Significance
The diverse environment of cellular membranes presents unique challenges in deciphering the roles that lipids play in modulating membrane protein structure and function. Here, we developed a native mass spectrometry approach to monitor binding of different lipid types to membrane proteins. We discovered that specific lipid−protein interactions can allosterically modulate the binding of lipids of different types. We also determined the structure of AmtB bound to cardiolipin, and mutation of residues involved in binding this lipid abolishes the observed allosteric effect. Our findings are of particular significance as they contribute to our general knowledge of how lipids modulate protein structure and function and how membrane proteins may recruit, through allostery, their own lipid microenvironment.
Keywords: native mass spectrometry, lipids, membrane proteins, lipid−protein interactions, allostery
Abstract
Membrane proteins interact with a myriad of lipid species in the biological membrane, leading to a bewildering number of possible protein−lipid assemblies. Despite this inherent complexity, the identification of specific protein−lipid interactions and the crucial role of lipids in the folding, structure, and function of membrane proteins is emerging from an increasing number of reports. Fundamental questions remain, however, regarding the ability of specific lipid binding events to membrane proteins to alter remote binding sites for lipids of a different type, a property referred to as allostery [Monod J, Wyman J, Changeux JP (1965) J Mol Biol 12:88–118]. Here, we use native mass spectrometry to determine the allosteric nature of heterogeneous lipid binding events to membrane proteins. We monitored individual lipid binding events to the ammonia channel (AmtB) from Escherichia coli, enabling determination of their equilibrium binding constants. We found that different lipid pairs display a range of allosteric modulation. In particular, the binding of phosphatidylethanolamine and cardiolipin-like molecules to AmtB exhibited the largest degree of allosteric modulation, inspiring us to determine the cocrystal structure of AmtB in this lipid environment. The 2.45-Å resolution structure reveals a cardiolipin-like molecule bound to each subunit of the trimeric complex. Mutation of a single residue in AmtB abolishes the positive allosteric modulation observed for binding phosphatidylethanolamine and cardiolipin-like molecules. Our results demonstrate that specific lipid−protein interactions can act as allosteric modulators for the binding of different lipid types to integral membrane proteins.
Studies over the past four decades have shed light on the crucial role of specific lipid−protein interactions in modulating membrane protein structure and function (1–12). Early studies, dating back to the 1980s, have demonstrated that the lipid environment influences the function of the nicotinic acetylcholine receptor from Torpedo wherein cholesterol plays a critical role in promoting function (10–12). Phosphatidylinositol 4,5-bisphosphate, a minor component of the cytoplasmic leaflet, is required for the activation of all inward rectifying potassium channels (13–16), and the sensitivity toward this lipid can be modulated by anionic lipids (17). Additionally, we have recently shown that the activity of the bacterial water channel aquaporin Z (AqpZ) can be modulated threefold by cardiolipin, a lipid interaction that was found by native mass spectrometry (MS) (7). Furthermore, lipids have recently been shown to allosterically modulate protein activity, and protein−ligand and protein−protein interactions (18–22). Despite the subset of examples from the literature presented above, very little is actually known about how lipids influence, on the molecular level, the structure and function of membrane proteins, which will advance our understanding of how lipids participate in a multitude of physiological processes.
Over the past two decades, native MS has emerged as a powerful biophysical technique to study protein structure, dynamics, and ligand interactions (23–30). Native MS affords the ability to analyze intact protein complexes and preserve noncovalent interactions in the mass spectrometer for analysis (28–32). Although MS has been applied to soluble proteins for nearly three decades (31), recent advances have led to the ability to preserve noncovalent interactions and maintain intact, folded membrane proteins in the gas phase, providing invaluable information on subunit stoichiometry, nucleotide, drug, peptide, and lipid binding (7, 33–37). Importantly, the detergent micelle protects membrane proteins from solution into the mass spectrometer (38, 39). In the case of membrane protein−lipid complexes, minimal activation is applied to gently remove the detergent micelle while preserving both native-like structure and noncovalently bound lipids (7, 40–45). When using charge-reducing detergents, such as C8E4, the minimal activation applied to strip the weakly bound detergent micelle from the membrane protein is not sufficient to dissociate lipids bound to membrane proteins, whereas, at much higher activation, lipids can readily dissociate from the membrane protein complex (45). Recently, binding thermodynamics for protein−ligand interactions, including membrane protein−lipid and protein−protein interactions, have been determined using native MS coupled to a temperature-controlled source (18, 42). Importantly, binding thermodynamics determined using other biophysical techniques, such as isothermal titration calorimetry and surface plasmon resonance, are in agreement with those obtained using native MS (18, 42, 46). Taken together, protein−ligand equilibrium and thermodynamic parameters that are in agreement with other biophysical techniques can be obtained by native MS.
