Significance
This study addresses why splenic B lymphocytes show differential cell proliferation and death decisions, and whether these may be predictable. Biology provides examples of both stochastic decision making and highly deterministic developmental programs. Prior studies of B lymphocytes suggested these cells make stochastic decisions, but the key experiment—long-term tracing of individual cell lineages—has not been done. Overcoming the technical challenges, we found that B cell fate decisions are largely nonstochastic. Using a mathematical model, we found that they are predictable, as long as the state of the molecular network in founder cells is known. That allowed us to identify the molecular determinants of proliferative fate decisions, which potentially constitute novel drug targets and biomarkers for B cell-mediated diseases.
Keywords: B cells, math model, heterogeneity, systems biology, proliferation
Abstract
Rapid antibody production in response to invading pathogens requires the dramatic expansion of pathogen-derived antigen-specific B lymphocyte populations. Whether B cell population dynamics are based on stochastic competition between competing cell fates, as in the development of competence by the bacterium Bacillus subtilis, or on deterministic cell fate decisions that execute a predictable program, as during the development of the worm Caenorhabditis elegans, remains unclear. Here, we developed long-term live-cell microscopy of B cell population expansion and multiscale mechanistic computational modeling to characterize the role of molecular noise in determining phenotype heterogeneity. We show that the cell lineage trees underlying B cell population dynamics are mediated by a largely predictable decision-making process where the heterogeneity of cell proliferation and death decisions at any given timepoint largely derives from nongenetic heterogeneity in the founder cells. This means that contrary to previous models, only a minority of genetically identical founder cells contribute the majority to the population response. We computationally predict and experimentally confirm nongenetic molecular determinants that are predictive of founder cells’ proliferative capacity. While founder cell heterogeneity may arise from different exposure histories, we show that it may also be due to the gradual accumulation of small amounts of intrinsic noise during the lineage differentiation process of hematopoietic stem cells to mature B cells. Our finding of the largely deterministic nature of B lymphocyte responses may provide opportunities for diagnostic and therapeutic development.
How similar cells give rise to distinct fates is a fundamental question with different answers in different biological contexts. Cell fates are established deterministically for some cell types or organisms, such as Caenorhabditis elegans (1). In other cases, cell fates are established seemingly stochastically, such as for the development of competence by the bacterium Bacillus subtilis (2) or the generation of alternative color vision photoreceptors in Drosophila melanogaster (3), and are thereby independent of cellular history (4). Here, we examined whether B lymphocyte proliferation decisions are the result of stochastic or deterministic fate decisions, and whether molecular network determinants may be identified.
B lymphocytes are an essential component of the adaptive immune response and source of antibody-producing cells. In response to invading pathogens, B lymphocytes rapidly proliferate, differentiate into antibody-producing cells, and produce antigen-specific antibodies, which are essential for an effective immune response. B cells genetically diversify by rearranging the Ig locus to produce a diverse antibody repertoire and, therefore, diverse B cell receptor (BCR)-antigen affinities, which control mitogenic signals.
While genetic heterogeneity arising from BCR diversification has the potential to be a source of heterogeneity of B cell fate, BCR-antigen affinity is a poor predictor of B cell proliferative expansion (5), and snapshot flow-cytometry measurements reveal a high degree of cell-to-cell generational heterogeneity even in response to BCR-independent stimuli (6). This led to the notion that B cell fate decision-making is highly stochastic. Indeed, direct measurement of division times at single-cell resolution revealed a highly variable first division (7, 8), consistent with a stochastic decision-making process. Based on these observations, Hodgkin et al. (9) developed a phenotypic model of lymphocyte proliferation using probability distributions of division and death times. The Cyton model has shown remarkable ability to fit dye dilution measurements by flow cytometry and derive corresponding cell biological parameters (such as division and death times) (9–13).
Whereas a key assumption of the Cyton model is the independent stochastic decision-making of each cell at each generation, direct observation of sibling cell behavior revealed correlations in cell fate decisions and division times (8, 10, 11, 14). This has prompted revisions of the model to consider heritability. Thus, lymphocyte population dynamics models have been proposed that structure cell decisions by age (9, 15, 16) or division number (17) (or technical aspects; refs. 18 and 19). However, the degree to which fate decisions are nonstochastic remains unclear (20). Recently developed approaches combining multiple division-tracking dyes revealed that clonal populations were all of a similar generation at given time-points during the proliferative expansion phase (21). To mathematically account for these results, one recent study proposed a distributed division destiny time that is inherited through cell division, controlled in part by the proto-oncoprotein Myc and a separate “time-to-die” mechanism (22).
Prior studies therefore provide the basis for considering the molecular mechanisms underlying B cell decision-making and, thereby, quantify the degree of inheritance versus intrinsic noise. Generally, progeny cells are thought to inherit proteomic networks that mediate decisions (23, 24), Indeed, direct observation of protein abundances indicated that the mixing time of inherited proteins exceeds two generations (more than 40 h) (25). However, in studies of TRAIL-induced death, the concordance of cell fates among siblings decayed rapidly (with a half-life of 1.5 h) (23). Blocking protein synthesis slowed this loss of concordance, indicating a substantial role for intrinsic gene expression noise (26). To what degree gene expression noise or other sources of intrinsic molecular variability affect phenotypic heterogeneity of B cell decision-making remains to be determined.
In the present study, we addressed the molecular underpinnings of the heterogeneity of cell fate decisions during B cell expansion and examined the roles of heritability and intrinsic noise. To obtain accurate, longitudinal, single-cell lineage information, we established an experimental workflow for long-term live cell microscopy of primary B cells and a computational workflow for image analysis and data processing. Resulting data were then used to parameterize a multiscale mechanistic model that accounts for B cell proliferation as a function of the interplay of cell cycle and apoptosis molecular networks, each receiving inputs from a multidimeric NF-κB signaling model (24). Using an iterative systems biology approach, we could thereby quantify the degree of heritability and stochasticity in cell fate decisions, identify the sources of phenotypic cell-to-cell heterogeneity, and then computationally predict and experimentally confirm cell-intrinsic determinants of proliferative capacity.
Results
B Lymphocyte Proliferative Decisions Are Predictable.
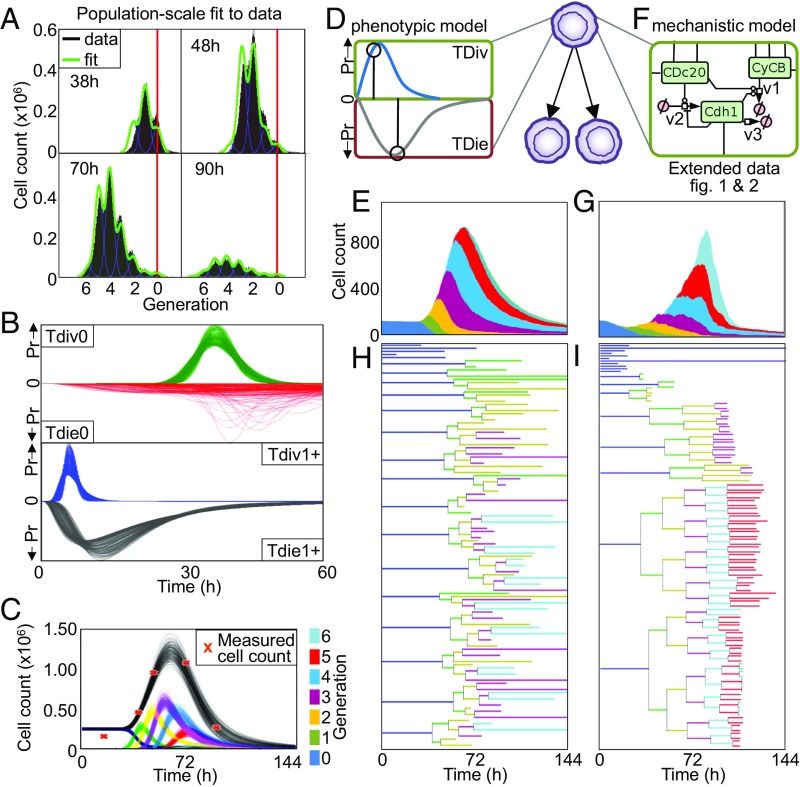
To study the heterogeneity of underlying B cell fate choices, we circumvented the complexity of differential BCR-antigen affinities by using the BCR-independent stimulus, Toll-like receptor 9 ligand, CpG, in dye dilution assays with Cell Trace Red (CTR) monitored at several time points by flow cytometry (Fig. 1A). The diversity of cellular responses within the population could be determined by fitting the probability distributions of the established nonheritable phenotypic-scale Cyton model to the data (9, 13), revealing that cell division and cell death times of proliferating cells vary between ∼4 and 12 h and 8 and 48 h, respectively (Fig. 1B). These distributed timings underlie the successive peaks of individual generations while the maximum total cell count was reached at about 72 h (Fig. 1C). We extended the Cyton model to a single-cell probabilistic formulation (referred to as the phenotypic model), in which each simulation sampled from these distributions of division and death times (Fig. 1D), and showed that it also recapitulated the total and generation-specific population dynamics (Fig. 1E). An alternative, multiscale model (referred to as the mechanistic model) of single-cell fate decision-making as a function of the molecular networks that control cell cycle, cell-survival/death, and the NF-κB signaling network (Fig. 1F and SI Appendix, Figs. S1 and S2) (24) also adequately recapitulated the total and generation-specific population dynamics (Fig. 1G).
