Significance
Transporters isomerize between conformations to shuttle cargo across membranes, but the mechanism is not understood. Double electron–electron resonance measurements on the sodium-dependent sugar transporter (vSGLT) were used to explore the conformational state of the transporter under specific ligand conditions. Although sugar transport by vSGLT is driven by sodium gradients, vSGLT adopts an inward-open conformation irrespective of the presence of sodium. In the presence of sodium and galactose, the transporter transitions to an occluded conformation. We propose that the cell’s negative membrane potential aids in driving vSGLT toward the outward-facing state to bind sugar and begin the transport cycle. These findings could be applicable to other transporters whereby the inherent cellular membrane potential is integrated into the transport cycle.
Keywords: vSGLT, DEER, sugar, transport, conformation
Abstract
Sodium-dependent transporters couple the flow of Na+ ions down their electrochemical potential gradient to the uphill transport of various ligands. Many of these transporters share a common core structure composed of a five-helix inverted repeat and deliver their cargo utilizing an alternating-access mechanism. A detailed characterization of inward-facing conformations of the Na+-dependent sugar transporter from Vibrio parahaemolyticus (vSGLT) has previously been reported, but structural details on additional conformations and on how Na+ and ligand influence the equilibrium between other states remains unknown. Here, double electron–electron resonance spectroscopy, structural modeling, and molecular dynamics are utilized to deduce ligand-dependent equilibria shifts of vSGLT in micelles. In the absence and presence of saturating amounts of Na+, vSGLT favors an inward-facing conformation. Upon binding both Na+ and sugar, the equilibrium shifts toward either an outward-facing or occluded conformation. While Na+ alone does not stabilize the outward-facing state, gating charge calculations together with a kinetic model of transport suggest that the resting negative membrane potential of the cell, absent in detergent-solubilized samples, may stabilize vSGLT in an outward-open conformation where it is poised for binding external sugars. In total, these findings provide insights into ligand-induced conformational selection and delineate the transport cycle of vSGLT.
Membrane bilayers are essential components for the compartmentalization of all cells and organelles, generating an impermeable boundary for ions and water-soluble molecules. To facilitate the selective transfer of hydrophilic solutes between compartments, cells employ membrane-embedded channels and transporters. Transporters are widely believed to follow an alternating-access mechanism (1) where the substrate binding site alternates between multiple states to deliver its cargo from one side of the membrane to the other. There is now a wealth of biophysical and structural data from a diverse set of transporters—occupying different folds and conformations—supporting the alternating-access mechanism (2–4), but the process by which they transition between states and how their ligands influence transporter equilibria varies among transporters.
The SLC5 family of human solute carriers is composed of 12 members, which are responsible for the cotransport of Na+ with glucose, galactose, myoinositol, choline, and anions. The sodium-dependent glucose transporters (SGLTs) are the most well-characterized members of the SLC5 family, where SGLT1 is responsible for absorption of dietary sugars in the brush border of intestinal enterocytes. SGLT1 and SGLT2 aid in maintaining systemic glucose concentration through glucose reabsorption from the kidney’s glomerular filtrate back to the bloodstream. In addition to their roles in normal physiology, SGLTs are the target of novel drugs for the treatment of type II diabetes. These drugs inhibit the absorption and reabsorption of glucose, thereby lower plasma glucose levels and glycosylated hemoglobin, while modestly reducing weight and systolic blood pressure (5, 6).
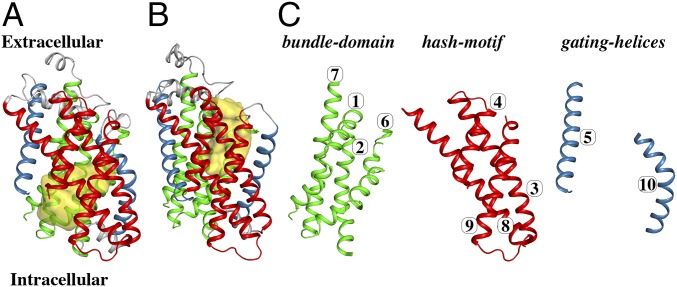
The structural data for SGLTs was largely generated from the Vibrio parahaemolyticus sodium galactose cotransporter (vSGLT), which shares 60% sequence similarity to human SGLT1. vSGLT displays an inverted repeat motif in which TM1–TM5 and TM6–TM10 are related by a ∼153° rotation parallel to the membrane plane (7, 8). This 5-TM inverted-repeat (5TMIR) architecture has been seen in other transporters (7, 9), including the Drosophila dopamine transporter (10) and the human serotonin transporter (11). A cornerstone of the 5TMIR topology is the spatial arrangement between the first two TMs (TM1–2 and TM6–7) and the last three TMs (TM3–5 and TM8–10), of each inverted repeat, frequently referred to as the “bundle” and “scaffold” domains, respectively. The “rocking-bundle” mechanism represents a minimalist solution to achieve alternating access by transitioning between outward- and inward-facing conformations through rigid-body motion of the bundle domain around the scaffold domain (12), yet there have been a number of variations on this theme (3).
X-ray structures of vSGLT were solved in an inward-occluded (7) and an inward-open conformation (8) (Fig. 1A), providing atomic-resolution details of these inward-facing states and the corresponding release of substrates; however, data for outward-facing conformations remain absent. Other proteins displaying the 5TMIR fold have been captured in both outward- and inward-facing conformations, suggesting that they represent comparable energetic states (2, 4, 13). This concept has further been explored using in-solution measurements such as double electron–electron resonance (DEER) and single-molecule fluorescence resonance energy transfer that clearly showed isomerization events that are compatible with both outward- and inward-facing conformations (14, 15). For Mhp1, a Na+/hydantoin transporter, solution studies demonstrated a dynamic equilibrium where Na+ and substrate together shift the equilibrium toward the outward-facing state, and this study reinforced the rocking-bundle model (14). For LeuT, a more complicated pattern emerged where ligand induced stabilization of certain populations that deviated from both the crystal structures and the rocking-bundle mechanism (16). Despite having a high structural identity, these different transport mechanisms highlight the diversity of the 5TMIR fold.
Fig. 1.
Overall architecture of the 10-TM core of vSGLT. (A) A cartoon representation of the crystal structure of vSGLT in the inward-open conformation (PDB ID code 2XQ2). The conserved 10-TM core is divided into TMs 3, 4, 8, and 9 constituting the hash motif (shown in red), TMs 1, 2, 6, and 7 constituting the bundle domain (shown in green), and two gating helices TMs 5 and 10 (shown in blue). The extracellular cavity connecting the ligand binding site to the intracellular space is shown in yellow. (B) An outward-facing homology model of vSGLT generated from the outward-facing structure of the Proteus mirabilis sialic-acid transporter (PDB ID code 5NV9) displays rigid-body movements between the bundle domain and hash motif, accompanied with TM10 opening, compared with the inward-facing structure. Color code is the same as in A, and the extracellular cavity is shown in yellow. (C) The structural motifs of the inward-open conformation are shown separately with the TM numbering.
In this work, shifts in the conformational ensemble of vSGLT in detergent micelles were monitored by DEER experiments where thiol reactive paramagnetic spin labels were attached to engineered cysteine residues. We observed the transporter in a state of dynamic equilibrium, sampling a number of conformations. Using defined conditions in which Na+ or Na+/galactose were either present or absent, the equilibrium could be shifted to favor a specific conformation. Comparison of the resulting distance distributions to structures of vSGLT in inward-facing conformations and to a model of vSGLT in the outward-facing conformation (Fig. 1B) show that vSGLT favors the inward-facing conformation in the absence and presence of Na+, while Na+/galactose shifts the equilibrium toward the outward-facing or occluded conformation. The substrate-dependent conformational transition could be inhibited by mutation of functionally relevant residues. This pattern of structural changes largely agrees with the rocking-bundle mechanism and is consistent with the ligand-dependent conformational equilibrium of Mhp1.
Results
vSGLT’s alternating-access mechanism was monitored by probing the individual subdomains, which are roughly associated with the rocking-bundle mechanism, using DEER spectroscopy. The “bundle domain” is formed by TMs 1, 2, 6, and 7 and the scaffold domain has been further subdivided into the “hash motif” (TMs 3, 4, 8, and 9) and the “gating helices” (TMs 5 and 10) (Fig. 1C). To explore the relationship between DEER distance distributions and observed conformational changes in a structural framework, models for both the inward- and outward-facing conformations are required. Currently, there is no structure of vSGLT in the outward-facing conformation. Thus, a homology model based on the outward-facing structure of the Proteus mirabilis sialic-acid transporter (SiaT) [Protein Data Bank (PDB) ID code 5NV9] was constructed (Fig. 1B). SiaT is a member of the solute sodium symporter family with a 2:1 ion:ligand stoichiometry. vSGLT and SiaT share a 24% sequence identity and 46% sequence similarity (SI Appendix, Fig. S1) with a root-mean-squared deviation of 3.0 Å between the core structures. The best model, from the program MODELLER (17), was chosen by considering the discrete optimized protein energy (DOPE) score (18) together with visual inspection of the sugar binding site rotamers and extracellular loops.
