Significance
Identifying proteins selectively associated with a genomic locus provides an important entry point toward understanding how a specific gene is regulated. Over the years, there have been several reports describing targeted chromatin-purification methods. However, none has been widely adopted due to the complexity and investment required for such protocols. Here, we present an adaptable chromatin purification system, CLASP, that capitalizes on the versatility of purified dCas9 RNA/protein complexes. We deployed CLASP to purify and identify proteins associated with telomere sequences in human cells as a proof of concept. Next, we targeted a different genomic locus, the Drosophila melanogaster histone cluster, and identified several regulators of the essential histone locus and validated their functional association with genes within the locus.
Keywords: CRISPR/Cas9, reverse ChIP, histone regulation, gene expression
Abstract
Eukaryotic gene regulation is a complex process, often coordinated by the action of tens to hundreds of proteins. Although previous biochemical studies have identified many components of the basal machinery and various ancillary factors involved in gene regulation, numerous gene-specific regulators remain undiscovered. To comprehensively survey the proteome directing gene expression at a specific genomic locus of interest, we developed an in vitro nuclease-deficient Cas9 (dCas9)-targeted chromatin-based purification strategy, called “CLASP” (Cas9 locus-associated proteome), to identify and functionally test associated gene-regulatory factors. Our CLASP method, coupled to mass spectrometry and functional screens, can be efficiently adapted for isolating associated regulatory factors in an unbiased manner targeting multiple genomic loci across different cell types. Here, we applied our method to isolate the Drosophila melanogaster histone cluster in S2 cells to identify several factors including Vig and Vig2, two proteins that bind and regulate core histone H2A and H3 mRNA via interaction with their 3′ UTRs.
To fully understand the molecular mechanisms governing multiple steps in gene expression, including transcription and posttranscriptional regulation for a given gene, one must first identify the various protein factors involved in the process. Over the past 30 y, great progress has been made by conventional biochemical fractionation in isolating and characterizing some of the major regulators of gene expression, such as components of the basal-transcription machinery, activators/coactivators, chromatin-remodeling complexes, and RNA-processing proteins, as well as factors influencing mRNA stability. Nevertheless, we still lack a detailed and comprehensive understanding of the coordinated molecular mechanisms controlling gene expression for the majority of genes (1).
Genome-wide survey techniques, such as ChIP coupled to high-throughput sequencing (ChIP-seq), have substantially increased the scope of discovery in molecular biology. ChIP-seq allows the precise mapping and identification of many potential DNA-binding sites for a given regulatory protein in a cell population of interest. This unbiased genome-wide identification of protein DNA-binding sites provides researchers the ability to test regulatory functions at enriched sequences and, in doing so, to begin to understand the function of select regulatory proteins within the cell (2). Although ChIP-seq is a powerful molecular tool in studying site-specific DNA-interacting regulators, it suffers from some significant shortcomings. Eukaryotic gene expression requires the coordinated activity of tens, if not hundreds, of proteins working in concert to ensure proper cell type-specific gene regulation (3). Finding available and highly specific antibodies for each individual putative regulatory protein necessary for ChIP-seq experiments is challenging and remains a significant roadblock to studying many as yet undiscovered genomic control factors. Furthermore, ChIP-seq requires prior knowledge that the protein of interest may have regulatory functions within the nucleus. These challenges have made the discovery of a more complete “regulome” responsible for various stages of gene-expression control at specific genomic loci a difficult and experimentally arduous process.
One of the many essential loci for which our understanding of gene regulation remains stubbornly incomplete is the canonical histone gene locus, which exists as highly repetitive clusters of unique sequence in eukaryotic genomes (4). As eukaryotic cells progress through the cell cycle, the doubling of the DNA content requires the rapid and coordinated synthesis of the linker histone H1 and the core canonical histone proteins H2A, H2B, H3, and H4, needed to efficiently package the newly synthesized DNA into histone-bound chromatin (5). Mirroring DNA replication, histone protein synthesis is a tightly regulated process wherein histone mRNA levels increase by 35-fold as the cell enters S phase but is quickly degraded once this cell-cycle phase has completed (6). The finely tuned maintenance of core histone levels throughout the cell cycle is crucial for proper gene regulation and cell health. For example, dysregulation of histone production leads to abnormal chromosomes and potential interference of histone methyltransferases and deacetylases (7–9). Some aspects of canonical histone gene expression are well described, such as the role of stem loop-binding protein (SLBP) in splicing and degradation, multi-sex combs (Mxc) in recruiting pre-mRNA–processing subunits, and the distinct roles of TATA box-binding protein-related factor 2 (TRF2) and TATA box-binding protein (TBP) in regulating H1 versus H2A histone transcription, respectively (10–12). However, the details of many other regulatory steps remain unknown. For example, what are the transcription factors (TFs) and chromatin regulators responsible for initiating transcription at the beginning of S phase? What is the mechanism of histone mRNA stability and degradation? Are there other proteins besides TRF2 that differentially regulate linker histone H1 from the core histone genes?
One approach to address these questions and to gain a more complete picture of the protein ensemble operating at the histone cluster (HisC) is to perform a reverse-ChIP analysis, where one isolates specific regions of the genome and characterizes the diverse repertoire of proteins associated with them (13–16). Over the last decade or so, multiple attempts at developing reverse-ChIP methodologies have been made by coupling some form of chromatin purification to mass spectrometry (17–22). However, these methods often require substantial effort in building complex transgenic cell lines de novo, and most lack robustness in identifying functionally relevant gene-specific regulators targeted to the gene locus of interest. Even proteomics of isolated chromatin segments (PICh), a more versatile reverse-ChIP method that uses a biotinylated oligo to hybridize with the region of interest, is relatively inefficient at isolating specific chromatin, suffers from a lack of adaptability and requires the user to substantially reoptimize the probe when targeting different sequences, thereby diminishing its usability to quickly target and troubleshoot multiple loci (23–25). Thus, a need still exists for a locus-specific proteome purification method that is robust, scalable for high-throughput target screening, and easily adaptable to different cell types.
