Significance
The construction of an organism with an altered genetic code negatively impacted its fitness. We evolved this organism for ∼1,100 generations in the laboratory to recover fitness and learn what changes would accumulate during evolutionary trajectories toward faster growth rates. We observed several selective mutations that helped alleviate insufficient translation termination or that corrected for unintended mutations that accumulated when we originally altered the genetic code. Further observed mutations were generally adaptive in a nonrecoded background. This work bolsters our understanding of the pliability of the genetic code and will help guide future efforts seeking to recode genomes. Finally, it results in a useful strain for nonstandard amino acid incorporation in numerous contexts relevant for research and industry.
Keywords: adaptive evolution, recoded genome, synthetic biology, genetic code expansion, nonstandard amino acids
Abstract
Efforts are underway to construct several recoded genomes anticipated to exhibit multivirus resistance, enhanced nonstandard amino acid (nsAA) incorporation, and capability for synthetic biocontainment. Although our laboratory pioneered the first genomically recoded organism (Escherichia coli strain C321.∆A), its fitness is far lower than that of its nonrecoded ancestor, particularly in defined media. This fitness deficit severely limits its utility for nsAA-linked applications requiring defined media, such as live cell imaging, metabolic engineering, and industrial-scale protein production. Here, we report adaptive evolution of C321.∆A for more than 1,000 generations in independent replicate populations grown in glucose minimal media. Evolved recoded populations significantly exceeded the growth rates of both the ancestral C321.∆A and nonrecoded strains. We used next-generation sequencing to identify genes mutated in multiple independent populations, and we reconstructed individual alleles in ancestral strains via multiplex automatable genome engineering (MAGE) to quantify their effects on fitness. Several selective mutations occurred only in recoded evolved populations, some of which are associated with altering the translation apparatus in response to recoding, whereas others are not apparently associated with recoding, but instead correct for off-target mutations that occurred during initial genome engineering. This report demonstrates that laboratory evolution can be applied after engineering of recoded genomes to streamline fitness recovery compared with application of additional targeted engineering strategies that may introduce further unintended mutations. In doing so, we provide the most comprehensive insight to date into the physiology of the commonly used C321.∆A strain.
Billions of years of evolution have given rise to diverse organisms that share a universal genetic code. The ability to recode genomes to contain fewer than the full set of 64 triplet codons has proven useful for enabling multivirus resistance, enhanced incorporation of nonstandard amino acids (nsAAs), and capability for biocontainment by synthetic auxotrophy. The first genomically recoded organism, Escherichia coli C321.ΔA (1–3), was generated by using multiplex automatable genome engineering (MAGE) (4) to replace all 321 UAG stop codons with UAA and delete the associated class I peptide release factor 1 (RF1—encoded by prfA), which recognizes UAA/UAG codons. Genome synthesis and assembly methods are currently being used to construct additional recoded genomes, including a 57-codon E. coli genome (5) and a synthetic yeast genome (6, 7). Given their common aim of UAG codon reassignment, these efforts would benefit from greater characterization of C321.ΔA under diverse conditions.
Previous genome engineering efforts directed toward the goal of UAG codon reassignment resulted in strains that exhibited large fitness deficits upon removal of RF1. RF1 was originally considered to be essential, and only conditionally lethal mutants were described (8). In 2010, RF1 deletion was enabled by conversion of seven UAG codons in essential genes to UAA and introduction of a UAG suppressor (9). Two subsequent studies showed that RF1 could be deleted if class I peptide release factor 2 (RF2—encoded by prfB), which recognizes UAA/UGA codons (10), was corrected in one of two ways: by removing RF2 autoregulation (11) or by introducing a PrfB_T246A variant (12). However, the resulting strains were severely growth-impaired given the presence of numerous unassigned UAG codons where ribosomes would presumably stall. More recently, RF1 deletion was enabled after conversion of 95 of the 273 UAG codons in E. coli BL21(DE3), which natively contains the PrfB_T246A variant (13); nevertheless, this strain displayed inferior growth in minimal media. C321.ΔA and its derivatives are the only strains that contain neither apparent UAG codons nor RF1, enabling UAG reassignment to nsAAs without competition from off-target sites or from RF1.
C321.ΔA has been widely adopted as a workhorse for nsAA incorporation. nsAAs broaden the repertoire of biological chemistry in living systems for diverse purposes, such as photocrosslinking (14), functionalization (15), structure determination (16), fluorescence (17, 18), metal binding (19), biosensing (20), and immobilization (21). nsAAs also augment protein function, such as the affinity and pharmacodynamic properties of therapeutically relevant proteins (22–24) and the catalytic properties of industrially relevant enzymes (25–27). Many of these potential applications for nsAAs, such as live cellular imaging, metabolic engineering using simple carbon sources, and industrial-scale protein expression, depend on the ability to culture cells in defined media. However, C321.ΔA exhibits a fitness deficit compared with its nonrecoded ancestor, which is due at least in part to off-target hitchhiker mutations that accumulated during recoding (4). This fitness deficit is exacerbated in defined media.