Fig. 1.
Phenotypic and mechanistic cell fate models of lymphocyte expansion predict similar cell population dynamics but distinct single-cell lineage trees. (A) Histogram of cell generations revealed by a dye dilution timecourse (38, 48, 70, and 90 h) of CTR-labeled, CpG-stimulated B cells. Plots indicate undivided founder cells (generation “0,” red line), total cell count (black histogram), and best fit of the probabilistic Cyton model using computational tool FlowMax (13) (green). (B) Probability density plots of log normal distributions of division and death time for founder cells (Tdiv0/Tdie0) and subsequent generations (Tdiv1+/Tdie1+) following FlowMax fitting to the data in A. (C) Line graphs of total cell count (black) and individual generations (indicated colors) over time following FlowMax fitting from A, compared with measured cell counts (orange crosses). (D) Schematic of the single-cell phenotypic cell-fate model in which cell fate is determined by independently sampling division and death times at each generation from appropriate distributions, the shortest of which determines cell fate. (E) Stacked area plot of total cell counts of the probabilistic single-cell model with the contribution of each generation marked with the same color as used in C. (F) Schematic of the mechanistic single-cell model in which molecular signaling networks determine cell fate and are inherited during cell division (24). (G) Stacked area plot of total cell counts of the mechanistic single-cell model with the contribution of each generation (as in E). (H) Lineage tree generated using the probabilistic single-cell model. (I) Lineage tree generated using the mechanistic single-cell model.
Although the two models (phenotypic and mechanistic) account for dye dilution flow cytometry data, they are based on fundamentally different assumptions. The phenotypic model assumes that each cell at each generation makes a stochastic decision about its division and death times. The mechanistic model assumes that cellular decisions are predictably determined by a molecular network, whose constituents are naturally inherited from their predecessor; in this case, variable cell fates are then largely due to preexisting differences in the molecular networks of founder cells. Given these different assumptions, we sought to produce distinguishable predictions by plotting respective lineage trees. We found the lineage trees were fundamentally distinct: The phenotypic model yields many similar looking lineage trees, with a range of terminal generations within each single lineage (Fig. 1H); the mechanistic model yields highly diverse lineage trees but with all progeny within a lineage sharing the same terminal generation (Fig. 1I). The fundamental difference in the lineage trees generated by each modeling approach is a reflection of the degree of inheritance within each lineage. The phenotypic model (without inheritance) assumes division and death mechanisms reset with each division and generates lineage trees consistent with a mechanistic model in which signaling molecule concentrations are resampled from a wide distribution with each division (SI Appendix, Fig. S3A). Conversely, the recently reformulated phenotypic model with heritable division destiny (22) generates lineage trees consistent with the mechanistic model with perfect inheritance (SI Appendix, Fig. S3B).
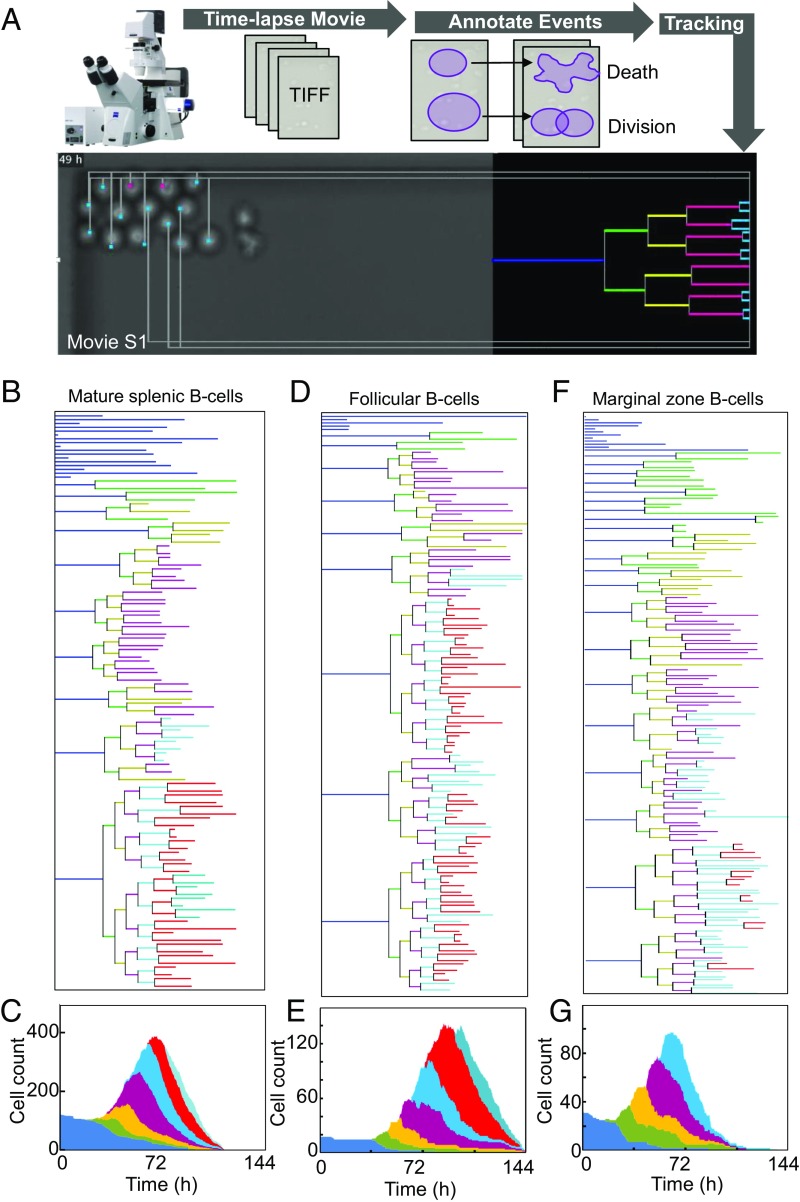
To quantify what degree of heritability best reflects B cell fate choices during population expansion, we developed a long-term time-lapse microscopy workflow coupled to accurate single-cell tracking (Fig. 2A and Movie S1). The resulting lineage trees showed a high degree of regularity as each founder cell gave rise to progeny of generally equal terminal generations (Fig. 2 B and C). However, founder cells differed dramatically in their proliferative capacity or the terminal generation of the lineage tree. Such intralineage regularity and interlineage heterogeneity was also observed within specific B cell subtypes (Fig. 2 D–G and SI Appendix, Fig. S4). In sum, mathematical models of B cell proliferative decisions must contain heritable mechanisms to accurately predict lineage data.
Fig. 2.
Lymphocyte lineage trees produced by long-term live cell microscopy. (A) Schematic of the live-cell time-lapse microscopy pipeline. See Movie S1 for annotated example of single-cell lineage tree construction. (B) Representative lineage tree for CpG-stimulated splenic mature B cells. (C) Stacked area plot of total cell count of the tracked cells with the contribution of each generation marked with similar color used in Fig. 1C. (D) Representative lineage tree for CpG-stimulated Follicular B cells. (E) Stacked area plot of total cell count of the tracked cells with the contribution of each generation marked with similar color used in Fig. 1C. (F) Representative lineage tree for CpG-stimulated splenic marginal zone B cells. (G) Stacked area plot of total cell count of the tracked cells with the contribution of each generation marked with similar color used in Fig. 1C. TIFF, tagged image file format.
Heterogeneity Between Lineages Exceeds Intrinsic Noise.
