Significance
Protein structure refinement by direct global energy optimization has been a longstanding challenge in computational structural biology due to limitations in both energy function accuracy and conformational sampling. This manuscript demonstrates that with recent advances in both areas, refinement can significantly improve protein comparative models based on structures of distant homologues.
Keywords: protein structure prediction, homology modeling, energy function, protein conformational search, protein structure refinement
Abstract
Proteins fold to their lowest free-energy structures, and hence the most straightforward way to increase the accuracy of a partially incorrect protein structure model is to search for the lowest-energy nearby structure. This direct approach has met with little success for two reasons: first, energy function inaccuracies can lead to false energy minima, resulting in model degradation rather than improvement; and second, even with an accurate energy function, the search problem is formidable because the energy only drops considerably in the immediate vicinity of the global minimum, and there are a very large number of degrees of freedom. Here we describe a large-scale energy optimization-based refinement method that incorporates advances in both search and energy function accuracy that can substantially improve the accuracy of low-resolution homology models. The method refined low-resolution homology models into correct folds for 50 of 84 diverse protein families and generated improved models in recent blind structure prediction experiments. Analyses of the basis for these improvements reveal contributions from both the improvements in conformational sampling techniques and the energy function.
The number of protein families for which computational models with reasonable accuracy can be built has steadily increased in the current structure- and sequence-rich era (1). Homology-modeling methods can in some cases produce models with sufficient accuracy for the inference of structure–function relationships, but in many cases starting models contain significant errors. Increasing the accuracy of such models is the goal of protein structure refinement and for the last decade has been a grand challenge for structural biology (2–8).
Structural averaging of molecular dynamics (MD) simulation trajectories (9, 10) and modeling with strong restraints to a high-resolution reference model (4, 5) have been shown to consistently improve model accuracy when starting models are close to the native structure. However, when starting models contain significant errors, the conformational phase space exceeds by orders of magnitude what can be explored using such methods, and little or no accuracy increase is observed (2). In contrast, coarse-grained conformational search and unrestrained simulations can sample more extensively but suffer from inaccuracy in energy functions and thus more often degrade than improve model quality (3, 8, 11). Because of the stringent and often conflicting requirements of energy function accuracy and extensive sampling, the substantial improvement of distant comparative models remains an outstanding challenge.
Here we describe a protein structure refinement method based on large-scale energy optimization that improves structure models with significant errors such as comparative models built from structures of distant homologs with sequence identity less than 30%. Study of the basis for this improved performance reveals contributions from improvements in both the sampling methodology and the energy function.
Results
Approach Summary.
In devising a refinement method capable of improving models far from the native structure, we were guided by the following considerations. First, large-scale structural changes are on a time scale that is likely too long for all-atom MD simulations to currently access, and an effective approach should involve moves that introduce discrete (rather than continuous) structural changes to enable energy barrier hopping. Second, since such moves will be generally unfavorable in standard all-atom representations, a lower-resolution coarse-grained model is appropriate for sampling. Third, since a coarse-grained model is necessarily less accurate than an all-atom model, the overall refinement trajectory should be guided by an all-atom energy function. Finally, since structural changes may need to occur at multiple noninteracting regions, an iterative refinement approach should improve success.
We implemented these considerations in a refinement method within the Rosetta modeling suite (12) that carries out large-scale sampling of the energy landscape using a two-stage protocol. The first stage introduces structural variation in the starting model, generating a population of diverse structures in different low-energy minima. The second stage utilizes an evolutionary algorithm to guide this model population toward the lowest all-atom energy (and hence likely more accurate) state. The second stage consists of 50 iterations; in each iteration, new structures are sampled, and the population is updated favoring lower all-atom energy structures while maintaining the structural diversity. For evaluation of the effectiveness of the protocol, we consider both the lowest-energy structures from each of the five largest clusters at the end of the iteration and a single model generated through structural averaging of models close to the lowest-energy cluster representative followed by constrained minimization (SI Appendix provides details); we refer to the former as “cluster representatives” and the latter as the “refined structure” in the remainder of the text.
To generate models at the diversification stage and at every iteration of the evolution stage, multiscale Monte Carlo (MC) modeling implemented in RosettaCM (13) was used with sampling carried out in a coarse-grained representation and model evaluation in an all-atom representation. The conformational search utilizes a “broken-chain” kinematics setup of the protein chain that allows large structural changes in internal coordinates to local regions [or regions predicted as unreliable (14)] without disruption of the remainder of the structure (13). Two types of structural perturbations are applied during MC search: “mutation” replaces backbone torsions with those from a generic fragment library (15), and “crossover” replaces the Cartesian coordinates of the backbones of a segment with those from another structure in the current pool of models (only mutation operators are used at the diversification stage) (13). Distance restraints are employed during sampling in the coarse-grained representation (but not in the subsequent all-atom model evaluation) to control the amount of structural variation and are weighted based on their frequency in the sampled population—frequently violated restraints are down-weighted. At the diversification stage and at each iteration at the evolution stage, after the coarse-grained conformational search, side-chains are built onto the backbones, and iterative side-chain and backbone optimization is carried out in an all-atom representation using the recently improved implicit solvent energy function (16). The energy of a model following all-atom optimization determines whether it is accepted into or rejected from the evolving population at each iteration of the evolution stage. Details of the method can be found in Methods.