Here, we present a reverse-ChIP method, CLASP (Cas9 locus-associated proteome), that takes advantage of the RNA-mediated DNA-targeting capability of Cas9 to efficiently and adaptably isolate specific genomic regions and their associated protein factors. By using purified recombinant catalytically inactive Cas9 (dCas9)–guide RNA ribonucleoprotein (RNP) complexes, CLASP does not require specialized cell lines and can be easily prepared with different guide RNAs to target multiple loci in any cell line or tissue. As a test of this reverse-ChIP platform, we have employed this CLASP to identify factors involved in Drosophila melanogaster HisC gene expression. Our newly established method to purify the chromatin of the H2A/H2B promoter generated a list of chromatin-associated proteins through mass spectrometry. Further characterization of a subset of these potential HisC-associated factors identified proteins that regulate H2A mRNA expression and revealed how a set of related RNPs can modulate H2A expression.
Results
Development and Validation of an in Vitro dCas9 Purification Method.
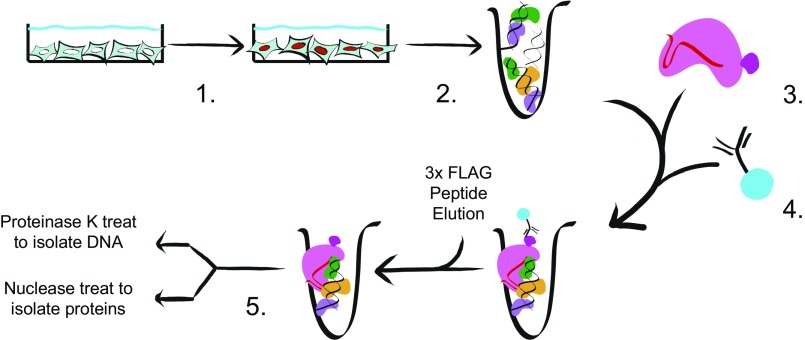
To identify potential regulators of histone transcription, we first developed a method of reverse ChIP that utilizes the D10A/H840A nuclease-deficient Cas9 (dCas9) RNP. dCas9 has proved a versatile tool in several contexts, including fluorescent imaging and epigenetic studies (26–29). Given that recombinant dCas9 binds in vitro targets with high stability and specificity, we employed it as a RNA-guided, DNA-targeting protein for an in vitro chromatin purification scheme that avoids the need for establishing customized transgenic cell lines (30). The workflow of CLASP is as follows: (i) chemically crosslink the cells of interest; (ii) isolate chromatin and shear it to desired size; (iii) add the RNP complex consisting of recombinant dCAS9-3×FLAG and single-guide RNAs (sgRNAs); (iv) enrich for RNP-bound chromatin using anti-FLAG immunoprecipitation (IP); and (v) isolate chromatin for protein identification via mass spectrometry (Fig. 1).
Fig. 1.
Layout of the CLASP. Graphic depiction of the CLASP method. (1) Cells of interest are crosslinked with a crosslinker of choice. (2) Small fragments of chromatin are generated by mechanical shearing of the fixed cells. (3) Recombinant dCas9-3×FLAG loaded with the chosen guide RNA is added to the chromatin mixture. (4) Anti-FLAG antibody conjugated to resin is added to RNP/chromatin and washed; enriched chromatin is eluted with 3×FLAG peptide. (5) Through the use of either Proteinase K or nuclease treatment, enriched DNA or protein samples can be isolated for downstream applications.
We first validated the method by targeting telomere sequences in HeLa cells. These sequences are relatively abundant, comprising 0.01–0.07% of the genome, and many proteins that bind them are well characterized (23, 31). Dot blot assays verified that telomeric DNA was enriched compared with nontargeting dCas9 IP, which targets a random sequence within the Escherichia coli genome and is not found in humans (Fig. S1A). Both Western blotting and multidimensional protein identification technology (MudPIT) mass spectrometry results showed that proteins associated with the telomeric sequence such as TPP1, TRF2, and RAP1 were enriched after telomere-targeted purification. These results demonstrate that CLASP is suitable for targeted chromatin isolation (Fig. S1 B and C).
Purification of the Drosophila HisC.
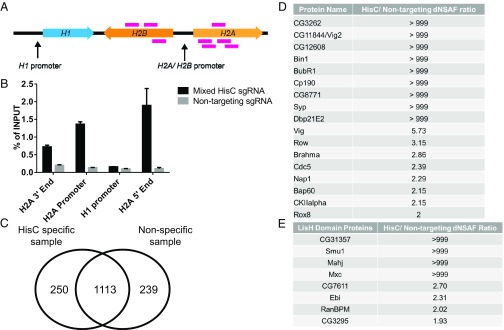
To identify potential regulators of the D. melanogaster HisC specifically associated with the H2A/H2B gene, we designed a series of guide RNAs that tiled the ∼5,000-bp unique sequences adjacent to the H2A/H2B promoter (Fig. 2A). The guide RNAs were designed specifically to bind outside the H2A/H2B promoter itself to avoid any potential dCas9 steric hindrance with promoter-associated factors (Fig. 2A). To rule out nonspecific dCas9 binding, the same nontargeting guide RNA used for the telomeric DNA pulldown was also used for the Drosophila nontargeting sample as a negative control (Table S3).
Fig. 2.
Purification and identification of proteins associated with the Drosophila HisC. (A) Graphic representation of the eight different genomic targets of the sgRNA used in the mixed HisC pool. (B) qPCR analysis of the HisC IP sample vs. the nontargeting IP sample. Each sample was prepared from two billion synchronized S2 cells fixed with 1% formaldehyde for 15 min. A dCas9-targeted ChIP sample loaded with a mix of HisC-targeting guide RNAs showed significant enrichment of the H2A/H2B promoter region and the 5′ end of the H2A gene promoter. (C) Venn diagram of the total proteins found only in HisC-specific IP samples, nontargeting IP samples, and within both samples. Results are from the first HisC pull-down sample. (D) Table of proteins that showed a dNSAF ratio ≥2 between the first HisC-specific sample and nontargeting sample and that have an association with nucleic acids within the DAVID database. (E) Table of proteins that were identified as having a LisH domain from the second HisC-specific pulldown and their relative abundance ratio in HisC-specific samples and nontargeting samples. A ratio of >999 indicates that the protein was not identified in the nontargeting sample in D and E.