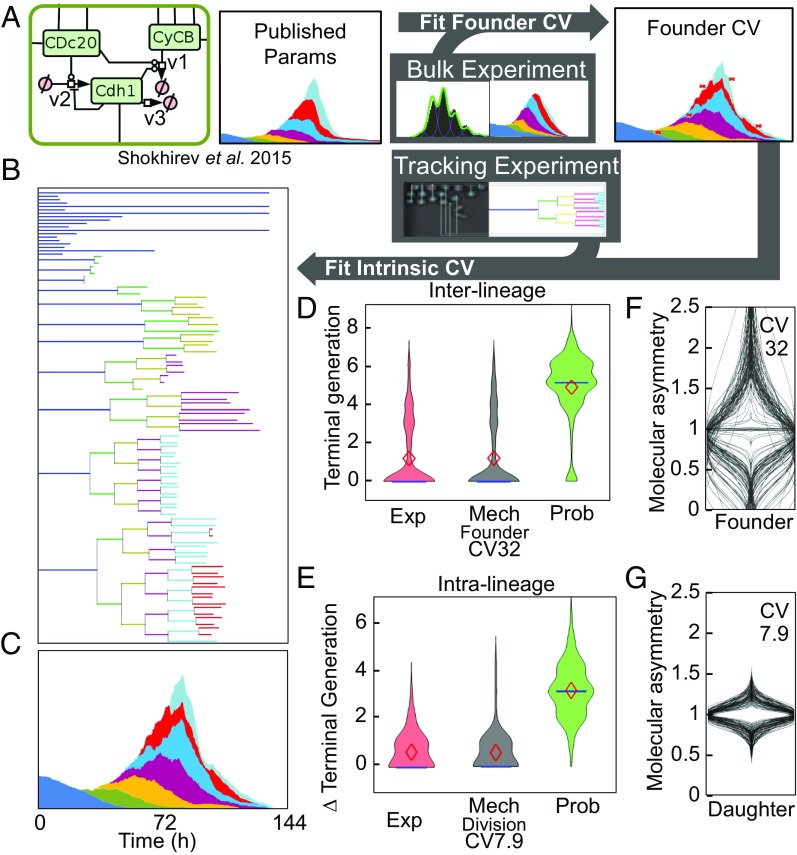
To quantify the relative contribution of interlineage (preexisting) and intralineage (intrinsic) heterogeneity, we used cell lineage data to parameterize these two sources of heterogeneity in the mechanistic model (Fig. 3A) such that simulated cell lineages (Fig. 3B) and generation-specific population dynamics (Fig. 3C) provide optimal fits to the experimental data. We considered two metrics in particular: interlineage differences in the terminal generation (Fig. 3D) and intralineage differences in terminal generation (Fig. 3E). Whereas the phenotypic model that lacks inheritance (probabilistic model) showed distributions that are quite distinct from the experimental result, the mechanistic model enabled tuning of preexisting heterogeneity and intrinsic noise to fit the experimental data. We first determined how widely biochemical parameters within molecular networks would need to be distributed between founder cells to optimally account for the observed distributions in terminal generations (SI Appendix, Fig. S5), and found it to correspond to an average coefficient of variation (CV) of 32% (Fig. 3F). Then we determined the width of the distribution of perturbations added to progeny molecular networks at each division that optimally accounted for differences in terminal generation within lineages and found it to correspond to a CV of 7.9% (Fig. 3G and SI Appendix, Figs. S6 and S7). The resulting, parameterized heritable mechanistic model produces lineage trees and generational distributions (Fig. 3 B and C) that closely match those observed by live cell tracking (Fig. 2 B–G).
Fig. 3.
Quantification of extrinsic and intrinsic cell-to-cell variability by interpreting lymphocyte lineage trees produced by long-term live cell microscopy. (A) Schematic of the optimization approach used to quantify the level of cell-to-cell variability between founder cells and within lineages. All parameters are taken from ref. 24 and the CV between founder cells and between cells within lineages are optimized using particle swarm optimization to minimize the distance between the live-cell tracking and the simulation. (B and C) Representative lineage tree and cell counts generated by the mechanistic model with 32% founder-cell CV and 7.9% CV of asymmetric division of the daughter cells. (D) Violin plot of the distribution, mean (♦), and median (―) of the maximal terminal generation obtained from 125 founder cells in experimentally tracked cells (Exp), mechanistic model simulations with indicated extrinsic CV (Mech), and the single-cell probabilistic model (Prob). (E) Violin plot, mean (♦), and median (―) of the distribution of ∆ (difference between the maximum and minimum) terminal generation within each lineage in 125 experimentally tracked lineages (Exp), mechanistic simulations with 31% extrinsic CV and indicated intrinsic CV (Mech), and the single-cell probabilistic model (Prob). (F) Violin plots of the distributions of each molecular species concentration within 125 simulated founder cells required to recapitulate the experimental lineage tree. Median CV for all metabolites: 32%. (G) Violin plots of the distributions of each molecular species concentration in the 100 simulations with the lowest distance between the ∆ terminal generation distribution of experimentally tracked cells and the model (optimum CV 7.9%).
The model predicts an average 32% CV in founder cell metabolites, however, due to molecular network architecture, not all signaling molecules are predicted to have the same magnitude of heterogeneity. We generated individual predictions for three key molecules (Bcl-2, Myc, and CycD) that showed distinct CVs (SI Appendix, Fig. S8). Using intracellular antibody staining followed by flow cytometry, an experimental technique that introduces an unknown amount of technical noise, we obtained higher CVs, but found, remarkably, that the relative CV associated with Myc and CyclD normalized to the lowest observed (associated with Bcl2) was accurately predicted (SI Appendix, Fig. S8C).
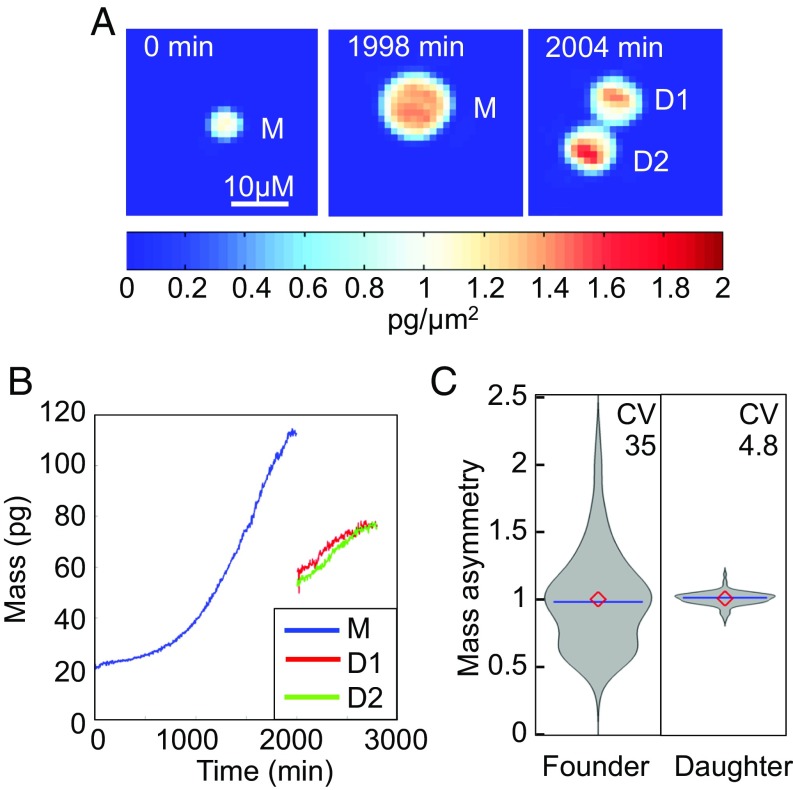
As modeling the experimentally observed lineage trees suggested that the symmetry of division must be tightly controlled (CV 7.9%), while signaling network states are widely distributed between founder cells (CV 32%), we asked whether this heterogeneity is also manifest in measurements of cell mass. Cell mass may be measured precisely by adapting quantitative phase microscopy, which measures the phase shift of light as it passes through and interacts with the material inside of cells (27), to live cell-tracking experiments. We quantified mass distributions in founder cells (at 5 h after stimulation) and in daughter cells following division (Fig. 4 A and B) and found that founder cell masses were widely distributed (CV ∼ 35%) but that cell division was generally highly symmetric with daughter cell masses having a CV of 4.8% (Fig. 4C). While it is unclear how cell mass asymmetry relates to B cell fate divergence during the proliferative expansion phase, our model simulations employ a shuffling (at each division) of protein concentrations (SI Appendix, Fig. S6) or kinetic rate constants (SI Appendix, Fig. S7) (that are mediated by enzymes not explicitly represented in the model). The concordance between molecular network distributions and mass distributions may suggest that they are the result of the same sources of heterogeneity and tightly controlled within lineages but distributed among founder cells.
Fig. 4.
Validation of extrinsic and intrinsic cell-to-cell variability predictions by live cell interferometry. (A) Representative quantitative phase measurement of a single founder cell (M) and the corresponding daughter cells (D1 and D2) at 0 min, 1,998 min, and 2,004 min after the start of imaging. The color code represents area density [mass (pg)/area (μm2)] with blue color as lower area density and red as higher area density. (B) Biomass over time for the annotated cells. (C) Violin plot of the distribution of biomass ratios for 188 founder cells at 5 h after stimulation and daughter cells resulting from 93 division events tracked by live cell quantitative phase microscopy.
Proliferative Fates Are Distinct Due to Nongenetic Heterogeneity of Founder Cells.