A simple comparison of the outward-facing model with the inward-facing structure (PDB ID code 3DH4) reveals a rigid-body movement between the bundle domain and hash motif, accompanied with a dramatic 16-Å opening of the extracellular portion of TM10 (gating helix) that exposes the extracellular vestibule (Fig. 1). Thus, these putative conformational changes were analyzed using site-directed spin labeling (19) of these domains and DEER measurements under various ligand conditions that shift the equilibrium of the transporter. To verify that the site-directed spin labeling procedures did not alter the transporter’s function, all mutants were assayed for galactose uptake into proteoliposomes (SI Appendix, Fig. S2) and were found to be of comparable activity to “wild-type” vSGLT (A423C). Molecular-dynamics (MD) simulations with dummy spin-labels (MDDS) (20, 21) were performed on the inward-facing structure and outward-facing model for each engineered site. These short MD simulations predict the spin-pair distance distributions between dummy spin-labels that are computationally attached to models, enabling the direct comparison with experimental DEER distance distributions. Additionally, the MDDS analysis provides an important guideline to assess the influence of ligands on the conformation of vSGLT.
vSGLT Forms Dimers in Micelles.
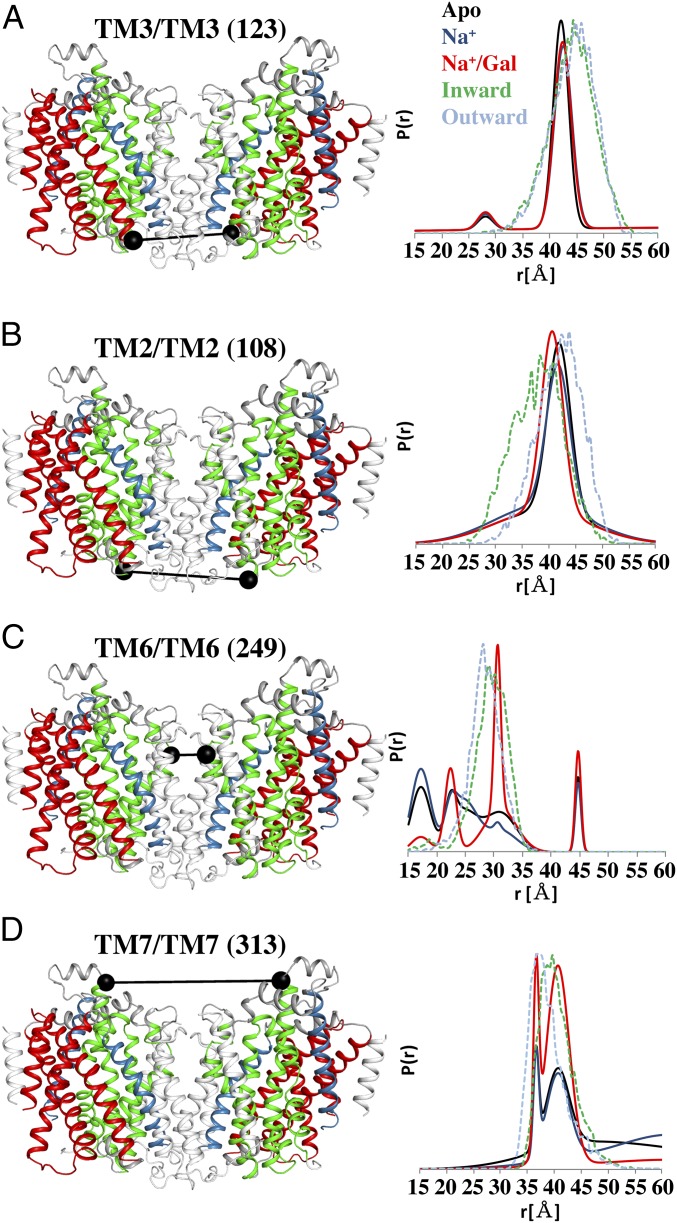
Freeze-fracture studies on heterologously expressed SGLT1 in Xenopus laevis oocytes (22) and vSGLT reconstituted into proteoliposomes (23) show a monomeric state. However, multiple crystal structures of vSGLT have been reported (7, 8), and all of them show the same dimeric packing, which is an important consideration when designing and interpreting paired distance distributions for DEER. To determine whether the dimeric arrangement observed in the crystal packing is maintained in detergent micelles, four single-cysteine constructs were labeled and measured. DEER-derived distance distributions for the four single-cysteine mutants are presented in Fig. 2. On the intracellular face of vSGLT, the intermolecular distance distributions for TM3 (residue 123 of the hash motif) (Fig. 2A) and TM2 (residue 108 of the bundle domain) (Fig. 2B) clearly demonstrate a distance distribution with a single peak at ∼42 Å, which agrees well with the distances calculated from the dimeric crystal packing of vSGLT and the MDDS calculations. A lack of ligand-induced changes in the distance distribution probabilities indicates that pair distance is unchanged during transport, but these positions may still reside on portions of the protein that undergo concerted movements.
Fig. 2.
DEER of single-cysteine constructs confirms a dimeric arrangement of vSGLT. Cartoon representations of the crystallographic vSGLT dimer are shown with the same color code as in Fig. 1, with an addition of the nonconserved helices in gray. Labeling sites used for DEER spectroscopy are marked with black spheres connected with black lines. A and B report the crystallographic location and corresponding distance distributions of sites on the intracellular side, whereas C and D show extracellular sites. Distance distributions represented as the probability of a distance [P(r)] versus distance (r) is shown for each pair. Data were obtained for the Apo (black), Na+-bound (blue), and Na+- and galactose-bound (red) intermediates. MDDS-derived distance distributions for the inward-facing structure (green dash) and the outward-facing model (cyan dash) correlate with the primary peaks in the DEER data.
A different pattern emerges for single-cysteine mutants on the extracellular side of TM6 and TM7 of the bundle domain. TM6 (residue 249) reports a dynamic region, which adopts a number of conformations in the Apo (black trace) and Na+ (blue trace) solutions. However, in the presence of Na+ and galactose (red trace), longer and more uniform distance components are observed, corresponding to the helix moving away from the dimer interface (Fig. 2C). Similarly, for TM7 (residue 313), distance components between 35 and 45 Å together with a broad, long-range component (>50 Å) are observed in the Apo (black trace) and Na+ (blue trace) traces. Under Na+ and galactose conditions (red trace), the long-range distance component is suppressed and the distance populations between 35 and 45 Å are increased, indicative of stabilizing a well-defined conformation (Fig. 2D). We attribute these long-distance components to an inward-open conformation in which TM7 helices move away from each other but do not adopt a well-defined conformation. It appears that the extracellular side, upon Na+ and galactose binding, adopts a restricted set of conformational states between protomers in the dimer. In addition, these data definitively show that vSGLT forms a stable dimer in detergent micelles, which is a critical parameter in designing intramolecular double-cysteine mutants and interpreting data from DEER spectra as the intermolecular distances—associated with the dimer—can convolute interpretation of the DEER spectra.
Conformational Changes of the Intracellular Side.