Refinement Performance Evaluation.
We first establish that the method described in the previous section can improve starting models distant from the native structure. This is a nontrivial property: Because of the high dimensionality of the search space, there are many more ways to make a model worse than to make it better. We then evaluate the factors determining success in this endeavor.
To benchmark the approach, we identified 44 proteins from previous CASP (critical assessment of techniques for structure prediction) (17) and CAMEO (continuous automated evaluation of models) (18) experiments, with diverse topologies and size ranging from 60 to 200 amino acids. In all 44 cases, the best homology models had substantial structural errors (SI Appendix, Table S1). For CASP targets, we selected as starting models the best models submitted by any server group [analogous to the CASP refinement category (3)]; for CAMEO targets we selected the best Robetta server (19, 20) model. The benchmark-set targets cover a broad range of starting model quality and sequence identity to homologs of known structures (SI Appendix, Fig. S1). The challenge of improving these models is analogous to that in CASP refinement experiments, where model generation already uses all available information from homologous structures [input models are generally more accurate than any single template (17)]; thus improving their accuracy consistently is a nontrivial challenge.
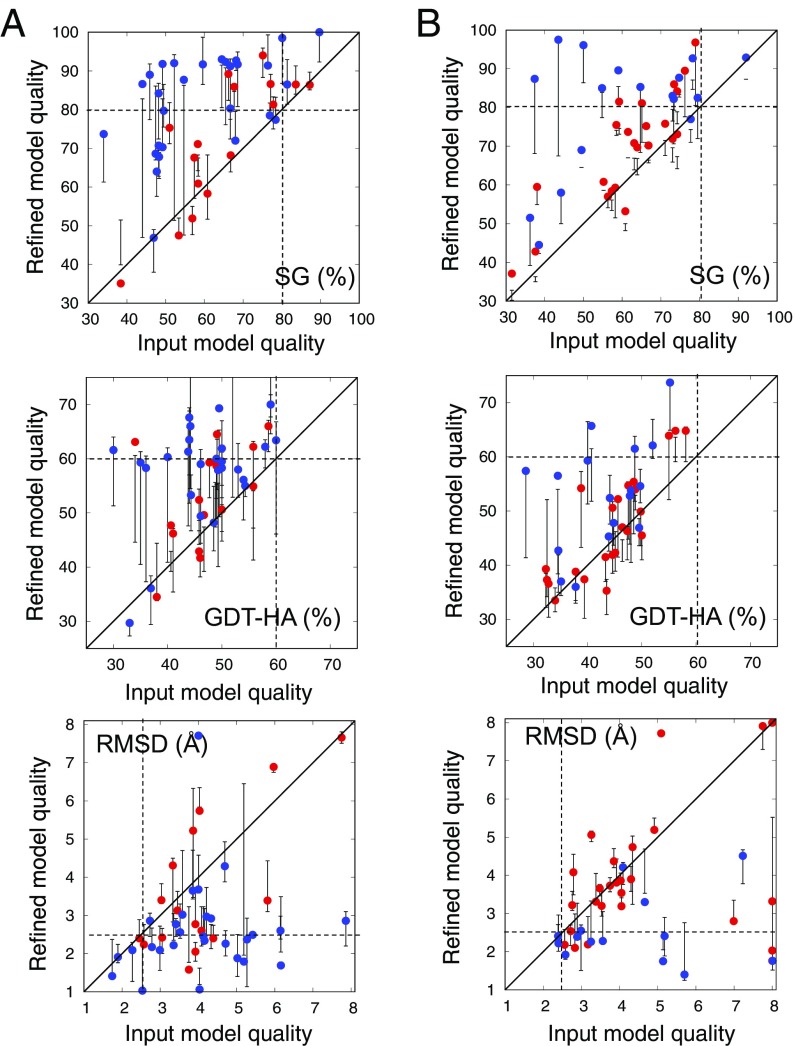
The results on the benchmark set show that the approach can significantly improve the input structures. The quality of the refined structure is compared with the input structures according to three different model backbone quality metrics in Fig. 1A (side-chain accuracy is given in SI Appendix, Fig. S2). We define a model as having a “correct fold” if two of three metrics are better than the thresholds shown as dashed lines in Fig. 1A. With this definition, in over 44 cases, refinement increases the number of correct folds from two to 24, and for 32 of the cases at least one of the cluster representatives had the correct fold. These improvements are greater than those of the best submissions in previous CASP refinement challenges in 75% of cases (SI Appendix, Table S2). (To be fair, the earlier predictions, unlike ours, were completely blind, but our automated approach had no knowledge of native structures.) Repeated refinement calculations did not in general yield improved results; in most cases the runs converged on similar structures (SI Appendix, Fig. S3), and in the remainder, model selection was a nontrivial challenge. An important application of model refinement is increasing suitability for solving X-ray crystal structures by molecular replacement (MR). Refinement improved the MR log likelihood gain (LLG) (21) by greater than 15 units for 8 of 10 CASP targets for which diffraction data were available (SI Appendix, Table S3). MR is generally successful if the LLG is greater than 60; of the 10 targets, the number with LLG > 60 increased from one before refinement to four after refinement.