Since HisC is transcriptionally active only during S phase, we synchronized Drosophila S2 cells with the two-block method of Ponasterone A and hydroxyurea, followed by a 2.5-h release (5, 11). This synchronization method was efficient in generating S-phase cells: Up to 80% of the total population of cells could be isolated in S phase while not significantly affecting cell viability (Fig. S2).
Using a combination of eight different sgRNAs (Fig. 2A and Table S3), we performed a HisC-targeted CLASP starting with two billion synchronized S2 cells. qPCR analysis indicated that we achieved a significant enrichment of the promoter and the 5′ end of the H2A histone gene while avoiding off-target enrichment of other regions of HisC, including the H1 promoter (Fig. 2B). After targeted chromatin purification and trichloroacetic acid (TCA) precipitation to isolate proteins, we used MudPIT mass spectrometry to analyze the specific and nonspecific proteins isolated by CLASP. With this initial sample, we identified 250 proteins unique to HisC-specific IP, 1,113 proteins common to both specific and nontargeting samples, and 239 proteins unique to the nontargeting IP sample (Fig. 2C).
Identification of Potential Histone Gene-Expression Regulators.
To narrow the list to a manageable number of potential candidates for functional validation, we utilized the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics database and identified proteins associated with nucleic acids that also had a relative abundance (distributed normalized spectral abundance factor; dNSAF) ratio of 2 or greater over the nontargeting IP sample (32, 33). Using these two criteria, we identified 17 potential regulators associated with the canonical histone genes with the first HisC purification (Fig. 2D).
A replicate CLASP purification of the HisC also identified proteins with the Lis1 homology (LisH) motif. This result stood out because the LisH domain is found in Mxc, a protein that has been previously described as a transcriptional regulator of the Drosophila HisC (12). In addition, the LisH domain has been shown to be important for Mxc function at the HisC (34). As expected, Mxc was one of the proteins enriched in the HisC-targeted sample with a LisH domain. In addition to Mxc, 7 of the 18 total LisH motif-containing proteins in the Drosophila proteome were also enriched, suggesting that additional proteins with LisH domains could play a role at the HisC (Fig. 2E).
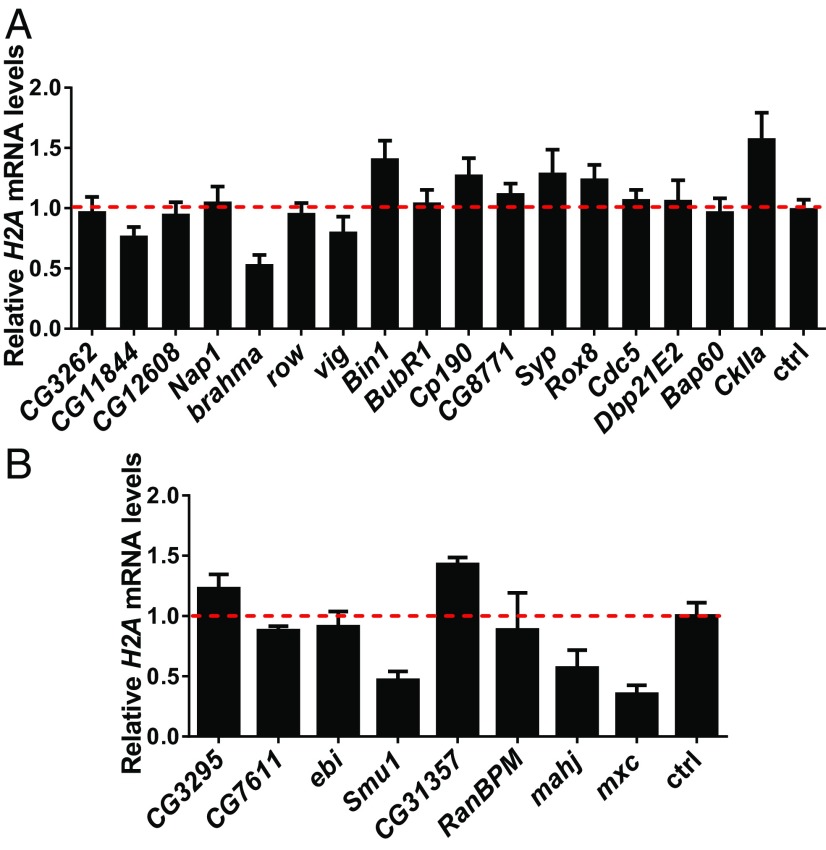
To validate whether some of the identified proteins have a functional effect on H2A/H2B gene expression, we performed dsRNA-mediated RNAi in S2 cells, isolated total RNA, and measured H2A mRNA levels relative to a reference gene via reverse-transcription qPCR. Of the 17 genes identified, multiple gene knockdowns altered the expression of H2A. However, we were interested in positive regulators of the histone locus, so we focused on the loss of CG11844, brahma, and vig, which all had a negative effect on histone expression, suggesting they function either to activate H2A transcription or to prevent its mRNA degradation (Fig. 3A). Interestingly, CG11844 is also known as “vig2,” a paralog of vig in D. melanogaster (35). The dsRNA-knockdown assay was also performed on the eight LisH domain-containing proteins. Not surprisingly, mxc knockdown showed a reduction in H2A mRNA levels. Interestingly, knockdown of Smu1 and mahj mRNA also showed significant reduction in H2A mRNA levels, suggesting a potential, hitherto undocumented, role for these two factors in regulating histone gene expression (Fig. 3B).