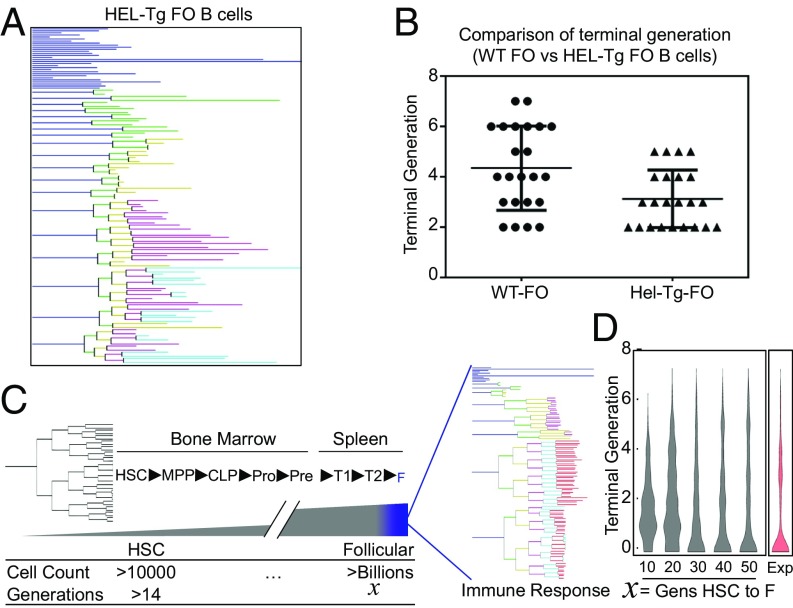
We wondered whether differences in the molecular networks in founder cells that determine cell fate may be a consequence of genetic BCR diversity. (Although the TLR9 stimulus CpG is, of course, a BCR-independent stimulus, distinct BCRs may produce distinct signaling histories and, hence, molecule networks.) To address this question, we utilized BCR-transgenic mice in which >90% of B cells carry the same BCR specific for the hen egg lysosome (HEL) antigen (28). Both B cell subtypes showed differences in terminal generation between founder cell lineages that are largely indistinguishable from nontransgenic controls, which is evident from dye dilution assay (SI Appendix, Fig. S9) and single-cell lineage tracking (Fig. 5 A and B). This indicates that heterogeneity in the molecular networks of founder cells has nongenetic origins.
Fig. 5.
Nongenetic heterogeneity is the source of cell-to-cell heterogeneity. (A) Lineage trees of HEL transgenic follicular B cells (HEL-Tg FO B cells). (B) The terminal generation reached by each dividing founder cell of wild-type (WT) FO B cell and HEL-Tg FO B cell. (C) Schematic depicting the origins of the splenic founder B lymphocyte, and the potential accumulation of intrinsic noise at each division. Hematopoietic stem cells (HSC) differentiate into multipotent progenitor cells (MPP), MPPs differentiate into common lymphoid progenitors (CLP), CLPs differentiate into progenitor B cells (Pro), Pro cells differentiate into precursor B cells (Pre), Pre cells differentiate into transitional 1 B cells (T1), T1 cells differentiate into transitional 2 B cells (T2), and T2 cells differentiate into follicular B cell (F). HSC to Pre cell differentiation occurs in bone marrow, and T1 to F differentiation occurs in spleen. (D) Violin plot of the distribution of terminal generations reached by 125 simulated founder cells (gray) with the indicated number of partitioning steps in which the molecular network is distributed (CV 7.2%) before stimuli and experimentally (Exp) tracked lineage information (red).
Many potential sources of nongenetic differences in founder cells are possible including, but not limited to, distinct histories of migration from the bone marrow, residence times in microenvironments, and prior antigen exposures. One possible source of the nongenetic differences in the state of molecular networks in mature splenic B cells is simply the accumulation of intrinsic noise through successive generations in each founder cell’s history (Fig. 5C). Based on our estimates of intrinsic noise and measurements of division asymmetry, we found that more than 30 generations would be required to distribute the molecular network sufficiently to explain the distribution of terminal generations observed by lineage tracking (Fig. 5D). This number is compatible with estimates of the number of divisions separating hematopoietic stem cells and splenic B cells (29–31). Thus, while distinct founder cell histories are more than likely, they may not be a required driver of founder cell heterogeneity.
The Population Response Is Dominated by Founder Cells with Low Propensity for Apoptosis.
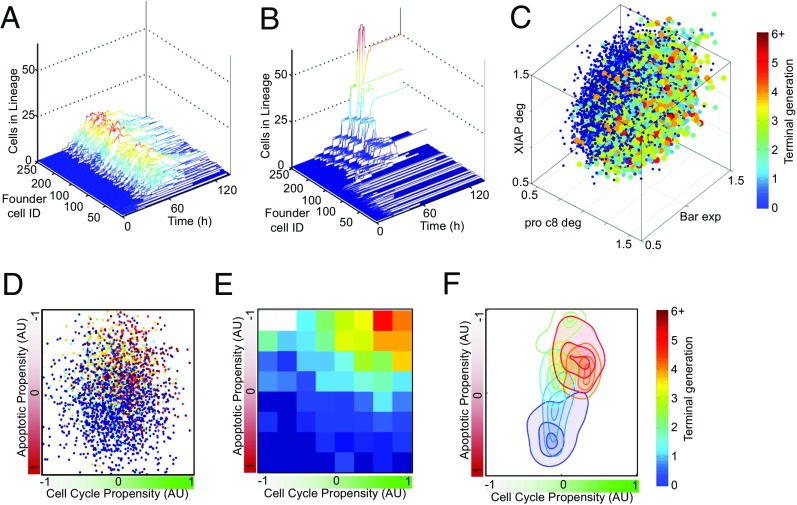
A consequence of the nonheritable phenotypic model, in which differences in terminal generation are predominantly determined by intrinsic noise during proliferation, is a prediction that many founder cells contribute similarly to population responses (Fig. 6A). In contrast, the mechanistic model, in which cell fate differences are predominantly driven by heritable differences in the molecular networks of founder cells, predicts that a remarkably small proportion of founder cells contribute to the expanded population. The lineages of these founder cells survive for a disproportionate number of generations, resulting in a large number of descendant cells that dominate the population response (Fig. 6B). Given the largely deterministic nature of cell fate decisions characterized by the mechanistic model, it ought to be possible to predict which founder cells will contribute highly to the population and undergo many divisions. Using multivariate statistical regressions, we sought molecular network components that are associated with founder cells that are dominant contributors to the overall population response. Although no single reaction rate was a reliable predictor of proliferative capacity, in principle, knowledge of the founder cell molecular network could predict its proliferative capacity (R2 ∼ 0.6) (SI Appendix, Fig. S10A). While each individual network component is a poor predictor, combinations such as low procaspase 8, high Bar, and high XIAP are predictive of high proliferative capacity (Fig. 6C and SI Appendix, Fig. S11). Indeed, classifying the state of founder cell molecular networks, in terms of propensity for apoptosis and cell cycle activity (Fig. 6 D–F), demonstrated that low apoptotic propensity was a strong indicator of founder cell terminal generation with nondividing cells. Cells which did not divide could be separated from cells that divided three or more times by difference in the state of the founder cell’s apoptosis regulatory network (Fig. 6F).
Fig. 6.
Identifying nongenetic determinants of founder B cell proliferative capacity. (A and B) Line plots of the number of descendent cells from 250 founder cells over time (0–120 h). Descendant cell numbers generated using the single-cell probabilistic model (A) and descendant cell numbers generated using the mechanistic model (B), colored to indicate the contribution of each founder cell’s descendant relative to the maximum. (C) Three-dimensional scatterplot of the terminal generation (color code) reached by 20,000 founder cells with distributed kinetic parameters against the values for the first three nonredundant predictors identified by LASSO [XIAP degradation rate (XIAP deg), procaspase 8 degradation rate (pro c8 deg), and bifunctional apoptosis regulator expression rate (BAR exp)]. (D) Scatter plot of the terminal generation reached by single-cell simulations. The position of each cell is determined by the state of five key signaling molecules in each of the apoptosis and cell cycle networks of each founder cell. The color code for terminal generation is shown on the right. (E) Heatmap of the data in D binned into eight equally spaced bins for each module’s activity. (F) Density contour map for the data in D. Contours are drawn at 0.85, 0.95, and 0.99, indicating the location of the highest density regions for each generation in the space created by the apoptosis and cell-cycle signaling network states.
Key Molecular Regulators in Founder Cells Are Identifiable.
