Significance
Optimization of the engineered pathway for production of D-glucaric acid, a precursor to nylons and detergents, requires strategies that manage pathway competition with glycolysis as well as the stability and activity of the rate-controlling enzyme. To address both of these challenges, two orthogonal, autonomous, and tunable dynamic regulation strategies were layered to produce the highest reported glucaric acid titers in Escherichia coli K-12 strains. The implementation of two regulatory circuits harnesses synthetic biology tools to program and optimize cellular behavior and demonstrates the power of multiplexed dynamic control for strain optimization.
Keywords: metabolic engineering, synthetic biology, dynamic regulation
Abstract
Microbial production of value-added chemicals from biomass is a sustainable alternative to chemical synthesis. To improve product titer, yield, and selectivity, the pathways engineered into microbes must be optimized. One strategy for optimization is dynamic pathway regulation, which modulates expression of pathway-relevant enzymes over the course of fermentation. Metabolic engineers have used dynamic regulation to redirect endogenous flux toward product formation, balance the production and consumption rates of key intermediates, and suppress production of toxic intermediates until later in the fermentation. Most cases, however, have utilized a single strategy for dynamically regulating pathway fluxes. Here we layer two orthogonal, autonomous, and tunable dynamic regulation strategies to independently modulate expression of two different enzymes to improve production of D-glucaric acid from a heterologous pathway. The first strategy uses a previously described pathway-independent quorum sensing system to dynamically knock down glycolytic flux and redirect carbon into production of glucaric acid, thereby switching cells from “growth” to “production” mode. The second strategy, developed in this work, uses a biosensor for myo-inositol (MI), an intermediate in the glucaric acid production pathway, to induce expression of a downstream enzyme upon sufficient buildup of MI. The latter, pathway-dependent strategy leads to a 2.5-fold increase in titer when used in isolation and a fourfold increase when added to a strain employing the former, pathway-independent regulatory system. The dual-regulation strain produces nearly 2 g/L glucaric acid, representing the highest glucaric acid titer reported to date in Escherichia coli K-12 strains.
Microbes have been engineered as renewable biochemical reactors to produce a variety of value-added compounds, such as fuels and fragrances, from low-cost substrates. After a pathway is constructed in an appropriate microbial host, it must be optimized for product titer, yield, and selectivity to become an economically viable process. Static strain engineering techniques for maximizing pathway productivity include balancing enzyme expression levels (1) and deleting genes encoding enzymes of competing reactions (2). Recently, dynamic regulation has emerged as an optimization technique by implementing “process control” at the cellular level (3, 4). Inspired by naturally occurring regulatory processes that allow cells to adapt to environmental changes such as temperature, osmolarity, and carbon source, dynamic regulation can be used to repurpose native regulation mechanisms to control expression of relevant enzymes.
Dynamic control can either be implemented through pathway-specific or pathway-independent strategies. Pathway-specific regulation systems control the expression of pathway enzymes in response to changes in concentration of a relevant intermediate or byproduct. Implementation of such dynamic control requires a biosensor that can detect an input change (i.e., substrate or metabolite concentration) and then actuate a response as a result of detection (5). Thus, identification and characterization of an appropriate sensor is particularly important. For example, Farmer and Liao diverted carbon flux away from the undesired byproduct acetate toward the desired product lycopene by up-regulating phosphoenolpyruvate synthase and isopentenyl diphosphate isomerase to maintain sufficient glyceraldehyde-3-phosphate (G3P) and dimethylallyl diphosphate (DMAPP) pools upon sensing acetyl phosphate (6). Zhang et al. (7) used the acyl-CoA sensor FadR to repress ethanol biosynthesis genes, adhB and pdc, until sufficient accumulation of fatty acids had occurred, preventing buildup of the toxic intermediate to improve fatty acid ethyl ester production by threefold. Xu et al. (8) increased fatty acids production by twofold using a malonyl-CoA sensor FapR to balance the production (by acetyl-CoA carboxylase) and consumption (by fatty acids synthase) of malonyl-CoA.
Pathway-independent dynamic regulation strategies have also been used to improve pathway productivity. Media formulation is one option in which strains are engineered with genetic circuits that respond to concentrations of critical nutrients in the medium. By tuning the concentration of these nutrients in the culture medium, the triggering time of the internal circuit can be controlled (9). These strategies, however, may lead to unknown global metabolic effects on the cell, as concentrations of critical compounds must be modulated. In addition to media formulation, quorum sensing (QS) circuitry has also been utilized to institute pathway-independent dynamic control, as it does not rely on pathway metabolites or substrates (10–12). The QS device constructed in the K.L.J.P. laboratory was used to knock down an enzyme involved in central carbon metabolism to boost titers of glucaric acid from a heterologous pathway. Essential pathways cannot be knocked out of the genome, but they can be dynamically down-regulated to arrest growth once a desired cell density is reached (10–14).
We sought to develop and apply both pathway-specific and pathway-independent strategies to explore the synergies that could be achieved by layering multiple dynamic regulation devices in the same strain, specifically in the context of glucaric acid production in Escherichia coli. A heterologous pathway for bioproduction of glucaric acid, a precursor to nylons and detergents, has previously been constructed in E. coli (15). In this pathway, glucose is imported into the cell and converted to glucose-6-phosphate (G6P) by the native bacterial phosphotransferase system. G6P is converted to myo-inositol (MI) by myo-inositol-1-phosphate synthase (MIPS) from Saccharomyces cerevisiae and endogenous phosphatases. MI is then converted to glucuronic acid by myo-inositol oxygenase (MIOX) from Mus musculus. Finally, glucaric acid is produced by uronate dehydrogenase (Udh) from Agrobacter tumefaciens. We identified two points in this pathway where dynamic control may be beneficial: the branch point at G6P, where MIPS competes with native glycolytic enzymes for flux of G6P; and the key intermediate MI, which builds up in the culture due to the low stability and activity of the enzyme MIOX (15).
