Significance
Penicillin-binding proteins (PBPs) are synthases that build the bacterial cell wall. They are one of our most important antibiotic targets. Several years ago, it was shown that a major PBP synthase of the Gram-negative bacterium Escherichia coli requires activation by LpoB. This outer-membrane lipoprotein is narrowly conserved in Gram-negative species whereas its target PBP synthase is broadly distributed. Here, we show that Pseudomonas aeruginosa and other Gram-negative bacteria use the distinct lipoprotein called LpoP to activate their PBPs via a mechanism similar to LpoB-PBP activation in E. coli. Our results therefore indicate that it may be possible to develop broad-spectrum inhibitors of Gram-negative PBP activation despite their use of diverse PBP regulators.
Keywords: penicillin, peptidoglycan, cell wall, PBP, antibiotic
Abstract
Penicillin-binding proteins (PBPs) are synthases required to build the essential peptidoglycan (PG) cell wall surrounding most bacterial cells. The mechanisms regulating the activity of these enzymes to control PG synthesis remain surprisingly poorly defined given their status as key antibiotic targets. Several years ago, the outer-membrane lipoprotein EcLpoB was identified as a critical activator of Escherichia coli PBP1b (EcPBP1b), one of the major PG synthases of this organism. Activation of EcPBP1b is mediated through the association of EcLpoB with a regulatory domain on EcPBP1b called UB2H. Notably, Pseudomonas aeruginosa also encodes PBP1b (PaPBP1b), which possesses a UB2H domain, but this bacterium lacks an identifiable LpoB homolog. We therefore searched for potential PaPBP1b activators and identified a lipoprotein unrelated to LpoB that is required for the in vivo activity of PaPBP1b. We named this protein LpoP and found that it interacts directly with PaPBP1b in vitro and is conserved in many Gram-negative species. Importantly, we also demonstrated that PaLpoP-PaPBP1b as well as an equivalent protein pair from Acinetobacter baylyi can fully substitute for EcLpoB-EcPBP1b in E. coli for PG synthesis. Furthermore, we show that amino acid changes in PaPBP1b that bypass the PaLpoP requirement map to similar locations in the protein as changes promoting EcLpoB bypass in EcPBP1b. Overall, our results indicate that, although different Gram-negative bacteria activate their PBP1b synthases with distinct lipoproteins, they stimulate the activity of these important drug targets using a conserved mechanism.
The cell wall forms a protective shell around bacteria that prevents osmotic rupture of the cytoplasmic membrane (1). This exoskeleton layer is made of the heteropolymer peptidoglycan (PG), which consists of glycan strands with the repeating unit N-acetylmuramic acid (MurNAc)–β-1–4-N-acetylglucosamine (GlcNAc). Attached to the MurNAc sugars is a pentapeptide that is used to form peptide cross-links between adjacent glycans, generating a matrix-like structure that surrounds the cell. Two enzymatic reactions are needed to make PG: polymerization of glycans by a PG glycosyltransferase (PGTase) and cross-linking by a transpeptidase (TPase) (1). These activities have long been known to be catalyzed by the penicillin-binding proteins (PBPs), the targets of penicillin and related beta-lactam drugs (2). Synthetic PBPs are found in two varieties: the class A enzymes (aPBPs) that possess both PGT and TP activity and class B PBPs (bPBPs) that have only TP active sites.
For many years, the aPBPs and related proteins with monofunctional PGT active sites were the only known cell-wall polymerases. However, the SEDS (shape, elongation, division, sporulation) protein RodA was recently shown to also function as a PGTase (3). Given the broad conservation of SEDS proteins and their known associations with bPBPs, it has been proposed that SEDS-bPBP complexes represent a second type of bifunctional PG synthase in bacteria (3, 4).
Although the identities of the major PG synthases in bacteria have been established, what has remained less clear is how the activities of these enzymes are regulated to control when and where PG biogenesis takes place. Understanding this regulation is important because it will not only shed light on the fundamental process of cell morphogenesis, but also will likely provide insights into new ways of disrupting PG biogenesis for antibiotic development. Currently, PG synthesis regulation is most well understood for the aPBPs of the model Gram-negative bacterium Escherichia coli. Several years ago, the outer-membrane lipoproteins EcLpoA and EcLpoB were found to be required for the activation of aPBPs in this organism (5, 6). EcLpoA specifically activates EcPBP1a whereas the unrelated lipoprotein EcLpoB was found to activate EcPBP1b (5, 6). Both lipoproteins interact directly with their cognate aPBP to form a transenvelope complex (7–9), and, when added to in vitro reactions, they modulate the PGT and TP activities of their target synthases (5, 6, 10). LpoA was found to be relatively well-conserved throughout the Gammaproteobacteria (6). On the other hand, LpoB and the UB2H regulatory domain of EcPBP1b to which it binds appeared to be largely restricted to the Enterobacteriaceae despite the relatively broad distribution of PBP1b sequences (6). However, subsequent studies of Vibrio cholerae and Shewanella oneidensis, which are members of the Vibrionaceae and Shewanellaceae, respectively, identified LpoB and UB2H-containing PBP1b homologs in these organisms (11, 12), suggesting a wider distribution than previously appreciated. Indeed, using a less stringent similarity cutoff than was used previously (6), we found that many PBP1b proteins from organisms outside of the Enterobacteriaceae, including the opportunistic pathogen Pseudomonas aeruginosa, contain a UB2H domain (SI Appendix, Fig. S1) but not a recognizable homolog of LpoB. This observation suggested that PBP1b enzymes from other Gram-negative bacteria may be controlled by an alternative activator. We therefore investigated PBP1b regulation in P. aeruginosa and found that PaPBP1b activity relies on a distinct lipoprotein that we have named LpoP. Our results indicate that, although this protein is unrelated to EcLpoB, it appears to stimulate PaPBP1b via a mechanism that is similar to EcLpoB activation of EcPBP1b.