Fig. 1.
Performance of refinement protocol on benchmark set. (A) Benchmark set1—44 proteins from CASP and CAMEO rounds up to September 2015. The starting homology models were generated by multiple different servers. (B) Benchmark set2—40 proteins from CAMEO rounds since October 2015. The starting homology models were generated by the Robetta server (19, 20). In each panel, model quality is compared between input models (x axis) and refined models (y axis) using three different model accuracy metrics. (Top) SphereGrinder (SG); (Middle) GDT-HA; (Bottom) rmsd (SI Appendix). For SG and GDT-HA, higher values are better, and the native structure has a value of 100. Models with values better than the thresholds indicated by the dashed lines (SG > 80, rmsd < 2.5 Å, and GDT-HA > 60) for two of the three metrics are considered “correct folds.” Points represent the single refined model; the error bars represent the range of model qualities of the five cluster representatives. Blue, proteins with less than 120 residues; red, proteins with 120 or more residues. The refinement protocol consistently improves input models in both benchmarks.
To further benchmark the refinement protocol, we applied it to a second benchmark set (set2, listed in SI Appendix, Table S4) consisting of 40 recent (since October 2015) CAMEO targets. As input we selected comparative models generated for CAMEO by the Robetta server. The sequence and structure databases used in the refinement procedure were chosen to be identical to those used by Robetta during the CAMEO model generation process; this ensures that any improvement in model quality is a result of the protocol and not the availability of additional sequence or structure information. The results were qualitatively similar to those with the first benchmark set (Fig. 1B); the small decrease in performance likely reflects increases in average protein size and the use of Robetta in model generation (the models start out in energy minima of the Rosetta energy function). Over benchmark set1 and set2 combined, the refined structure has the correct fold by the definition above for 40 of 84 targets, and one of the cluster representatives, for 50 of 84 targets.
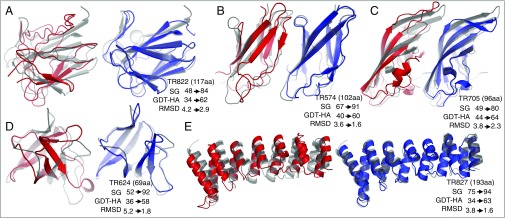
It is instructive to consider several specific examples of the structural changes that occur during refinement. The most dramatic improvement in the first benchmark was for TR822 (Fig. 2A), in which refinement recovers not only the native β-strand pairing pattern but also improves the locations of these strands, increasing model accuracy by over 30%. The input structure for TR624 and TR827 (Fig. 2 D and E) has roughly correct topology but with large deviations in secondary structure orientations, likely originating from significant sequence changes in homologous structures, which are largely corrected by the energy-guided refinement. The TR574 and TR705 (Fig. 2 B and C) cases show that the approach can improve both secondary structure and loop regions.
Fig. 2.
Examples of structural improvements brought about by refinement. (A) TR822, (B) TR574, (C) TR705, (D) TR624, and (E) TR827. Native, input structures, and refined models are shown in gray, red, and blue cartoon representations, respectively.
The refinement protocol was further tested in a completely blind setting on 43 targets from the tertiary structure prediction and refinement categories of the latest CASP (CASP12). Excluding membrane proteins and oligomers with extensive subunit interfaces (four targets in total), for 29 of the remaining 39 cases, accuracy was improved for the best of the five cluster representatives with increases of more than 20% in five cases (SI Appendix, Fig. S4). Nevertheless, the performance was generally less consistent compared with that of the benchmark; large proteins (over 200 residues) and oligomers—excluded in the benchmark—were rarely improved and sometimes degraded in quality (SI Appendix, Fig. S5). Our large-scale sampling approach likely fails in these cases due to the very large size of the conformational space; as noted above, there are many more ways to move away from any point in a high-dimensional space than toward it. The other approaches with more consistent results in CASP12 were quite a bit more conservative, staying relatively close to the input structure, and hence both the improvements and the decreases in model quality had smaller overall magnitudes (22).
Contributions to Successful Refinement.
The CASP12 results show there is still considerable room for improvement of the refinement method, particularly for larger proteins. To understand the origin of successful refinement and to guide further methods development, the set of targets for which the sampling problem is tractable (monomeric proteins with <200 residues) can be used as a laboratory to systematically investigate the contributions to successful refinement. We carried out a series of control experiments on benchmark set1, replacing aspects of the search strategy and the energy function with alternatives one at a time to isolate the factors responsible for the structural improvement. To make the comparisons simpler and quantitative, we use a single metric—SphereGrinder (SG) (23)—which best captures large-scale structural improvements (3, 8, 11, 22); over the benchmark set the SG values are correlated with both the rmsd and the GDT-HA (global distance test-high accuracy) (24) (SI Appendix, Fig. S6).