At the G6P branch point, glucaric acid production competes with essential glycolytic pathways, which cannot be knocked out. Previous work has demonstrated that dynamically redirecting carbon flux from glycolysis to glucaric acid production at intermediate points in the fermentation can notably improve endpoint titers (11, 16). We utilized a previously implemented QS-based switch to autonomously trigger down-regulation of the glycolytic enzyme Pfk-1 at different points over the course of the fermentation (11) and thereby siphon carbon into glucaric acid production. Briefly, the circuit relies on constitutive production of a small signaling molecule, 3-oxohexanoylhomoserine lactone (AHL), which is sensed by a protein activator, EsaRI70V. EsaRI70V activates its cognate promoter, PesaS, in the absence of AHL. Upon sensing a certain concentration of AHL, EsaRI70V is released from PesaS, thereby deactivating gene expression from PesaS. By instituting differential rates of AHL production, the switching times of the circuit can be modulated. This strategy is pathway independent, as the QS circuit does not rely on any intermediates of the glucaric acid pathway and represents one route to improving product titers.
In addition to the G6P branch point, the key intermediate, MI, represents another point where dynamic regulation can improve production. Previous work has shown that once MIOX is expressed, its activity declines rapidly during the fermentation (15). Thus, delaying Miox expression until its substrate, MI, accumulates sufficiently may delay the loss in activity and result in higher MI conversion (17). A pathway-specific strategy was required to directly control production of the rate-controlling enzyme MIOX as a function of its substrate concentration. To implement this, a MI biosensor was constructed to sense the key intermediate, with Miox transcription as the output. In this system, the MI-responsive transcriptional regulator, IpsA, from Corynebacterium glutamicum (18) represses an engineered promoter driving Miox expression until sufficient production of MI, at which point MI-bound IpsA is released from the DNA and Miox transcription proceeds. The timing of Miox expression is modulated by expressing ipsA at varying levels. At the optimal Miox expression time, glucaric acid production increased 2.5-fold compared with the unregulated Miox control. Thus, this strategy represents a separate route to improving glucaric acid production that is orthogonal to the pathway-independent QS strategy described above.
To simultaneously address the challenges at both intermediates G6P and MI, the pathway-specific MIOX control was layered over the pathway-independent Pfk-1 control. When both control strategies were implemented, the optimal combination of Pfk-1 down-regulation and Miox expression timing resulted in the production of nearly 2 g/L glucaric acid, compared with 0.4 g/L glucaric acid produced by the strain with only Pfk-1 down-regulation, at both the 250-mL shake flask and 3-L bioreactor scales.
Results and Discussion
Construction and Characterization of the Myo-Inositol Biosensor.
To connect Miox expression to the concentration of MI, we desired a biosensor to detect MI and regulate transcription of Miox. The ligand-responsive transcription factor, IpsA, natively regulates MI and cell wall biosynthesis in C. glutamicum (18). IpsA has been shown to bind with high specificity to MI, as well as to multiple promoter regions in the C. glutamicum genome (18). In the absence of MI, IpsA binds to DNA sites, such as the promoter region of the gene encoding MIPS and recruits RNA polymerases to activate transcription. In the presence of MI, however, IpsA binds to MI and dissociates from DNA, preventing polymerase recruitment and deactivating gene expression (18). IpsA was also found to up-regulate cg0044, a putative ABC transporter, and down-regulate inositol catabolism genes, O-succinylbenzoic acid-CoA ligase (menE), and iron homeostasis factors (18). However, initial attempts to directly import these IpsA-regulated promoters into E. coli yielded no expression.
Thus, we engineered hybrid promoters that contained −35 and −10 regions recognizable by the native E. coli σ70 RNA polymerase and an IpsA DNA binding site to permit IpsA-mediated transcription (Fig. 1A and SI Appendix, Fig. S1). In this system, IpsA represses transcription from the hybrid promoter, presumably by blocking access to RNA polymerase. Binding of MI to IpsA causes the complex to unbind from DNA and transcription to proceed. This system may be used to exercise MI-inducible control of expression over any gene placed downstream of the hybrid promoter.
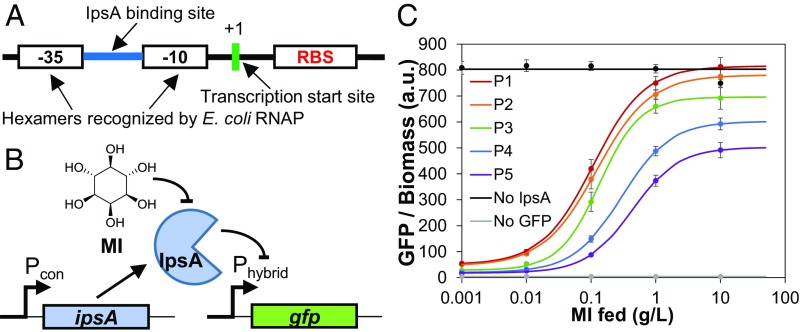
Fig. 1.
Construction and characterization of the myo-inositol (MI) biosensor. (A) Architecture of the hybrid promoter. See SI Appendix, Fig. S1 for alternate hybrid promoter configurations. (B) Biosensor circuit, where a constitutive promoter expressing ipsA represses the hybrid promoter driving gfp expression, and gfp is derepressed in the presence of MI. (C) GFP fluorescence normalized to biomass in MG1655 (DE3) cells harboring the sensor or control plasmids as a response to MI added to LB medium. Measurements were taken 24 h postinoculation. Individual points with error bars represent the mean ± 1 SD of three replicate cultures. Data points were fitted to a dose–response transfer function to yield smooth curves. P1–P5 represent the different promoters (corresponding to Pcon in A) driving ipsA expression (SI Appendix, Table S2). No IpsA, sensor-less plasmid expressing gfp from the hybrid promoter; no GFP, empty vector.