Results
Identification and Characterization of Synthetic Lethal Partners of PaPBP1a and PaPBP1b.
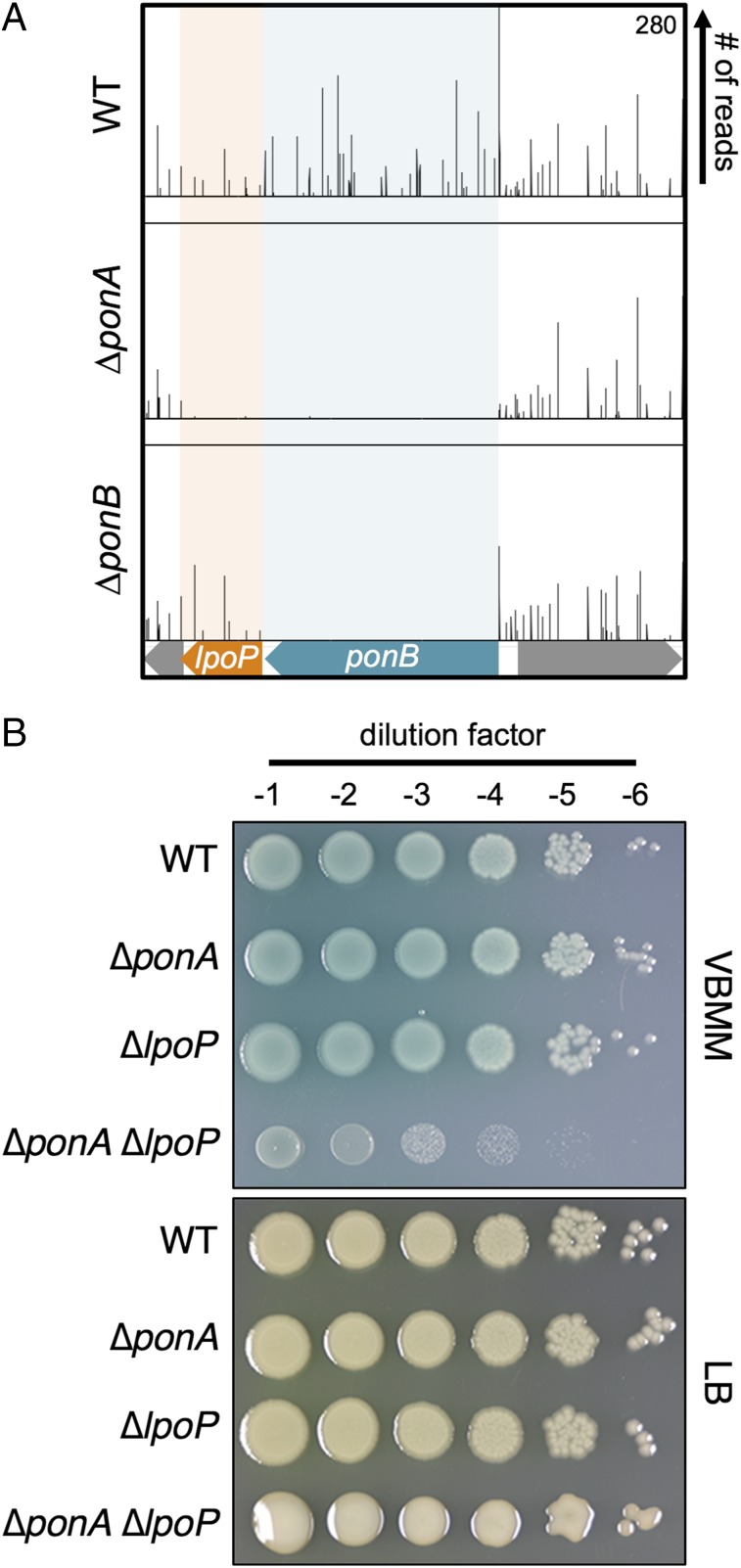
We previously identified the EcLpoA and EcLpoB factors using a synthetic lethal screen in E. coli based on a plasmid-loss phenotype (5). Because EcPBP1a and EcPBP1b are known to form a synthetic lethal pair, we reasoned that critical activators of one aPBP could be identified as synthetic lethal partners of the other. We employed a similar logic to investigate aPBP regulation in P. aeruginosa but used transposon sequencing (Tn-Seq) analysis (13) to identify synthetic lethal relationships. Transposon mutagenesis was performed on wild-type P. aeruginosa PAO1 as well as on derivatives deleted for PaponA (encoding PaPBP1a) or PaponB (encoding PaPBP1b). As part of the mutagenesis procedure, transposon mutants were selected on Vogel-Bonner minimal medium (VBMM) containing gentamicin, resistance to which is encoded in the transposon. Genomic DNA was then harvested from each of the resulting pooled transposon libraries and the transposon-chromosome junctions were mapped by Illumina sequencing. As expected from previous studies, both the PaponA and PaponB genes were readily inactivated by insertions in the wild-type background (14). However, the PaponA gene was devoid of insertions in the ΔPaponB strain, indicating that it had become essential in this background (SI Appendix, Fig. S2A). The gene for PalpoA displayed a Tn-Seq profile similar to PaponA, which is consistent with it functioning as a PaPBP1a activator (SI Appendix, Fig. S2A). Analogously, PaponB displayed a Tn-Seq pattern consistent with it being essential when PaPBP1a was inactivated (Fig. 1A). The downstream PA4699 gene also appeared to be synthetically lethal with ΔponA, suggesting that it might encode a PaPBP1b activator (Fig. 1A). Based on the results presented below, we have renamed this gene PalpoP for (lipoprotein activator of PBP1b).