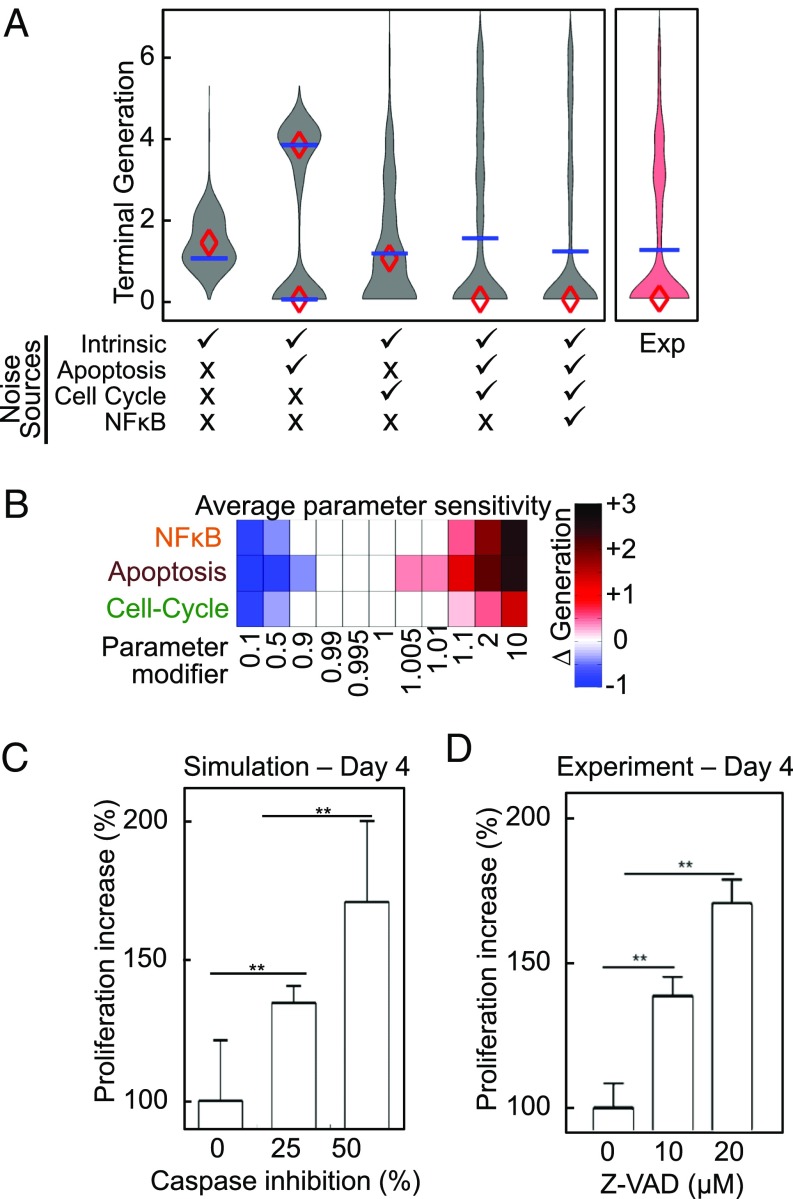
To characterize how nongenetic heterogeneity within the signaling networks of founder cells affects their proliferative capacity, we compared the distributions of terminal generations resulting from simulations containing only intrinsic noise with those that included distributed founder cell molecular network states within the apoptosis and cell cycle networks (Fig. 7A). We found that distributing the components of the apoptosis control network resulted in a bimodal distribution of proliferative capacities, including not only cells with a higher propensity to die curtailing even a first division, but also founder cells of lineages with much higher terminal generations. Additionally, distributing the components of the cell cycle network could result in a further increase, but also decrease in terminal generations (Fig. 7A and consistent with Fig. 6A).
Fig. 7.
Identifying and targeting the molecular determinants of lymphocyte proliferation. (A) Violin plots, mean (♦), and median (―) of the distributions of terminal generations reached in a simulation of 250 founder cells with identical starting molecular network states and subsequent asymmetric division (intrinsic noise), with the addition of distributed starting state of the apoptosis network, cell cycle network, NF-κB network, and combinations of these. Experimentally (Exp) tracked terminal generation is shown in red. (B) Sensitivity analysis heatmap of change in terminal generation reached by the mean founder cell when parameters within each indicated network are scaled by the indicated parameter modifiers across two orders of magnitude. (C) Bar graph of the proliferation increase predicted by the mechanistic simulation at day 4 following caspase inhibition. One hundred twenty-five founder cells were simulated with caspase inhibition reactions scaled as indicated, and Cdh1 positivity was used as a marker for proliferating cells. **P ≤ 0.01 by Student’s t test. (D) Bar graph of experimentally measured proliferation increase following inhibition of caspase activity using Z-VAD (10 and 20 μM) at day 4. Ki67 expression was used as a marker for proliferating cells. Data shown is from simulation and biological triplicates at the 4-d timepoint following CpG stimulation. **P ≤ 0.01 by Student’s t test.
A sensitivity analysis found that many reaction rates within the signaling network controlling cell fate could perturb the terminal generation of stimulated lymphocytes; however, terminal generation was predicted to be most sensitive to the state of the apoptosis control network in founder cells (Fig. 7B and SI Appendix, Fig. S11). The sensitivity predictions are consistent with B cell proliferation control by cRel and IκBε seen in previous experiments (24, 32). Perturbations targeting the cell-cycle network (such as Myc overexpression) are predicted to increase proliferation (SI Appendix, Fig. S12A), and the computational model recapitulated increases in total cell number and mean division number reported recently (SI Appendix, Fig. S12B) (22). The prediction that terminal generation was most sensitive to small perturbations within the apoptosis control network such as those controlling the level and activity of caspases (SI Appendix, Fig. S9), rather than cell growth and cell cycle control networks, was surprising as inhibition of the mitochondrial amplification loop was not predicted by the model to substantially alter proliferation, a result also confirmed in a recent experimental study of Bcl-2 overexpression (22, 24). However, the sensitivity analysis predicted that inhibition of the central apoptotic caspase cascade would significantly enhance proliferation (SI Appendix, Figs. S11 and S12D).
To generate a testable quantitative prediction of the degree of increased proliferation resulting from caspase inhibition, we simulated the effect of a 25% and 50% reduction in caspase activity, predicting a dose-dependent increase in the fraction of proliferating cells at late time points (Fig. 7C). To test this prediction experimentally, naïve B cells were treated with pan-caspase inhibitor Z-VAD (33, 34) before CpG stimulation; indeed, we found increased B cell proliferation at late time points in a dose-dependent manner (Fig. 7D), based on an increased fraction of cells positive for the proliferation marker Ki67 (SI Appendix, Fig. S13). Examining why survival signals would enhance proliferative capacity in more detail, we found that while growing B cells are protected from cell death in early divisions, as previously documented (24), during later divisions, cell death can occur even in growing cells, resulting in censoring of subsequent generations (SI Appendix, Fig. S14). Thus, cell death regulation emerges as a critical determinant of proliferative capacity of founder cells during the B cell immune response.
Discussion
In this study, we showed that the diversity of B cell proliferative fate decision-making is largely the result of nongenetic differences between founder cell molecular network states. Indeed, utilizing a mechanistic mathematical model of the molecular interaction networks that determine cell proliferation decisions, we were able to identify predictive markers of a high proliferative capacity and potential drug targets that control lymphocyte proliferation with high sensitivity.
The nonheritable phenotypic models of lymphocyte proliferation have demonstrated great utility in quantifying cell-biological parameters (such as division and death time) from snapshot flow cytometry data. These models were based on the assumption that cells at each generation engage in stochastic decision-making process (7, 8, 10, 12, 24, 35). However, recent studies revealed concordance between recently divided siblings (14) and a recent revision of the phenotypic model proposed to assign division destiny times to founder cells that is then inherited to descendants through the lineage. The present study extends these findings by demonstrating that heritability of phenotypic cell fate from one generation to the next can be understood as a function of the heritability of the underlying molecular networks. Leveraging the availability of sufficient high-quality lineage data (complete lineage trees with no missing branches), we could quantitatively distinguish between the heritability vs. seeming stochasticity of cell fate decisions through the lineage. Using the molecular mechanistic model of B cell fate decisions, we could then the quantify the contributions of heterogeneity in the molecular networks of the starting founder cell population (“extrinsic noise”) vs. randomization of abundances of molecular network components at each division (“Intrinsic noise”) and how each noise source accounts for the heterogeneity of B cell fates in the population. We found that the former with a CV of 32% substantially exceeded the latter with a CV of 7.9%.
Our findings are surprising in the context of previous studies quantifying the heritability of fate decisions by HeLa cells treated with the apoptosis-inducing TNF receptor superfamily ligand TRAIL (23). These studies also found cell fate concordance of recently divided siblings but reported a decay of this concordance with a half-life of 1.5 h (a longer half-life when gene expression was blocked) (23). Including such a substantial degree of intrinsic noise into our B cell model would produce lineage trees indistinguishable from the nonheritable models (Fig. 1H and SI Appendix, Fig. S3A), but not account for the experimentally observed lineages. Our study therefore revealed an unexpected degree of heritability, suggesting that intrinsic noise is tightly controlled during lymphocyte proliferation. This may indicate that gene expression noise itself is limited or that topological characteristics of the network minimize the effect of that noise as, for example, observed for the MyD88 pathway in macrophages (36) or the chemotaxis network of Escherichia coli (37). This is supported by the high degree of symmetry in the division mass (CV ∼ 5%). Given that even this small asymmetry of division nevertheless may contribute to randomization of cell fate decisions, we surmise that the gene expression noise in proliferating B lymphocytes is remarkably small.
What may be the reasons for the discrepancies between HeLa cell apoptosis and B cell proliferation? Aside from invoking cell type differences, we wonder whether gene expression noise may be a function of whether cells are in a basal steady state or in a highly activated state in which protein production is maximized. The distinction between basal vs. activated states may also pertain to the covariance of gene expression, a mechanism proposed to limit phenotypic heterogeneity in T lymphocytes (38); it is possible that gene expression variation relevant to HeLa cell apoptosis control is largely independent, whereas there is a greater degree of covariance in the expression of genes regulating lymphocyte proliferation. We note that key regulators of lymphocyte proliferation are targets of the same stimulus-induced transcription factor, NF-κB.