For initial characterization of the biosensor, we placed gfp under control of the hybrid promoter and measured the GFP fluorescence output at a range of MI concentrations (Fig. 1B). Fluorescence measurements revealed a dose-dependent response to MI (Fig. 1C and SI Appendix, Fig. S1), with increasing MI concentrations leading to increased fluorescence. We investigated multiple hybrid promoter variants with varying IpsA binding site sequences and locations (SI Appendix, Fig. S1 and Table S1). The binding sites cg44 and cgA were obtained from the promoter regions of C. glutamicum genes cg0044 and cg3323, respectively. These binding sites were placed either between the −35 and −10 regions or downstream of the −10 region, as it has been shown that these locations lead to tight repression (19, 20). We found the hybrid promoter variant with binding site cgA placed between −35 and −10 to be very leaky (SI Appendix, Fig. S1). The remaining variants demonstrated similar dose–response curves (SI Appendix, Fig. S1), and the hybrid promoter with binding site cg44 placed between the −35 and −10 regions was selected for further characterization (Fig. 1). For additional tunability, the amount of IpsA in the circuit was modulated by utilizing promoters of different strengths to drive ipsA expression (SI Appendix, Fig. S2 and Table S2). As expected, increased expression of ipsA lowered the maximal GFP signal and reduced the leaky expression in the absence of any MI (Fig. 2 and SI Appendix, Fig. S3). The presence of multiple knobs for tunability allows flexibility in reaching similar GFP levels through different circuit architectures.
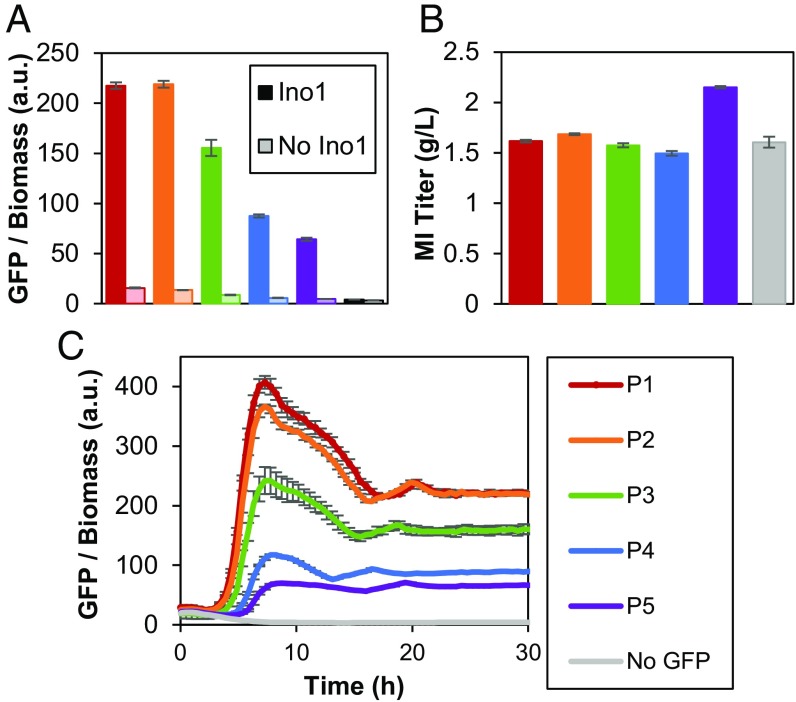
Fig. 2.
Sensor detection of MI produced from MIPS and phosphatase in MG1655 (DE3) cells. (A) GFP response at 48 h postinoculation. Expression of INO1 in darker bars; corresponding control (empty vector in place of INO1) in lighter bars. (B) Corresponding MI titers at 48 h. P1–P5 represent the different promoters driving ipsA expression. No GFP refers to a vector expressing ipsA from the tet promoter with a multiple cloning site in place of GFP. (C) GFP response over time with INO1 expressed. Error bars represent ±1 SD from the mean of three replicate cultures.
In addition to evaluating the circuit with exogenously added MI, we tested whether a response could be triggered from intracellularly produced MI. The enzyme MIPS (encoded by INO1), which facilitates the conversion of glucose to MI (15), was used to intracellularly produce MI from fed glucose. Compared with the control strain harboring an empty plasmid, cells expressing INO1 demonstrated a 16- to 55-fold increase in fluorescence, depending on the expression level of IpsA (Fig. 2A). Consistent with the trends observed with exogenously added MI in previous trials, increased ipsA expression decreased the endpoint fluorescence measurement, even while all strains produced ∼1.5 g/L MI (Fig. 2B). This trend suggests that a circuit with increased IpsA concentration requires more MI to achieve the same GFP output as a circuit with weaker ipsA expression. Furthermore, increased ipsA expression lengthened the amount of time required for a rise in fluorescence (Fig. 2C), suggesting that derepression of gfp is delayed with increasing repressor concentration, and that modulating ipsA expression controls the timing of switching on the reporter gene. This collection of genetic circuits (P1–P5) with a tunable range of production responses provided a variety of MIOX control circuits to test in the glucaric acid pathway.
Dynamic Regulation of the Glucaric Acid Pathway by Controlling MIOX Expression.
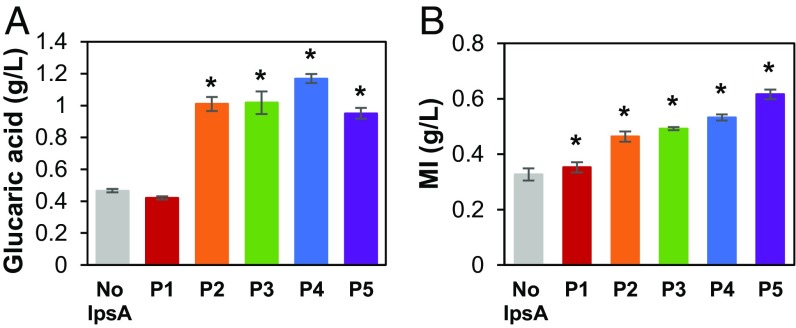
We implemented MI-dependent dynamic control of MIOX in the glucaric acid pathway by utilizing the circuit to control Miox expression in response to MI concentration. As MIOX is an unstable enzyme whose activity drops rapidly within the first 24 h of fermentation (15), we sought to control the timing of Miox expression to optimize the window of its maximal activity. Delaying Miox expression until a larger pool of MI has accumulated may boost enzyme activity and pathway productivity (17). In the IpsA-based circuit, the transcription rate of Miox is a joint function of MI and IpsA concentration, and expression of Miox is delayed until sufficient MI is produced. Glucaric acid production was evaluated in strains in which the genes gudD and uxaC, encoding for enzymes that metabolize glucaric acid and its intermediates, were deleted (2). These trials showed that delayed Miox expression improved glucaric acid titers up to 2.5 ± 0.084-fold compared with the constitutively expressed Miox control (Fig. 3A). These trends in production were consistent at 24 and 48 h (SI Appendix, Fig. S4). Strain P4 produced the highest titer, likely because the P4 circuit resulted in optimal timing of Miox expression. Strain P1 permitted expression of Miox too early in the fermentation, leading to MIOX instability and lower conversion, whereas P5 delayed Miox expression for too long, with insufficient time for conversion of MI. The trends in MI buildup correspond to the delays in Miox expression: increased ipsA expression allowed increased MI buildup before expression of Miox (Fig. 3B).