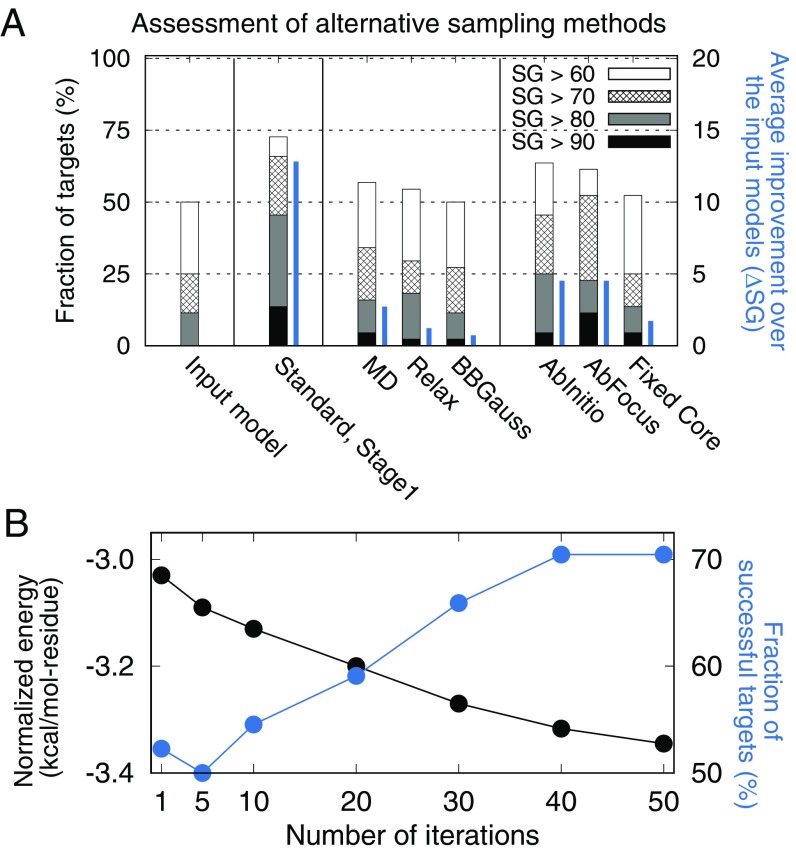
A first set of control experiments was carried out to elucidate the contribution from different aspects of the conformational sampling method (Fig. 3). To investigate the importance of the use of different resolution representations in our multiscale modeling approach (rather than using an exclusively all-atom representation), we carried out control calculations using as all-atom representation methods: (i) refinement by explicit water MD simulation with parameters optimized for high-resolution refinement (MD) (9), (ii) iterative all-atom optimization of protein core coupled with discrete side-chain optimization (Relax) (25, 26), and (iii) Monte Carlo sampling combining continuous backbone and side-chain movements (BBGauss) (27). The second and third controls use the same all-atom energy function as our multiscale approach. None of the control methods produced significant improvements in the starting models (Fig. 3A). In particular, while consistent improvements were obtained with refinement using explicit water MD simulation (MD), which is quite powerful for refinement of close-to-native models (3), they were quite small; only 7 of the 44 benchmark cases passed the “correct fold” threshold defined above. All-atom representations make it difficult to escape local energy minima around the input structure (SI Appendix, Fig. S7 A and B), due to the high probability of introducing unfavorable interactions accompanying any large perturbation to the structure. (The coarse-grained representation is much more tolerant of clashes.) Simulation parameters can be adjusted to encourage diversification for an exclusively all-atom representation approach, but focusing back in on the lowest-energy structure by annealing (7) or with unrestrained simulation (8) is challenging; even when sampling is successful, selecting one single representative from a massive simulation trajectory is a nontrivial challenge (8, 9). Incorporating restraints from physical intuition into replica-exchange MD simulation (28) has yielded good results for proteins with less than 100 residues (29); it is unclear how this approach will work for larger proteins or starting models with more substantial structural errors.
Fig. 3.
Contribution of conformational sampling protocol to refinement success. (A) Control experiments evaluating different sampling methods over the proteins in benchmark set1. The fraction of targets for which the best cluster representative after the diversification stage had an SG value above the thresholds is shown in the wide stacked bars (values on left axis), and the average improvement over the input structure is shown in narrow blue bars (values on right axis). MD, Relax, and BBGauss exclusively utilize an all-atom representation, while FixedCore, AbFocus, and AbInitio utilize different kinematic setups from the standard method. Distribution of per-target improvements over the input models and structural similarity to the input models for the methods are in SI Appendix, Fig. S7. (B) Dependence of model quality and all-atom energy on the number of iterations at the evolution stage. All-atom energies, shown in the black line, are normalized by dividing by the number of residues and averaged over 44 cases (values on left axis); the fraction of targets with SG > 80 at each iteration is shown in the blue line (values on right axis). As the number of iterations increases, the energy decreases, and the model quality increases.