Experimental evolution is a powerful method for directly observing rapid evolutionary change in the laboratory, and nonmodel organisms can quickly adapt to laboratory conditions after a period of sustained propagation (28). We describe here the adaptive laboratory evolution (ALE) of an organism containing a genome with fewer than 64 codons. By sequencing the whole genomes of two clonal isolates from more than 50 independent evolved populations, we present evidence that the conversion of 321 UAG codons into UAA codons, and especially the subsequent deletion of RF1, introduces a burden to E. coli K-12 cellular translation machinery under several industrially relevant defined media. Our evolutionary analysis reveals that point mutations in RF2 that are known to provide increased activity on UAA codons are selected for and recover much of the fitness loss. Furthermore, we observe that natural selection exhibits a variety of mechanisms to correct the most detrimental off-target mutations introduced during engineering of the recoded strain, including the introduction of premature stop codons (PSCs) in essential genes and the inactivation of a stress-related transcription factor.
Adaptive Evolution Achieves Robust Growth of Evolved Recoded Strains in Glucose Minimal Media
We seeded 14 independent populations for serial batch evolution of each of the following four strains: (i) “Parent” (ECNR2, which is the nonrecoded parent strain of C321.ΔA); (ii) “Recoded.ΔRF1” (C321.ΔA, a C321 derivative with RF1 removed); (iii) “Recoded.ΔRF1-v2” (C321.ΔA-v2, a C321.ΔA derivative containing engineered reversions to three off-target MAGE mutations) (29); and (iv) “Recoded” (C321, a C321.ΔA derivative with RF1 restored to its native locus). These strains contained inactivated mutS, which significantly increases MAGE efficiency but also results in a hypermutator phenotype. Hypermutators are known to arise naturally during long-term evolution (30), and we hypothesized that we might accelerate our ALE by using MutS− strains for seeding. Independent lineages were propagated for over 1,000 generations and subjected to genotyping, doubling time analysis, and storage at ∼70 to 100 generation intervals (SI Appendix, Fig. S1). We increased the dilution and passaging rate as growth rates increased so that strains did not evolve to survive long periods of stationary phase but instead experienced selective pressure primarily for exponential growth.
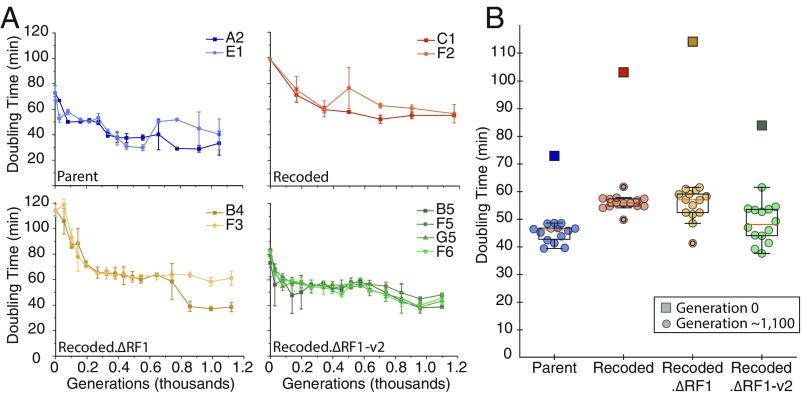
After ∼1,100 generations, we determined, through intermittent sampling of population doubling times, that the fastest populations from both the experimental and the control lines were approaching 30 min, which is close to the parental doubling time in rich media (Fig. 1). We therefore paused the evolution at this point and characterized and sequenced all evolved populations. Coarse samplings of the doubling time improvements over evolutionary time were run on all evolved populations (SI Appendix, Fig. S2). We then chose two populations from each strain (except for Recoded.∆RF1-v2, for which we chose four populations) that showed the greatest improvement to doubling time by the end of the evolution and measured the growth rate improvement over time more finely (Fig. 1). All strains showed marked improvement to their fitness in minimal media over the time course of the ALE, but the less fit strains experienced larger improvements in fitness (SI Appendix, Fig. S3). Faster adaption of less fit strains is not likely to be caused by an elevated mutation rate because our analysis of neutral mutations suggests that lineages are accumulating mutations at the same rate (SI Appendix, SI Materials and Methods). The fastest growing biological isolates from each experimental population converged upon a doubling time of between 35 and 45 min. We next sought to identify the causal mutations behind improvements to strain fitness with an analysis of whole genome sequencing data from two clonal isolates of each evolved population.
Fig. 1.
Representative trajectories showing changes in fitness during the evolution of the four E. coli strains discussed. (A, Top Left; Parent) Two lineages of nonrecoded ECNR2 (engineered from E. coli MG1655 K-12). (A, Top Right; Recoded) Two lineages of recoded C321.∆A (321 UAG→UAA and RF1−). (A, Bottom Left; Recoded.∆RF1) Four lineages of recoded C321.∆A-v2 (C321.∆A with engineered reversion of some off-target mutations that occurred during recoding). (A, Bottom Right; Recoded.∆RF1-v2) Two lineages of recoded C321 (C321.∆A with prfA gene restored). (B) Ancestral and final doubling time measurements sampled from all lineages of Parent (blue), Recoded (red), Recoded.∆RF1 (yellow), and Recoded.∆RF1-v2 (green).