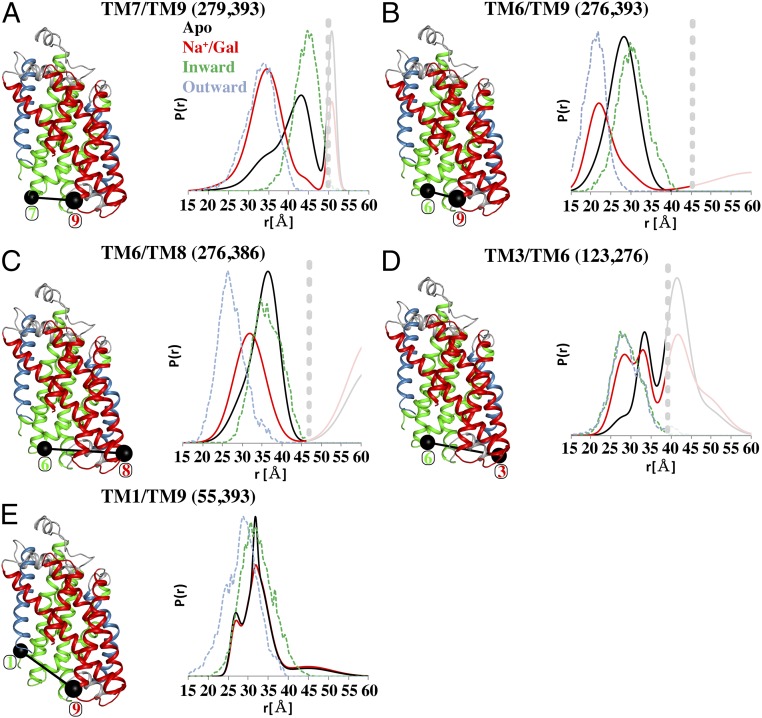
To investigate the mechanism of ligand-induced alterations at the intracellular face of vSGLT, we measured distances of doubly spin-labeled mutants. The sites for probe attachment were selected to investigate conformational changes within a single protomer (intramolecular distances), although long-distance components associated with the dimer arrangement were also evident. The hash motif and bundle domain are thought to transition as independent domains to facilitate alternating access in proteins with the 5TMIR fold (24). Monitoring movements between the bundle domain and hash motif revealed distinct Na+/Gal-induced changes in distance between probes (Fig. 3). TM7/TM9 (residues 279 and 393), TM6/TM9 (residues 276 and 393), and TM6/TM8 (residues 276 and 386) (Fig. 3 A–C, respectively) showed nearly indistinguishable distance distributions for the Apo (black traces) and Na+ conditions, but Na+/Gal (red traces) induces a decrease in average distance. This distance change is driven by enhanced population of short-distance components with a concomitant decrease of longer-distance components present in the equilibrium ensemble. In general, the traces of the Apo and Na+ conditions were similar for all measurements; therefore, from here onward, the Na+ traces are only presented in SI Appendix, Fig. S3.
Fig. 3.
Ligand-dependent conformational changes between TMs of the hash motif and bundle domain on the intracellular side of vSGLT. Since the distance distributions for the Apo and Na+ conditions overlap, the Na+ traces have been removed from the distance distribution plots for clarity but are included in SI Appendix, Fig. S3. The distance distributions originating from the dimeric arrangement are faded and enclosed to the right of horizontal broken lines. The effect of Na+/Gal binding to vSGLT results in a closing motions of TMs 7/9, 6/9, and 6/8 (A–C), with a good agreement between the Apo and Na+ conditions with the MDDS calculation for the inward-open structure and of the Na+/Gal condition with the outward-facing conformation. The distance distributions for TM3/TM6 are very wide with an equilibrium shift toward the short-distance regime for the Na+/Gal condition (D). TM1/TM9 do not display marked ligand-induced distance changes for both the DEER data and MDDS calculations (E).
MDDS calculations for these three pairs performed with the inward-facing conformation primarily overlap with the Apo and Na+-derived distance distributions, indicating vSGLT favors an inward-open conformation in the absence of galactose. Meanwhile, the MDDS calculations performed on the outward-facing model (in which the intracellular vestibule is closed) overlap well with the Na+/Gal distributions. Thus, the addition of Na+ and galactose appears to seal the intracellular vestibule, indicating a shift toward an inward-closed conformation.
Similarly, probes at TM3/TM6 (residues 123 and 276) report a multimodal distance distribution with intramolecular components ranging from 20 to 35 Å together with a larger intermolecular component likely arising from dimeric contribution of residue 123 (Fig. 2A). The presence of Na+/Gal increases the population of the shortest distance component, further indicating closure of the inner face. Interestingly, the MDDS analysis captured the breadth of the experimental distributions but failed to distinguish between the two intramolecular populations identified in the DEER analysis (Fig. 3D). In contrast, TM1/TM9 (residues 55 and 393) does not demonstrate ligand-induced changes in the distributions (Fig. 3E). This result highlights a significant deviation in the pattern of distance changes observed with Mhp1 where ligand binding drove a ∼10-Å decrease in distance between probes (14). Although the MDDS predicted strongly overlapping distances in both inward- and outward-facing conformations, a shift toward shorter distances was anticipated based on the outward-facing model. Given that this construct retained galactose uptake (SI Appendix, Fig. S2), either explicit relative motion is not required between TM1/TM9 or spin label rotamer preference obscured the movements. In support of the latter interpretation, the electron paramagnetic resonance (EPR) spectrum of this pair is consistent with rotational restriction of the probes in all states (SI Appendix, Fig. S3). Nevertheless, there is a global trend where Na+/Gal induces a shortening of distances between the hash motif and the bundle domain corresponding to closure of the intracellular vestibule.
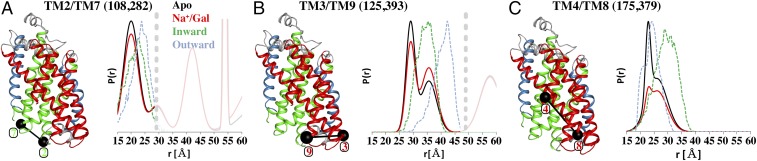
According to the rocking-bundle mechanism, the bundle domain and hash motif move as rigid bodies to facilitate the isomerization between outward- and inward-open conformations. In contrast to intracellular pairs within the bundle domain, subtle ligand-dependent movements within the hash motif were observed (Fig. 4). TM2/TM7 (residues 108 and 282) of the bundle domain is insensitive to the addition of Na+ or Na+/Gal, suggesting that these helices may move in a concerted manner (Fig. 4A). However, small differences between the Apo or Na+ states and Na+/Gal-bound form were apparent for TM3/TM9 (residues 125 and 393) in the hash motif (Fig. 4B). TM3/TM9 is the only intracellular pair where Na+/Gal induced an increase in the proportion of the longer distance population, and this increase is also reflected in the MDDS data for the outward-facing model, indicating that the longer distance is still compatible with inner-gate closure. Likewise, TM4/TM8 (residues 175 and 379) of the hash motif exhibits bimodal distance distributions indicative of equilibrium sampling of two conformations that are slightly modulated by ligand binding (Fig. 4C). Similar to other pairs, the distance distribution of TM4/TM8 is nearly identical for Apo (black trace) and Na+ states. Analysis of the Na+/Gal (red trace) state highlights a decrease in the population of the short-distance component with a concomitant increase in the apparent DEER signal background that is likely associated with changes in intermolecular dimer distances (SI Appendix, Fig. S3). Together, it appears that the bundle domain is rigid, while the hash motif exhibits limited ligand-induced conformational changes, which is a deviation from strict rigid-body movements.
Fig. 4.
Ligand-dependent conformational stabilization within the hash motif and the bundle domain on the intracellular face of vSGLT. TMs 2/7 of the bundle domain display rigid-body movements (A), while ligand-dependent equilibrium shifts that are not predicted to occur in the framework of the rocking-bundle mechanism are evident for helices of the hash motif (B and C).
Conformational Changes of the Extracellular Side.
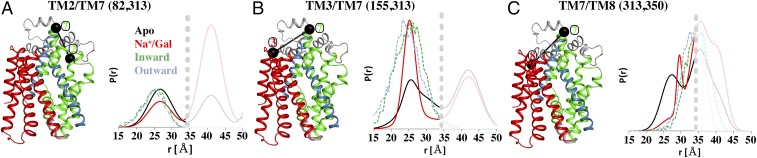
In a similar manner, ligand-induced alterations at the extracellular face of vSGLT were monitored using three pairs of engineered mutants (Fig. 5). TM2/TM7 (residues 82, 313) reports on the spatial relationship between two helices in the bundle domain. Under all conditions, a peak at ∼26 Å is observed in the DEER traces (solid traces), which corresponds to both the inward-facing structure (green dash) and outward-facing model (cyan dash) (Fig. 5A). The lack of discernable changes in distance between these sites supports concerted bundle domain movement. A long-distance peak centered at ∼41 Å and enhanced by Na+/Gal likely emanates from the TM7 (residue 313) intermolecular dimer distance (Fig. 2D). TM3/TM7 (residues 155, 313) that probes distance distributions between the bundle domain and hash motif, displays a more complex pattern, which is sensitive to Na+/Gal. The Apo and Na+ states have broad distance distributions in the 20–35 Å range, indicative of conformational heterogeneity. The binding of Na+/Gal shifts the equilibrium toward the shorter distance component while reducing the conformational flexibility (Fig. 5B). Broad multicomponent distance distributions were also characteristic of TM7/TM8 (313 and 350), including a prominent short distance component not observed in the MDDS analysis. Upon Na+/Gal binding, the average intramolecular distance increases as a consequence, in part, of suppression of this short distance component (Fig. 5C). As observed for TM2/TM7, the interdimer contribution from TM7 (Fig. 2D) dominates the range >35 Å. Taken together, the DEER analysis indicates that TM7 and TM2 move in a coordinated manner toward the dimer interface in the presence of galactose. This ligand induced change further suggests coupled movements between the hash motif and bundle domain helices on the extracellular side during the transport process.