Fig. 3.
dsRNA knockdown assays reveal potential histone gene expression regulators. (A) Proteins enriched by a dNSAF ratio of 2 and associated with nucleic acids are knocked down by dsRNA over 72 h. cDNA is synthesized with iScript reverse transcriptase with a mixture of poly-A and random hexamer primers. H2A mRNA levels for each knockdown were calculated relative to the nonspecific dsRNA control using Tub84b as a reference gene. Compared with the nonspecific knockdown, CG11844, vig, and brahma show a negative effect on H2A mRNA expression. (B) LisH domain-containing proteins that are enriched in the HisC sample are knocked down by dsRNA as described in A. Compared with the nonspecific knockdown, Smu1, mahj, and mxc all showed a negative effect on the mRNA expression of H2A. Data plotted are averages and SDs from three separate knockdowns.
Vig and Vig2 Are Histone RNA-Binding Proteins.
vig and vig2 were previously identified as members of the RNAi complex in D. melanogaster (36, 37). With most of the literature focused on their roles as part of the RNAi machinery, and one report on vig and vig2 knockouts affecting heterochromatin formation at the organismal level, previous studies gave little, if any, indication of potential Vig function at the Drosophila HisC (38). Visualizing Vig and Vig2 localization in S2 cells by immunofluorescence also did not inform how these proteins might be affecting histone gene expression, as both are enriched and evenly distributed in the cytoplasm (Fig. S3). However, the mammalian homolog of vig and vig2, SERBP1, has been previously described as encoding an RNA-binding protein that plays a role in regulating the stability of the RNA with which it interacts (39, 40). Given SERBP1 function in mammalian cells, we hypothesized that Vig and Vig2 may specifically interact with H2A mRNA to control its stability and regulate histone gene expression posttranscriptionally.
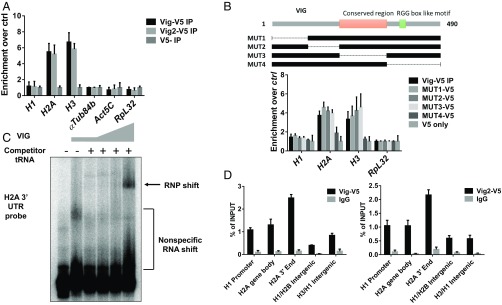
To test this hypothesis, we generated S2 cell lines stably overexpressing either V5-tagged Vig (Vig-V5) or Vig2 (Vig2-V5) and performed an RNP IP via the V5 tag. By isolating the RNAs enriched from each protein IP, we can test whether certain RNAs specifically interact with the proteins of interest. Indeed, when we perform RNP IPs with Vig and Vig2, they both bound and enriched for H2A and H3 mRNA but not H1 mRNA. These results are consistent with what was known regarding their mammalian homolog, SERBP1, and as anticipated by their association with the HisC locus revealed by our dCas9-targeted CLASP pulldown (Fig. 4A). Previous studies of SERBP1 identified an RGG box toward the C terminus of the protein as being necessary for binding to target mRNAs (39). To test whether this holds true for the Drosophila homologs, we generated deletion mutants for Vig spanning the entire protein and performed RNP IP to check whether histone mRNA binding was disrupted (Fig. 4B). As expected, only the deletion of the C terminus containing an RGG box-like motif led to a reduction in Vig binding to both H2A and H3 mRNA (Fig. 4B).
Fig. 4.
Vig and Vig2 bind specifically to histone mRNA. (A) Enrichment of HisC mRNA from V5 IPs using cells stably overexpressing Vig-5 and Vig2-V5 fusion protein. Compared with empty vector controls, Vig and Vig2 protein IPs significantly enrich for histone mRNA as opposed to reference genes such as Tub84b, Actin, and Rpl32. (B) Graphic depiction of deletion mutants made across the Vig protein. The deletion mutants are used for a transient transfection of S2 cells, and after 72 h the fusion protein is immunoprecipitated via the V5 tag. Reverse transcription is performed as described in A. The enrichment of core histone mRNA through Vig IP remains undisrupted for the majority of deletion mutants, with the exception of MUT4, which deletes the C terminus of Vig containing the putative RGG box motif. (C) Recombinant Vig protein is overexpressed and purified. Recombinant Vig is then added to a mixture of a P32 end-labeled H2A 3′ UTR probe with and without 100-fold excess of nonspecific competitor yeast tRNA. The mixture is run on a 6% acrylamide gel in 1× Tris–glycine–EDTA buffer and visualized by exposing on a phosphoimager screen. Recombinant VIG protein binds and shifts the H2A 3′ UTR probe in the presence of a large excess of nonspecific competitor RNA. (D) ChIP is performed on S2 cells stably expressing Vig-V5 and Vig2-V5. qPCR analysis of Vig-V5 and Vig2-V5 ChIP samples shows that both Vig-V5 and Vig2-V5 is enriched at the 3′ end of the H2A gene compared with other regions of the Drosophila HisC. All data plotted are averages and SDs from three separate pull-down experiments.
Vig Binds to the 3′ UTR of H2A mRNA.
Canonical histone mRNAs are structurally unique and highly regulated to ensure that histone production is tightly coupled to S phase (41). The 3′ UTR of canonical histone mRNAs contain an evolutionarily conserved stem loop that recruits SLBP, which plays important roles in both the maturation and degradation of histone mRNAs (42). To test which part of the histone mRNA Vig interacts with, we expressed Vig in E. coli, purified it with nickel-nitrilotriacetic acid agarose (Ni-NTA) and cation exchange chromatography, and used the recombinant Vig protein for RNA EMSAs (Fig. S4). When combined with a H2A 3′ UTR probe, Vig efficiently bound and shifted the 3′ UTR probe in the presence of nonspecific competitor RNA (Fig. 4C). In line with these results, ChIP experiments at the HisC also showed enrichment of Vig and Vig2 at the 3′ end of the H2A gene body (Fig. 4D), providing further evidence for Vig interaction with the 3′ UTR of the H2A mRNA.