As B cell expansion is largely determined by the highly heterogeneous proliferative capacity of mature B cells, we sought to identify the underlying molecular determinants. We focused our studies on the molecular networks that are engaged regardless of the mitogenic stimulus, that is the IKK–NF-κB signaling network, the cell cycle control network, and the apoptosis control network. Among these, expression variation that determined apoptotic susceptibility was found to be the most predictive of proliferative capacity (Figs. 6 and 7). However, when including receptors and their proximal signaling pathways, we may indeed find other prominent molecular sources of phenotypic heterogeneity. Indeed, in studies of T cell responses, interleukin-2 receptor α (IL-2Rα) was identified as a significant predictor of cell fate, with its expression varying across five orders of magnitude. In contrast, TLR9 (the cognate receptor for CpG) shows an expression variability of one order of magnitude (38–41), insufficient to be a key determinant in the stimulation conditions employed in our studies. In the absence of one key determinant of phenotypic heterogeneity, a regression analysis of high-throughput simulations of the mechanistic model suggests that many individual molecules contribute to the cell’s overall propensity for proliferation and survival (Fig. 6 C–F). Measuring expression variation directly is challenging as the functionally relevant variability in expression (e.g., 32% CV) is comparable to the technical variability of single-cell assays such as flow-cytometry or fluorescence microscopy using antibodies or fluorescent reporters (39).
The mechanisms by which the distinct molecular network states of founder cells are acquired in vivo likely involve a combination of intrinsically stochastic processes and distinct environmental exposure histories for each cell. In the absence of any other sources of variability, the small degree of intrinsic noise quantified here may be sufficient to establish heterogeneous founder cell states only after more than 30 generations (Fig. 5D). Low abundance components such as mitochondria have recently been identified recently as a source of variability in TRAIL-induced apoptosis (42). Heterogeneous chromatin states may also contribute to the gene expression heterogeneity among founder cells. For example, in T lymphocytes, chromatin states have been found to contribute to IL-4 expression variability and maintain its state over ∼3 d (43). This relatively long maintenance of chromatin state could represent a cellular memory of distinct exposure histories that contributes to founder-cell heterogeneity but is unlikely to contribute to intralineage heterogeneity.
What may be the physiological consequences of nongenetic differences in splenic founder B cell molecular networks? Our work suggests the hypothesis that founder cell molecular heterogeneity contributes significantly to noise in lymphocyte clonal selection based on antigen-antibody affinity; as such, it might increase the diversity of the antibody repertoire. Antibody diversity is critical for an effective immune response (44) and a key indicator of immune health during aging (45). Our results therefore suggest that nongenetic heterogeneity of a splenic B cell plays an important physiological role in immune system function.
Following the proliferative expansion studied here, single lymphocytes can produce distinct cellular progeny, such as memory or plasma B cells (10, 46, 47). These cell fate decisions are also not primarily stochastic but are thought to reflect the uneven impact of the germinal center microenvironment that fosters asymmetric segregation of signaling molecules, transcription factors, and growth factors into daughter cell (46). Thus, despite the great degree of phenotypic heterogeneity, rigorous characterization of molecular interaction networks along with cellular interactions renders both B lymphocyte proliferation and differentiation decisions surprisingly predictable. Indeed, the origins of the phenotypic heterogeneity, that are thus identifiable, are of critical relevance to achieving the ambitious goals of personalized precision medicine.
Methods
B Cell Isolation and Culture.
Spleens were harvested from 10- to 12-wk-old female C57BL/6 mice and Hemizygote BCR transgenic mice (The Jackson Laboratory, stock no. 002595). Homogenized spleens were incubated with anti-CD43 magnetic beads for 15 min at 4–8 °C, washed with MACS buffer [Phosphate buffer saline, (pH 7.4), 0.5% BSA, and 2 mM EDTA (pH 8)] and pass-through LS columns (Miltenyi Biotech). The purity of B cells was >95% based on B220 staining.
Marginal zone (MZ) and Follicular (FO) B cells were isolated using magnetic sorting followed by fluorescence activated cell sorting (FACS). Homogenized spleens were incubated with MZ and FO B cell biotin-antibody mixture for 5 min at 4–8 °C followed by incubation with Streptavidin microbeads for 10 min at 4–8 °C, then washed with MACS buffer and passed through the LS column. The eluted B cell population was enriched with MZ and FO B cells. MZ and FO B cells were purified by FACS sorting. Briefly, the enriched B cell population was stained with B220-eF450 (48-0452-82; eBioscience), CD23-PE (101607; Biolegend), CD21-APC (123411; Biolegend), CD93-FITC (11-5892-81; eBioscience), CD3e-FITC (11-0031-82; eBioscience), CD11b-FITC (11-0112-82; eBioscience) and Ly-6C-Alexa 488 (53-5932-82; eBioscience). The stained cells were gated on B220+CD93−CD3e−CD11b−Ly-6C− population to further exclude immature B cell and any residual non-B cell population present in the enriched B cell population. MZ and FO B cells were sorted based on the expression CD21 and CD23 on B220+ cells (48, 49). The purity of MZ and FO B cells were ∼98% (SI Appendix, Fig. S3).
B cells were grown in fresh media with 1% penicillin streptomycin solution (Mediatech Inc.), 5 mM l-glutamine (Mediatech Inc.), 20 mM Hepes buffer (Mediatech Inc.), 1 mM MEM nonessential amino acid (Sigma), 1 mM Sodium pyruvate (Sigma), 10% FBS, and 55 µM 2-ME (Fisher Scientific).
CTR Labeling and Cell Biological Parameter Estimation.
Purified B cells were labeled with CTR (1 μM) and stimulated with 250 ng/mL CpG ODN 1668 (tlrl-1668-1; Invivogen). The cells were grown in 48-well tissue-culture plates (2 × 105 cells per mL) at 37 °C, 5% CO2 containing humidified chamber for a period of 5 d. Stimulated B cells were collected at different time points and stained with 7 aminoacitomycine D (7AAD) (420404; Biolegend) to exclude dead cells. B cell proliferation and survival were measured in C6 Accuri Flow Cytometry (BD Bioscience) and 100 μL of volume counted in each time point. After manual gating on viable (7AAD-negative) B cells, FlowMax software was used to construct CTR histograms (13). All assays were performed in duplicate. The fluorescence of the undivided peak was manually identified for each time point, and maximum-likelihood fcyton model parameter distributions were identified from 500 best fit solutions.
Quantitative Phase Microscopy Mass Measurement.
Quantitative phase microscopy measures the phase shift of light as it passes through and interacts with the material inside of cells. Purified naïve B cells were stimulated with CpG (250 ng/mL) and grown in poly-l-lysine (PLL)-treated microgrid array (MGA-250; Microsurface) in an environment-controlled microscope as described previously. Images were acquired on an Axio Observer Z1 inverted microscope (Carl Zeiss Microscopy GmbH) using a SID4-Bio quadriwave lateral shearing interferometry (50) camera (Phasics) and an LD Plan Neofluor objective at 40×, 0.6 N.A. Images were acquired with illumination from a 660-nm collimated LED (Thorlabs) every 3 min for 5 d.
Z-VAD Treatment and Proliferation of B Cells.
Proliferation of B cells was measured by Ki67 expression. Ki67 is expressed in the nucleus of cells that are in active cell cycle, thus Ki67 was used as a proliferation marker. (51–53). Purified naïve B cells were pretreated with Z-VAD(OMe)-FMK (10 and 20 μM) (BML-P416-0001; Enzo Life Science International) for 90 min followed by stimulation with CpG (250 ng/mL) for 4 d. Cells were harvested at 4 d, washed twice with PBS, stained with fixable viability dye eF520 (65-0867-14; eBioscience) for 30 min in 4 °C, washed with PBS, and permiblization and staining with Ki67-PerCp-eFluoro 710 (46-5698-82; eBioscience) was performed using Fix and Perm cell permeabilization kit (# GAS003; ThermoFisher) as recommended. Briefly, cells were fixed with reagent A for 15 min at room temperature (RT), washed with PBS, permeabilized, and stained with reagent B for 20 min at RT followed by washing with PBS. Cell proliferation was measured by AccuriC6, and data were analyzed with FlowJo. Cell proliferation was determined by an increase of cell fluorescence index over the naïve B cells. The data are representative of three biological replicates.
Time-Lapse Microscopy.
Purified naïve B cells and MZ and FO B cells were stimulated with CpG (250 ng/mL) and grown in a microgrid array (MGA-250; Microsurface) in an environment-controlled microscope. Images were acquired on an Axio Observer Z1 inverted microscope (Carl Zeiss Microscopy GmbH) with a Pln Apo 20×, 0.8 N.A. objective to a Coolsnap HQ2 CCDcamera (Photometrics) using ZEN imaging software (Carl Zeiss Microscopy GmbH). Environmental conditions were maintained at 37 °C, 8.5% CO2 with a heated enclosure, and CO2 controller (Pecon). Brightfield images were taken every 3 min for 5 d. Microgrid arrays were coated with PLL before incubating cells to reduce the movement of the cells during image acquisition to facilitate lineage tracking.