Fig. 5.
Ligand-dependent conformational stabilization of the extracellular face of vSGLT. TM2/TM7 of the bundle domain display rigid-body movements (A), whereas TM3/TM7 (B) and TM7/TM8 (C) that probe the spatial relationship between the hash motif and bundle domain display ligand-induced changes. For TM3/TM7, sugar binding induces a shortening of the distances between the pair, but for TM7/TM8 ligand binding induces longer distances. These movements were not predicted by our models as evident by the MDDS calculations for inward-facing conformation (green dash) and outward-facing conformation (cyan dash).
Conformational Changes of Gating Helices.
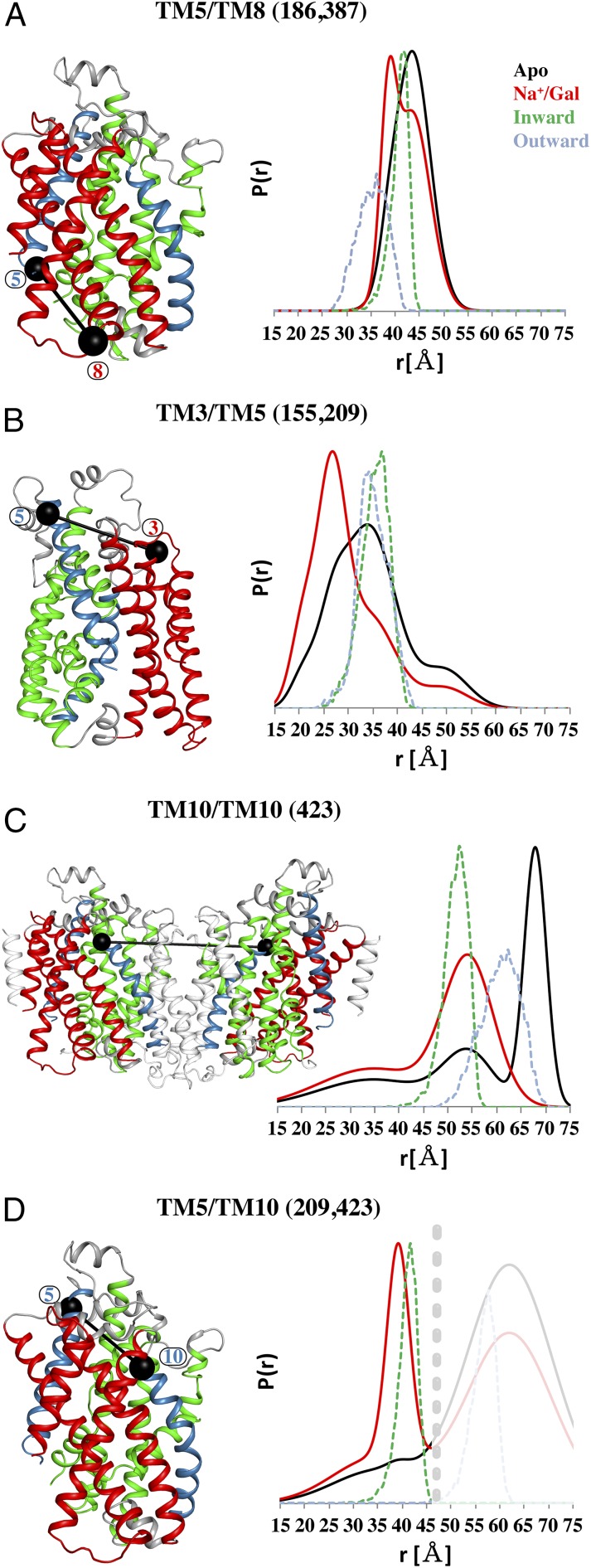
The gating helices (TM5 and TM10), in conjunction with the hash motif and bundle domain, are thought to work in concert to govern the opening and closing of the extracellular and intracellular vestibules (12). Specifically, ligand binding induces a dramatic bending of the extracellular portion of TM10, which prevents the ligands from escaping to the extracellular milieu. Conversely, the intracellular portion of TM5 bends to open the intracellular vestibule facilitating substrate release (25). The DEER-derived distance distributions at the intracellular face between TM5 and TM8 (residues 186 and 387) in the hash motif suggest a shift toward shorter distances in the Na+/Gal condition while retaining a component corresponding to the Apo and Na+ conditions (Fig. 6A). This movement is similar to MDDS calculations for this pair, once again suggesting that an inward-facing state predominates in the Apo and Na+-only conditions and binding of galactose leads to closure of the intracellular face. We also monitored the extracellular motions of gating helix TM5 with respect to TM3 in the hash motif, even though TM5 is thought to primarily play a role in intracellular gating (residues 155, 209). Interestingly, we observed a striking Na+/Gal-induced decrease in distance between TM3/TM5, which was not predicted by the inward- or outward-facing models (dashed traces) (Fig. 6B). Collectively, the data indicates that TM5 is involved in global movements associated with modulating access to the sugar binding site on both sides of the transporter.
Fig. 6.
Conformational changes of gating helices. On the intracellular side, the shorter distance induced for the Na+/Gal condition of TM5/TM8 indicates that sugar binding induces a gate closure that seals the ligand from the intracellular cavity. This shorter distance agrees well with the MDDS calculation based on the outward-open conformation (cyan dash) (A). TM3/5 on the extracellular side display a very broad distance distribution for the Apo and Na+ states. Ligand binding induces a narrowing of the distribution toward shorter distances that was not predicted by our models (B). The intermolecular distance distributions for TM10 on the extracellular side are very wide for the Apo and Na+ states, and a clear shift toward shorter distances is observed for the Na+/Gal condition (C). The same trend of Na+/Gal-induced occlusion is seen for TM5/TM10 (D).
Monitoring the conformational changes on the extracellular portion of gating helix TM10 (residue 423) by DEER spectroscopy proved to be difficult. The phase memory time was too short in our experimental conditions to completely parameterize the long spin label distances (>50 Å) between dimer-related residue 423 and the nonspecific intermolecular background, leading to approximations of the shape and width of individual distance components. Nevertheless, a clear change in the spin echo decay of the Na+/Gal sample indicated a shift toward shorter distances as seen in the resulting distance distributions (Fig. 6C and SI Appendix, Fig. S3). Importantly, continuous wave (CW)-EPR spectra revealed that the local environment of the spin label at residue 423 is dramatically different between the Apo and Na+ states compared with the Na+/Gal state. Indeed, Na+-dependent galactose binding substantially reduces spin label mobility, which is consistent with burial of the label into the protein core (SI Appendix, Fig. S3). Furthermore, we observed a Na+/Gal-dependent decrease in distance between TM5 and TM10 (209 and 423) (Fig. 6D), suggesting coordinated movement of both gating helices on the extracellular side. The short-distance component that arises from Na+/Gal binding aligns well with predicted distances in the inward-facing model, suggesting that the gating helices predominately adopt a conformation consistent with a fully occluded state upon substrate binding.
Nonfunctional Mutants Alter Conformational Equilibria.