Fig. 3.
Production of (A) glucaric acid and (B) MI from MI-controlled expression of Miox in MG1655 (DE3) ΔgudD ΔuxaC at 72 h. IpsA is expressed from promoters P1–P5. No IpsA, constitutive (unregulated) Miox expression. Error bars represent ±1 SD from the mean of three replicate flasks. *P < 0.01 compared with the no IpsA control by two-tailed t test.
To confirm that repression of Miox by IpsA is relieved upon the production of MI, we performed qRT-PCR with two of the dynamically regulated strains to demonstrate changes in Miox mRNA levels over time (SI Appendix, Fig. S5). Two control strains were implemented to demonstrate static expression levels of MIOX: “No IpsA,” where Miox is constitutively transcribed from the hybrid promoter, and “no Ino1,” where MI is not produced, leading to constant repression of Miox by ipsA expressed from promoter P5. As expected, strains with promoters P2 and P5 expressing ipsA exhibited up to ∼20- and 50-fold increases in Miox transcript levels, indicating significant derepression of Miox over time, while static strains referred to as no IpsA and no Ino1 did not exhibit any changes in Miox transcript levels. The strain with P5 exhibited a larger fold increase than P2 likely because the stronger P5 promoter is less leaky and has greater repression of Miox at initial times, with a P2 to P5 transcript level ratio of 5.45 ± 1.89 at the initial time point.
Although we found improved growth with IpsA present when only INO1 was expressed (SI Appendix, Fig. S6), it is unlikely that the improvement in glucaric acid titer is due to the boost in growth, as we did not observe significant growth benefits with the full pathway (SI Appendix, Fig. S7). As a control, we tested glucaric acid production with a circuit variant in which the promoter driving Miox did not contain an IpsA binding site (but is otherwise identical to the hybrid promoter) and observed no benefit in titer or growth (SI Appendix, Fig. S8). This indicates that the improvements in production from the biosensor circuit are not simply due to expression of ipsA or other circuit components.
Layered Dynamic Regulation to Substantially Increase Glucaric Acid Production.
While product titers were improved by instituting the MI biosensor strategy independently, we sought to implement even more precise regulation of the glucaric acid pathway by layering the QS-based Pfk-1 control system on top of the IpsA-based MI biosensor in the same strain (Fig. 4). The QS-based system autonomously down-regulates Pfk-1 at intermediate points in the fermentation to siphon carbon flux from glycolytic pathways to the glucaric acid pathway, while the IpsA-based MI biosensor delays transcription of Miox until threshold concentrations of MI have accumulated. Thus, we combined two tunable, orthogonal control systems, a pathway-independent and a pathway-dependent system, to synergistically boost pathway productivity. When acting independently in different strains, each system improved glucaric acid titers by two- to threefold (11), suggesting that the combination of multiple pathway controls could boost titers even higher than either system alone.
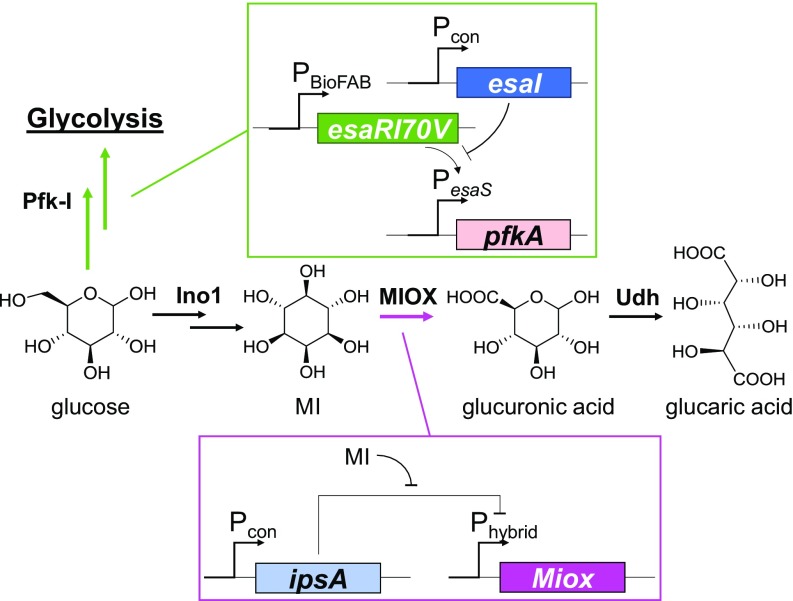
Fig. 4.
Schematic of layered dynamic regulation in the glucaric acid pathway. The initial branch point at glucose-6-phosphate (G6P) is controlled using a quorum sensing system, where pfkA placed under control of the PesaS promoter is responsive to the AHL (produced by EsaI) concentration, corresponding to the cell density. Pfk-1 activity is knocked down at sufficiently high cell densities. Different levels of esaI expression, achieved by varying the strength of constitutive promoter Pcon, alters the timing of pfkA knockdown. Miox is controlled via a hybrid promoter (Fig. 1A) regulated by the MI-specific sensor IpsA.