A Broad Mutational Analysis of Evolved Populations
We ran all genome sequencing data through two parallel analytical pipelines (SI Appendix, SI Materials and Methods). Because of the hypermutator mutS− phenotype, our evolved strains accumulated mutations much more quickly than WT E. coli. The lineages averaged between 40 and 55 mutations per clonal population after ∼1,100 generations. Evolved Parent lines had the fewest mutations on average (∼40 mutations per clone) while Recoded.∆RF1 lines had the most mutations (∼55 mutations per clone). The rates of mutation accumulation roughly correlated to the initial fitness of the ancestor strains (SI Appendix, Fig. S4). These rates are higher than rates normally seen in ALE with WT strains that have not acquired a hypermutator phenotype (2 to 5 mutations per 1,000 generations) (31–33) but roughly correspond to rates seen in populations of E. coli that evolved similar hypermutator phenotypes (mutT−) during long-term laboratory evolution in the Lenski laboratory (62 mutations per 1,000 generations) (34). A summary of key mutations is shown in Table 1.
Table 1.
A subset of the most frequently mutated genes that were characterized in this study or are otherwise of interest
| Gene | ECNR2 | C321 | C321.∆A | C321.∆A-v2 | Total | Brief remarks | Ref(s). |
| fimH | 1 | 1 | 2 | 11 | 15 | Important for biofilm formation in M9 + glucose and LB | 53 |
| fis | — | 4 | 6 | — | 10 | 32 | |
| flu | 14 | — | 5 | 13 | 32 | Important for biofilm formation in M9 + glucose but not in LB | |
| folA | — | 10 | 5 | 2 | 17 | Hitchhiker mutation to promoter during C321 recoding | 48 |
| gltB | — | 4 | 3 | — | 7 | Highly expressed in biofilms in Gram-negative bacteria | 32 |
| kup | 1 | 4 | 2 | — | 7 | 32 | |
| mdtJ | — | — | 1 | 4 | 5 | Knockout led to increased biofilm mass in minimal media | |
| ompT | 5 | 1 | — | 1 | 7 | Highly expressed in biofilms in Gram-negative bacteria | |
| oxyR | — | — | 5 | 12 | 17 | Null mutation up-regulates flu | |
| prfB | 1 | — | 6 | 7 | 14 | Release factor 2, discussed in text | |
| prfC | — | — | 3 | 3 | 6 | Release factor 3, discussed in text | |
| purL | 1 | 4 | 2 | — | 7 | Purine biosynthesis is important for biofilm formation in many Gram-negative bacteria | |
| purT | — | 3 | 1 | 7 | 11 | Purine biosynthesis is important for biofilm formation in many Gram-negative bacteria | |
| pykF | 9 | 4 | 1 | 2 | 16 | 31, 32 | |
| pyrE | 16 | 7 | 6 | 8 | 37 | Mutation in MG1655 founder strain | 31, 33, 54 |
| rph | 2 | 4 | 2 | 8 | 16 | Mutation in MG1655 founder strain | 31, 33, 54 |
| rpoB | 7 | 5 | 2 | — | 14 | Commonly seen in adaptation to minimal media | 31, 33, 54 |
| rpoC | 10 | 7 | 4 | 2 | 23 | Commonly seen in adaptation to minimal media | 33, 54 |
Numbers represent unique instances of observed mutations to the evolved lineages. Em dashes indicate zero observations. Refs. refer to works in which the indicated mutations have been previously observed during evolutionary studies.
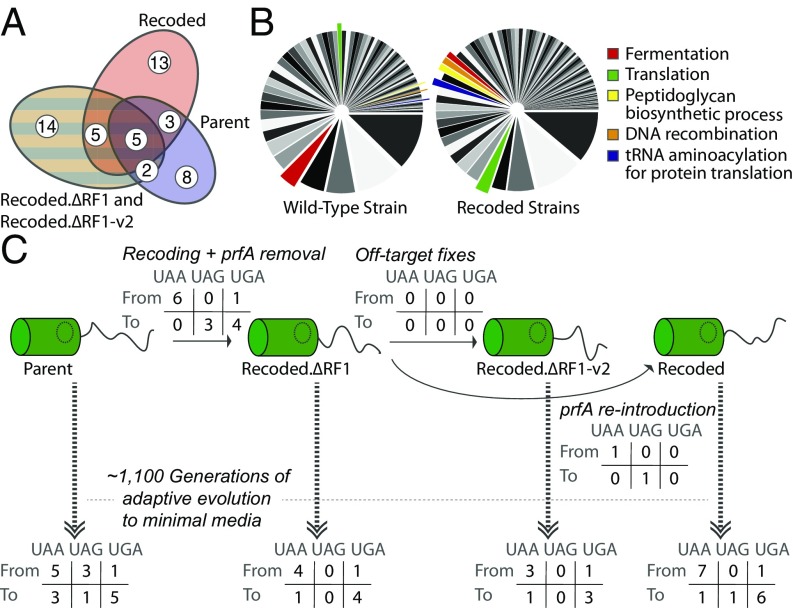
We observed a notable overlap in multiply hit genes (at least five observed mutations) between lineages (Fig. 2A). Some of these gene targets that were observed to affect all (or most) populations are discussed in SI Appendix. In several cases, our study provides further insight into the evolutionary profiles of these genes in general adaptation of MG1655-derived E. coli strains to minimal media (SI Appendix, SI Discussion). In the remainder of the text, we focus on mutations observed disproportionately in recoded strains.