Cell Tracking.
Cell tracking was performed on phase-contrast images using semiautomated software (24). A representative example of cell tracking and lineage tree generation can be seen in Movie S1 (created in MATLAB). Briefly, founder cells were annotated in the first image, and approximate linear paths were manually drawn for each identified founder cell. While PLL coating and low cell density enables many frames to be approximated to a linear path at early time points, the temporal resolution of cell path annotations necessarily increased in later time points. Founder cells were tracked until a division or death event was manually identified and annotated. Death events were recognized by morphological changes in cell morphology consistent with apoptotic blebbing. Division events were manually annotated such that each daughter cell stores the cell ID of its mother cell, enabling complete lineage reconstruction. The cell tracking software was further developed to report terminal generation, Δ terminal generation, generation-specific cell count, total cells within lineage, and generation-specific SD of division times for each founder cell. To aid comparison of computational models and live-cell microscopy, the experimental cell tracking software was modified to take model-generated lineage information as input and perform identical statistical analysis and lineage tree generation for both in vitro and in silico experiments.
Measurement of Heterogeneity in Intracellular Protein Expression.
Naïve mature B cells were isolated from the spleen as described above. Cells were washed twice with PBS, stained with fixable viability dye eF660 (65-0864-14; eBioscience) for 30 min in 4 °C, washed with PBS, and permeabilization and staining (with primary antibody followed by secondary antibody) were performed using Fix and Perm cell permeabilization kit (GAS003; Thermo Fisher) as recommended. Cell permeabilization and staining protocol was followed as described above with little modification in staining time. Cells were stained 15 min with primary antibody and 15 min with secondary antibody. Cell proliferation was measured by CytoFLEX flow cytometry (Beckman Coulter), and data were analyzed with FlowJo. Protein abundances were graphed by fluorescence histogram, and the CV was determined. CycD1 (sc-718; SantaCruz), Myc (ab32072; Abcam), and Bcl-2 (2870S; Cell Signaling) were chosen as these are key regulators of cell cycle and cell survival.
Mathematical Modeling.
Single-cell cyton model.
A single-cell computational model of the Cyton assumptions was created using MATLAB. Parameters describing the distributions of division and death times (Tdiv and Tdie, respectively) in first generation and subsequent generation cells, along with proliferative fate parameters (pF) from FlowMax were used as input parameters into the model to recapitulate CTR population distributions. Ten thousand founding cells were simulated with identical parameter distributions. To simulate the experimental time course each initial cell sampled from both division and death distributions created by the input parameters and the lowest of the two sampled times determined the fate of the cell. To recapitulate the proportion of cells progressing to the next generation (pF) as identified using FlowMax, every cell sampled from a uniform distribution and determined if the value was greater than the appropriate progression factor for the cells generation; only if the sampled value was less than the progression factor was a division time sampled, otherwise the cell would die with the sampled Tdie. Cells which achieved a division fate triggered the introduction of two new cells that then independently sampled from the distributions corresponding to division 1+ events.
Mechanistic mathematical model.
Mechanistic computational modeling was performed as described previously (24). Briefly, established ordinary differential equation models of the NF-κB signaling network (32), apoptotic control network (54), and cell cycle control network (55, 56) were combined into single-cell simulations (SI Appendix, Figs. S1 and S2). One hundred twenty-five starting cells were used for all simulations except where indicated, for example to generate data for identification of fate predictors in which 20,000 starting cells were simulated to improve statistical power. All parameters were maintained as published previously with the exception of CVs as indicated in Fig. 3 and SI Appendix, Figs. S4–S6. Cleaved PARP (cParp) and cadherin-1 (Cdh1) concentration thresholds triggered cell death and division, respectively (for parameters, see Dataset S1). Division events triggered the addition of two new daughter cells, which inherited reaction rate parameters and metabolite concentrations from the mother cell. The timing of the cell cycle in individual cellular agents is determined by cyclin expression and resultant dynamics in the established mammalian cell-cycle module that, in turn, controls the timepoint at which cdh1 crosses the previously defined threshold (24, 55). Similarly, cell death timings are determined by the dynamics within the established apoptosis module that determines death when cParp exceeds the published threshold (24, 54). The computational modeling code is available in SI Appendix.
Founder cell variation.
To distribute the state of starting cells, all initial conditions were sampled from lognormal distributions, and synthesis and degradation rate constants were sampled from a normal distribution with mean value from the established model and CV indicated. An initial simulation phase was run to enable individual cells to obtain a steady state after which IKK activity was transiently increased recapitulating TLR9 activation. The CV for the distribution from which founder cell parameters were sampled was fit using particle swarm optimization (max velocity: 2%, maximum CV: 50, minimum CV: 0, population size: 20) (SI Appendix, Fig. S4D) in which each particle is a simulation with 125 founder cells distributed with a chosen CV and scored by the Euclidian distance of each generation and total cell count time courses to the experimentally measured data shown in Fig. 2A.
Intrinsic noise.
Simulations of asymmetric distribution were performed by sampling the concentration of metabolites inherited by daughter cells. A division ratio was sampled with a mean of 1 and CV as indicated. The initial condition of all metabolites in one daughter cell were scaled uniformly by the sampled division ratio and inversely scaled in the other daughter cell. The CV for the distribution from which daughter cell parameters were sampled was fit using particle swarm optimization (maximum velocity: 1%, maximum CV: 30, minimum CV: 0, population size: 20) (SI Appendix, Fig. S5C) in which each particle is a simulation with 125 founder cells distributed with a chosen CV and scored by the Chi-Square statistic comparing the distribution of intralineage ∆ terminal generation between the simulated results and the single-cell tracked data (Fig. 3E). Simultaneous particle swarm optimization of both intrinsic and extrinsic noise was also performed to account for potential confounding effects between intrinsic and extrinsic noise; however, a similar optimal CV for both values was found (SI Appendix, Fig. S5D). For simulations in which noise was added with division events, parameters inherited by daughter cells were sampled from with mean value inherited from the mother cell and CV as indicated (SI Appendix, Fig. S6). Estimates of intrinsic noise accumulation were generated by repeated sampling of the distribution of all metabolite concentrations before simulation.
Sensitivity analysis.
Sensitivity analysis of terminal generation was performed on a simulation without division-introduced variation. Scaling factors were sampled at 10 points logarithmically chosen from 10-fold lower and higher than the standard parameter value. A founder cell was simulated for each scaled parameter value, leading to a simulation of 1,680 distinct founder cells. The terminal generation reached by each founder cell was stored to quantify the effect of perturbations in each parameter on terminal generation.
Statistical analysis.
To identify the best predictors of terminal generation, a simulation with distributed founder cell parameters was performed with 20,000 founder cells. Parameter values, initial (steady-state) metabolite concentrations (i.e., SI Appendix, Fig. S4A; x1,1, x2,1, x3,1), and terminal generation were stored for each founder cell. Least absolute shrinkage and selection operator (LASSO) regression was performed using either founder cell parameter values initial (steady-state) metabolite concentrations (i.e., SI Appendix, Fig. S4A; x1,1, x2,1, x3,1) as predictors and maximum depth for each founder cell as observations using MATLAB’s statistical toolbox (1∙10−6 relative convergence threshold for coordinate descent algorithm and 50-fold cross-validation). One hundred regularization coefficients λ were fitted, and cross-validation mean square error was plotted for each λ penalty values.
Caspase inhibition simulation.
One hundred twenty-five founder cells were simulated with distributed molecular networks. All caspase inhibition reactions were scaled as indicated, and Cdh1 positivity was used as a marker for proliferating cells.
Founder cell module analysis.
One hundred twenty-five single-cell simulations were run with founder cells distributed as described previously but with distributions of kinetic rates restricted to reactions only in apoptosis, cell-cycle or NF-κB regulatory modules, or combinations thereof.
To visualize the space of founder-cell networks and the relation to terminal generation, 5,000 single-cell simulations were run with distribution of both cell cycle and apoptosis networks in founder cells. Scores for “apoptotic propensity” were generated by identifying five chemical species within the apoptosis network that were significantly different between founder cells that divided one or more times and those that died (phosphorylated caspase 8, Bar, phosphorylated caspase 3, phosphorylated caspase 6, and XIAP). The concentration of these metabolites in founder cells was then normalized to unit variance with negative regulators of terminal generation inverted to match positive regulators, and the average of the five values was the founder cell’s apoptotic propensity score. An identical approach was taken for cell cycle activity scores of chemical species showing significant differences within the cell cycle network of founder cells that divided to late terminal generation (3+) compared with those that did not (Cdh1, cyclin A, IEP, Exopolyphosphatase, and Growth).
Supplementary Material
Acknowledgments
We thank laboratory members Yi Liu and Diane Lefaudeux, as well as Dinesh Rao [University of California, Los Angeles (UCLA)], Roy Wollman (UCLA), and Ken Duffy (Hamilton Institute) for critical discussions and/or reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715639115/-/DCSupplemental.