One of the greatest modern challenges in membrane protein structural biology is understanding how the chemically diverse environment of the biological lipid membrane modulates membrane protein structure and function. Overwhelmingly, there are, at present, over 40,000 biologically relevant structures in the LIPID Metabolites and Pathways Strategy structure database (47). This striking number is over sevenfold more than the predicted human membrane proteome (48). As a step toward understanding how this diverse environment influences membrane protein structure and function, we report a native MS approach designed to determine the allosteric nature of heterogeneous lipid binding events to integral membrane proteins. We selected the Ammonia channel (AmtB) from Escherichia coli, as it has been previously characterized by native MS, a biophysical technique that preserves noncovalent interactions in the mass spectrometer (28, 29, 31, 32). AmtB forms a trimeric complex of ∼127 kDa with each subunit containing 11 transmembrane helices (49). Native MS studies have revealed that binding of individual lipids of similar type both stabilizes the channel and exhibits unique thermodynamic signatures (7, 40, 42, 45). In particular, from our previous studies, we identified a specific binding site for phosphatidylglycerol (PG), a lipid found to stabilize the channel (7). Mutation of residues involved in PG binding to AmtB resulted in decreased stabilization and different thermodynamic signatures compared with the wild-type channel, supporting the idea of a PG-specific binding site (7, 42). We build upon these previous findings and methods to study the molecular interaction of AmtB with a heterogeneous mixture of lipids—here a mixture of two lipid types or lipid pairs.
Results
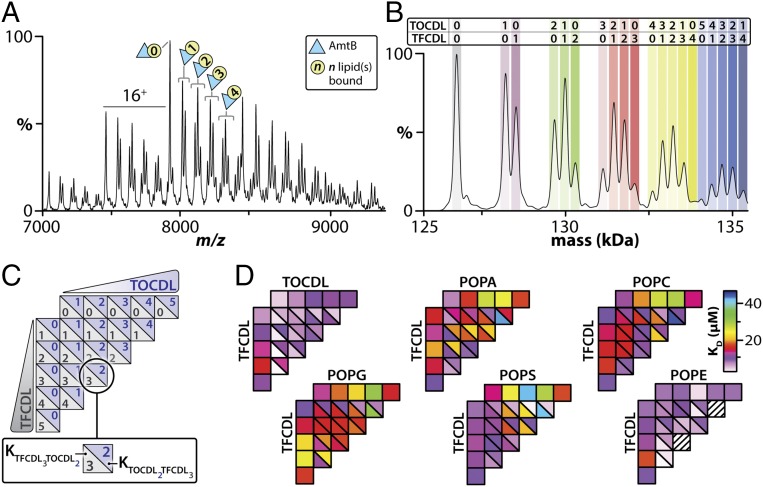
Our first objective for studying heterogeneous lipid binding events to AmtB by native MS was to identify lipids such that multiple binding events for each lipid and combinations thereof could be resolved in the mass spectrum. Using the experimental mass spectral resolution from our previous study as a guide (42), we first considered lipids found in the biological membrane of E. coli (50) and simulated mass spectra for a number of lipid pairs, wherein a lipid pair consists of a mixture of two lipids that differ in chemical composition that can bind to AmtB. From these simulations, we observed significant peak overlap due to insufficient mass resolution (SI Appendix, Fig. S1A). We then simulated mass spectra for nearly 200 commercially available lipid pairs, yielding greater than 19,000 possible combinations (SI Appendix, Fig. S1B). From this dataset, we identified as a top candidate a fluorescent-labeled cardiolipin molecule, TopFluor cardiolipin (TFCDL, 1,1′,2,2′-tetraoleoyl cardiolipin[4-(dipyrrometheneboron difluoride)butanoyl]) (SI Appendix, Fig. S1C), which had the least peak overlap with lipids found in the biological membrane. The modified headgroup of TFCDL provides the necessary mass shift such that peaks can be resolved for this lipid in combination with six other lipids: phosphatidic acid (PA), phosphatidylethanolamine (PE), PG, phosphatidylserine (PS), and phosphatidylcholine (PC) containing 1-palmitoyl-2-oleoyl (PO, 16:0–18:1) tails, and cardiolipin (TOCDL, 1,1′,2,2′-tetraoleoyl-cardiolipin). With the exception of TFCDL and POPC, the remaining five lipids are present in the biological membrane of E. coli (50). In addition to their physiological relevance, the selected lipids for our study have identical acyl chains, with the exception of TFCDL and TOCDL, which contain four oleoyl (18:1) tails (SI Appendix, Fig. S2). TFCDL and TOCDL differ only in their headgroups, where TFCDL contains a BODIPY moiety, a fluorophore, covalently attached to the 3′ hydroxyl in the TOCDL phosphoglycerol headgroup (SI Appendix, Fig. S2).