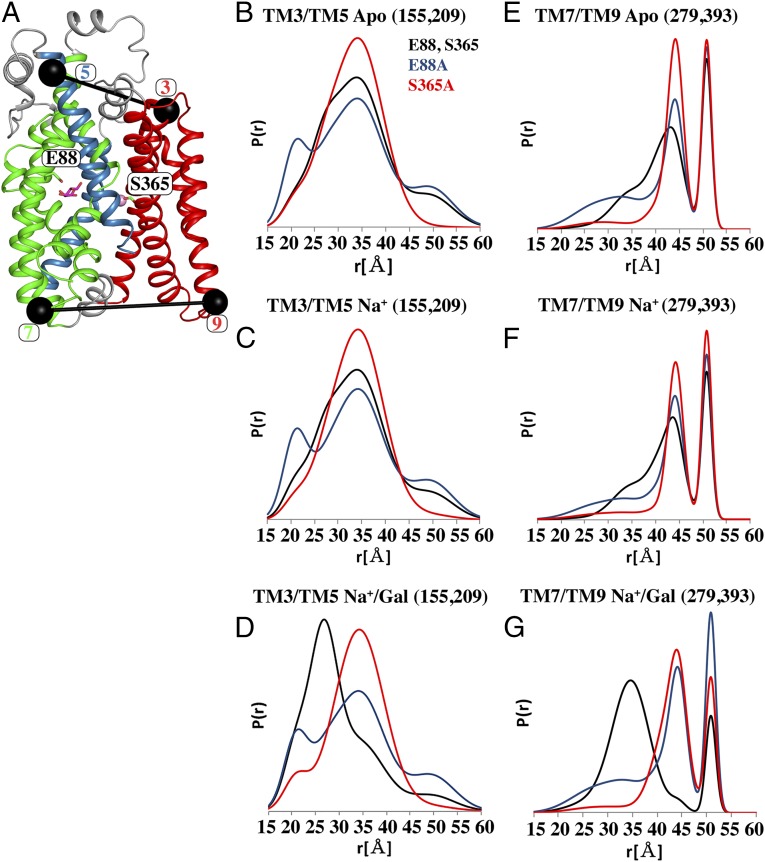
Earlier structural and functional studies revealed the precise coordination of galactose and Na+ by vSGLT. Mutations of a galactose-coordinating residue (E88A) and a Na+-coordinating residue (S365A) abolished Na+-dependent galactose uptake (7). To further investigate how these nonfunctional mutants influence conformational changes in the transporter, we performed DEER measurements on two spin-labeled mutants, 155/209 and 279/393, in the background of E88A and S365A (Fig. 7A). CW-EPR spectra of the spin-labeled background mutants and the corresponding functional constructs show subtle changes in the line shapes, suggesting only minor changes to local packing due to the inactivating mutations (SI Appendix, Fig. S3).
Fig. 7.
Nonfunctional mutants display impaired formation of the Na+/Gal state. DEER analysis for TM3/TM5 and TM7/TM9 was performed on the background of the active constructs (black), a Na+ site inactivating mutation, S365A (red), and a sugar site inactivation mutant, E88A (blue). A cartoon representation of the labeling sites (black spheres connected by black lines), galactose (in sticks), Na+ (pink sphere), and the two side chains involved in ligand and sodium binding (sticks) are shown in A. Ligand-dependent distance distributions under the Apo, Na+, and Na+/galactose conditions are shown for TM3/TM5 (B–D), respectively, and for TM7/TM9 (E–G). Disruption of the Na+ or sugar binding sites alters the conformational distributions sampled by vSGLT in all solution states and prevents the formation of the Na+/galactose-bound conformation sampled by the active protein.
As previously described, TM3/TM5 (residues 155 and 209) and TM7/TM9 (residues 279 and 393) capture features of the Na+/galactose-induced conformational transition on the extracellular (Fig. 6B) and intracellular sides (Fig. 3A), respectively. Notably, disruption of the Na+ or sugar binding sites alters the conformational distribution sampled by vSGLT in all states for both pairs. In the Apo and Na+ conditions, disrupting sugar binding with the E88A mutation increases sampling of short-distance components that are populated in the wild-type background upon galactose binding (Fig. 7 B, C, E, and F). Thus, the E88A mutation appears to shift the equilibrium toward the outward-facing or occluded intermediate. In contrast, mutation of the Na+ site (S365A) reduces sampling of specific distance components, even those associated with the putative outward-facing conformation (Fig. 7 B–G). Despite the disparate structural effects of these two distinct mutations, both E88A and S365A largely suppress the formation of the Na+/galactose-bound conformation observed in the wild type (Fig. 7 D and G). Not only do these results emphasize that occupation of an intact Na+ site is required to facilitate the Na+/galactose-driven conformational change, but they also uncover the relationship between the ensemble of conformations and the integrity of the Na+ and sugar binding sites.
MD Simulations.
MD simulations of the inward-facing structure (PDB ID code 3DH4) and the outward-facing model of vSGLT were performed to probe the dynamics of the transporter in these dissimilar conformations. Specifically, we simulated three dimer systems embedded in detergent micelles to closely match the experimental conditions. The systems were composed of two inward-facing transporters, two outward-facing transporters, and a mixed dimer with one protomer in each state. Fully atomistic simulations were carried out for 50 ns on each system, and the combined results from inward-facing protomers and outward-facing protomers resulted in 150 ns of aggregate of analysis for each conformation (Materials and Methods).
Root-mean-square fluctuation (RMSF) calculations performed on these simulations corroborate many of the DEER findings. Namely, in both conformations the bundle domain maintains a higher degree of rigidity compared with the hash motif and gating helices (SI Appendix, Fig. S4). Globally, the outward-facing protomers show a higher degree of flexibility than the inward-facing protomers, and highlight the conformational freedom of the extracellular gating helix TM10. One intriguing exception to the rocking-bundle mechanism is seen for the intracellular portion of TM6. This part of the protein was not sampled by DEER, and the MD simulations show that it has a high degree of flexibility that decouples it from the rest of the bundle domain. The involvement of the intracellular part of TM6 in opening of the intracellular vestibule and its decoupling from the bundle domain were also observed for LeuT (16, 26).
Discussion
Crystal structures of occluded, outward- and inward-facing states from a number of 5TMIR fold proteins display numerous common structural characteristics (4, 24), yet there are clear differences between individual transporters that are most likely due to variance in the type of transported substrate, driving ion, and stoichiometry of driving ion(s). To construct a mechanistic view of the transport process, we complemented static snapshots of vSGLT—generated by crystallography—with biophysical and simulation data. The current structural data for vSGLT consist of a galactose bound inward-occluded conformation, a substrate-free inward-open conformation, and an outward-facing model from a closely related structure (SiaT). The switch between the inward-facing conformations is associated with release of galactose to the intracellular milieu, and it involves a 13° bending of the intracellular half of TM1 and a 6° relative rigid-body shift of the hash motif and bundle domain from each other (8). Our ligand-induced DEER measurements sampled broader structural changes that are attributed to isomerization events between the inward- and outward-facing conformations.
The majority of the DEER data for vSGLT, under the three conditions employed, underscores a dynamic equilibrium between distinct conformations. As observed for Mhp1 (14), there were no significant distance changes between the Apo and Na+ conditions, and the protein adopts a predominantly inward-facing state. Meanwhile, concurrent binding of Na+ and ligand dramatically shifted the equilibrium to an outward-facing or occluded state. In contrast, Na+ binding alone to LeuT drives the protein toward an outward-facing state (27, 28). To date, all Na+-driven transporters using 5TMIR fold have a conserved Na2 site, which is formed by side- and main-chain interactions from TM1 and TM8 and is in close proximity to the ligand binding site. It is notable that both Mhp1 and vSGLT have a 1:1 ligand:ion stoichiometry, while LeuT has a nonconserved Na1 site that is coordinated directly to the bound substrate, giving it a 1:2 stoichiometry. Unlike LeuT with its added Na1 site, addition of sodium does not change the equilibrium between inward-facing and outward-facing states of both vSGLT and Mhp1. Thus, it is possible that Na+—occupying the Na2 site—does not stabilize an outward-facing state, and that the role of an additional Na+ at the Na1 site drives the outward-facing state. Nonetheless, Tavoulari et al. (29) recently demonstrated that both Na+ sites in LeuT are required to form the outward-facing state, emphasizing the functional variation and complexity in the 5TMIR family.
The data presented here are consistent with both the Apo and Na+-only conditions biasing the equilibrium toward the inward-open conformation, whereas the presence of sodium and galactose shifts the equilibrium toward an outward-open or occluded conformation. Specifically, on the intracellular side, there is good agreement between the experimental DEER data of the Apo and sodium conditions with the MDDS calculations derived from the inward-open crystal structure, while the Na+/Gal condition correlates well with the outward-facing model on the intracellular side, indicative of a closed inner gate (Figs. 3 A–C, 4C, and 6A). On the extracellular side, our data are less conclusive due to the limited number of stable constructs that we were able to produce, which is potentially related to the high degree of conformational flexibility on the extracellular face revealed by the MD simulations. However, a number of constructs (Figs. 5B and 6 B–D) showed distinct Na+/galactose-induced reduction in distances that hint at occlusion of the vSGLT binding pocket from both sides. The most substantial difference between the outward-facing and occluded states of Mhp1 and vSGLT is the lack of movement of TM10 in Mhp1 that obstructs the extracellular substrate permeation pathway in vSGLT.