Fig. 2.
Next-generation sequencing results and variants of interest from independent evolved populations. (A) Venn diagram showing distribution and quantity of multiply hit genes from each individual strain. (B) Gene ontology results showing all categories of mutations found in nonrecoded Parent (Left) or in recoded strains (Right). Sector size corresponds to the number of mutations observed to genes with a particular GO term. Highlighted in red are GO terms of interest. (C) Results from analysis of changes in stop codon usage in evolved lineages.
We compared the gene ontology (GO) of mutations found in the Parent lines against those found in recoded lines (Fig. 2B). There were four GO terms for which there was either a threefold enrichment in Parent lines or recoded lines, and for which at least 10 mutations were seen in the enriched line(s). The recoded lines were enriched for mutations in genes associated with translation, tRNA aminoacylation for protein translation, and DNA recombination while the Parent lines were enriched for mutations in genes involved in fermentation. The enrichments for mutations associated with translational machinery in recoded lines is consistent with the change in stop codon repertoire and removal of RF1 from these populations.
Stop Codon Identity in Evolved Strains Displays Differential Selection by Population Phenotype
As the recoded strains have a globally altered stop codon repertoire, we sought to examine how ALE differentially affected stop codons in each evolved population. We hypothesized that RF1 removal from recoded strains may put selective pressure on stop codons during adaptive evolution because of literature related to translation termination (SI Appendix, SI Discussion). We examined all genes in sequenced clones for stop codon reassignment (SCR) (SI Appendix, SI Materials and Methods). In recoded strains lacking RF1, we did not observe any appearances of UAG codons during evolution except in one instance in a pseudogene, indicating a robustness of the recoding performed to remove UAG stop codons from the strain to long-term laboratory passaging.
Two trends stood out to us in analyzing SCR (Fig. 2). First, in all populations, genes with UAA codons experienced more SCR than those with UGA codons. WT MG1655 had UAA:UGA:UAG codon ratios of ∼10 to 5 to 1, respectively. In this study, however, we saw that genes with UAA codons see about four to five times the rate of SCR as genes with UGA codons (about twice the expected ratio P < 0.1). Most observed SCR was due to either frameshift mutations near the 3′ end of the protein-coding region or read-through of termination codons by way of their mutation to sense codons (SI Appendix, Table S1). These two causes should show a roughly equal propensity to mutate to any of the three termination codons, but we observed a bias toward UGA stop codons. Although MutS− strains exhibited an excess of transitions over transversions, the SCR mutations characterized consisted entirely of frameshifts and transitions, and thus mutational bias was not responsible for the trend we observed. Together these observations suggest the possibility of an adaptive preference for UGA over UAA or UAG in minimal media, but further study would be needed to confirm this observation. The second notable trend in SCR is that, at every transition between ECNR2 and the various C321 derivative strains, there was at least one mutation toward a UAG codon, except in those cases where RF1 was absent. This seems to indicate a fitness cost of mutation toward a termination codon in a strain missing its cognate release factor. Lastly, notable, perhaps for its absence, is that no significant difference in SCR was noticed in recoded lineages compared with the WT ECNR2 lineage, even in the absence of RF1. This suggests that stop codon reassignment is not an easily accessible source for fitness gains to the recoded C321 strains.
Selective Mutations Occur in Translational Machinery: prfB and prfC
Three mutations observed repeatedly were found in the prfB and prfC genes. PrfB encodes RF2, and prfC encodes release factor 3 (RF3), which is a ribosome-dependent GTPase that stimulates release of RF1 and RF2 from the ribosome after termination (35). In the recoded lineages missing prfA (Recoded.∆RF1 and Recoded.∆RF1-v2), we observed independently arising missense mutations to the remaining release factor: prfB. The dominant mutation, observed in 31 of 54 sequenced clones from prfA-lineages, was PrfB_T246A, which is a revertant to the PrfB sequence present in most non–K-12 E. coli strains (36). This mutation has been characterized in vitro (36) and in vivo (37, 38), and it is known to increase net RF2 activity on UAA codons by roughly fivefold in vivo (38). Another mutation observed in independent lineages was PrfB_E170K. Based on previously targeted mutagenesis studies of prfB, PrfB_E170K appeared to also have increased termination efficiency for UAA codons (39, 40). We also observed five separate missense mutations to prfC in the recoded lineages, but, in contrast to prfB, for which mutations only appeared in the recoded lines missing prfA, prfC mutations appeared also in the recoded C321 line with prfA reintroduced. The most frequently observed mutation to prfC was PrfC_A350V, which arises independently in at least one population from all three recoded lines. To our knowledge, no one has studied the effect of the A350V mutation although this position was one of many targeted for mutagenesis in a previous study (41). During a study of strains containing temperature-sensitive RF1 and RF2 variants, prfC mutations appeared at positions 96, 118, 399, and 440, each of which suppressed growth defects (42). The previous observation of RF3 mutations during defective translation termination lends support to the notion that the A350V mutation may also alleviate impaired termination. Note that, when translation termination is impaired in other ways, such as by deletion of ribosomal modification machinery, mutations in prfB and in prfC are also known to arise, but not at the positions observed in our study (SI Appendix, SI Discussion). Overall, in our study, three RF mutations were found to occur independently across multiple populations: PrfB_E170K, PrfB_T246A, and PrfC_A350V. None of these mutations were found in evolved Parent populations. Furthermore, no sequenced clones were found to contain more than one of these three mutations, and a functional RF1 seems to be enough to epistatically shield prfB from selectional mutation.