As a step toward understanding integral membrane protein−lipid interactions in the chemically diverse environment of the lipid bilayer, we performed systematic titrations of mixtures of TFCDL with one of the six “light” lipids (POPA, POPC, POPE, POPG, POPS, or TOCDL), which are lighter in mass compared with TFCDL, with detergent solubilized AmtB followed by recording their native mass spectra at a fixed temperature (42) (Fig. 1A and SI Appendix, Fig. S3). The mole fraction for each combination of lipids bound was determined after deconvoluting the mass spectra from the titration series (Fig. 1B). Notably, we did not observe lipid binding patterns consistent with dimers or multimers of lipids binding to AmtB. The free concentration of the two different lipids was back-calculated, and an equilibrium coupled binding model (SI Appendix, Fig. S4) was fit to the experimental mole fraction data, equating to roughly 750 data points per replicate (SI Appendix, Figs. S5 and S6 and Movie S1–S6). This allowed for the determination of the equilibrium dissociation constants (Kd) for AmtB binding up to a total of five lipids of either TFCDL, light lipid, or combinations thereof (Fig. 1 C and D and SI Appendix, Fig. S7). We first examined the Kd values for sequential binding of the light lipid and found the values to be similar to those from our previous study (42), where only one lipid type was titrated. The sequential binding of TFCDL displayed negative cooperativity for the binding of up to four molecules, followed by positive cooperativity for the fifth binding event. In contrast, binding of AmtB to both TFCDL and a light lipid molecule revealed defined and unique states with higher equilibrium affinity constants for each lipid pair. Mixed species containing both TFCDL and POPA generally possessed a higher Kd than those for sequential binding of the lipid components alone. Moreover, POPC, POPG, and TOCDL demonstrated a preference for binding multiple light lipids and one TFCDL. In the case of POPE, the lowest Kd observed occurred for a species containing four TFCDL and one POPE molecule. In contrast, the lowest Kd observed for POPS occurred when four POPS and one TFCDL were bound to AmtB.
Fig. 1.
Native MS enables the determination of equilibrium binding constants for heterogeneous lipid binding events to AmtB. (A) Native mass spectrum of AmtB at a concentration of 2 µM in the presence of 11 µM TFCDL and 11 µM TOCDL. (B) Deconvolution of the mass spectrum shown in A using UniDec (58). The apo and lipid bound states of TFCDL and TOCDL to AmtB are labeled. (C and D) Equilibrium binding constants for AmtB binding TFCDL and TOCDL, POPA, POPC, POPG, POPS, or POPE. The legend shown in C illustrates the number of TFCDL and TOCDL lipids bound to AmtB per box and the respective Kd value plotted. Hatched lines represent species that were not observed, leading to erroneous values in the fitting routine. Reported are the averages from repeated measurements (n = 3).