Control experiments on the components of the input model diversification stage highlight the importance of the kinematic setup of the protein chain and the set of structural perturbations incorporated into coarse-grained sampling. While keeping the energy function unchanged, the performance of the standard approach was compared with that of three alternatives. AbInitio resembles Rosetta de novo modeling (30) in which the protein is treated kinematically as a continuous chain, and sampling is uniform across the protein chain. AbFocus is the same as AbInitio, except that five times more intensive sampling is carried out in the regions predicted to be unreliable. FixedCore resembles RosettaCM (13) and other local reconstruction-based refinement methods (4, 5) in using error estimation and a broken-chain kinematics setup, but mutations are only allowed in unreliable regions. These alternatives produce either smaller (FixedCore) or lower-consistency improvements (AbInitio and AbFocus) (Fig. 3A; see SI Appendix, Fig. S7 A and B for more details). When the evolution stage is carried out using a first-generation pool generated from any of the three approaches, the resulting models are quite a bit worse.
A final series of control experiments was used to explore the robustness of the approach to the details of the global optimization control logic. The evolution protocol was varied by changing structural operators, parent-selection logic, or population update logic, keeping the remaining components identical to the standard approach (SI Appendix, SI Methods and Fig. S7C). Improvement over the starting population occurs in all cases, implying that our results are not the outcome of overfit search parameters. The only exception—where performance is significantly poorer—results from eliminating protocol iteration and instead generating the same amount of total models repeatedly from the first-generation structures: improvements originate from propagation through multiple iterations rather than a single huge improvement in one fortunate MC simulation. The optimal number of iterations at the evolution stage—generating ∼100 new structures at each iteration—was found to be ∼40 iterations; while the all-atom energy continues to drop beyond this point the structures do not change much, as they are in deep energy minima (Fig. 3B).
Energy Function Accuracy Is Critical for Structure Refinement by Large-Scale Sampling.
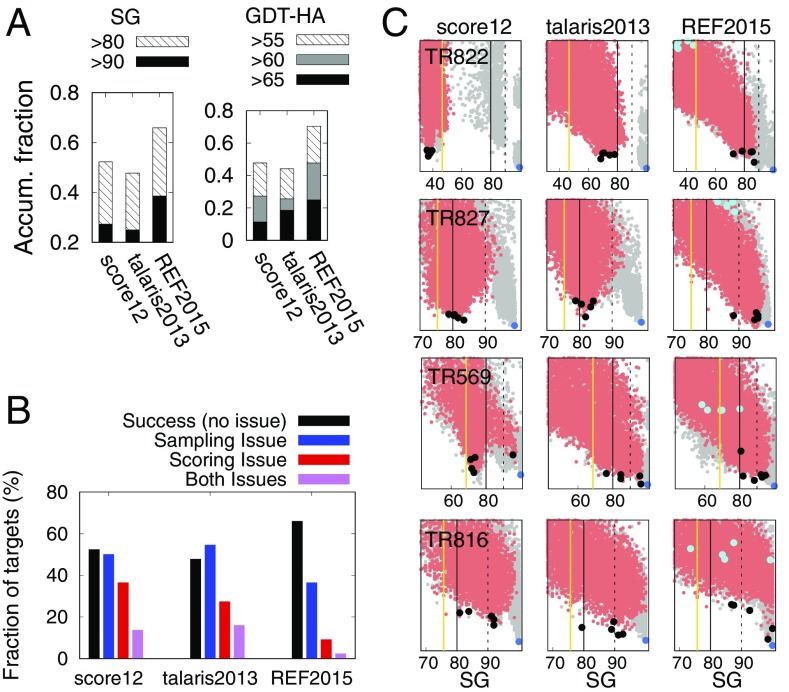
To address the role of energy function improvement in refinement success, a second set of control experiments was carried out using different all-atom energy functions. We pick three energy functions historically used as standard all-atom energy functions in Rosetta: score12 (31), talaris2013 (referred to as ElecHBv2 in ref. 32), and REF2015 (referred to as opt-nov15 in ref. 16), listed in order of development. REF2015, used in this study, is the current default energy function in Rosetta; all nonbonded atomic-level parameters were fully reoptimized to simultaneously reproduce small-molecule thermodynamic properties as well as protein properties. Fig. 4A shows a remarkable improvement in model quality with the energy function improvements from talaris2013 to REF2015: energy function improvement was clearly essential to the success of our global optimization-based refinement method.
Fig. 4.