The Contribution of Mutated RF Alleles to Fitness Depends on Media Composition
We reconstructed these RF mutations individually in ancestral strains and tested their effects on fitness using two approaches: doubling time analysis and head-to-head competition. We also examined the effect of each mutation in a wide range of relevant media conditions. Media composition has been shown to influence how impaired translation termination affects growth rate (SI Appendix, SI Discussion). In general, translational termination defects often show increased severity in poorer media conditions due potentially to two factors: (i) Slower growing E. coli cells produce lower levels of release factors and fewer ribosomes, meaning more demand for release and recycling on fewer molecules (43), and (ii) growth in minimal media or on low quality carbon sources necessitates expression of a broader set of genes (44, 45), which more often contain weak termination codons than highly expressed genes (46). We therefore decided to ask whether adaptive fitness improvements to RFs would be applicable across media types.
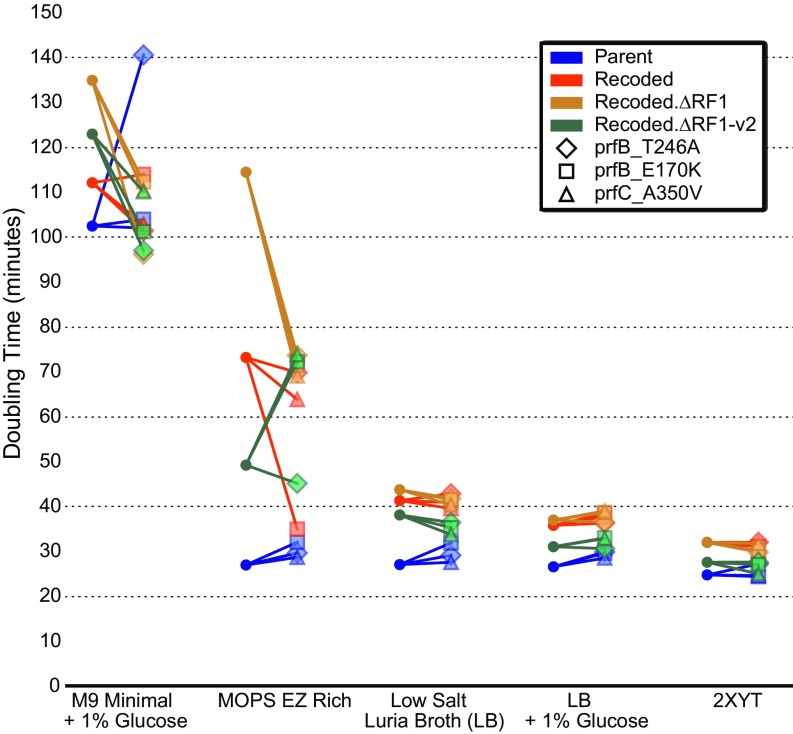
Although we performed ALE only in M9+glucose, we measured doubling times for strains grown in two defined media (M9+glucose and Mops EZ Rich) and three complex media (LB, LB+glucose, and 2XYT). We display the results in two formats for clarity (Fig. 3 and SI Appendix, Fig. S5), and, to complement this analysis, we performed head-to-head competition assays against a fluorescent reference strain (SI Appendix, Fig. S6). Several trends emerged from these data. First, in agreement with past observation, we saw that doubling times were much larger for ancestral recoded strains than for ECNR2 across media types. Next, in support of the idea that RF stress is more acute in poorer media conditions, we observed that allelic fitness variants have little effect in rich media but offer large improvements to recoded strain fitness in defined media. Furthermore, in accordance with RF variants being the most acutely selected variant type in this study, we observed a decreasing difference in fitness between recoded and nonrecoded strains as media became richer, implying that RF stress is a key driver of fitness loss in minimal media for the recoded strains. Overall, some RF mutations were beneficial in defined media but neutral in complex media; the mutation that consistently appeared most beneficial to recoded strains was PrfB_T246A, which is corroborated by its greater frequency of occurrence during evolution.
Fig. 3.
Effects of individually reconstructed release factor mutations and media context on growth rate of ancestral strains. Different media compositions are shown on the x axis, arranged from poorer carbon sources in defined media (leftmost) to rich and complex media (rightmost). The three release factor mutations investigated are PrfB_T246A, PrfB_E170K, and PrfC_A350V.