Fig. 1.
Genetic connections between PaponA and genes encoding PaPBP1b and PaLpoP. (A) Shown are Tn-Seq profiles for the P. aeruginosa ponB locus from analyses performed in the PAO1 WT strain and derivatives lacking either PBP1a (∆ponA) or PBP1b (∆ponB). Individual lines above the locus map represent unique transposon insertion sites, and the height of each line reflects the number of sequencing reads at that site. Colored boxes indicate genes with significantly fewer reads in either mutant compared with the WT. (B) Cells of PAO1 (WT), PA104 (ΔponA), PA550 (ΔlpoP), and PA685 (ΔponA ΔlpoP) were grown overnight in LB at 30 °C, washed twice with VBMM, and normalized for cell density (OD600 = 1). Samples were serially diluted, and 5 μL of each dilution (10−1–10−6) was spotted onto VBMM agar before overnight incubation at 37 °C. Note that the ΔponA ΔlpoP colonies on LB are mucoid, indicating that they may be induced for an envelope stress response that results in extracellular polysaccharide production.
To validate the Tn-Seq results, mutant strains deleted for the aPBP and/or Lpo genes were constructed. As expected, all of the deletion mutants lacking a single aPBP or Lpo factor were viable (Fig. 1 and SI Appendix, Fig. S2B). Similar to previous results (14), we were also unable to construct a double ∆PaponA ∆PaponB mutant on either minimal VBMM or rich medium. The synthetic lethal phenotype resulting from simultaneous aPBP inactivation was further confirmed by constructing a strain with its sole copy of PaponB under control of the isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible toplac-dn1 promoter (Ptoplac-dn1) and deleting PaponA in this background. Growth of the resulting strain was IPTG-dependent, indicating that at least one aPBP is required for P. aeruginosa viability (SI Appendix, Fig. S3).
Unlike the dual aPBP deletion strain, construction of ∆PaponA ∆PalpoP and ∆PaponB ∆PalpoA double strains was possible even though the mutants are also predicted to be defective for both aPBPs. These double mutants displayed a conditional growth phenotype. On rich medium, their growth approached that of the wild-type strain (Fig. 1B and SI Appendix, Figs. S2B and S4). However, in accordance with the Tn-Seq results, the double mutants displayed severe growth defects in the minimal VBMM medium used for the analysis (Fig. 1B and SI Appendix, Figs. S2B and S4). These phenotypes could be complemented by the expression of the missing lpo gene from a plasmid, indicating that the growth defect is indeed due to the loss of Lpo protein function and not an effect of the deletion allele on nearby genes (SI Appendix, Fig. S5).
The ability to construct the ∆PaponA ∆PalpoP and ∆PaponB ∆PalpoA mutants suggests that the aPBPs of P. aeruginosa retain residual activity in the absence of their cognate activator. Furthermore, PaPBP1b appears to possess more residual activity than PaPBP1a, given the greater severity of the ∆PaponB ∆PalpoA growth defect relative to that of ∆PaponA ∆PalpoP cells on VBMM. In support of the aPBPs retaining activity in the absence of their cognate Lpo factor, overproduction of PaPBP1a or PaPBP1b suppressed the growth defect of ∆PaponB ∆PalpoA or ∆PaponA ∆PalpoP strains on VBMM, respectively (SI Appendix, Fig. S5). The medium dependence of the growth phenotype suggests either that this residual (Lpo-independent) activity is elevated in rich medium relative to VBMM or that low levels of aPBP activity are poorly tolerated on VBMM relative to LB. These results differ from E. coli, in which the inactivation of one aPBP combined with its noncognate Lpo activator is strictly lethal in both minimal and rich medium (5). It is not clear whether this reflects a greater dependence of E. coli aPBPs on their Lpo factors for activity or that E. coli is just more sensitive to reduced aPBP activity than P. aeruginosa. In any case, the mutant analysis in P. aeruginosa confirms the Tn-Seq results, and the severe growth defect observed when PaLpoP is inactivated in strains lacking PaPBP1a implicates PaLpoP as a potential activator of PaPBP1b.
PaLpoP Primary Structure and Phylogenetic Distribution.