The QS-based Pfk-1 control system was previously developed in our laboratory to institute autonomous, dynamic control at the G6P branch point, where carbon flux is split between native metabolic pathways and the heterologous glucaric acid production pathway. Upon buildup of the cognate N-acyl homoserine lactone (AHL), the circuit is triggered and leads to down-regulation of Pfk-1, thereby limiting flux through glycolysis. The carbon flux is therefore redirected through MIPS into glucaric acid production. Expression of Pfk-1 is controlled by the PesaS promoter, which is active in the absence of AHL. Upon AHL buildup, the promoter is deactivated, and transcription of its downstream gene is arrested. The triggering time can be tuned by modulating the AHL production rate by varying the expression of the AHL synthase, EsaI. A variety of expression rates were obtained by instituting a library of promoter and ribosome binding site (RBS) variants upstream of esaI. Stronger esaI expression leads to a higher AHL production rate and a faster down-regulation of Pfk-1 activity (11). All circuit parts were genomically integrated, leading to a series of strains, each with a distinct promoter–RBS combination driving esaI expression (L31GA, L19GA, and L24GA; SI Appendix, Table S3). Similarly, timing of Miox expression was tunable in the MI biosensor circuit through the varied expression levels of IpsA.
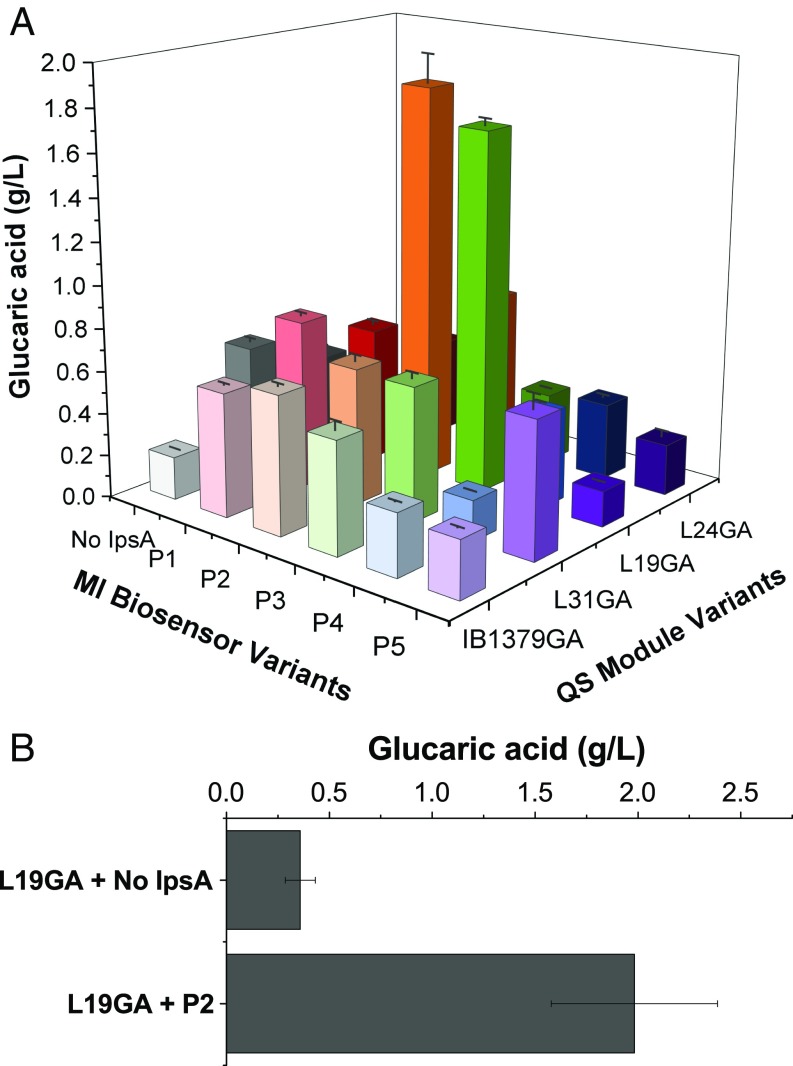
To identify the optimal combination of Pfk-1 knockdown and Miox expression timing when both devices are present in the same strain, we tested each EsaI expression level with each IpsA expression level. Strains contained uxaC, gudD, zwf, and pfkB knockouts to prevent product consumption and removal of carbon flux through other pathways (SI Appendix, Table S3). At the 250-mL shake flask scale, strain “L19GA + P2” produced 1.84 g/L glucaric acid, the highest titer reported thus far in the E. coli K-12 strains. This is a 4.3 ± 0.051-fold increase in titer over the best-performing quorum sensing strain without Miox regulation, “L19GA + no IpsA,” which produced 0.43 g/L glucaric acid and a 9.1 ± 0.69-fold increase over the completely static strain “IB1379GA + no IpsA” (no pfkA knockdown or Miox delay), which produced only 0.20 g/L glucaric acid (Fig. 5A and SI Appendix, Table S4). In our previous work, strain L19GA (without IpsA) produced 0.68 g/L glucaric acid (11), slightly more than strain L19GA + no IpsA in this work. However, different expression vectors for the pathway genes were used due to the need to accommodate and be consistent with the MIOX regulatory circuit developed in this work. These differences, perhaps including the additional expression of ipsA, are the most likely causes of the discrepancy in titers. Generally, we found that intermediate Pfk-1 knockdown times, along with intermediate Miox expression times, led to improved titers. To ensure that IpsA itself did not render growth and production benefits, we also expressed ipsA with Miox driven by a promoter that was not regulated by IpsA and observed no boost in titer over the control strain not expressing ipsA (SI Appendix, Fig. S9). Strain L19GA + P2 was chosen for scale-up in a 3-L bioreactor with dissolved oxygen (DO) and pH control. This strain produced 1.98 g/L glucaric acid, corresponding to 20% yield by mass, while strain L19GA + no IpsA only produced 0.4 g/L glucaric acid (Fig. 5B).
Fig. 5.