Selective Mutations Occur in Genes Not Associated with Translational Machinery
In addition to the RF mutations described above, we observed six genes of interest that were enriched for mutation in recoded lines as opposed to Parent. The first was folA, which encodes the essential dihydrofolate reductase. We observed 18 mutations in or around folA, including PSCs or mutations in the promoter region, which has an off-target hitchhiker mutation from the recoding of Recoded.ΔRF1 (C49765T). The four most frequently arising mutations appeared in the folA promoter region near the hitchhiker mutation. Additional mutations included four separately arising PSCs and many low-frequency mutations later in the gene or to the 5′ end of folA, in a region likely to impact its mRNA structure (47). Interestingly, after alternating the passaging of Recoded.ΔRF1 in LB and minimal media for just 12 growth cycles, Monk et al. (48) similarly observed one instance of a PSC in folA and two mutations in the promoter region. Monk et al. predicted that these mutations up-regulate folA and lead to an increase in metabolic flux through this enzyme. However, we viewed the PSC mutations in folA as a likely signature for down-regulation of FolA translation. Indeed, nonsense mutations in E. coli genes have been known to not completely abolish corresponding enzyme activity, instead allowing residual expression of roughly 10−4 of WT levels (49). To investigate this hypothesis, we cloned the native and Recoded.ΔRF1 folA promoter sequences upstream of a gene encoding a fluorescent reporter protein and measured fluorescence over a 24-h time course in M9+glucose and LB media (SI Appendix, Fig. S7). We measured fluorescence of the reporter protein rather than measuring transcript levels because we did not expect transcripts that contain PSCs to be fully translated. Strains expressing the fluorescent reporter under control of either the C321.ΔA folA promoter or a moderately strong constitutive promoter exhibited a high level of fluorescence in both media while fluorescence was essentially undetectable for strains expressing the reporter under control of the native folA promoter. This result indicates that the PSCs in the folA gene significantly decrease FolA translation, which compensates for overexpression caused by the hitchhiker mutation. Given that signal from the native folA promoter was low under both media conditions, it is likely that folA expression in Recoded.ΔRF1 is globally burdensome and that it should be corrected through engineering or evolution for all envisioned applications.
Five other genes of interest that were highly mutated in recoded strains were fimH, oxyR, purT, mdtJ, and fis. We investigated their contribution to fitness by allelic reconstruction in both the Recoded.∆RF1-v2 and Recoded.∆RF1-v2.PrfB_T246A backgrounds to determine if their fitness effects would be additive with RF2 mutation. Four of these genes were involved in biofilm formation (SI Appendix, SI Discussion), and the fis gene encoded a multipurpose transcription factor and nucleoid regulator. With one exception (fimH), these genes all contained PSCs or frameshift mutations. Therefore, in these cases we introduced PSCs ∼30 bases downstream of the start codon during allele reconstruction whereas, in fimH, we tested the two most frequent missense mutations. When we compared the fitness of the reconstituted variants by competition assay with the unaltered strains, Recoded.∆RF1-v2 was significantly improved by mdtJ, oxyR, and purT knockouts whereas the fitness effects in Recoded.∆RF1-v2.PrfB_T246A were muted, with no mutations showing significant fitness improvements (SI Appendix, Fig. S8). This seems to indicate that the fitness benefit of these alleles may be masked by RF2 improvement.
Evolved Recoded Strains Show High Specificity for nsAA Incorporation in Defined Media
The absence of UAG termination codons and RF1 permits dedicated reassignment of UAG to an nsAA. To evaluate the utility of our evolved recoded strains for nsAA incorporation, we isolated clonal populations from three evolved Recoded.∆RF1-v2 strains and cotransformed them with plasmids harboring an orthogonal translation system and a reporter protein. If the orthogonal translation system is sufficiently specific for an nsAA, then nsAA incorporation is proportional to full-length reporter protein formation based on suppression of UAG codons in the gene encoding the reporter protein (50).
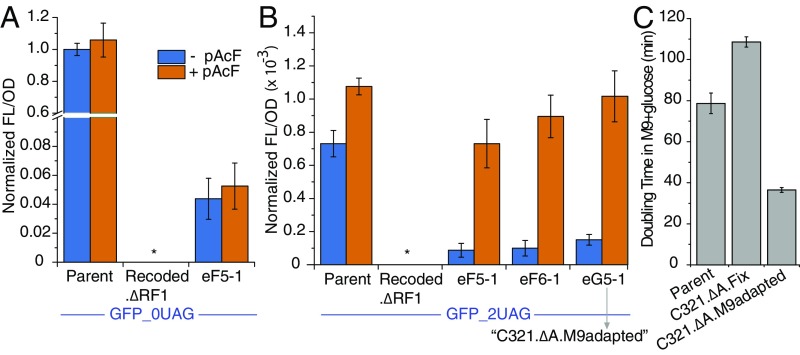
We observed that nsAA incorporation [as measured by fluoresence (FL) normalized by optical density at 600 nm (OD), or FL/OD] was unaffected by the addition of p-acetyl-phenylalanine in ECNR2 and an evolved recoded strain expressing the 0-UAG reporter (the ancestral Recoded.ΔRF1 strain did not observably grow during the 24-h observation period) (Fig. 4 A and B). Notably, FL/OD of the evolved recoded strain was ∼20-fold lower than that of Parent. We observed substantially lower FL/OD using the 2-UAG reporter, consistent with competition for UAG suppression with RF1 in Parent and the limited activity of previously developed orthogonal translation systems even in the absence of RF1. Interestingly, FL/OD in the presence of p-acetyl-phenylalanine did not vary significantly between Parent and the evolved recoded strains, but, in the absence of p-acetyl-phenylalanine, FL/OD was lower in the recoded strains. This result demonstrates that our evolved recoded strains not only grow and incorporate nsAAs in minimal media but also exhibit improved dynamic range for nsAA incorporation applications. We comment more on how to improve protein expression in evolved strains in SI Appendix, SI Discussion.