PaLpoP is a 259-aa protein that, similar to LpoB, consists of an N-terminal signal sequence with a lipobox predicted to target it to the outer membrane (residues 1–20) immediately followed by an intrinsically disordered domain (residues 21–189). Despite these similarities at the N terminus, the C termini of LpoB and PaLpoP are strikingly different. PaLpoP harbors two tandem tetratricopeptide repeats (TPRs) at the C terminus (residues 190–255) (SI Appendix, Fig. S6). TPR domains are a widely distributed helix-turn-helix structural motif typically involved in mediating protein–protein interactions (15). The C terminus of LpoB, on the other hand, consists of a distinct globular domain composed of a beta sheet surrounded by alpha helices (8). In terms of phylogenetic distribution, LpoP is typically found in families of the Gammaproteobacteria that lack an LpoB homolog, but retain a PBP1b homolog (SI Appendix, Fig. S7A). Unlike LpoB, which is often encoded at a distinct locus from PBP1b, the LpoP-coding sequence tends to be closely linked to the PBP1b-coding gene and is often found immediately downstream (SI Appendix, Fig. S7B). Interestingly, a few species were found to encode both LpoB and LpoP homologs. Further inspection of the genomes of several of these organisms revealed that they encode two PBP1b homologs. Overall, the genetic and phylogenetic analyses support a role for LpoP in PBP1b function.
PaLpoP Interacts Directly and Specifically with PaPBP1b.
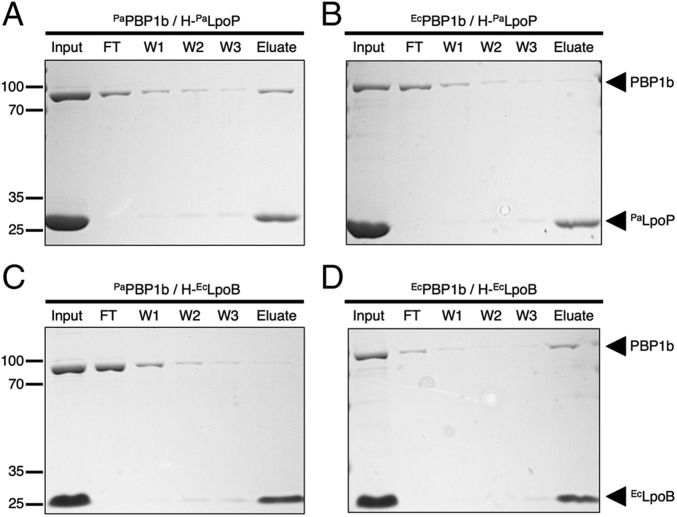
We next investigated the ability of PaLpoP to interact with PaPBP1b in vitro. Soluble versions of PaLpoP and EcLpoB were affinity-purified with an N-terminal histidine-tag (His-Tag) that replaced the signal sequence. These proteins were mixed with purified preparations of untagged PaPBP1b and EcPBP1b solubilized in detergent, and interactions were assessed using a pull-down assay with metal affinity resin. As expected, based on the genetic analysis, PaPBP1b but not EcPBP1b could be pulled down with PaLpoP (Fig. 2). Conversely, EcLpoB pulled down EcPBP1b but not PaPBP1b (Fig. 2). We therefore conclude that PaLpoP and PaPBP1b interact directly and specifically.
Fig. 2.
PaLpoP directly and specifically interacts with PaPBP1b. (A–D) H-PaLpoP (A and B) or H-EcLpoB (C and D) was incubated with PaPBP1b (A and C) or EcPBP1b (B and D) for 60 min at room temperature in binding buffer (20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 0.1% Triton-X-100). Ni-NTA agarose beads were then added to each reaction, followed by incubation for 2 h at 4 °C with rotation. The beads were pelleted by centrifugation and washed three times with binding buffer containing 30 mM imidazole, and the proteins retained on the resin were eluted with sample buffer containing EDTA (100 mM). Proteins in the initial reaction (input), initial supernatant (FT), wash supernatants (W1, W2, and W3), and elution (Elute) were separated on a 12% SDS polyacrylamide gel and stained with Coomassie Brilliant Blue. All proteins were present in the initial binding reaction at a concentration of 4 μM. Positions of molecular mass markers (numbers in kDa) are given to the left of each panel.
PaPBP1b-PaLpoP Can Functionally Substitute for E. coli aPBP Activity.