Modeling of vSGLT’s gate helices predicts a high degree of flexibility on the intracellular side of TM5 (TM5i) and the extracellular side of TM10 (TM10e). For Mhp1, both Apo and Na+ conditions displayed a very broad distance distribution, indicative of conformational disorder. However, upon ligand binding, a narrower distribution consistent with the outward-facing or occluded conformations was observed (14). For vSGLT, TM5i shows nearly identical distributions for the Apo and Na+ conditions that are consistent with simulation data obtained from the inward-facing structure of vSGLT (8). For intracellular pairs involving TM5, Na+/Gal binding increases the population of shorter distances that are in equilibrium with the longer-distance components, indicating closure of the intracellular gate. However, this inner closure is not as pronounced as it is for Mhp1 (14). A possible explanation for more limited conformational changes in vSGLT may be due to an additional helix [TM(−1)], which is absent in Mhp1 and may restrict TM5i movements (SI Appendix, Fig. S5). Instead, vSGLT shows more structural freedom for the intracellular portion of TM1 and TM6, as also observed for LeuT (26), and reflected in our simulation data (SI Appendix, Fig. S4). On the extracellular side, TM10e is predicted to have a large 16-Å gating motion that is supported by previous fluorescence data (30). The EPR measurements confirm that TM10e is highly dynamic, sampling different environments in a ligand-dependent manner (SI Appendix, Fig. S3). It is clear that, although there are many conserved features between proteins with the 5TMIR fold, there is significant variation in the conformational freedom of the 10-TM core segments.
Previous studies have shown the importance of the integrity of both Na+- and sugar-binding sites for transport (7, 8). The conformational states of our sugar and Na+ site mutants, probed at TM7/TM9 and TM3/TM5 (Fig. 7), further demonstrate that binding of both ion and sugar are necessary for inducing the isomerization between states. Importantly, these results also establish a striking correlation between the sampled conformational landscape and the biochemical architecture of ion and substrate binding sites. Indeed, variations in conformational sampling imposed by nonfunctional background mutations corresponded to impaired formation of the Na+/Gal state.
It is somewhat surprising that vSGLT is biased toward the inward-facing conformation in both the Apo and Na+ conditions, since under physiological conditions external Na+ binding presumably stabilizes the outward conformation, preparing the transporter for acquiring sugar from the external environment. However, there are two crucial factors that are not present in detergent-solubilized samples, which may bias the energetics to favor the outward-facing state—membrane potential and endogenous lipids. Nearly all cells maintain an intrinsic transmembrane potential in the −40- to −80-mV range, which acts as a power source for a variety of cellular processes. Indeed, hyperpolarizing potentials have been shown to favor the outward-facing conformation of hSGLT1 expressed in oocytes (31, 32).
We used electrostatic calculations (33) to determine vSGLT’s gating charge between the inward- and outward-facing states and arrived at a value of 0.7 |e|, and we also predict the voltage dependence of ion binding to the Na2 site from both sides of the membrane. A kinetic scheme for vSGLT derived from the hSGLT1 model with these calculated electrostatic parameters (Materials and Methods) shows that a negative membrane potential increases the probability of the outward-facing state from 10 to 70% (SI Appendix, Table S3). Thus, while such small gating charges may not imbue transporters with the switch-like behavior observed in voltage-gated channels (34), they can play a role in stabilizing key states along the transport pathway, which may enhance substrate uptake.
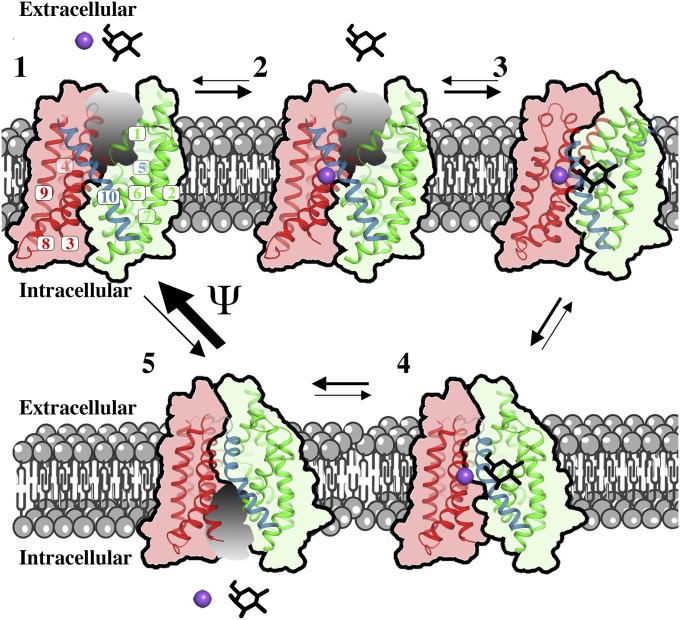
It is thus intriguing to incorporate this energetic potential with our vSGLT structures, the outward-facing model, DEER data, and computational and functional studies into a unifying model that provides a feasible depiction of the transport process in vSGLT (Fig. 8). In this scheme, the transporter inherently favors the outward-facing state in the presence of a negative membrane potential (state 1) and infrequently isomerizes to the inward-facing conformation (state 5). This isomerization principally follows the rocking-bundle mechanism with a few deviations such as uncoupled movements of helices belonging to the hash motif (TM3/TM9, TM4/TM8). Sodium binding to the outward-facing conformation (state 2) precedes sugar binding (35, 36), but together they would overcome the membrane potential and promote stochastic transitions of the transporter to the occluded state (state 3), primarily by a rigid-body motion between the hash motif and bundle domain with major involvement of TM10 in substrate occlusion. The isomerization to the inward-open state (state 4) is governed by rocking-bundle movements with TM5 opening. Sodium and substrate are released to the intracellular space in a stochastic manner (13), and this results in an empty inward-facing conformation (state 5) that is driven to the outward-facing state by the membrane potential, completing the transport cycle. Additional work will be required to determine which specific residues are responsible for the voltage sensing.
Fig. 8.
Sugar transport mechanism. Under Apo conditions, vSGLT mainly resides in the inward-facing conformation (step 5) and infrequently isomerizes to the outward-facing conformation (step 1). We hypothesize that, under physiological conditions, the membrane potential drives the conformation of vSGLT toward the outward-facing conformation. This isomerization principally follows the rocking-bundle mechanism with a few deviations such as uncoupled movements of helices belonging to the hash motif (TM3/TM9 and TM4/TM8). On the extracellular side, sodium binding to the outward-facing conformation (step 2) precedes sugar binding, after which the transporter assumes an occluded conformation (step 3) through rigid-body motions of the hash and bundle domains as well as major structural changes in TM10 to occlude the substrate. Before sodium and substrate release, SGLT switches to the inward-open conformation (step 4) and sugar and sodium are released to the intracellular space in a stochastic manner. Inward release results in the Apo inward-facing conformation (step 5) that enables the transport process to reinitialize.
This work utilizes DEER spectroscopy to describe the influence of ligand and sodium on the conformational equilibrium of vSGLT. vSGLT has a number of commonalities with Mhp1, likely related to their similar 1:1 Na+/ligand stoichiometry. Sodium alone induces minimal changes in stability of the inward- and outward-facing conformation of the transporters. Concurrent binding of sodium and substrate stabilizes an outward-open or occluded conformation. This transition largely follows the rocking-bundle mechanism. This is in stark contrast to LeuT, which has a 2:1 Na+/ligand stoichiometry, where sodium binding alone stabilizes the outward-facing conformation by a mechanism that deviates significantly from the rocking-bundle mechanism (16). The 5TMIR family is diverse in terms of substrate, type of driving ion, and ion stoichiometry, and our work further highlights that different family members utilize unique mechanisms to carry out transport in this central class of transporters.
Materials and Methods
DNA Construct and Mutagenesis.
The coding region of vSGLT followed by a glycophorin helix and a 6×histidine tag at the C terminus was cloned into the IPTG-inducible T5 controlled pJExpress401 plasmid (DNA2.0). All mutagenesis was performed utilizing the QuikChange site-directed methodology (Agilent Technologies) with primers obtained through Eurofins (MWG Operon). Each mutation reaction was verified by oligonucleotide sequencing (Laragen).
Protein Expression, Purification, and Labeling.
Overnight TB cultures supplemented with 50 μg/mL kanamycin were inoculated with XL1 Blue Escherichia coli cells (Agilent Technologies) harboring the vSGLT pJExpress401 plasmid and incubated at 37 °C, 220 rpm. The following day, fresh TB kanamycin cultures were inoculated to a density of A600 = 0.05 and further propagated to a density of 1.6 at 37 °C, 220 rpm. IPTG (Gold Biotechnology) at 0.75 mM was used for protein induction, and the temperature was reduced to 33 °C. After an incubation of 3 h, cultures were pelleted by centrifugation and either frozen or resuspended and passed through an EmulsiFlex-C3 cell extruder (Avestin). Broken cells were subjected to a 35-min centrifugation at 15,000 × g and the clear supernatant was further spun down for 60 min at 302,000 × g to isolate membranes.