Fig. 4.
Characterization of select evolved recoded clones for applications featuring nsAAs. (A) Expression of a control GFP reporter containing no UAG codons in various strains in the presence and absence of the nsAA p-acetyl-phenylalanine (pAcF). (B) nsAA incorporation assay based on expression of a GFP reporter containing two UAGs in various strains in the presence and absence of pAcF. Evolved recoded clone G5-1 was selected as C321.∆A.M9adapted for deposit in the Addgene repository. (C) Doubling time measurements for C321.∆A.M9adapted and related strains in glucose minimal media.
Evolved recoded clone G5-1, which showed the highest nsAA-dependent protein expression from among the evolved ancestors, was selected as “C321.∆A.M9adapted” for deposit in the Addgene repository (cat. no. 98568). Finally, we compared the doubling times of Parent, a previously engineered strain, C321.∆A.opt (29), and C321.∆A.M9adapted from the present study. C321.∆A.M9adapted exhibited faster growth than both Parent and C321.∆A.opt in glucose minimal media (Fig. 4C).
Collectively, our results show that, while genome engineering strategies and laboratory evolution are both useful tools, they each present tradeoffs that suggest different use cases and a clear order of operations. Genome engineering strategies facilitate rational genetic changes to known targets; however, these approaches can also introduce undesired changes (1), and they may also suffer from the problem of unforeseen epistatic interactions in varied genetic backgrounds. In contrast, laboratory evolution enables natural selection to guide both steps of target identification and change making at once, and it is the most logical tool for improving the phenotype of fitness. It is particularly beneficial when applied after large-scale genome engineering to evaluate the robustness of engineered alterations and to correct unwanted alterations. Evolution performed in specific contexts, however, can result in specialized strains that are less fit in other contexts (33). Evolution is also limited in the total number of traits, and the simultaneous number of traits, that it can be used to enhance because of the selection requirement (51). Thus, these strategies should be used in concert, and, in this work, our final strain, C321.∆A.M9adapted, is indeed the product of ALE being used to further improve on strain fitness after some rational engineering of the recoded strain for correction of predicted high-impact hitchhiker mutations.
Conclusions
Here, we overcame one of the principal shortcomings of the recoded E. coli lineage, evolving a derivative strain that grows robustly in both rich and minimal media, and show that adaptive evolution is a useful tool for the recovery of fitness loss from laboratory engineering. After only ∼1,100 generations of adaptation to minimal media, independent recoded lineages recovered nearly all of their fitness loss from the recoding of 321 stop codons and the correlated off-target mutational load. We anticipate that the deposited strain C321.∆A.M9adapted will be of significant interest to researchers interested in a fast-growing recoded derivative that thrives in a broad range of media types. Particularly compelling use cases for C321.∆A.M9adapted include recombinant protein expression in defined media, cellular microscopy, and metabolic engineering.
Materials and Methods
The following four strains were each passaged in 14 independent populations for ∼1,100 generations in defined M9 minimal media plus 1% glucose plus biotin plus carbenicillin: ECNR2 (4), C321.∆A (1), C321.∆A-v2 (29), and C321 (prfA+). C321.∆A-v2 is a variant of C321.∆A in which some off-target mutations were corrected using MAGE. C321 was constructed by scarlessly reintroducing the prfA gene into its WT locus in C321.∆A using recombineering and CRISPR-Cas9–based selection (52). In brief, the prfA gene along with flanking homology was amplified from ECNR2 to generate a linear DNA cassette for recombination. C321.∆A was transformed with plasmids containing inducible Cas9 and guide RNA containing the junction sequence at the site of insertion. C321.∆A was heat-shocked to induce the lambda red system as in standard recombineering/MAGE protocols and transformed with the prfA cassette for homologous recombination. All these strains were derived from E. coli K-12 MG1655. For more details, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Alex Leffell (Massachusetts Institute of Technology) for experimental help when things were busy; Seth Shipman, George Chao, Erkin Kuru, and Gabriel Filsinger (Harvard University) for helpful conversations; and Gleb Kuznetsov and Daniel Goodman (Harvard University) for coaching us through the variant analysis in Millstone and for providing us with many strains and resources. Computational resources for this work were provided by the Amazon Web Services Cloud Credits for Research Program. This project was graciously funded by US Department of Energy Grant DE-FG02-02ER63445.
Footnotes
Conflict of interest statement: G.M.C. is the founder of Gen9, ReadCoor, EnEvolv, and GRO Biosciences and has related financial interests in ReadCoor, EnEvolv, and GRO Biosciences. For other potential conflicts, please see arep.med.harvard.edu/gmc/tech.html.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. CP025268) and the Addgene repository (accession no. 98568).