Contribution of all-atom energy function improvements to refinement success. Full refinement calculations on benchmark set1 were carried out using three different energy functions—score12 (29), talaris2013 (30), and REF2015 (16)—and results are shown for the best of the five cluster representatives. (A) Stacked bars show fractions of targets with SG (Left) and GDT-HA (Right) values above the thresholds indicated in the legends. Refinement with REF2015 produces better structures than with score12 or talaris2013. (B) Distribution of refinement outcomes using the different energy functions. Success—SG of lowest-energy structure sampled >80 and energy gap of greater than 0.1 kcal/mol-residue between this structure and the lowest-energy structure with SG < 80 (e.g., all four cases in C with REF2015). Sampling issue—no structures sampled with SG > 80 (e.g., TR822 with score12 in C). Scoring issue—lowest-energy structure (including structures from the native biased simulations) with SG > 80 have energy gaps to the structures with SG < 80 of less than 0.1 kcal/mol-residue (e.g., TR569 with score12). (C) Full energy landscapes for cases with large differences between energy functions. Model quality is on the x axis (in SG), and energy is on the y axis; analyses with other metrics are in SI Appendix, Fig. S8. Yellow line represents the input model quality; red dots represent the entire set of structures sampled by standard approach; cyan dots represent the five models with lowest energy at the beginning of evolution stage (for REF2015 only); black dots represent the final five cluster representatives. Gray dots on the background are native biased simulations; the global energy minimum is in blue dots.
A more detailed view of how improvements in the energy function guide sampling is provided by energy landscape exploration analysis using a priori knowledge of the native conformation. While the energy function-based model discrimination improves from score12 to talaris2013 to REF2015 (red bars in Fig. 4B), as we progress from talaris2013 to REF2015, the improvement in energy function in addition leads to improvement in sampling (blue bars in the figure), which is not the case in the step from score12 to talaris2013. There is an enhanced driving force guiding sampling to the native structure in REF2015 that likely originates primarily from an improved Lennard-Jones (LJ) model that captures the energetics of hydrophobic core formation more accurately (SI Appendix, Table S5). For most of the cases shown in Fig. 4C (SI Appendix, Fig. S8, for other metrics), while the native conformation is one of the lowest-energy states in the entire landscape in all energy functions, REF2015 is the only one with sufficient gradient to guide the iterative sampling process toward the native structure starting from an inaccurate input structure; this driving force is important in challenging refinement problems, as less sampling is needed to converge on the correct structure. The optimization of REF2015 utilized in addition to small-molecule thermodynamic data, an evaluation metric measuring both the shape of the folding funnel (the difference in energy between the native structure and nearby structures) and discrimination power (the difference in energy between the native structure and far away structures), and the improvement in the gradient toward the native state, perhaps resulting from the former, appears to be the dominant source of improvement in refinement.
Discussion
We have demonstrated that protein structure models can be improved by energy-guided large-scale sampling and traced how success in refinement depends on the sampling protocol and energy function. In previous studies this concept—exploring the energy landscape with iterative multiscale modeling—was limited either by strongly restraining input structures (4–6), carrying out only a few iterations (33), or using experimental data to guide the search (34). The major stumbling block was that without restraints on the input structure or experimental data to guide the search, inaccuracies in the energy function would cause structures to drift away rather than toward the native structure (33). As noted above, because of the high dimensionality of the space, there are far more ways to degrade a model than to improve it. The folding funnels (examples shown in Fig. 4C) show that with recent improvements, the Rosetta implicit solvent energy function can have sufficient accuracy to guide sampling into the native energy basin. Best results are obtained when the improved energy function is coupled with a robust and rapid all-atom relaxation method (26) using an iterative multiscale representation approach; thorough relaxation of perturbed coarse-grained models is critical, otherwise close-to-native models could be rejected from the structural pool. Advances in sampling techniques developed in Rosetta for homology modeling (13) and experimental data-guided search (35), including local error estimation, broken-chain kinematics, and multiscale modeling, clearly facilitate exploring the complicated energy landscape. The refinement protocol can readily incorporate additional structural information for more complex or larger proteins. For example, a simpler version of the protocol was recently used in the computation of protein structures using sequence coevolution information (35). The all-atom energy function and coevolution restraints were optimized together, considerably improving the input models built by de novo modeling or coevolution pattern search to an accuracy where functional insights become possible.
The failures of the approach on specific targets reveal areas for improvement. The assumption behind the approach is that the global energy minimum is located near the monomeric native conformation and that this minimum can be discovered through large-scale energy-guided sampling. The approach will fail if the global energy minimum is not a monomer (SI Appendix, Fig. S5A), as was the case for a number of CASP12 targets whose biological units were homo- or hetero-oligomers. Insufficient sampling is another cause of failure (SI Appendix, Fig. S5 B and C): refinement attempts failed to improve targets with starting models with totally different folds, with over 200 residues and/or complex topologies, and with significant sequence registration errors in secondary structures. Of these causes of failures, sequence registration errors are perhaps the most tractable issue to address (SI Appendix, Fig. S5C). The problem of refining larger proteins is more challenging due to the exponential increase in the size of the search space with increasing chain length—consistent success in refinement of larger proteins may ultimately require significant increases in computing power, a more advanced sampling strategy, or further energy function improvements so there is a stronger guiding energy gradient further from the native structure.