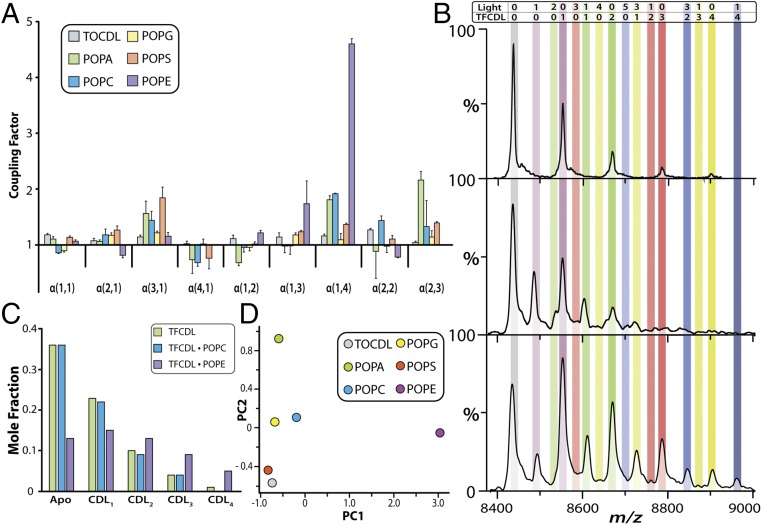
To gain further insight into the trends observed above, we calculated the coupling factor (α), which informs the magnitude of positive, neutral, or negative modulation of ligand affinity (51), for each quadrant in our equilibrium coupled model (Fig. 2A and SI Appendix, Fig. S7B). Given the sensitivity of our native MS approach, we determined coupling factors that spanned a range of values, including positive, neutral, and negative allosteric modulation (Fig. 2A). The allosteric modulation for the TOCDL and TFCDL pair displayed largely neutral allosteric modulation, which is anticipated given the cardiolipin-like nature of TFCDL and, in essence, behaves as a homogeneous lipid pair. POPG, which is essentially half of a CDL molecule, demonstrated a pattern similar to TOCDL where no significant allosteric modulation was observed. POPA contained the largest number of α values less than 1, indicating negative allostery or a decrease in ligand affinity. For POPA, there is a twofold reduction in Kd when binding a mixture of TFCDL and POPA compared with binding of four POPA molecules. The largest positive allosteric modulation we observed was for POPE and TFCDL, which steadily increased along the direction of binding one POPE and multiple TFCDL molecules, peaking at a total of five bound lipids in this study (Fig. 1).
Fig. 2.
Different lipid pairs binding to AmtB exhibit distinct coupling factors. (A) Coupling factors (α) for AmtB binding x light lipid and y TFCDL species (αx,y). Reported are the mean and SEM from repeated measurements (n = 3). (B) The 15+ charge state region is shown for AmtB titrated with TFCDL (Top), TFCDL and POPC (Middle), and TFCDL and POPE (Bottom). The mole ratio of TFCDL to AmtB is 3:1, and POPC and POPE are at a mole ratio of 1.5 and 1, respectively, to AmtB. The number of TFCDL and POPC or POPE bound is shown as in Fig. 1B. (C) Mole fraction for TFCDL bound to AmtB determined from deconvolution of mass spectra shown in B. (D) PCA of the coupling factors for different lipid pairs binding to AmtB.
Coinciding with the calculation of coupling factors, we inspected individual mass spectra recorded for the titration of different lipid pairs. AmtB in the presence of TFCDL at a 4:1 molar ratio (TFCDL:AmtB) binds up to four TFCDL molecules (Fig. 2B). The addition of POPC at a molar ratio of 3:1.5:1 (TFCDL:POPC:AmtB) did not enhance TFCDL binding (Fig. 2B). In contrast, AmtB in the presence of 3:1 TFCDL:POPE resulted in a dramatic enhancement of TFCDL binding, with up to four TFCDL molecules bound to AmtB (Fig. 2 B and C). In summary, native MS captures snapshots of solution equilibria and clearly demonstrates the potent allosteric effect observed for the POPE and TFCDL pair binding to AmtB.