Protein purification commenced by a homogenization of the membrane pellet and samples were adjusted to 2% (wt/vol) n-dodecyl-β-d-maltoside (DDM) (Inalco) and gently stirred for 1–3 h. Soluble material was isolated by a 1-h 53,300 × g centrifugation and the protein was purified using Ni2+ affinity on a HisTrap HP column and size-exclusion chromatography using a Superdex 200 column (GE Healthcare).
For spin labeling, fractions containing purified vSGLT were pooled and incubated with 2 mM DTT for 30 min on ice to ensure thiol reduction. Before labeling, DTT was removed by injecting the sample over a HiTrap 5-mL desalting column (GE Healthcare) into 50 mM Hepes, pH 7.4 (KOH), 6% glycerol (vol/vol), and 1 mM DDM. The sample was labeled with two rounds of 20-fold molar excess 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate (MTSSL) (Enzo Life Sciences) per engineered cysteine at 4 °C in the dark over a 4-h period. After a third addition of spin label, the sample was incubated on ice for 12 h (37). Unreacted spin label was removed by desalting. Samples were then concentrated using an Amicon Ultra (100,000 molecular-weight cutoff) for DEER and EPR analysis.
Transport Assays.
MTSSL-labeled vSGLT constructs were reconstituted in a buffer containing 150 mM KCl, in 0.5% DM, and mixed at a protein/lipid ratio of 1:200 with lipids. Reconstitution was initiated by adding 5 mg/mL SM-2 Bio-Beads and incubating with agitation at 4 °C overnight. The proteoliposomes were collected and washed twice by ultracentrifugation with 150 mM KCl, 10 Hepes/Tris, pH 7.5, 0.1 mM CaCl2, and 0.1 mM MgCl2. After the final resuspension, the proteoliposomes underwent three cycles of freeze–thaw in liquid nitrogen and either stored at −80 °C or were directly used for sugar uptake. Transport activity of the mutants was measured by uptake of 50 μM 14C d-galactose, for 18 min at 22 °C in the presence or absence of a 150 mM Na+ gradient or 150 mM K+ replacing Na+ in triplicates for each condition, and collected through 0.3-μm filters (Millipore). Uptake levels were measured by scintillation counting and corrected for protein content in the proteoliposomes. All mutants were assayed for galactose uptake into proteoliposomes (SI Appendix, Fig. S2) and were found to be of comparable activity to wild-type vSGLT (A423C).
Homology Modeling of the Outward-Facing Conformation.
The outward-facing conformation of vSGLT was modeled primarily on the SiaT structure from P. mirabilis (PDB ID code 5NV9). A consensus sequence alignment was determined by comparing a sequence-based approach using EMBOSS stretcher (38) and a structure-based alignment between vSGLT (chain A of PDB ID code 3DH4) and SiaT using MatchMaker (39). The two alignments agree throughout much of the sequence, but a final hand alignment was determined by accounting for large structural differences where the structure-based procedure fails while also closing gaps in the transmembrane domains. With this alignment, 100 outward-facing models were constructed with MODELLER, version 9.15 (40), using SiaT as a template structure for TM segments −1–12 and the inward-facing apo structure of vSGLT (PDB ID code 2XQ2) as a template for TM13 and the N-terminal glycophorin A TM segment used in the DEER experiments. The model with the best DOPE score (18) (no. 1 of 100 models) was selected for MDDS analysis and MD simulations.
CW-EPR, DEER Spectroscopy, and Analysis.
EPR spectra were collected at 23 °C on a Bruker EMX spectrometer (X-band, 9.5 GHz) at an incident power of 10 mW and 1.6 Gauss modulation amplitude. Distance measurements were carried out on a Bruker E580 pulsed EPR spectrometer at Q-band frequency (34 GHz) employing a standard four-pulse protocol at 83 K (41). Pulse lengths were 10–12 ns (π/2) for the probe pulse and 40 ns for the pump pulse. The frequency separation was 63 MHz. To ascertain the role of Na+, samples were subjected to 150 mM NaCl, while for the Na+/Gal condition samples were subjected to 150 mM NaCl and 20 mM galactose. Samples for DEER analysis were cryoprotected with 25% (wt/vol) glycerol and flash frozen in liquid nitrogen. DEER signals obtained under different conditions for the same spin-labeled pair were analyzed globally with home-written software operating in the Matlab (MathWorks) environment (42). The fitting routine assumes that the distance distribution P(r) is a sum of Gaussians. The number of Gaussians to sufficiently describe P(r) was statistically determined by F test between fits of increasing Gaussian components. Comparison of the experimental distance distributions with the inward-facing crystal structure and outward-facing model using MDDS was facilitated by the DEER Spin-Pair Distributor at the CHARMM-GUI (21, 43, 44) website.
MD Simulations.
Three systems were constructed consisting of dimers with both transporters inward facing (taken directly from the 3DH4 dimer structure), both outward facing (using the SiaT-based homology model), and a mixed dimer. The dimer orientation in the outward–outward and mixed simulations were based on the 3DH4 dimer orientation. Each protomer had Na+ bound in the Na2 site, but only the inward-facing protomers were galactose bound. Next, the inward–inward dimer was embedded in a DDM detergent belt previously equilibrated around the LeuT monomer. The LeuT-DDM system was superposed onto each vSGLT protomer in the dimer and all clashing DDM molecules were removed. This system was then solvated and neutralized with 150 mM NaCl in a rectangular box of dimension 178 × 145 × 154 Å3, resulting in a final system size of ∼375,000 atoms. Simulations were carried out using the CHARMM36 parameter set (CHARMM22 with CMAP corrections for the protein) (45, 46), CHARMM force field for pyranose monosaccharides for galactose (47), and CHARMM36 for lipids (48) using the TIP3P water model (49) and NAMD 2.10 (50) as the MD engine. The system was first minimized using conjugate gradient for 10,000 steps followed by 25 ps of dynamics in a constant volume and temperature (NVT) ensemble at 310 K with the Na2 site Na+, ligand core, and protein backbone heavy atoms restrained with a 10 kcal·mol−1·Å−2 harmonic force constant. The temperature was maintained at 310 K using Langevin dynamics with a 1 ps−1 damping coefficient. Restraints decreased to 7.5 kcal·mol−1·Å−2 and simulated for an additional 25 ps. Next, we switched to a constant pressure and temperature ensemble using the Langevin piston barostat with a 200-fs piston period and 100-fs piston decay constant to maintain the pressure at 1 bar. Constraints were gently reduced over the next 1.2 ns, and the system was then allowed to equilibrate for 5 ns without restraints before production runs, which lasted 50 ns. Hydrogen bond lengths were constrained with SHAKE, and a 1-fs time step was employed in the initial equilibration stages, followed by a 2-fs time step throughout. Particle mesh Ewald was employed for long-range electrostatics using an interpolation order of 6, and van der Waals interactions were smoothly switched to zero between 11 and 12 Å. The other two dimer simulations were constructed by taking the final DDM configuration from the end of the production run and transferring them to the other dimer structures and removing clashing lipids.
RMSF values were computed by combining trajectories of the inward- and outward-facing protomers from each system. Bundle domain RMSF values were calculated by first superposing TM1, 2, 6, and 7, the hash motif RMSF values were calculated by first superposing TM3, 4, 8, and 9, and the RMSF values of the gating helices were computed by superposing TM1–10.
Kinetic Model.
Kinetic equations were derived from the five-state stochastic release model recently developed for hSGLT1 (13) (see SI Appendix, Table S1, for all base rate constants). We adapted the hSGLT1 model to vSGLT, which only binds 1 Na+, based on the cartoon model in SI Appendix, Fig. S6. While the vSGLT model is not calibrated from experimental current recordings, as the hSGLT1 model is, we based the model on three ideas. First, only a single Na+ binds, so ion binding rates depend on the external/internal [Na+] raised to the first power, rather than the second power. Second, we borrowed all rate constants directly from the hSGLT1 model, except for those that involved Na+ binding. For the vSGLT model, we reduced the rate constants involving sodium binding (k12 and k54) by a factor of 10, which keeps the overall rate similar in both systems since internal/external Na+ is on the order of 0.1 M. Third, the voltage dependence of all of the kinetic transitions (εij) was determined directly from electrostatic calculations carried out on the inward-facing and outward-facing models, as described in Gating Charge Calculations. The final set of parameters used in the vSGLT kinetic modeling are presented in SI Appendix, Table S2. The differential equations for hSGLT1 can be found in ref. 13, and the vSGLT equations are as follows:
where i indicated intracellular values and o represents extracellular values, and [Na] and [G] are the sodium and galactose molar concentrations, respectively. The rate constants depend on membrane voltage according to the following:
where the zero superscript indicates the base rate constants shown in SI Appendix, Tables S1 and S2, and (εij − εji) is the equivalent charge movement from state i to j determined from the electrostatic calculations. We assume that the voltage dependence of the charge transfer influences the forward and reverse rates equally.