Discussion
Since its initial biochemical characterization in 2012, Cas9 has become a Swiss Army knife of molecular biology (43). Here, we add to its versatility by coopting Cas9 as the targeting agent for a sequence-specific, in vitro chromatin purification method, CLASP. This versatile and convenient method allowed us to purify the HisC and identify five potential regulators of H2A gene expression, which we then validated by measuring loss-of-function effects on H2A transcript level in Drosophila cells (Figs. 2 and 3).
Our most striking result was finding both Vig and Vig2 at the H2A/H2B gene region of the HisC, elucidating their previously unrecognized function in D. melanogaster. Vig was originally identified as a part of the RNAi complex, and studies in D. melanogaster focused on its role in gene silencing (36, 37). Even less was known about the molecular function of Vig2, as publications were limited to a possible role in affecting global heterochromatin formation, along with Vig, and to the identification of Vig2 in the structure of the D. melanogaster 80S ribosome (38, 44). The identification of SERBP1 as a mammalian homolog of Vig and Vig2 gave us a hint that these proteins might be binding and regulating histone mRNA (40). With the CLASP data in hand, we indeed found that both Vig and Vig2 bind and regulate canonical histone mRNA by binding to its 3′ UTR (Fig. 4A). The preferential targeting of Vig and Vig2 to H2A but not H1 mRNA is consistent with the differential timing of active transcription for these two genes during S phase, with H1 being transcribed throughout the S phase, while core histone genes are highly transcribed only toward the beginning of S phase (45). Thus, Vig and Vig2 might be needed to protect the early transcribed H2A mRNA against degradation throughout the S phase. With more and more potential mRNA-binding partners being discovered for SERBP1 and its homologs, it is also possible that these proteins act to maintain mRNA stability for a wide range of targets.
The identification of Mxc in our dCas9 HisC CLASP experiment served as an informative positive control, as it was previously described as a regulator of the Drosophila HisC. Unexpectedly, along with Mxc, seven other LisH domain-containing proteins out of 18 in the Drosophila proteome were enriched in the HisC-specific CLASP dataset. Knockdown of two potential regulators containing LisH motifs, Smu1 and Mahj, had a significant effect on H2A mRNA levels similar to the Mxc knockdown (Fig. 3B). The LisH domain has been shown to be crucial for Mxc recruitment to the HisC, and because of its prominent role in dimerization, it is proposed that the LisH domain mediates Mxc assembly into an oligomeric network that provides a scaffold onto which other components of the HisC assemble (46–48). As both Mahj and Smu1 contain LisH domains, it will be interesting in future studies to determine whether these domains help recruit LisH proteins to the HisC.
Another interesting candidate that had an effect on histone gene expression was brahma. Being part of the SNF2/SWI2 nucleosome remodeling complex, Brahma is known to be crucial for RNA polymerase II transcription and euchromatin maintenance (49, 50). Our preliminary data suggest that Brahma’s wide-ranging effect also involves regulation of the HisC, as knockdown of Brahma causes a decrease in steady-state mRNA levels of H1, H2A, and H3 genes (Fig. S5). Because Brahma’s potential regulation of the entire HisC is reminiscent of a previously reported HERS-mediated repression of the same region, it is tempting to speculate that they play opposite roles to ensure the proper formation of active and repressive chromatin at the histone locus during cell-cycle progression (51).
Although we were gratified that CLASP uncovered several functionally relevant regulators of the HisC locus, we were surprised that none of the proteins selectively associated with HisC turned out to be classic sequence-specific DNA-binding TFs expected to control histone gene expression. We considered two potential explanations for this finding. First direct measurements by single-molecule tracking reveal typical RNA polymerase II TFs with cognate site DNA-binding residence times on the order of a few seconds to a minute (52). Given the time scale of typical cross-linking protocols and the striking recent report on the artifacts of chemical cross-linking TF–DNA complexes, it seems likely that capturing bound TFs may pose a rather difficult challenge (53). A further compounding problem is that TFs are often expressed at very low levels, and our CLASP procedure may not be sensitive enough to isolate and unambiguously detect such rare regulatory proteins. Although no classic sequence-specific TFs were identified, several components of the general transcription machinery were detected. Specifically, RPB1 and TFIIF were enriched in the HisC-specific pull-down samples, while TFIIB was found to be depleted (Dataset S2). The lack of TFIIB at HisC is consistent with our previous finding through imaging analysis and provides further evidence that HisC may require a distinct preinitiation complex (45). Additional improvements in the efficiency and sensitivity of CLASP will be required to more comprehensively survey the full spectrum of the proteome associated with specific loci in the metazoan genome.
In conclusion, by developing CLASP, a versatile and experimentally tractable in vitro dCas9-targeted chromatin purification and locus-specific proteome isolation method, we identified regulators of the D. melanogaster HisC and determined their likely mechanism of modulating histone mRNA expression. In addition, the CLASP approach greatly increases our ability to experimentally screen for suitable reverse-ChIP regions within eukaryotic genomes (54). This advantage also means that customized stable cell lines overexpressing Cas9 are no longer necessary and that purified Cas9/RNPs can be applied to chromatin purification from any cell line that can be grown in sufficient quantities. Although our analysis of the D. melanogaster HisC takes advantage of the HisC’s intrinsic repetitiveness to increase the effective target concentration, single-locus chromatin purification and identification of regulatory proteins have recently been reported (19, 22). We believe that, in the future, improvements of CLASP by coupling with in vitro biotinylation of an attached tag by recombinant BirA could provide a powerful approach to probe multiple single-copy genomic targets to identify interesting regulatory proteins (55).
Materials and Methods
CLASP of D. Melanogaster HisC from S2 Cells.