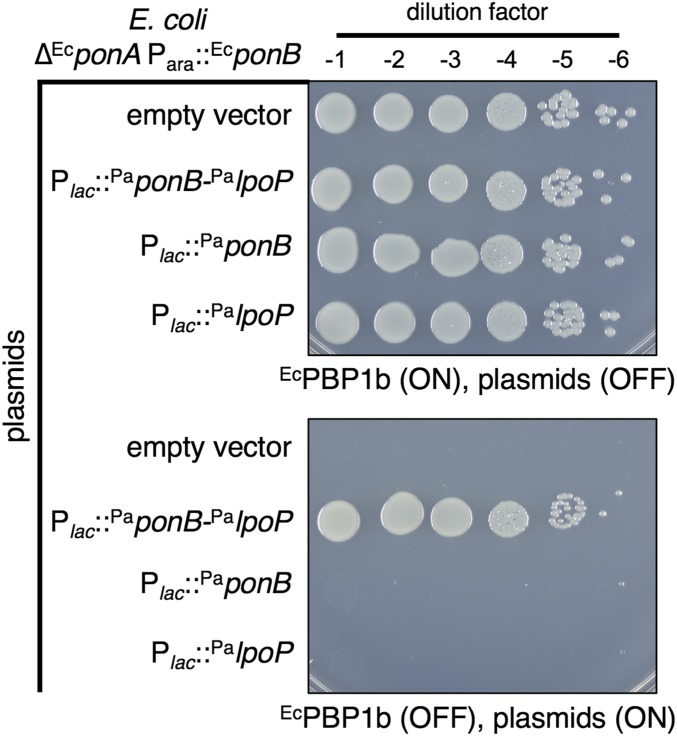
To determine whether PaLpoP is required to promote PaPBP1b activity, we tested the ability of PaPBP1b to supply aPBP activity in a heterologous E. coli system with or without coexpression of PaLpoP. For this experiment, we took advantage of an E. coli strain harboring an arabinose-inducible copy of its native EcponB gene (Para::EcponB) that is also deleted for EcponA. In the absence of arabinose, the strain lyses due to deficient aPBP activity and displays a severe plating defect (Fig. 3 and SI Appendix, Figs. S8 and S9). Expression of the PaponB-PalpoP operon from an IPTG-inducible lactose promoter (Plac) restored growth and viability to the strain on arabinose-free medium supplemented with IPTG (Fig. 3 and SI Appendix, Fig. S8). By contrast, expression of PaponB or PalpoP alone was insufficient to prevent lysis and restore growth, with the growth defect of the strains resembling that of the empty vector control (Fig. 3 and SI Appendix, Figs. S8 and S9). Importantly, PaPBP1b was produced at comparable levels whether or not it was coexpressed with PaLpoP (SI Appendix, Fig. S10). We conclude that PaPBP1b is capable of substituting for the E. coli aPBPs in a manner dependent on PaLpoP.
Fig. 3.
PaPBP1b-PaLpoP complements the loss of aPBP activity in E. coli. Cells of E. coli MM10 (∆EcponA Para::EcponB) and its derivatives harboring the integrated plasmid expression constructs pMT116 (empty vector), pNG66 (Plac::PaponB-PalpoP), pNG68 (Plac:: PaponB), or pNG51 (Plac::PalpoP) were grown overnight in M9 medium with 0.2% arabinose at 37 °C. Cells were washed twice with M9 medium lacking a carbon source, normalized for cell density (OD600 = 1), and serially 10-fold diluted (10−1–10−6). Five microliters of each dilution was spotted onto M9 agar with or without the arabinose or IPTG inducers as indicated. Plates were incubated overnight at 37 °C and photographed.
To determine whether LpoP factors from other organisms also promote the activity of their cognate PBP1b, we tested the function of Acinetobacter baylyi proteins in the E. coli system. As with the P. aeruginosa proteins, we found that AbPBP1b was produced but only capable of supplying aPBP activity to E. coli when it was coexpressed with AbLpoP (SI Appendix, Figs. S10 and S11A). Notably, AbLpoP was unable to complement the PaLpoP defect of P. aeruginosa mutants deleted for PaponA and PalpoP, suggesting that the LpoP-PBP1b functional pairs must be species-specific (SI Appendix, Fig. S11B). We conclude that LpoP proteins are likely to generally function as cofactors for PBP1b proteins in organisms that express them.
Evidence That EcLpoB and PaLpoP Activate Their Cognate PBP1b via a Similar Mechanism.
To investigate the mechanism by which PaLpoP promotes PaPBP1b activity in vivo, we selected for mutants that bypass the PaLpoP requirement for PaPBP1b function. For this investigation, we took advantage of the observation that the growth phenotype of the ∆PaponA ∆PalpoP mutant increases in severity when plated on VBMM at 42 °C such that growth is all but eliminated (SI Appendix, Fig. S12). This growth defect allowed us to plate ∆PaponA ∆PalpoP cells on VBMM at 42 °C to select for spontaneous suppressors. Twelve independent suppressors were isolated and subjected to whole-genome sequencing along with the parental strain. The majority of the suppressors (9 of 12) harbored a single missense change within the PaponB-coding region. An additional strain had a mutation that mapped upstream of PaponB (SI Appendix, Table S1). We suspected that this mutation increases PaponB expression and therefore did not study this mutant further. The final two suppressors had mutations mapping to a single gene not directly involved in PG synthesis. These suppressors will be described and characterized as part of a separate report.