See Commentary on page 2853.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715530115/-/DCSupplemental.
References
- 1.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 6.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 8.Rydén SM, Isaksson LA. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 9.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DBF, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DBF, et al. Release factor one is nonessential in Escherichia coli. ACS Chem Biol. 2012;7:1337–1344. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukai T, et al. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci Rep. 2015;5:9699. doi: 10.1038/srep09699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J, et al. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 17.Summerer D, et al. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci USA. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J Am Chem Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 19.Xie J, Liu W, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angew Chem Int Ed Engl. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 20.Niu W, Guo J. Novel fluorescence-based biosensors incorporating unnatural amino acids. Methods Enzymol. 2017;589:191–219. doi: 10.1016/bs.mie.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Ravikumar Y, Nadarajan SP, Hyeon Yoo T, Lee C-S, Yun H. Incorporating unnatural amino acids to engineer biocatalysts for industrial bioprocess applications. Biotechnol J. 2015;10:1862–1876. doi: 10.1002/biot.201500153. [DOI] [PubMed] [Google Scholar]
- 22.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 23.Liu CC, Choe H, Farzan M, Smider VV, Schultz PG. Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry. 2009;48:8891–8898. doi: 10.1021/bi9011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H, et al. Optimized clinical performance of growth hormone with an expanded genetic code. Proc Natl Acad Sci USA. 2011;108:9060–9065. doi: 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson JC, Duffy SP, Hess KR, Mehl RA. Improving nature’s enzyme active site with genetically encoded unnatural amino acids. J Am Chem Soc. 2006;128:11124–11127. doi: 10.1021/ja061099y. [DOI] [PubMed] [Google Scholar]
- 26.Ugwumba IN, et al. Improving a natural enzyme activity through incorporation of unnatural amino acids. J Am Chem Soc. 2011;133:326–333. doi: 10.1021/ja106416g. [DOI] [PubMed] [Google Scholar]
- 27.Windle CL, et al. Extending enzyme molecular recognition with an expanded amino acid alphabet. Proc Natl Acad Sci USA. 2017;114:2610–2615. doi: 10.1073/pnas.1616816114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll SM, Xue KS, Marx CJ. Laboratory divergence of Methylobacterium extorquens AM1 through unintended domestication and past selection for antibiotic resistance. BMC Microbiol. 2014;14:2. doi: 10.1186/1471-2180-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuznetsov G, et al. Optimizing complex phenotypes through model-guided multiplex genome engineering. Genome Biol. 2017;18:100. doi: 10.1186/s13059-017-1217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 31.LaCroix RA, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 33.Conrad TM, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wielgoss S, et al. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc Natl Acad Sci USA. 2013;110:222–227. doi: 10.1073/pnas.1219574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikuni O, et al. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinçbas-Renqvist V, et al. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 38.Mora L, Heurgué-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J Biol Chem. 2007;282:35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura Y, Uno M, Toyoda T, Fujiwara T, Ito K. Protein tRNA mimicry in translation termination. Cold Spring Harb Symp Quant Biol. 2001;66:469–475. doi: 10.1101/sqb.2001.66.469. [DOI] [PubMed] [Google Scholar]
- 40.Uno M, Ito K, Nakamura Y. Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc Natl Acad Sci USA. 2002;99:1819–1824. doi: 10.1073/pnas.032457599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe Y, Nakamura Y, Ito K. A novel class of bacterial translation factor RF3 mutations suggests specific structural domains for premature peptidyl-tRNA drop-off. FEBS Lett. 2010;584:790–794. doi: 10.1016/j.febslet.2009.12.048. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura K, Ito K, Kawazu Y, Mikuni O, Nakamura Y. Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J Mol Biol. 1996;258:588–599. doi: 10.1006/jmbi.1996.0271. [DOI] [PubMed] [Google Scholar]
- 43.Adamski FM, McCaughan KK, Jørgensen F, Kurland CG, Tate WP. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J Mol Biol. 1994;238:302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- 44.Tao H, Bausch C, Richmond C, Blattner FR, Conway T. Functional genomics: Expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M, et al. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- 46.Brown CM, Stockwell PA, Trotman CNA, Tate WP. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990;18:2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monk JW, et al. Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli. ACS Synth Biol. 2017;6:45–54. doi: 10.1021/acssynbio.6b00192. [DOI] [PubMed] [Google Scholar]
- 49.Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunjapur AM, et al. Engineering posttranslational proofreading to discriminate nonstandard amino acids. Proc Natl Acad Sci USA. 2018;115:619–624. doi: 10.1073/pnas.1715137115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold FH. Design by directed evolution. Acc Chem Res. 1998;31:125–131. [Google Scholar]
- 52.Reisch CR, Prather KLJ. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci Rep. 2015;5:15096. doi: 10.1038/srep15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilar C, et al. Genetic changes during a laboratory adaptive evolution process that allowed fast growth in glucose to an Escherichia coli strain lacking the major glucose transport system. BMC Genomics. 2012;13:385. doi: 10.1186/1471-2164-13-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.