References
- 1.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 2.Kuchina A, et al. Temporal competition between differentiation programs determines cell fate choice. Mol Syst Biol. 2011;7:557. doi: 10.1038/msb.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins ED, et al. Quantal and graded stimulation of B lymphocytes as alternative strategies for regulating adaptive immune responses. Nat Commun. 2013;4:2406. doi: 10.1038/ncomms3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deenick EK, Gett AV, Hodgkin PD. Stochastic model of T cell proliferation: a calculus revealing IL-2 regulation of precursor frequencies, cell cycle time, and survival. J Immunol. 2003;170:4963–4972. doi: 10.4049/jimmunol.170.10.4963. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins ED, Markham JF, McGuinness LP, Hodgkin PD. A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proc Natl Acad Sci USA. 2009;106:13457–13462. doi: 10.1073/pnas.0905629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins ED, Turner ML, Dowling MR, van Gend C, Hodgkin PD. A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc Natl Acad Sci USA. 2007;104:5032–5037. doi: 10.1073/pnas.0700026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy KR, et al. Activation-induced B cell fates are selected by intracellular stochastic competition. Science. 2012;335:338–341. doi: 10.1126/science.1213230. [DOI] [PubMed] [Google Scholar]
- 11.Marchingo JM, et al. T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science. 2014;346:1123–1127. doi: 10.1126/science.1260044. [DOI] [PubMed] [Google Scholar]
- 12.Kan A, et al. Stochastic measurement models for quantifying lymphocyte responses using flow cytometry. PLoS One. 2016;11:e0146227. doi: 10.1371/journal.pone.0146227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shokhirev MN, Hoffmann A. FlowMax: A computational tool for maximum likelihood deconvolution of CFSE time courses. PLoS One. 2013;8:e67620. doi: 10.1371/journal.pone.0067620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinjyo I, et al. Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat Commun. 2015;6:6301. doi: 10.1038/ncomms7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard S, Pujo-Menjouet L, Mackey MC. Analysis of cell kinetics using a cell division marker: Mathematical modeling of experimental data. Biophys J. 2003;84:3414–3424. doi: 10.1016/S0006-3495(03)70063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyrien O, Chen R, Zand MS. An age-dependent branching process model for the analysis of CFSE-labeling experiments. Biol Direct. 2010;5:41. doi: 10.1186/1745-6150-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Boer RJ, Perelson AS. Estimating division and death rates from CFSE data. J Comput Appl Math. 2005;184:140–164. doi: 10.1007/s11538-006-9094-8. [DOI] [PubMed] [Google Scholar]
- 18.Banks HT, et al. Estimation of cell proliferation dynamics using CFSE data. Bull Math Biol. 2011;73:116–150. doi: 10.1007/s11538-010-9524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luzyanina T, et al. Numerical modelling of label-structured cell population growth using CFSE distribution data. Theor Biol Med Model. 2007;4:26. doi: 10.1186/1742-4682-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner SL, Adams WC. Lymphocyte fate specification as a deterministic but highly plastic process. Nat Rev Immunol. 2014;14:699–704. doi: 10.1038/nri3734. [DOI] [PubMed] [Google Scholar]
- 21.Marchingo JM, et al. T-cell stimuli independently sum to regulate an inherited clonal division fate. Nat Commun. 2016;7:13540. doi: 10.1038/ncomms13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzel S, et al. A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat Immunol. 2016 doi: 10.1038/ni.3598. [DOI] [PubMed] [Google Scholar]
- 23.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shokhirev MN, et al. A multi-scale approach reveals that NF-κB cRel enforces a B-cell decision to divide. Mol Syst Biol. 2015;11:783. doi: 10.15252/msb.20145554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 26.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zangle TA, Teitell MA. Live-cell mass profiling: An emerging approach in quantitative biophysics. Nat Methods. 2014;11:1221–1228. doi: 10.1038/nmeth.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan TG, et al. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J Exp Med. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fulcher DA, Basten A. B cell life span: A review. Immunol Cell Biol. 1997;75:446–455. doi: 10.1038/icb.1997.69. [DOI] [PubMed] [Google Scholar]
- 30.Höfer T, Busch K, Klapproth K, Rodewald H-R. Fate mapping and quantitation of hematopoiesis in vivo. Annu Rev Immunol. 2016;34:449–478. doi: 10.1146/annurev-immunol-032414-112019. [DOI] [PubMed] [Google Scholar]
- 31.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 32.Alves BN, et al. IkappaBepsilon is a key regulator of B cell expansion by providing negative feedback on cRel and RelA in a stimulus-specific manner. J Immunol. 2014;192:3121–3132. doi: 10.4049/jimmunol.1302351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva EM, et al. Caspase inhibition reduces lymphocyte apoptosis and improves host immune responses to Trypanosoma cruzi infection. Eur J Immunol. 2007;37:738–746. doi: 10.1002/eji.200636790. [DOI] [PubMed] [Google Scholar]
- 34.Schrantz N, et al. Role of caspases and possible involvement of retinoblastoma protein during TGFbeta-mediated apoptosis of human B lymphocytes. Oncogene. 1999;18:3511–3519. doi: 10.1038/sj.onc.1202718. [DOI] [PubMed] [Google Scholar]
- 35.Polonsky M, Chain B, Friedman N. Clonal expansion under the microscope: Studying lymphocyte activation and differentiation using live-cell imaging. Immunol Cell Biol. 2016;94:242–249. doi: 10.1038/icb.2015.104. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Z, Taylor B, Ourthiague DR, Hoffmann A. Distinct single-cell signaling characteristics are conferred by the MyD88 and TRIF pathways during TLR4 activation. Sci Signal. 2015;8:ra69. doi: 10.1126/scisignal.aaa5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kollmann M, Løvdok L, Bartholomé K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 38.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cotari JW, Voisinne G, Dar OE, Karabacak V, Altan-Bonnet G. Cell-to-cell variability analysis dissects the plasticity of signaling of common γ chain cytokines in T cells. Sci Signal. 2013;6:ra17. doi: 10.1126/scisignal.2003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feinerman O, et al. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Mol Syst Biol. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tkach KE, et al. T cells translate individual, quantal activation into collective, analog cytokine responses via time-integrated feedbacks. eLife. 2014;3:e01944. doi: 10.7554/eLife.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos LC, et al. 2017. Origins of fractional control in regulated cell death. bioRxiv:10.1101/201160.
- 43.Mariani L, et al. Short-term memory in gene induction reveals the regulatory principle behind stochastic IL-4 expression. Mol Syst Biol. 2010;6:359. doi: 10.1038/msb.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shroeder HW. The evolution and development of the antibody repertoire. Front Immunol. 2015;6:33. doi: 10.3389/fimmu.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ademokun A, et al. Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell. 2011;10:922–930. doi: 10.1111/j.1474-9726.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnett BE, et al. Asymmetric B cell division in the germinal center reaction. Science. 2012;335:342–344. doi: 10.1126/science.1213495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thaunat O, et al. Asymmetric segregation of polarized antigen on B cell division shapes presentation capacity. Science. 2012;335:475–479. doi: 10.1126/science.1214100. [DOI] [PubMed] [Google Scholar]
- 48.White HN, Meng QH. Recruitment of a distinct but related set of VH sequences into the murine CD21hi/CD23- marginal zone B cell repertoire to that seen in the class-switched antibody response. J Immunol. 2012;188:287–293. doi: 10.4049/jimmunol.1101264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turchinovich G, et al. Programming of marginal zone B-cell fate by basic Kruppel-like factor (BKLF/KLF3) Blood. 2011;117:3780–3792. doi: 10.1182/blood-2010-09-308742. [DOI] [PubMed] [Google Scholar]
- 50.Bon P, Maucort G, Wattellier B, Monneret S. Quadriwave lateral shearing interferometry for quantitative phase microscopy of living cells. Opt Express. 2009;17:13080–13094. doi: 10.1364/oe.17.013080. [DOI] [PubMed] [Google Scholar]
- 51.Iwasaki H, Arai F, Kubota Y, Dahl M, Suda T. Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood. 2010;116:544–553. doi: 10.1182/blood-2009-08-240903. [DOI] [PubMed] [Google Scholar]
- 52.Soares A, et al. Novel application of Ki67 to quantify antigen-specific in vitro lymphoproliferation. J Immunol Methods. 2010;362:43–50. doi: 10.1016/j.jim.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ringhoffer S, Rojewski M, Döhner H, Bunjes D, Ringhoffer M. T-cell reconstitution after allogeneic stem cell transplantation: Assessment by measurement of the sjTREC/βTREC ratio and thymic naive T cells. Haematologica. 2013;98:1600–1608. doi: 10.3324/haematol.2012.072264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loriaux PM, Tesler G, Hoffmann A. Characterizing the relationship between steady state and response using analytical expressions for the steady states of mass action models. PLOS Comput Biol. 2013;9:e1002901. doi: 10.1371/journal.pcbi.1002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conradie R, et al. Restriction point control of the mammalian cell cycle via the cyclin E/Cdk2:p27 complex. FEBS J. 2010;277:357–367. doi: 10.1111/j.1742-4658.2009.07473.x. [DOI] [PubMed] [Google Scholar]
- 56.Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.