Sampling problems also occur in the very close-to-native regime where the MC moves in the coarse-grained representation may be too coarse to achieve the small changes required to improve the structure. This is evident in the smaller improvements in the GDT-HA metric during refinement, which penalizes deviations as small as 0.5 Å from the native structure (the other two metrics are tolerant to deviations of this magnitude). The GDT-HA values of many of the refined models are around 60% or less, indicating room for improvement, which will likely be necessary for greater success in molecular replacement (SI Appendix, Table S3). Combining with more continuous MD simulation for higher-resolution refinement, already demonstrated in previous CASPs to be effective in consistent refinement of close-to-native models (9), is a promising direction. While we have demonstrated that implicit solvent models are suitable for refinement from distant starting structures, closer to the native state explicit water molecules can become important in determining the precise conformations of loops; such effects will be missed by our current approach but could perhaps be captured by incorporating explicit solvent MD simulations at a final stage.
Methods
Sampling Operators.
Two types of sampling operators are used: a mutation operator and a crossover operator. The sampling operators first set up a “star fold tree” kinematics for propagating changes to the starting conformation in a coarse-grained representation by breaking the chain at the beginning of each unreliable region and loop with more than three residues, as defined by a secondary structure assignment software DSSP (36). There are then three sampling stages: (i) stage1, a MC simulation in internal coordinates in which the degrees of freedom are the internal coordinates of the resulting disconnected chains, and the rigid body transforms between them; (ii) stage2, a MC simulation in Cartesian coordinates with moves consisting of local segment replacements and minimization in Cartesian space, and strong restraints between the termini of each chain segment to close the chain breaks; (iii) stage3, all-atom refinement (26).
In the mutation operator, the primary MC moves are fragment insertions in the loops or unreliable regions (see Diversification Stage section below), but also to the other parts at a lower probability (10% of the frequency of the unreliable regions) to further increase structural diversity. Stage1 consists of 12,000 MC-attempted three or nine residue fragment replacements as in Rosetta de novo structure prediction (backbone torsion angles of randomly selected segments are replaced with those of the fragment), and stage2, 700 attempted nine residue fragment insertions in Cartesian coordinates made by superimposing the N and C-terminal residues of a randomly selected fragment on the first and last residues of a randomly selected nine residue insertion site. The fragments are obtained by the standard fragment-picking method developed for de novo modeling (15) from a July 2011 database. Fragments from the target itself or its homologs are excluded for targets whose native structure is deposited to Protein Data Bank before 2011.
In the crossover operator (used at the evolution stage), the MC moves consist not only of fragment insertions but also chunk replacements. A chunk replacement substitutes one or more different chain segments of the conformation with corresponding segments from the five selected members in the current pool of the evolution stage. Using more than two parents differs from typical crossover operations in evolutionary algorithms; however, we expect this “grouped crossover” increases sampling efficiency. Chunk replacements comprise 10% of the 12,000 MC attempts in stage1 (the remaining 90% are fragment insertion) and 20% of the 700 MC attempts in stage2 (the remaining 80% are local fragment superpositions).
Diversification Stage.
The diversification stage begins by estimating residue-level local errors in backbones and identifying unreliable regions based on structural fluctuations in short MD simulations (20 trajectories of 20 ps) (37). The residues are sorted based on the fluctuations in the MD simulations, and those with the largest fluctuations are considered unreliable. The fraction of residues that are selected as unreliable is a function of both protein size and target difficulty, ranging from 10% for easy targets over 200 residues to 50% for hard targets under 100 amino acids (SI Appendix).
The diversification stage consists of multiple independent applications of the mutation operator to the input model. Two types of restraints are used to constrain sampling (the restraints are solely for guiding the coarse-grained sampling; they do not affect the all-atom modeling or model selection). In restrained_sampling, all residue pairs in the structure are linearly restrained to the values in the input model. In permissive_sampling, the restraints are weighted based on the estimated residue-level error. Weights on residue pairs in which one or both are in unreliable regions are set to near zero and are weaker than in the restrained_sampling case even in the reliable regions to allow for generating more diverse structures (SI Appendix). The 1,000 and 2,000 independent samples are generated using restrained_sampling and permissive_sampling, respectively. For very small proteins under 70 residues, Rosetta de novo structure calculations are also carried out and are filtered by structural similarity to the input models [TM-score (38) > 0.5].
Each of the populations of models are then clustered, and the lowest-energy members of each cluster are identified (the cluster representatives). The five lowest-energy cluster representatives from the restrained_sampling runs and the 45 lowest-energy cluster representatives from the permissive_sampling runs are then combined, giving a total pool size of 50. When de novo models are included for proteins under 70 residues, 1, 9, and 40 cluster representatives are selected from the restrained_sampling, permissive_sampling, and de novo populations, respectively.
Evolution Stage.
The evolution stage starts from the 50 selected models from the diversification stage and proceeds in a series of iterations, maintaining a pool of 50 structures. At each iteration, 10 members of the pool are selected as seeds, and for each, 6 mutation operations and another 6 crossover operations (with different combinations of randomly picked parents other than seed) are carried out to generate a total of 120 trial structures. From the original 50 parents and the newly generated 120 trial structures, 50 models are selected for the next iteration based on all-atom energy and divergence from the other pool members. After 15–25 iterations, the unreliable regions and restraints are updated according to the structural variation in the current population. Details of the evolution stage are described in SI Appendix.
