51:160–167. doi: 10.1002/art.20236. [DOI] [PubMed] [Google Scholar]
- 3.Mogil JS. Pain genetics: Past, present and future. Trends Genet. 2012;28:258–266. doi: 10.1016/j.tig.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell KJ. Curiouser and curiouser: Genetic disorders of cortical specialization. Curr Opin Genet Dev. 2011;21:271–277. doi: 10.1016/j.gde.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Simner J, Carmichael DA. Is synaesthesia a dominantly female trait? Cogn Neurosci. 2015;6:68–76. doi: 10.1080/17588928.2015.1019441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron-Cohen S, Wyke MA, Binnie C. Hearing words and seeing colours: An experimental investigation of a case of synaesthesia. Perception. 1987;16:761–767. doi: 10.1068/p160761. [DOI] [PubMed] [Google Scholar]
- 7.Simner J, et al. Synaesthesia: The prevalence of atypical cross-modal experiences. Perception. 2006;35:1024–1033. doi: 10.1068/p5469. [DOI] [PubMed] [Google Scholar]
- 8.Rothen N, et al. Psychophysiological evidence for the genuineness of swimming-style colour synaesthesia. Conscious Cogn. 2013;22:35–46. doi: 10.1016/j.concog.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Rouw R, Scholte HS. Increased structural connectivity in grapheme-color synesthesia. Nat Neurosci. 2007;10:792–797. doi: 10.1038/nn1906. [DOI] [PubMed] [Google Scholar]
- 10.Tomson SN, Narayan M, Allen GI, Eagleman DM. Neural networks of colored sequence synesthesia. J Neurosci. 2013;33:14098–14106. doi: 10.1523/JNEUROSCI.5131-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouw R, Scholte HS. Neural basis of individual differences in synesthetic experiences. J Neurosci. 2010;30:6205–6213. doi: 10.1523/JNEUROSCI.3444-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nunn JA, et al. Functional magnetic resonance imaging of synesthesia: Activation of V4/V8 by spoken words. Nat Neurosci. 2002;5:371–375. doi: 10.1038/nn818. [DOI] [PubMed] [Google Scholar]
- 13.Newell FN, Mitchell KJ. Multisensory integration and cross-modal learning in synaesthesia: A unifying model. Neuropsychologia. 2016;88:140–150. doi: 10.1016/j.neuropsychologia.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Janik McErlean AB, Banissy MJ. Color processing in synesthesia: What synesthesia can and cannot tell us about mechanisms of color processing. Top Cogn Sci. 2017;9:215–227. doi: 10.1111/tops.12237. [DOI] [PubMed] [Google Scholar]
- 15.Simner J, Bain AE. A longitudinal study of grapheme-color synesthesia in childhood: 6/7 years to 10/11 years. Front Hum Neurosci. 2013;7:603. doi: 10.3389/fnhum.2013.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron-Cohen S, Harrison J, Goldstein LH, Wyke M. Coloured speech perception: Is synaesthesia what happens when modularity breaks down? Perception. 1993;22:419–426. doi: 10.1068/p220419. [DOI] [PubMed] [Google Scholar]
- 17.Grossenbacher PG, Lovelace CT. Mechanisms of synesthesia: Cognitive and physiological constraints. Trends Cogn Sci. 2001;5:36–41. doi: 10.1016/s1364-6613(00)01571-0. [DOI] [PubMed] [Google Scholar]
- 18.Galton F. Inquiries into Human Faculty and Its Development. Macmillan; London: 1883. [Google Scholar]
- 19.Tomson SN, et al. The genetics of colored sequence synesthesia: Suggestive evidence of linkage to 16q and genetic heterogeneity for the condition. Behav Brain Res. 2011;223:48–52. doi: 10.1016/j.bbr.2011.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asher JE, et al. A whole-genome scan and fine-mapping linkage study of auditory-visual synesthesia reveals evidence of linkage to chromosomes 2q24, 5q33, 6p12, and 12p12. Am J Hum Genet. 2009;84:279–285. doi: 10.1016/j.ajhg.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregersen PK, et al. Absolute pitch exhibits phenotypic and genetic overlap with synesthesia. Hum Mol Genet. 2013;22:2097–2104. doi: 10.1093/hmg/ddt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher JE, Aitken MR, Farooqi N, Kurmani S, Baron-Cohen S. Diagnosing and phenotyping visual synaesthesia: A preliminary evaluation of the revised test of genuineness (TOG-R) Cortex. 2006;42:137–146. doi: 10.1016/s0010-9452(08)70337-x. [DOI] [PubMed] [Google Scholar]
- 23.Novich S, Cheng S, Eagleman DM. Is synaesthesia one condition or many? A large-scale analysis reveals subgroups. J Neuropsychol. 2011;5:353–371. doi: 10.1111/j.1748-6653.2011.02015.x. [DOI] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niccolai V, Jennes J, Stoerig P, Van Leeuwen TM. Modality and variability of synesthetic experience. Am J Psychol. 2012;125:81–94. doi: 10.5406/amerjpsyc.125.1.0081. [DOI] [PubMed] [Google Scholar]
- 26.Battle A, Brown CD, Engelhardt BE, Montgomery SB. GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration & Visualization—EBI; Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis & Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213, and erratum (2018) 553:530. [Google Scholar]
- 27.Sunkin SM, et al. Allen brain atlas: An integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 29.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 30.Banissy MJ, et al. Grapheme-color and tone-color synesthesia is associated with structural brain changes in visual regions implicated in color, form, and motion. Cogn Neurosci. 2012;3:29–35. doi: 10.1080/17588928.2011.594499. [DOI] [PubMed] [Google Scholar]
- 31.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward J, et al. Atypical sensory sensitivity as a shared feature between synaesthesia and autism. Sci Rep. 2017;7:41155. doi: 10.1038/srep41155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes James EA, Simner J, Baron-Cohen S, Treffert Darold A, Ward J. Is synaesthesia more prevalent in autism spectrum conditions? Only where there is prodigious talent. Multisens Res. 2017;30:391–408. doi: 10.1163/22134808-00002558. [DOI] [PubMed] [Google Scholar]
- 36.Basu SN, Kollu R, Banerjee-Basu S. AutDB: A gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blockus H, Chédotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 40.Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE. Myo10 in brain: Developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 41.Quax TE, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Presnyak V, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fasen K, Elger CE, Lie AA. Distribution of α and β integrin subunits in the adult rat hippocampus after pilocarpine-induced neuronal cell loss, axonal reorganization and reactive astrogliosis. Acta Neuropathol. 2003;106:319–322. doi: 10.1007/s00401-003-0733-y. [DOI] [PubMed] [Google Scholar]
- 44.Bradshaw AD, et al. Integrin alpha 2 beta 1 mediates interactions between developing embryonic retinal cells and collagen. Development. 1995;121:3593–3602. doi: 10.1242/dev.121.11.3593. [DOI] [PubMed] [Google Scholar]
- 45.Lein PJ, Higgins D, Turner DC, Flier LA, Terranova VP. The NC1 domain of type IV collagen promotes axonal growth in sympathetic neurons through interaction with the alpha 1 beta 1 integrin. J Cell Biol. 1991;113:417–428. doi: 10.1083/jcb.113.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard J, Rothen N, Coolbear D, Ward J. Enhanced associative memory for colour (but not shape or location) in synaesthesia. Cognition. 2013;127:230–234. doi: 10.1016/j.cognition.2012.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.