Two billion synchronized S2 cells fixed in 1% formaldehyde (16% MeOH-Free; Polysciences) for 15 min were resuspended in lysis buffer [50 mM Hepes (pH 7.9), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100], incubated on ice for 10 min, spun down, and washed with 10 mM Tris⋅HCl (pH 8.1), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA. The cell pellet was rinsed with shearing buffer [0.1% SDS, 1 mM EDTA, 10 mM Tris (pH 8.1)]. Cells were sheared with a Covaris sonicator until HisC genomic DNA was visualized to be ∼150 bp by Southern blots. Chromatin was adjusted to NMNT buffer [10 mM Tris (pH 7.9), 500 mM NaCl, 5 mM MgCl2, 0.05% Nonidet P-40]. Chromatin was cleared with a hard spin (14,000 × g for 10 min) at 4 °C and then diluted to 10 mL of NMNT buffer per 5 × 108 cells in starting material. Then 0.16 mg of dCas9-3×FLAG was incubated with guide RNA at a 1:5 molar ratio for 1 h at 37 °C, was added to chromatin mix, and was incubated at RT overnight. M2 agarose resin (Sigma) was added to RNP/chromatin mix at a ratio of 50 uL of resin per 500 million S2 cells in starting material and incubated at RT for 2 h. Resin was spun down at 2,200 rpm and washed four times with NMNT buffer using 500 uL of wash volume per 20 uL of resin. Then 0.32 mg/mL of 3×FLAG peptide in 0.1 M NaCl NMNT buffer was added to resin to elute for 2 h at RT with shaking. The eluted sample was separated from resin and used for subsequent experiments.
CLASP of Telomere Sequences from HeLa Cells.
For CLASP of telomere sequences from HeLa cells, the same protocol as described above for HisC pulldown was used, except for the following changes: 500 million HeLa cells fixed at 1% formaldehyde for 15 min were used per pulldown, chromatin was sheared to ∼800 bp as visualized by agarose gel, and 0.18 mg of dCas9-3×FLAG fusion protein was used per 500 million HeLa cells in the starting material.
Purification of Recombinant Vig and dCAS9-3×FLAG Fusion Protein.
dCAS9-3×FLAG fusion protein was cloned into pET302 NT-His vectors (Thermo Fisher) and transformed into BL21-Codon Plus RIPL-competent cells (Agilent). Bacterial cultures were induced at OD 0.6 for incubation at 18 °C overnight with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cell pellets were lysed in lysis buffer [500 mM NaCl, 50 mM Hepes (pH 7.5), 5% glycerol, 10 mM 2-mercaptoethanol, 1% Triton X-100, 10 mM imidazole, and protease inhibitors]. Lysates were frozen at −80 °C overnight and sonicated. Sonicated lysates were cleared by ultracentrifugation and incubated with Ni-NTA resin overnight at 4 °C. Resin was then washed with 20× resin volume of 250 mM NaCl wash buffer [250 mM NaCl, 50 mM Hepes (pH 7.5), 5% glycerol, 10 mM 2-mercaptoethanol, and 25 mM imidazole] and eluted with 250 mM NaCl wash buffer + 250 mM imidazole. Peak elution fractions were pooled and applied to a POROS HS20 column (Applied Biosystems) and subjected to a linear gradient from 0.25 M to 1 M NaCl. Eluted fractions were analyzed by SDS/PAGE followed by PageBlue staining (Thermo Fisher). Peak fractions were pooled and dialyzed to 200 mM NaCl, 50 mM Hepes (pH 7.5), 5% glycerol, and 1 mM DTT. Samples were aliquoted and flash frozen for storage in −80 °C.
Recombinant Vig protein was purified in a similar fashion except the lysis buffer was composed of 25 mM Hepes (pH 7.5), 1 M NaCl, 10% glycerol, 0.05% Nonidet P-40, 0.4% Triton X-100, 0.08 mg/mL lysozyme, and 0.5 mM PMSF. Ni-NTA was washed with 40 resin volumes of 0.5 M NaCl lysis buffer and then with 10 resin volumes of 0.2 M NaCl lysis buffer. Proteins were eluted with 250 mM Imidazole in 0.2 M NaCl lysis buffer. Peak elution fractions were pooled, applied to a POROS HS20 column (Applied Biosystems), and subjected to a linear gradient from 0.2 M to 1 M KCl. Eluted fractions were analyzed by SDS/PAGE followed by PageBlue staining (Thermo Fisher). Peak fractions were pooled and dialyzed to 100 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 10% glycerol, 0.01% Nonidet P-40, and 1 mM DTT. Samples were aliquoted and flash frozen for storage in −80 °C.
RNA Isolation, Reverse Transcription, and Real-Time PCR Analysis.
Total RNA was extracted and purified using TRIzol reagent (Life Technologies) according to the manufacturer’s protocol. cDNA synthesis was performed with 1 μg of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad) and was diluted 10-fold. Real-time PCR analysis was carried out with SYBR Select Master Mix for CFX (Life Technologies) using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Gene-specific primer sequences are provided in Supporting Information.
In Vitro sgRNA Transcription and Purification.
The 19-bp targeted DNA sequence was inserted into the middle of a 58-bp primer behind a T7 promoter sequence (5′-TTAATACGACTCACTATAGNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGC-3′). The custom primer was then used with a reverse template (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′) in a DNA polymerase extension reaction to generate a dsDNA template. The dsDNA template was used with the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs) to generate ssRNA ∼100 bases in length. The reaction was DNase treated, and full-length RNA was purified by isolating the correct length after running on a denaturing polyacrylamide gel with 8 M urea.
MudPIT Mass Spectrometry and Analysis.