We next performed a principal component analysis (PCA) on the average coupling factors for different lipid pairs binding to AmtB, to identify patterns (Fig. 2D) (52). The lipid pair that exhibited the largest allosteric modulation, TFCDL and POPE, was separated the farthest from the five other light lipids by the first principal component (PC1). In contrast, the remaining lipids clustered around −0.5 for PC1 but were separated by the second principal component (PC2) into three groups: POPA; POPC and POPG; and TOCDL and POPS. POPA displayed the greatest negative allosteric modulation, providing an explanation for its separation from the other groups on the PC2 axis.
To test the specificity of TFCDL and POPE binding to AmtB, we acquired native mass spectra of AqpZ, a tetrameric integral membrane protein from E. coli, bound to TFCDL and mixtures of lipids (SI Appendix, Fig. S8). AqpZ in the presence of TFCDL at a 3:1 molar ratio (TFCDL:AqpZ) bound up to two TFCDL molecules. The addition of POPE at a molar ratio 3:3:1 (POPE:TFCDL:AqpZ) did not enhance TFCDL binding as observed for AmtB. The mass spectrum for AqpZ in the presence of 3:3 TFCDL:POPC was similar to the mixture for TFCDL and POPE. These results provide additional support for the positive allosteric modulation observed for AmtB binding TFCDL and POPE.
To gain structural insight into the potent allosteric effect observed for TFCDL and POPE, we set out to determine the crystal structure of AmtB in this lipid environment. Although we screened different ratios of AmtB:TFCDL:POPE, the best diffracting crystals grew in a 1:3:1.5 molar ratio. The crystals were light brown, in accord with an enrichment of TFCDL and quenching of the fluorophore (SI Appendix, Fig. S9A). The 2.45-Å resolution structure of AmtB is composed of one subunit in the asymmetric unit cell (Fig. 3). One TFCDL lipid was resolved in the electron density, which had strong positive density (σ > 10) in the difference density map (Fo − Fc) after molecular replacement (SI Appendix, Fig. S9F). TFCDL is located on the periplasmic facing side of AmtB, and, through crystallographic symmetry, the lipid is bound to each subunit within the trimeric assembly (SI Appendix, Fig. S9B), which is distinct from the site determined for PG (7). We did not observe electron density for POPE, likely owing to the small molar ratio present in solution. The overall structure of AmtB bound to TFCDL displayed an all-atom RMSD of 0.5 ± 0.1 Å compared with the other AmtB structures deposited in the Protein Data Bank (PDB). Despite this similarity, structural rearrangements were observed for a number of residues that form distinct contacts with TFCDL. Hydrogen bonds were formed to both the sidechain and backbone amide of N73 from the phosphate moiety on the TFCDL phosphoglycerol bridge (Fig. 3B). An additional hydrogen bond is formed to H156 by the sn2 ester linkage of TFCDL where the indole ring is precisely positioned through interaction with the sidechain of E357 on a neighboring helix. This H156·E357 interaction is unique among the other 21 structures for AmtB currently in the PDB. In comparison with the structure of AmtB bound to PG (PDB 4NH2), E357 rotates ∼150° and shifts ∼3.6 Å toward H156 (SI Appendix, Fig. S9E). Three of the four 18:1 tails of TFCDL were resolved and are arranged within the transmembrane spanning surface of AmtB, forming an interface area of 655 Å2 for the AmtB·TFCDL molecular interaction (Fig. 3C). The boron-dipyrromethane (BODIPY) moiety attached to the headgroup of TFCDL is positioned away from the protein such that the neutral moiety is packed against the lipid tails, where it does not form direct contacts with AmtB (SI Appendix, Fig. S9D). The covalent modification of TFCDL removes the ability for an additional hydrogen bond donation that would occur in CDL; however, the position of this ester group is not in proximity to form any potential hydrogen bonds with AmtB.