Glucaric acid production from strains with layered dynamic regulation at 72 h. (A) Glucaric acid titer as a function of Pfk-1 switching time (QS module variants) and Miox expression time (MI biosensor circuit variants) at the 250-mL shake flask scale. Error bars represent ±1 SD from the mean of three replicate flasks. (B) Glucaric acid production at the 3-L bioreactor scale for strains L19GA + P2 and L19GA + no IpsA. Error bars represent ±1 SD from the mean of two replicates. No IpsA, constitutive (unregulated) Miox expression; IB1379GA, wild-type pfkA expression (no knockdown of pfkA).
The combination of the two orthogonal regulation strategies in one pathway demonstrates synergistic effects, as the titer from the composite strain was greater than when either strategy was evaluated alone. The Pfk-1 switch shunts flux toward MIPS and away from glycolysis, improving MI production. Addition of the second layer expresses Miox at an optimal time, further boosting titers. The use of orthogonal, tunable controllers allowed the identification of optimal switching times for each independent device. While many combinations of MIOX and Pfk-1 regulation systems showed improved production over the completely static, unregulated strain “IB1379GA + no IpsA” (Fig. 5A), only two strains produced over 1.5 g/L glucaric acid, demonstrating that tuning is required for optimization. For the pfkA down-regulation switch, strain L19GA was the highest-producing strain for both single dynamic regulation (11) and layered regulation. However, the optimal Miox expression switch varied by strain background: promoter P4 for the single regulation system and P2 for the layered regulation system. As pfkA down-regulation affects MI production and therefore the timing and dynamics of Miox expression, the MIOX control switch is more sensitive to differences in strain background. In the layered control system, pfkA and Miox are expressed only when necessary, as we move closer toward the concept of “just-in-time” transcription (21) in heterologous pathways, a widespread phenomenon that exists in native pathways such as amino acid biosynthesis (21, 22), arabinose utilization (23), and metal resistance mechanisms (24).
Conclusions
Dynamic regulation is a metabolic engineering strategy for improving production in pathways with toxic intermediates, essential competing reactions, or flux imbalances. The layered dynamic control system employed here mimicked natural regulatory systems by up-regulating a downstream enzyme only upon sufficient production of its intermediate, and down-regulating an essential competing enzyme upon sufficient cell growth. The combinatorial search through the two systems suggests that the timing of induction and repression must be carefully tuned to identify the optimal producer. This work demonstrates the potential of using multiple, independently controlled regulation strategies to implement process control at the cellular level.
Materials and Methods
Myo-Inositol Sensor and Glucaric Acid Pathway Plasmid Construction.
All primers used for PCR amplification of genes and promoters are found in SI Appendix, Table S5. All plasmids used in this study are found in SI Appendix, Table S6.
The hybrid promoter cg44 (TTTACAgtctttattgattcagtTATTATgctagcacgtgcaatttttaaaattaaaggcgttacccaac agaggagaaatactag) was constructed by placing the IpsA binding site (underlined) from C. glutamicum gene cg0044 (18) in between the −35 and −10 sites (capitalized) of promoter Bba_J23101 from the Registry of Standard Biological Parts (25). The transcription start site is denoted in red and the RBS in green. To construct pHH-cg44-GFP, the hybrid promoter cg44 and gfp were inserted into a modified form of the plasmid pHHD01K (26, 27), which includes the rrnB terminator (ccaggcatcaaataaaacgaaaggctc agtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctc) downstream of the gene encoding tetR. Briefly, gfp was amplified from pSB1A2-GFP (17) with primers pHH-GFP-R and cg44-RBS-GFP-F1. To incorporate the full length of the hybrid promoter cg44 sequence, overlap extension PCR was employed by adding primer cg44-RBS-GFP-F2 to the reaction. The backbone of the modified pHHD01K was amplified with primers cg44-pHH-A and GFP-pHH-B. The backbone and insert containing gfp were annealed and amplified using circular polymerase extension cloning (CPEC) (28). All other hybrid promoter variants pHH-cgX-GFP, where cgX is the name of the hybrid promoter (SI Appendix, Fig. S1 and Table S1), were constructed by CPEC after PCR amplifying the insert containing gfp using primers GFP-C-R and cgX-F and backbone using primers GFP-C-F and cgX-R.
The gene encoding IpsA was obtained by PCR from C. glutamicum genomic DNA with primers EcoRI-IpsA-F and BamHI-IpsA-R, digested with EcoRI and BamHI, and ligated into the multiple cloning site of pHH-cgX-GFP to construct pHH-cg44-GFP-Ptet-ipsA. To create the tunable set of sensor plasmids, the tet promoter was replaced with promoters P1–P5 (SI Appendix, Table S2) from the Registry of Standard Biological Parts (25). Promoters P1, P2, and P5 were inserted via CPEC using primers Px-ipsA-F and Px-pHH-R, where x = 1, 2, 5. Promoters P3 and P4 were amplified by PCR of oligos Xho1-Px-F and Kpn1-Px-R (x = 3, 4), digested with XhoI and KpnI, and ligated into the digested pHH-cg44-GFP-Ptet-ipsA backbone.
For dynamic control of MIOX, the restriction sites SacI and NotI were first added to replace gfp in pHH-cg44-GFP-P2-ipsA via CPEC. The backbone was amplified by PCR using primers NotI-pHH-F and SacI-pHH-R. The insert was amplified by PCR using primers pHH-NotI-Cm-R and pHH-SacI-Cm-F. The backbone and insert were annealed and amplified by CPEC. Next, Miox was cloned by restriction digest into the SacI and NotI sites. Miox was PCR amplified with SacI-MIOX-F and NotI-MIOX-R from pTrc-MIOX (15), digested, and ligated in place of GFP to form pHH-cg44-MIOX-P2-ipsA. To construct the dynamic control plasmids containing the remaining ipsA promoters P1, P3–P5, the entire promoter plus ipsA sequence was digested out of the pHH-cg44-GFP-Px-ipsA (x = 1, 3, 4, 5) using enzymes XhoI and SbfI, and ligated into the backbone of pHH-cg44-MIOX-P2-ipsA.