The TCA-precipitated proteins were urea-denatured, reduced, alkylated, and digested with recombinant endoproteinase Lys-C (Promega) and modified trypsin (Promega) (56, 57). In addition, one telomere reverse-ChIP sample was digested with Asp-N followed by GluC (Roche) after denaturation with 8 M urea, reduction with 5 mM Tris(2-carboxyethyl)phosphine (TCEP), and cysteine carbamidomethylation. Peptides generated by LysC/trypsin or AspN/GluC were loaded onto a 100-μm fused silica (Polymicro Technologies) capillary column packed with 3 cm of 5-μm reverse-phase C18 resin (Aqua; Phenomenex), 4 cm of 5-μm strong cation exchange resin (Luna; Phenomenex), and 8 cm of reverse-phase C18 resin. The loaded microcapillary column was placed in-line with a Quaternary Agilent 1100 series HPLC pump and a LTQ linear ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (Thermo Scientific). Ten-step MudPIT mass spectrometry was performed on the ionized peptides as described (56). MS/MS spectra were interpreted using SEQUEST (v. 27.9) on the human dataset (58) or ProLuCID (v. 1.3.3) on the fly dataset (59) and were searched against a nonredundant protein D. melanogaster database [National Center for Biotechnology Information (NCBI) 20 February 2013] containing 160 usual contaminants (human keratins, IgGs, and proteolytic enzymes). For the telomere samples, the spectra were searched against the human database (NCBI 25 March 2015) containing 160 usual contaminants and the dCas9 protein sequence. To estimate false discovery rates (FDRs), the amino acid sequence of each nonredundant protein was randomized. Peptide/spectrum matches were sorted and selected using DTASelect (60) with the following criteria set: spectra/peptide matches were retained only if they had a Delta CN score (DeltCN) of at least 0.8, and minimum cross-correlation score of 1.8 for singly, 2.0 for doubly, and 3.0 for triply charged spectra. Additionally, the peptides had to be minimum of seven amino acids in length and fully tryptic (except for the AspN/GluC-digested sample). Peptide hits from multiple runs were compared using CONTRAST (60). The dNSAFs were used to estimate relative protein levels (61). Mass spectrometry data will also be available after publication from the Stowers Original Data Repository at https://www.stowers.org/research/publications/libpb-1230.
DAVID Bioinformatics Analysis.
GenInfo identifiers were taken from MudPIT mass spectrometry results and converted to UniProt identifiers using UniProt ID mapping (www.uniprot.org/uploadlists/). UniProt identifiers were inputted into DAVID Bioinformatics Resources 6.8 (david.ncifcrf.gov/tools.jsp), and UP_KEYWORDS, GOTERM_BP_DIRECT, GOTERM_CC_DIRECT, GOTERM_MF_DIRECT, and INTERPRO annotations were used for functional clustering of the gene list.
Drosophila S2 Cell Culture and Synchronization.
S2 cells were cultured in M3BPYE medium supplemented with 5% heat-inactivated FBS. Two confluent T150 flasks of Drosophila S2 cells were dissociated from the flask and cultured in a Wheaton double-sided arm spinner flask (Fisher) with 75 mL of M3BPYE medium with 5% heat-inactivated FBS. Cells were kept growing in suspension at a density of 1 million to 3 million cells/mL. To synchronize, 0.2 nM of Ponasterone A (Sigma) was added to the suspension culture. After 24 h, the S2 cells were spun down at 800 × g for 5 min, washed once with 1× PBS, and then resuspended in fresh medium containing 1.5 mM hydroxyurea (Sigma). After 18 h, the cells were spun down, washed with 1× PBS, and resuspended in fresh medium only. Cells were collected after 2.5 h in fresh medium.
S2 Cell Immunofluorescence.
Eighteen-millimeter coverslips were cleaned with methanol and ethanol washes and then were incubated with 0.01% poly-lysine solution in water for 15 min. Cells were grown on poly-lysine–treated coverslips until ∼70% confluency and then were fixed with 4% paraformaldehyde in 1× PBS for 10 min. The fixed samples were washed in 1× PBS, permeabilized with 0.5% Triton X-100 in 1× PBS, and blocked with 3% BSA in 1× PBS. Primary antibody was added to the samples in 1× PBS with 0.1% Triton X-100 and incubated at 4 °C overnight. Samples were washed and incubated with Alexa Fluor 555 or Alexa Fluor 647 (Thermo Fisher) secondary antibody for 1 h at room temperature. The samples were then washed, briefly incubated with 300 nM DAPI, and prepped with ProLong Gold mounting medium (Thermo Fisher) for confocal imaging.
dsRNA Preparation and Drosophila S2 RNAi Knockdown Assays.
dsRNA templates were generated by placing a T7 promoter in front of PCR primers against an exon region of the targeted gene and performing PCR. The resulting template was visualized and isolated by the QIAquick Gel Extraction Kit (Qiagen). One hundred nanograms of template DNA were used with the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs). The reaction mixture was treated with DNaseI and purified using TRIzol reagent (Life Technologies) according to the manufacturer’s protocol. Resulting RNA was resuspended in water, heated to 65 °C for 30 min, and slowly cooled to RT to anneal and make dsRNA. S2 cells were resuspended in serum-free M3BPYE medium and cultured with dsRNA for 30 min at RT; 10% FBS M3BPYE (Sigma S8398) was added to get a final concentration of 3.75% FBS. Cells were incubated at 27 °C for 72 h before TRIzol extraction for total RNA. Control dsRNA was made from the pBluescript sequence.
Vig and Vig2 IP and RT-qPCR.
S2 cells stably expressing Vig-V5 and Vig2-V5 were dissociated from flasks, spun down, and lysed in lysis buffer [20 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5% Nonidet P-40, 800 units RnaseIN/mL]. Lysates were incubated on ice and then spun down at 4 °C to clear insoluble particles. One hundred microliters of supernatant were taken and added to anti-V5 agarose beads (Sigma) that had been blocked with 5% BSA and were resuspended in 900 uL of 1× NT2 buffer [50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40, 0.04 units RnaseIN/mL]. The mixture was rocked overnight at 4 °C, and then resin was washed with 1× NT2 buffer adjusted to 200 mM NaCl. Twenty units of DnaseI (New England Biolabs) were added to the washed resin in 1× NT2 buffer with 150 mM NaCl and incubated at 37 °C for 30 min. SDS was added to mixture to get a 0.1% final concentration, and the mixture was treated with 2.5 uL of Proteinase K (Thermo Fisher) at 56 °C for 1 h. RNA was isolated by using TRIzol reagent (Life Technologies) according to the manufacturer’s protocols. cDNA synthesis was performed with 50 ug of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad) and was diluted 10-fold. Real-time PCR analysis was carried out with SYBR Select Master Mix for CFX (Life Technologies) using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Gene-specific primer sequences are provided in Supporting Information.