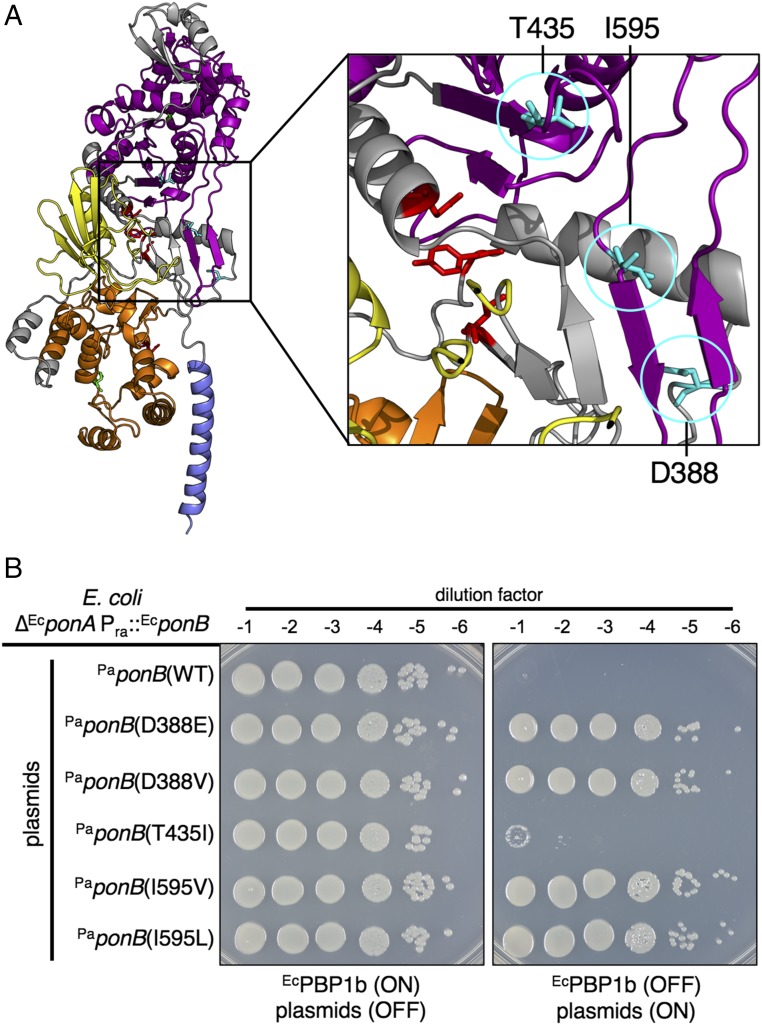
The missense mutations in PaponB resulted in two different changes each at codons D388 and I595, resulting in the production of the protein variants PaPBP1b(D388E), PaPBP1b(D388V), PaPBP1b(I595V), and PaPBP1b(I595L). The other unique change was at codon T435 to generate PaPBP1b(T435I). In the predicted structure of PaPBP1b based on the solved structure of EcPBP1b (16), these substitutions map to the region of the protein that lies between the PGT and TP domains and just behind the UB2H domain (Fig. 4A). Notably, amino acid substitutions in the same region of EcPBP1b were recently found to result in an EcLpoB bypass phenotype and to mimic LpoB activation of PGT activity (17). We therefore reasoned that the changes identified in PaPBP1b would similarly result in a synthase that functions without the need for PaLpoP. Accordingly, production of these PaPBP1b variants in P. aeruginosa ∆PaponA ∆PalpoP cells restored their growth on VBMM at 42 °C to a greater extent than the wild-type protein, the overexpression of which alone had a mildly suppressive effect (SI Appendix, Fig. S13). Furthermore, with the exception of PaPBP1b(T435I), the PaPBP1b variants were also capable of functioning in the E. coli system without coexpression with PaLpoP. This result contrasts starkly with PaPBP1b(WT), which was completely dependent on PaLpoP coexpression to substitute for the E. coli aPBPs (Fig. 4B and SI Appendix, Fig. S14). Based on the relatedness of the cofactor bypass mutants identified in PaPBP1b to those previously described for EcPBP1b, we conclude that PaLpoP functions as an activator of PaPBP1b and that it activates its cognate synthase via a similar mechanism despite being unrelated to EcLpoB in sequence and predicted structure.
Fig. 4.
Amino acid substitutions in PaPBP1b bypass the PaLpoP requirement. (A) Shown is a model structure of PaPBP1b made with Swiss-Model using the highest scoring template: E. coli PBP1b in complex with acyl-aztreonam and moenomycin (Protein Data Bank ID code 5HLB) (16). Residues D388, T435, and I595 are indicated in cyan. Catalytic residues involved in PGT activity (E190) and TP activity (S469) are highlighted in green. Homologous residues to those of EcPBP1b previously implicated in EcLpoB bypass (17) are highlighted in red. Domains are colored as follows: blue: transmembrane; orange: glycosyltransferase; yellow: UB2H; purple: transpeptidase. (B) Cells of E. coli MM10 (∆EcponA Para::EcponB) harboring the plasmid expression constructs pNG117 [Plac::PaponB(WT)], pNG118 [Plac::PaponB(T435I)], pNG119 [Plac::PaponB(I595V)], pNG120 [Plac::PaponB(I595L)], pNG121 [Plac::PaponB(D388E)], and pNG122 [Plac::PaponB(D388V)] were grown overnight in M9 supplemented with 0.2% arabinose and tetracycline to maintain the plasmids. Cells were then treated and plated on the indicated medium as described in Fig. 3. Note that PaPBP1b(T435I) was tested for function at 37 °C and 42 °C, but failed to support growth at both temperatures.
Discussion
It has been several years since the original identification of the EcLpoA and EcLpoB outer-membrane lipoproteins in E. coli and their role in activating PG synthesis by their cognate aPBPs, EcPBP1a and EcPBP1b, respectively (5, 6). Of the two aPBPs, the control of EcPBP1b by EcLpoB is the most well understood. EcLpoB binds to the UB2H domain of EcPBP1b and via this binding event is thought to stimulate a conformational change in the synthase that activates its PGT domain to promote PG polymerization and cross-linking (8–10, 17). Based on this likely mechanism for PBP1b activation, it was surprising to find that many Gammaproteobacteria encode a PBP1b protein with a recognizable UB2H domain but do not have a corresponding LpoB homolog. This observation suggested that these organisms encode an alternative PBP1b activator and prompted us to look for it using P. aeruginosa as a model system. The search resulted in the identification of LpoP as a cofactor that promotes PBP1b activity.
A Common Mechanism for PBP1b Activation by Different Lipoproteins.