The plasmid pRSFDCarb-IN-Udh, encoding enzymes MIPS and Udh, was modified from pRSFDuet-IN-Udh (29), where the kanamycin resistance gene was swapped for carbenicillin resistance using CPEC. The bla gene encoding carbenicillin resistance was PCR amplified from pET-Duet with primers Carb-C-RSFD and Carb-N-RSFD. The pRSFDuet-IN-Udh backbone was PCR amplified with primers RSFD-Carb-C and RSFD-Carb-N. The insert and backbone were annealed and amplified via CPEC. All restriction enzymes, ligases, and polymerases were purchased from New England Biolabs.
Strain Construction.
All strains (SI Appendix, Table S3) were constructed as described in ref. 11.
Fluorescence Measurements.
MG1655 (DE3) cells were transformed with the sensor plasmids and grown in Luria Bertani (LB) broth with 50 µg/mL kanamycin. Overnight cultures were used to inoculate 1:100 vol/vol into 1-mL wells containing LB with 50 µg/mL kanamycin on a 48-well flower plate and cultured at 30 °C, shaking 1,200 rpm, and 80% humidity in the BioLector (m2p Labs). For variants with the tet promoter driving ipsA, 50 ng/µL anhydrotetracycline (aTc) was added to the medium. GFP fluorescence (excitation, 488 nm; emission, 520 nm) and biomass (excitation, 620 nm) were monitored continuously using LED optics for 24 h. Experiments were performed in triplicate, during three separate, identical incubation periods.
The promoter strengths of P1–P5 were measured by placing rfp under their control. MG1655 (DE3) cells were transformed with the plasmid series and grown in culture tubes containing 5 mL LB with 50 µg/mL kanamycin. Samples of 200 µL volume were removed 24 h postinoculation and washed twice with 0.1 M sodium phosphate buffer (30 mM NaH2PO4 and 61 mM Na2HPO4) and centrifuged 2,000 × g, 4 °C for 5 min. RFP fluorescence (excitation, 580 nm; emission, 610 nm) and optical density (absorbance at 610 nm) were measured using a Tecan Infinite 200 PRO plate reader.
Myo-Inositol Fermentation.
MG1655 (DE3) cells were transformed with the sensor plasmids and plasmid pTrc-Ino1. Cultures were grown in T12 medium [10 g/L D-glucose, 7.5 g/L yeast extract, 7.5 g/L soy peptone, 7 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 3 g/L (NH4)2SO4, 4 mM MgSO4] plus 50 µg/mL kanamycin and 100 µg/mL carbenicillin. Fermentation cultures were inoculated from overnight cultures at 1:100 vol/vol. Fermentation was carried out for 48 h in the BioLector (m2p labs, Baesweiler, Germany; 30 °C, 1,200 rpm, 1.1 mL initial working volume). INO1 expression was induced with 50 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) at inoculation. Biomass and GFP were monitored as described above. At 24 and 48 h postinoculation, 100 µL culture from each well was removed, centrifuged at 3,000 × g, 4 °C, 5 min, and the supernatant retained for titer measurements. Experiments were performed in triplicate.
Glucaric Acid Fermentation.
Strains MG1655 (DE3), IB1379GA, L19GA, L24GA, or L31GA (SI Appendix, Table S3) were transformed with the MIOX regulation plasmids and pRSFDCarb-IN-Udh. Overnight cultures were used to inoculate 30-mL starter cultures at ∼1:50 vol/vol in T12 medium with 10 g/L glucose. Starter cultures were incubated for 4–8 h, until midexponential phase, and used to inoculate fermentation cultures (T12 medium with 10 g/L glucose) at a target OD600 (optical density measured at 600 nm) of 0.05. The 50-mL fermentation cultures were incubated for 72 h in 250 mL baffled shake flasks at 30 °C, 250 rpm, and 80% humidity. INO1 and udh expression were induced with 100 µM IPTG upon inoculation of the fermentation cultures. The 1.4-mL samples were removed every 24 h for OD600 measurements and centrifuged at 10,000 × g, room temperature for 10 min to obtain supernatants for titer measurements. Experiments were performed in triplicate.
Bioreactor Fermentation.
Overnight cultures in T12 medium with 10 g/L glucose were used to inoculate 50-mL seed cultures in 250-mL baffled shake flasks and incubated at 30 °C, 250 rpm, 80% humidity for ∼8 h. Seed cultures were then used to inoculate a 3-L Labfors bioreactor (Infors AG, Bottmingen, Switzerland; 1 L working volume) at OD600 = 0.05. The pH was controlled at 7 using 4 M NaOH and 4 M HCl solutions. DO was controlled at 35% of maximum saturation by agitation rate (minimum 250 rpm) and constant air sparging at 1 L per minute. Batch fermentation was carried out for 72 h, with 5-mL samples removed every 24 h for optical density and titer measurements as described for shake flasks.
Glucaric Acid and Myo-Inositol Quantification.
Supernatants were analyzed by high performance liquid chromatography (HPLC) on an Agilent 1200 series instrument with an Aminex HPX-87H column. An isocratic, 25-min method with 5 mM sulfuric acid mobile phase at a flow rate of 0.6 mL/min was used. The column temperature was maintained at 65 °C, and the refractive index detector (RID) at 35 °C. The elution times for relevant species are as follows: glucaric acid (7.9 min), myo-inositol (9.5 min), glucose (9.1 min).
Quantification of mRNA Levels.
Miox transcript levels were quantified using quantitative RT-PCR of mRNA isolated from culture samples. Samples were collected at the equivalent of 1 OD600 per 1 mL, centrifuged at 845 × g for 10 min to pellet, resuspended in 600 µL RNA Protect (Qiagen), and stored at −80 °C until mRNA extraction. To lyse the cells, samples were thawed at room temperature, centrifuged at 3,000 rpm for 5 min, resuspended in 100 µL 1 mg/mL lysozyme in TE buffer (10 mM Tris⋅HCl at pH 8.0, 1 mM EDTA), and vortexed vigorously for 10 min. Total mRNA extraction of the lysed pellets was performed following the illustra RNAspin Mini Kit protocol (GE Healthcare Life Sciences). Extracted mRNA was treated with DNaseI (TURBO DNA-Free Kit; Life Technologies) to remove any genomic DNA. Purified mRNA concentrations were quantified with the Nanodrop 2000 (Thermo Fisher Scientific). cDNA synthesis was performed with the QuantiTect Reverse Transcription Kit (Qiagen), with 100 ng/µL total mRNA added per cDNA synthesis reaction. Quantitative PCR reactions (25 µL total) were set up as follows: 12.5 µL 2× Brilliant II SYBR qPCR High ROX Master Mix (Agilent Technologies), 0.5 µL each primer, 9.5 µL water, and 2 µL cDNA. Amplification was performed with the ABI 7300 Real Time PCR System (Applied Biosystems) at the following cycle conditions: initial incubation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 30 s and 60 °C for 1 min. Threshold cycle (Ct) values were determined automatically by the ABI System software. Fold differences in Miox transcript levels were calculated between each sample and its respective initial time point value.