Implementation.
All of the sampling operators in the study—for both diversification and evolution stages—are run using the HybridizeMover in Rosetta (13). All of the scripts and instructions required for running the protocol are available online; see SI Appendix for details.
Supplementary Material
Acknowledgments
Computing resources for this work are from Hyak supercomputer system at the University of Washington. This work was supported by the US National Institutes of Health (R01GM092802 to D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719115115/-/DCSupplemental.
References
- 1.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci USA. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feig M. Computational protein structure refinement: Almost there, yet still so far to go. WIREs Comput Mol Sci. 2017;7:e1307. doi: 10.1002/wcms.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modi V, Dunbrack RL., Jr Assessment of refinement of template-based models in CASP11. Proteins. 2016;84:260–281. doi: 10.1002/prot.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee GR, Heo L, Seok C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins. 2016;84:293–301. doi: 10.1002/prot.24858. [DOI] [PubMed] [Google Scholar]
- 5.Park H, DiMaio F, Baker D. CASP11 refinement experiments with ROSETTA. Proteins. 2016;84:314–322. doi: 10.1002/prot.24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xun S, Jiang F, Wu Y-D. Significant refinement of protein structure models using a residue-specific force field. J Chem Theory Comput. 2015;11:1949–1956. doi: 10.1021/acs.jctc.5b00029. [DOI] [PubMed] [Google Scholar]
- 8.Raval A, Piana S, Eastwood MP, Dror RO, Shaw DE. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 9.Mirjalili V, Feig M. Protein structure refinement through structure selection and averaging from molecular dynamics ensembles. J Chem Theory Comput. 2013;9:1294–1303. doi: 10.1021/ct300962x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirjalili V, Noyes K, Feig M. Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging. Proteins. 2014;82:196–207. doi: 10.1002/prot.24336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, et al. Lessons from application of the UNRES force field to predictions of structures of CASP10 targets. Proc Natl Acad Sci USA. 2013;110:14936–14941. doi: 10.1073/pnas.1313316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leaver-Fay A, et al. Rosetta3: An object-oriented software suite for the simulation and design of marcromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gront D, Kulp DW, Vernon RM, Strauss CEM, Baker D. Generalized fragment picking in Rosetta: Design, protocols and applications. PLoS One. 2011;6:e23294. doi: 10.1371/journal.pone.0023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H, et al. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction: Progress and new directions in round XI. Proteins. 2016;84:4–14. doi: 10.1002/prot.25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas J, et al. The protein model portal–A comprehensive resource for protein structure and model information. Database (Oxford) 2013;2013:bat031. doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ovchinnikov S, et al. Improved de novo structure prediction in CASP11 by incorporating coevolution information into Rosetta. Proteins. 2016;84:67–75. doi: 10.1002/prot.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunkóczi G, Wallner B, Read RJ. Local error estimates dramatically improve the utility of homology models for solving crystal structures by molecular replacement. Structure. 2015;23:397–406. doi: 10.1016/j.str.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovan L, et al. Assessment of the model refinement category in CASP12. Proteins. 2017;86:152–167. doi: 10.1002/prot.25409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopp J, Bordoli L, Battey JND, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69:38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 24.Antczak PLM, Ratajczak T, Blazewicz J, Lukasiak P, Blazewicz J. 2015. SphereGrinder–Reference structure-based tool for quality assessment of protein structural models. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), (IEEE, Washington, DC), pp 665–668.
- 25.Tyka MD, et al. Alternate states of proteins revealed by detailed energy landscape mapping. J Mol Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betancourt MR. Efficient Monte Carlo trial moves for polypeptide simulations. J Chem Phys. 2005;123:174905. doi: 10.1063/1.2102896. [DOI] [PubMed] [Google Scholar]
- 28.Perez A, MacCallum JL, Dill KA. Accelerating molecular simulations of proteins using Bayesian inference on weak information. Proc Natl Acad Sci USA. 2015;112:11846–11851. doi: 10.1073/pnas.1515561112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez A, Morrone JA, Brini E, MacCallum JL, Dill KA. Blind protein structure prediction using accelerated free-energy simulations. Sci Adv. 2016;2:e1601274. doi: 10.1126/sciadv.1601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 31.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 32.O’Meara MJ, et al. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J Chem Theory Comput. 2015;11:609–622. doi: 10.1021/ct500864r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyka MD, Jung K, Baker D. Efficient sampling of protein conformational space using fast loop building and batch minimization on highly parallel computers. J Comput Chem. 2012;33:2483–2491. doi: 10.1002/jcc.23069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovchinnikov S, et al. Protein structure determination using metagenome sequence data. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Park H, et al. Refinement of protein termini in template-based modeling using conformational space annealing. Proteins. 2011;79:2725–2734. doi: 10.1002/prot.23101. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.