Several lines of evidence support a model in which LpoP stimulates PBP1b activity via a mechanism similar to LpoB despite the lack of relatedness between the two lipoproteins. First, the phenotypes of LpoB and LpoP in their respective organisms are highly similar, with each showing a synthetic lethal/sick phenotype when inactivated in combination with their noncognate aPBP. Second, each lipoprotein also specifically interacts with its cognate PBP1b in vitro (5) and is either capable of stimulating the activity of its cognate PBP in vitro (EcLpoB) (5, 6, 8, 10) or in a heterologous host (PaLpoP). The strongest case for LpoB and LpoP activating their respective PBP1b synthases via a similar mechanism comes from the analysis of PBP1b variants that gain the ability to function without Lpo factor activation. Such variants were previously isolated in EcPBP1b (17). They were found to function in vivo without EcLpoB, and their in vitro PG polymerase activity in the absence of added EcLpoB resembled that of the wild-type protein following EcLpoB stimulation (17). Most of the amino acid changes in these activated variants were located in a region of the EcPBP1b structure (16) between the PGT and TP domain that sits behind UB2H (17). It is therefore reasonable to assume that the substitutions induce a conformational change in EcPBP1b that mimics EcLpoB binding to UB2H and that this change promotes PGT activation. Substitutions in PaPBP1b in very similar locations allow it to function in the absence of PaLpoP. The striking correspondence of the Lpo bypass changes in EcPBP1b and PaPBP1b strongly supports the idea that the two unrelated lipoproteins have converged evolutionarily to activate their cognate PBPs via similar mechanisms.
In the case of PaLpoP, its predicted unstructured region likely allows the protein to reach across the periplasm from the outer membrane to interface with PaPBP1b as has been proposed for EcLpoB (8). The TPR domains are the most likely candidates for the portion of PaLpoP that directly interfaces with PaPBP1b, and this region probably does so by binding the UB2H domain like EcLpoB. Consistent with this possibility, TPR domains are broadly distributed in biology to mediate protein–protein interactions (15) and are found in many regulators of cell-wall biogenesis, including LpoA (5–7, 18), NlpI (19, 20), and CpoB (21). Interestingly, the periplasmic protein CpoB in E. coli is thought to use its TPR domain to associate with EcPBP1b via its UB2H domain during cell division to coordinate EcPBP1b activity with that of the Tol-Pal system during cell division (21). P. aeruginosa encodes a CpoB homolog in addition to PaLpoP, suggesting that it may have two TPR-containing proteins that bind its PBP1b. However, this possibility and the role of PaCpoB in PaPBP1b control will require further investigation.
Support for an Autonomous Role for aPBPs in PG Biogenesis.
The recent demonstration of PG polymerase activity for the SEDS protein RodA (3) has raised the question of how SEDS and aPBP PG polymerases work together to build the cell wall. In a previous study, we began addressing this problem by comparing the subcellular localization dynamics of RodA and the aPBPs in E. coli and Bacillus subtilis (4). RodA is a component of the rod-shape–determining PG biogenesis system called the Rod system (22). This machine is organized by filaments of the actin-like MreB protein, which have previously been shown to localize in dynamic foci distributed throughout the cell cylinder that rotate circumferentially around the long axis of the cell (23–25). This rotational movement is blocked by PG synthesis inhibitors, suggesting that it reflects active PG assembly by the machinery. Similar directed motions had been observed for RodA and its bPBP partner in B. subtilis (23, 24), and we confirmed that the E. coli proteins also display such circumferential movement (4). On the other hand, no such motion was observed for fluorescent fusions to PBP1b in E. coli or the main aPBP in B. subtilis, PBP1 (4, 26). Instead, these aPBPs displayed undirected, diffusive movement (4, 26). Based on the distinct dynamics observed for Rod system components versus aPBPs, we proposed a model for cell-wall synthesis in which the aPBPs function largely outside of the Rod system during cell elongation rather than being part of the complex as has been commonly believed (22).
The observation reported here that heterologous LpoP-PBP1b pairs from both P. aeruginosa and A. baylyi can supply essential aPBP activity to E. coli also supports a largely autonomous role for aPBPs in PG biogenesis. P. aeruginosa and A. baylyi are relatively distant from Enterobacteriaceae among the Gammaproteobacteria, and their PBP1b proteins are only 43 and 37% identical to the E. coli protein, respectively. It is therefore unlikely that these heterologous PBP1b proteins with their distinct lipoprotein activators are capable of integrating productively within E. coli PG synthetic complexes like the Rod system and division machinery. Thus, the fact that PaPBP1b and AbPBP1b can stand in for the E. coli aPBPs further supports the notion that the critical function of the aPBPs in cell-wall synthesis does not require them to engage in multiprotein synthetic machineries. Such a “free agent” or at least semiautonomous role for the aPBPs would also explain the long-standing observation that two or more aPBPs in a given bacterium, like E. coli PBP1a and PBP1b, are often interchangeable and largely redundant.
Importantly, although our results suggest that aPBPs can function as largely independent PG synthases, we are not proposing that they interact only with their Lpo activators. Indeed, many coimmunoprecipitation studies have been performed previously to detect aPBP interaction partners. For example, several interactions between EcPBP1b and cell-division proteins have been identified, and these interactions have been shown to alter PG synthase activity in vitro (21, 27, 28). What remains to be determined is whether or not these interactions and their associated PBP activity changes are physiologically relevant. The aPBP swapping experiments using P. aeruginosa and A. baylyi proteins suggest that the formation of these complexes may not be required for cell-wall assembly under normal circumstances. However, the isolation of mutants specifically defective for aPBP interactions with the cell-division machinery and other PG biogenesis factors and an assessment of their resulting phenotypes is needed before definitive conclusions can be drawn.