Statistics.
When relevant, paired two-tailed Student t tests were performed to demonstrate statistically significant differences between data points. Error propagation was conducted to estimate the uncertainty on the fold changes in titer.
Supplementary Material
Acknowledgments
This work was supported by the US National Science Foundation through the Graduate Research Fellowship program (to A.G.), the Synthetic Biology Engineering Research Center (Grant EEC-0540879 to S.J.D., A.G., and K.L.J.P.), the Division of Molecular and Cellular Biosciences (Grant MCB-1517913 to A.G. and K.L.J.P.), and the Biotechnology Training Program of the National Institutes of Health (Grant T32GM008334 to S.J.D.). This work was further supported by the Institute for Collaborative Biotechnologies through Grant W911NF-09-0001 from the US Army Research Office. The content of the information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Footnotes
Conflict of interest statement: A.G. and K.L.J.P. are coinventors on a pending patent application that covers the quorum sensing-based dynamic control system.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716920115/-/DCSupplemental.
References
- 1.Xu P, Rizzoni EA, Sul SY, Stephanopoulos G. Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth Biol. 2017;6:148–158. doi: 10.1021/acssynbio.6b00187. [DOI] [PubMed] [Google Scholar]
- 2.Shiue E, Prather KLJ. Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport. Metab Eng. 2014;22:22–31. doi: 10.1016/j.ymben.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Venayak N, Anesiadis N, Cluett WR, Mahadevan R. Engineering metabolism through dynamic control. Curr Opin Biotechnol. 2015;34:142–152. doi: 10.1016/j.copbio.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Brockman IM, Prather KLJ. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol J. 2015;10:1360–1369. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu D, Evans T, Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 8.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Kessler W, van den Heuvel J, Rinas U. Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl Microbiol Biotechnol. 2011;91:1203–1213. doi: 10.1007/s00253-011-3407-z. [DOI] [PubMed] [Google Scholar]
- 10.Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng. 2015;30:7–15. doi: 10.1016/j.ymben.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Gupta A, Reizman IMB, Reisch CR, Prather KLJ. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35:273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bothfeld W, Kapov G, Tyo KEJ. A glucose-sensing toggle switch for autonomous, high productivity genetic control. ACS Synth Biol. 2017;6:1296–1304. doi: 10.1021/acssynbio.6b00257. [DOI] [PubMed] [Google Scholar]
- 13.Brockman IM, Prather KLJ. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab Eng. 2015;28:104–113. doi: 10.1016/j.ymben.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan SZ, Manchester S, Prather KLJ. Controlling central carbon metabolism for improved pathway yields in Saccharomyces cerevisiae. ACS Synth Biol. 2016;5:116–124. doi: 10.1021/acssynbio.5b00164. [DOI] [PubMed] [Google Scholar]
- 15.Moon TS, Yoon S-H, Lanza AM, Roy-Mayhew JD, Prather KLJ. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl Environ Microbiol. 2009;75:589–595. doi: 10.1128/AEM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reizman IMB, et al. Improvement of glucaric acid production in E. coli via dynamic control of metabolic fluxes. Metab Eng Commun. 2015;2:109–116. doi: 10.1016/j.meteno.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiue E. 2014 Improvement of D-glucaric acid production in Escherichia coli. Available at dspace.mit.edu/handle/1721.1/87132. Accessed August 12, 2017.
- 18.Baumgart M, et al. IpsA, a novel LacI-type regulator, is required for inositol-derived lipid formation in Corynebacteria and Mycobacteria. BMC Biol. 2013;11:122. doi: 10.1186/1741-7007-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collado-Vides J, Magasanik B, Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaslaver A, et al. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 22.Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleif R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 2000;16:559–565. doi: 10.1016/s0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
- 24.Merulla D, van der Meer JR. Regulatable and modulable background expression control in prokaryotic synthetic circuits by auxiliary repressor binding sites. ACS Synth Biol. 2016;5:36–45. doi: 10.1021/acssynbio.5b00111. [DOI] [PubMed] [Google Scholar]
- 25. Registry of Standard Biological Parts. Available at parts.igem.org/Main_Page. Accessed September 21, 2015.
- 26.Dhamankar H, Tarasova Y, Martin CH, Prather KLJ. Engineering E. coli for the biosynthesis of 3-hydroxy-γ-butyrolactone (3HBL) and 3,4-dihydroxybutyric acid (3,4-DHBA) as value-added chemicals from glucose as a sole carbon source. Metab Eng. 2014;25:72–81. doi: 10.1016/j.ymben.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Dhamankar HH. 2013 Microbial synthesis of 3,4-dihydroxybutyric acid, 3-hydroxybutyrolactone and other 3-hydroxyalkanoic acids. Available at dspace.mit.edu/handle/1721.1/87535. Accessed May 5, 2015.
- 28.Quan J, Tian J. Circular polymerase extension cloning. Methods Mol Biol. 2014;1116:103–117. doi: 10.1007/978-1-62703-764-8_8. [DOI] [PubMed] [Google Scholar]
- 29.Shiue E, Brockman IM, Prather KLJ. Improving product yields on D-glucose in Escherichia coli via knockout of pgi and zwf and feeding of supplemental carbon sources. Biotechnol Bioeng. 2015;112:579–587. doi: 10.1002/bit.25470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.