Fig. 3.
Crystal structure of AmtB Bound to TFCDL. (A) Top view of the periplasmic face of AmtB (Left) and parallel view (Right) to the transmembrane portion of the trimeric AmtB·TFCDL3 assembly. Structure is shown in surface representation, with secondary structure shown in the top view. TFCDL is shown as a surface representation, with the headgroup (phosphoglycerol bridge) colored light red and the acyl chains in white. (B) Molecular interactions formed between TFCDL and AmtB. Hydrogen bonds are shown as yellow dashed lines. (C) A large interacting surface (dark blue) is generated by TFCDL bound to AmtB. TFCDL is shown as sticks and colored as in A. The BODIPY moiety of TFCDL has been omitted for clarity but is shown intact in SI Appendix, Fig. S9.
In an effort to gain insight into the allosteric mechanism for TFCDL and POPE, we analyzed AmtB containing the His156 to Ala mutation (AmtBH156A), a residue that interacts with TFCDL and forms a distinct interaction with E357. Equilibrium binding constants for AmtBH156A interacting with TFCDL, POPC, or POPE were determined in a similar fashion to the lipid pairs investigated for the wild-type channel. Interestingly, the Kd for TFCDL in the presence of POPC or POPE significantly increased, nearly tripling in some cases (SI Appendix, Table S2). Equilibrium binding affinities for POPC and POPE bound to AmtBH156A were not affected by the H156A mutation, as these values were similar to the wild-type protein. Moreover, the potent allosteric modulation observed for the wild-type protein binding TFCDL and POPE was completely abolished for AmtBH156A (Fig. 4).
Fig. 4.
Mutation of His156 to Ala in AmtB (AmtBH156A) diminishes TFCDL binding and abolishes the allosteric effect for binding TFCDL and POPE. (A) Mass spectra for AmtBH156A with TFCDL, TFCDL and POPC, and TFCDL and POPE as labeled. (B) Zoomed regions for the AmtBH156A 15+ charge state illustrating reduced POPE and TFCDL binding compared with WT AmtB. The mole ratio of TFCDL to AmtBH156A is 3:1. POPC and POPE were analyzed at a mole ratio of 1.5 and 1 to AmtB, respectively. Shown as described in Fig. 2B.
Discussion
Allostery is a biological phenomenon that underlies the regulation of macromolecular structure and function and has been identified in numerous biological processes, such as cellular signaling, transcriptional control, and disease (53–56). Here, we have shown that this biological phenomenon extends to lipid−protein interactions using AmtB as a model membrane protein system. We observed differences for AmtB binding to lipid pairs that differ only in their headgroup composition (Fig. 1). The six light lipids, which differ from each other in molecular weight by only a few daltons (SI Appendix, Fig. S2), display markedly different equilibrium binding constants that can be captured using native MS. In addition, PCA of the coupling factors (Fig. 2D) reveals four distinct patterns for the six different lipid pairs, providing additional support for the specificity of lipid binding events to AmtB. The allostery observed for TFCDL and POPE appears to be specific, as binding enhancement for this lipid pair to AqpZ, a different integral membrane protein, was not observed. These findings elucidate the preferential binding of lipids to AmtB and, more importantly, highlight the role of allostery as the underlying mechanism of action that modulates the affinity of lipid binding.
The largest positive allosteric modulation observed was for POPE and TFCDL. This observation is most intriguing, as PE accounts for around 75% of the total lipid composition in E. coli (57). POPC, which differs from POPE by three methyl groups, shared some similarity to POPE, albeit with a decreased magnitude of allosteric modulation (Fig. 1). In comparison with POPE, the additional methyl groups on the choline headgroup of POPC could impose steric restrictions that weaken or abolish the interaction of the polar headgroup with AmtB. These observed allosteric results demonstrate the highly specific nature of lipid−protein interactions and illustrate the exquisite sensitivity of native MS for uncovering specific protein−lipid interactions.