Conclusion.
In this report, we describe the surprising finding that different Gram-negative species use distinct lipoprotein cofactors to stimulate the activity of their PBP1b PG synthases and that they do so via a common mechanism. Thus, despite the apparent plasticity in the choice of activators between species, it may be possible to target aPBP activation as a means of disrupting cell-wall synthesis for the treatment of infections with a range of problematic Gram-negative bacteria.
Materials and Methods
Media, Bacterial Strains, and Plasmids.
Cells were grown in either LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl, unless otherwise indicated) and minimal M9 medium supplemented with 0.2% casamino acids and 0.2% sugar, as indicated, or VBMM (29). Where appropriate, antibiotics were used at the following concentrations, unless otherwise specified: ampicillin (50 μg/mL), carbenicillin (200 μg/mL), chloramphenicol (25 μg/mL), gentamicin (15 μg/mL for E. coli, 50 μg/mL for P. aeruginosa), kanamycin (50 μg/mL), and tetracycline (5 μg/mL). The bacterial strains used in this study are listed in SI Appendix, Table S2. All P. aeruginosa strains used in the reported experiments are derivatives of PAO1, and E. coli strains are derivatives of MG1655. Plasmids used in this study are listed in SI Appendix, Table S3. Details for strain and plasmid constructions are provided in SI Appendix.
Transposon Sequencing.
P. aeruginosa PAO1 and its ∆ponA and ∆ponB derivatives were mutagenized with the minitransposon from pBTK30 (30) following conjugation from E. coli (see SI Appendix for details). Genomic DNA from thawed cell pellets of each mutant library was extracted, fragmented, poly(C)-tailed, purified, and sequenced using methods similar to those previously described (31). See SI Appendix for an in-depth description of the protocol.
Protein Purification.
E. coli and P. aeruginosa PBP1b, EcLpoB, and PaLpoP proteins were purified as His-tagged or His-SUMO–tagged fusions using previously published procedures (5, 17). See SI Appendix for details.
Supplementary Material
Acknowledgments
We thank all members of the T.G.B. and David Rudner laboratories for advice and helpful discussions; Stephen Lory and Simon Dove for help with P. aeruginosa methods and for providing strains and expert advice; and Ankur Dalia for providing A. baylyi genomic DNA. This work was supported by the National Institute of Allergy and Infections Diseases of the National Institutes of Health (Grants R33 AI111713 and R01 AI083365). C.F. was supported in part by a postdoctoral fellowship from the Swiss National Science Foundation (Project #P300PA_171535).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717925115/-/DCSupplemental.
References
- 1.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016;1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jean NL, et al. Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: Implications for PBP1A stimulation. Structure. 2014;22:1047–1054. doi: 10.1016/j.str.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan AJF, et al. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci USA. 2014;111:8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King DT, Lameignere E, Strynadka NCJ. Structural insights into the lipoprotein outer membrane regulator of penicillin-binding protein 1B. J Biol Chem. 2014;289:19245–19253. doi: 10.1074/jbc.M114.565879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupoli TJ, et al. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J Am Chem Soc. 2014;136:52–55. doi: 10.1021/ja410813j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dörr T, et al. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infect Immun. 2014;82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin J, Sun Y, Mao Y, Jin M, Gao H. PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis. Antimicrob Agents Chemother. 2015;59:3357–3364. doi: 10.1128/AAC.04669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Zhang Y-M, Davies C. Penicillin-binding protein 3 is essential for growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;61:e01651-16. doi: 10.1128/AAC.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Andrea LD, Regan L. TPR proteins: The versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Sung M-T, et al. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci USA. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markovski M, et al. Cofactor bypass variants reveal a conformational control mechanism governing cell wall polymerase activity. Proc Natl Acad Sci USA. 2016;113:4788–4793. doi: 10.1073/pnas.1524538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sathiyamoorthy K, Vijayalakshmi J, Tirupati B, Fan L, Saper MA. Structural analyses of the Haemophilus influenzae peptidoglycan synthase activator LpoA suggest multiple conformations in solution. J Biol Chem. 2017;292:17626–17642. doi: 10.1074/jbc.M117.804997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson CGM, Kajander T, Regan L. The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold. FEBS J. 2005;272:166–179. doi: 10.1111/j.1432-1033.2004.04397.x. [DOI] [PubMed] [Google Scholar]
- 20.Singh SK, Parveen S, SaiSree L, Reddy M. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci USA. 2015;112:10956–10961. doi: 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray AN, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. elife. 2015;4:e07118. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 25.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TK, Meng K, Shi H, Huang KC. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Commun. 2016;7:13170. doi: 10.1038/ncomms13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller P, et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 28.Bertsche U, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 29.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 30.Goodman AL, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Lai GC, Cho H, Bernhardt TG. The mecillinam resistome reveals a role for peptidoglycan endopeptidases in stimulating cell wall synthesis in Escherichia coli. PLoS Genet. 2017;13:e1006934. doi: 10.1371/journal.pgen.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.