The analysis presented here only relies upon the steady-state probability of the transporter in each state C1–C5. We determined these values by numerically solving the ordinary differential equations until steady state was reached. The equations were solved using the Rosenbrock stiff solver implemented in Berkeley Madonna. The steady-state probabilities for both models under different conditions are shown in SI Appendix, Table S3.
Gating Charge Calculations.
We computed the voltage dependence of each step of the cycle in SI Appendix, Fig. S6 using the electrostatics solver APBSmem (33). Briefly, for two different conformations (inward-facing versus outward-facing, outward-facing Na+ versus outward-facing empty, etc.), we computed the difference in the total interaction energy of all charges in the system with the membrane voltage between both states over a series of membrane potentials (51), and then we extracted the equivalent “gating charge” movement from the slope of the line fitted to the resulting energy curve. The fraction of the electric field through which a single equivalent charge moves from one state i to another j is Qij = (εij − εji) and we set εij = Qij/2 and εji = −εij.
In the vSGLT model above, the net charge movement from stages 2–4 are evenly split among all four rates (k23, k32, k34, k43). The influence of charge in each step is evenly split between the forward and backward rates. Additionally, k12 and k54 were reduced by 0.1 since the hSGLT1 model has an extra factor of 0.01 to 0.1 M in these steps for the second sodium binding event.
Supplementary Material
Acknowledgments
We thank Drs. Althoff, Bergdoll, and Ramswamy for fruitful discussions, Dr. Khelashvili (Cornell University) for providing snapshots of LeuT in a detergent micelle, and Mr. Grandfield for his help with performing transport assays. This work was supported by NIH Grants R01GM089740 (to M.G.), DK19567 (to E.M.W.), U54 GM087519 (to H.S.M.), and R01GM078844; and University of California, Los Angeles, Cardiovascular Theme Discovery Award (to J.A.). Computations were performed at the San Diego Supercomputing Center through the support of National Science Foundation Division of Molecular and Cellular Biosciences Grant MCB-80011 and the Extreme Science and Engineering Discovery Environment.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718451115/-/DCSupplemental.
References
- 1.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 2.Drew D, Boudker O. Shared molecular mechanisms of membrane transporters. Annu Rev Biochem. 2016;85:543–572. doi: 10.1146/annurev-biochem-060815-014520. [DOI] [PubMed] [Google Scholar]
- 3.Kazmier K, Claxton DP, Mchaourab HS. Alternating access mechanisms of LeuT-fold transporters: Trailblazing towards the promised energy landscapes. Curr Opin Struct Biol. 2017;45:100–108. doi: 10.1016/j.sbi.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez C, Koshy C, Yildiz O, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012;490:126–130. doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 5.Wright EM. Glucose transport families SLC5 and SLC50. Mol Aspects Med. 2013;34:183–196. doi: 10.1016/j.mam.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Whalen K, Miller S, Onge ES. The role of sodium-glucose co-transporter 2 inhibitors in the treatment of type 2 diabetes. Clin Ther. 2015;37:1150–1166. doi: 10.1016/j.clinthera.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468:988–991. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abramson J, Wright EM. Structure and function of Na(+)-symporters with inverted repeats. Curr Opin Struct Biol. 2009;19:425–432. doi: 10.1016/j.sbi.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman JA, Green EM, Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532:334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest LR, Rudnick G. The rocking bundle: A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adelman JL, et al. Stochastic steps in secondary active sugar transport. Proc Natl Acad Sci USA. 2016;113:E3960–E3966. doi: 10.1073/pnas.1525378113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazmier K, Sharma S, Islam SM, Roux B, Mchaourab HS. Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1. Proc Natl Acad Sci USA. 2014;111:14752–14757. doi: 10.1073/pnas.1410431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazmier K, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21:472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Šali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 18.Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 20.Roux B, Islam SM. Restrained-ensemble molecular dynamics simulations based on distance histograms from double electron-electron resonance spectroscopy. J Phys Chem B. 2013;117:4733–4739. doi: 10.1021/jp3110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam SM, Stein RA, McHaourab HS, Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: Application to T4 lysozyme. J Phys Chem B. 2013;117:4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eskandari S, Wright EM, Kreman M, Starace DM, Zampighi GA. Structural analysis of cloned plasma membrane proteins by freeze-fracture electron microscopy. Proc Natl Acad Sci USA. 1998;95:11235–11240. doi: 10.1073/pnas.95.19.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turk E, et al. Molecular characterization of vibrio parahaemolyticus vSGLT: A model for sodium-coupled sugar cotransporters. J Biol Chem. 2000;275:25711–25716. doi: 10.1074/jbc.M003127200. [DOI] [PubMed] [Google Scholar]
- 24.Vergara-Jaque A, Fenollar-Ferrer C, Kaufmann D, Forrest LR. Repeat-swap homology modeling of secondary active transporters: Updated protocol and prediction of elevator-type mechanisms. Front Pharmacol. 2015;6:183. doi: 10.3389/fphar.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimamura T, et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328:470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C, et al. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophys J. 2012;103:878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claxton DP, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat Struct Mol Biol. 2010;17:822–829. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavoulari S, et al. Two Na+ sites control conformational change in a neurotransmitter transporter homolog. J Biol Chem. 2016;291:1456–1471. doi: 10.1074/jbc.M115.692012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veenstra M, Lanza S, Hirayama BA, Turk E, Wright EM. Local conformational changes in the vibrio Na+/galactose cotransporter. Biochemistry. 2004;43:3620–3627. doi: 10.1021/bi0357210. [DOI] [PubMed] [Google Scholar]
- 31.Loo DDF, Jiang X, Gorraitz E, Hirayama BA, Wright EM. Functional identification and characterization of sodium binding sites in Na symporters. Proc Natl Acad Sci USA. 2013;110:E4557–E4566. doi: 10.1073/pnas.1319218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright EM, Loo DDF, Hirayama BA. Biology of human sodium glucose transporters. Physiol Rev. 2011;91:733–794. doi: 10.1152/physrev.00055.2009. [DOI] [PubMed] [Google Scholar]
- 33.Marcoline FV, Bethel N, Guerriero CJ, Brodsky JL, Grabe M. Membrane protein properties revealed through data-rich electrostatics calculations. Structure. 2015;23:1526–1537. doi: 10.1016/j.str.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoppa N, McCormack K, Tanouye M, Sigworth F. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- 35.Parent L, Supplisson S, Loo DD, Wright EM. Electrogenic properties of the cloned Na+/glucose cotransporter: II. A transport model under nonrapid equilibrium conditions. J Membr Biol. 1992;125:63–79. doi: 10.1007/BF00235798. [DOI] [PubMed] [Google Scholar]
- 36.Turk E, Gasymov OK, Lanza S, Horwitz J, Wright EM. A reinvestigation of the secondary structure of functionally active vSGLT, the vibrio sodium/galactose cotransporter. Biochemistry. 2006;45:1470–1479. doi: 10.1021/bi052160z. [DOI] [PubMed] [Google Scholar]
- 37.Claxton DP, Kazmier K, Mishra S, Mchaourab HS. Navigating membrane protein structure, dynamics, and energy landscapes using spin labeling and EPR spectroscopy. Methods Enzymol. 2015;564:349–387. doi: 10.1016/bs.mie.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 39.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinforma. 2002;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 42.Stein RA, Beth AH, Hustedt EJ. A straightforward approach to the analysis of double electron-electron resonance data. Methods Enzymol. 2015;563:531–567. doi: 10.1016/bs.mie.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 44.Islam SM, Roux B. Simulating the distance distribution between spin-labels attached to proteins. J Phys Chem B. 2015;119:3901–3911. doi: 10.1021/jp510745d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 47.Guvench O, et al. Additive empirical force field for hexopyranose monosaccharides. J Comput Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 50.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callenberg KM, et al. APBSmem: A graphical interface for electrostatic calculations at the membrane. PLoS One. 2010;5:e12722. doi: 10.1371/journal.pone.0012722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.