Propidium Iodide Stain and Cell-Cycle Analysis.
Cells were collected, resuspended in 1× PBS, and fixed with ice-cold 70% EtOH for at least 2 h. The samples were then washed with 1× PBS, resuspended into propidium iodide (PI)/Triton X-100 solution (0.1% Triton X-100, 0.2 mg/mL Rnase A, 0.02 mg/mL PI in 1× PBS), and incubated at 37 °C for 15 min. Fluorescence was detected using BD LSRFortessa (BD Biosciences).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited with the ProteomeXChange [accession nos. PXD008043 (Drosophila histone cluster) and PXD008044 (human telomere].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718844115/-/DCSupplemental.
References
- 1.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 3.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 4.Strausbaugh LD, Weinberg ES. Polymorphism and stability in the histone gene cluster of Drosophila melanogaster. Chromosoma. 1982;85:489–505. doi: 10.1007/BF00327345. [DOI] [PubMed] [Google Scholar]
- 5.Marzluff WF, Duronio RJ. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 6.Harris ME, et al. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 8.Singh RK, et al. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghule PN, et al. Fidelity of histone gene regulation is obligatory for genome replication and stability. Mol Cell Biol. 2014;34:2650–2659. doi: 10.1128/MCB.01567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 11.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White AE, et al. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol Cell Biol. 2003;23:9275–9282. doi: 10.1128/MCB.23.24.9275-9282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasinskas A, Hamkalo BA. Purification and initial characterization of primate satellite chromatin. Chromosome Res. 1999;7:341–354. doi: 10.1023/a:1009211929408. [DOI] [PubMed] [Google Scholar]
- 15.Vincenz C, Fronk J, Tank GA, Langmore JP. Nucleoprotein hybridization: A method for isolating active and inactive genes as chromatin. Nucleic Acids Res. 1991;19:1325–1336. doi: 10.1093/nar/19.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman JL, Langmore JP. Nucleoprotein hybridization: A method for isolating specific genes as high molecular weight chromatin. Biochemistry. 1985;24:7486–7497. doi: 10.1021/bi00346a068. [DOI] [PubMed] [Google Scholar]
- 17.Hamperl S, et al. Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. In: Stockert JC, Espada J, Blázquez-Castro A, editors. Methods in Molecular Biology. Humana; Totowa, NJ: 2014. pp. 329–341. [DOI] [PubMed] [Google Scholar]
- 18.Waldrip ZJ, et al. A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics. 2014;9:1207–1211. doi: 10.4161/epi.29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pourfarzad F, et al. Locus-specific proteomics by TChP: Targeted chromatin purification. Cell Rep. 2013;4:589–600. doi: 10.1016/j.celrep.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: A method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170:1028–1043.e19. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Déjardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antão JM, Mason JM, Déjardin J, Kingston RE. Protein landscape at Drosophila melanogaster telomere-associated sequence repeats. Mol Cell Biol. 2012;32:2170–2182. doi: 10.1128/MCB.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ide S, Dejardin J. End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nat Commun. 2015;6:6674. doi: 10.1038/ncomms7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng W, Shi X, Tjian R, Lionnet T, Singer RH. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci USA. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grolimund L, et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun. 2013;4:2848. doi: 10.1038/ncomms3848. [DOI] [PubMed] [Google Scholar]
- 32.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Terzo EA, et al. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol Biol Cell. 2015;26:1559–1574. doi: 10.1091/mbc.E14-10-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho AB, Vicoso B, Russo CAM, Swenor B, Clark AG. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA. 2015;112:12450–12455. doi: 10.1073/pnas.1516543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomari Y, Zamore PD. Perspective: Machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 38.Gracheva E, Dus M, Elgin SCR. Drosophila RISC component VIG and its homolog Vig2 impact heterochromatin formation. PLoS One. 2009;4:e6182. doi: 10.1371/journal.pone.0006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J Biol Chem. 2001;276:3341–3347. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- 40.Ahn JW, et al. SERBP1 affects homologous recombination-mediated DNA repair by regulation of CtIP translation during S phase. Nucleic Acids Res. 2015;43:6321–6333. doi: 10.1093/nar/gkv592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marzluff WF. Metazoan replication-dependent histone mRNAs: A distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 45.Guglielmi B, La Rochelle N, Tjian R. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Mol Cell. 2013;51:480–492. doi: 10.1016/j.molcel.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duronio RJ, Marzluff WF. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017;14:726–738. doi: 10.1080/15476286.2016.1265198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim MH, et al. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 2004;12:987–998. doi: 10.1016/j.str.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 48.Cerna D, Wilson DK. The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J Mol Biol. 2005;351:923–935. doi: 10.1016/j.jmb.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 49.Armstrong JA, et al. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 2002;21:5245–5254. doi: 10.1093/emboj/cdf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayama T, Shimojima T, Hirose S. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development. 2012;139:4582–4590. doi: 10.1242/dev.083246. [DOI] [PubMed] [Google Scholar]
- 51.Ito S, et al. Epigenetic silencing of core histone genes by HERS in Drosophila. Mol Cell. 2012;45:494–504. doi: 10.1016/j.molcel.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teves SS, et al. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016;5:e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horlbeck MA, et al. Nucleosomes impede cas9 access to DNA in vivo and in vitro. Elife. 2016;5:e12677. doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu CM, Zhou H, Zhang WF, Yang HM, Tang JB. Site-specific, covalent immobilization of BirA by microbial transglutaminase: A reusable biocatalyst for in vitro biotinylation. Anal Biochem. 2016;511:10–12. doi: 10.1016/j.ab.2016.07.026. [DOI] [PubMed] [Google Scholar]
- 56.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 57.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 58.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 59.Xu T, et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.