Complementary to our native MS results, the crystal structure of AmtB bound to TFCDL provides molecular details on specific lipid−protein interactions and insight into the mechanism of allostery (Fig. 3). Interestingly, the BODIPY moiety attached to the headgroup of TFCDL does not form direct contacts with AmtB. Instead, the fluorophore is positioned away from the protein such that the neutral moiety is packed against the lipid tails (SI Appendix, Fig. S9B). This result implies that TFCDL is an effective mimic of CDL, and the observed allosteric effect reported here for TFCDL and POPE is anticipated to hold if TFCDL is substituted with other CDL molecules. This is further supported by AmtBH156A, which harbors a point mutant engineered to disrupt CDL binding, that exhibits reduced binding affinity to TFCDL and abolishes the allosteric effect for the TFCDL·POPE pair (Fig. 4). Given the impact of the H156A mutation, we speculate that H156 fluctuates between two states observed crystallographically: (i) interacting with E357, as in our crystal structure, or (ii) not interacting with E357, as found in other AmtB structures. Lipid binding could allosterically modulate the H156·E357 interaction, which could order the specific CDL binding site and enhance the apparent binding affinity, providing a plausible mechanism for the allostery observed.
In closing, we demonstrate that lipid−protein interactions can, indeed, allosterically modulate remote binding sites for lipids of different type. In our study, all lipid pairs contained a TFCDL, and we foresee that higher mass resolution instruments (35) will enable the opportunity to study natural lipid pairs, such as POPE with other PO-type lipids, as well as more complex lipid mixtures. It is likely that interaction with additional lipids, beyond two different lipids and/or five total lipids bound, will reveal additional insight into protein−lipid interactions, such as amplification of the observed allosteric effects and the adoption of biologically useful conformations. We anticipate allosteric coupling of lipid−protein interactions to be a general phenomenon observed in other membrane proteins. Lastly, our data provide compelling evidence that allosteric protein−lipid interactions could be utilized in the biological membrane to effectively recruit a defined lipid microenvironment.
Methods
Protein Expression and Purification.
The Ammonia channel (AmtB) from E. coli was expressed (7, 42) and purified (18) as previously described. Detailed methods are provided in SI Appendix, Supporting Methods.
Preparation and Titration of Phospholipids.
Phospholipid samples were prepared as previously described (42). Lipid stocks were diluted such that both light and heavy lipids were mixed with AmtB to yield samples for native MS analyses (SI Appendix, Supporting Methods).
Native MS Analysis.
Native MS was performed on a Synapt G1 HDMS instrument (Waters Corporation) equipped with a 32k RF generator. Additional details are provided in SI Appendix, Supporting Methods.
Automated Data Processing.
To handle the large volume of data as well as interpretation of complex mass spectra, we developed a pipeline for automated data processing. In brief, MS data files were deconvoluted in batch using UniDec software (58) with the following settings: no smoothing, m/z range 6,000 to 12,500, charge range 10 to 25, mass sampling of 10 Da, and peak FWHM of 10. A custom Python script was written to import the zero-charge mass spectrum outputted from UniDec and extract the intensities for apo and various lipids bound species followed by conversion to mole fraction. For each titration experiment, the free light lipid can be back calculated as follows:
| [1] |
and the free concentration of the heavy lipid can be calculated as
| [2] |
where F represents the mole fraction of a particular species. The total protein ([P]total) was determined with the DC Protein Assay kit (Bio-Rad) using BSA as the standard. The extracted mole fraction data and free concentration of lipids were saved and used for determination of equilibrium binding constants.
Determination of Lipid Equilibrium Binding Constants.
Analysis of MS data obtained for AmtB and AmtB bound to different lipid species was performed using UniDec software packages (58) and in-house scripts written in Python programming language. The intensities of AmtB (P) and AmtB bound to light (L) or heavy (H) lipid species were converted to mole fraction for a given lipid titration. Apparent equilibrium binding constants (KA) for protein−lipid species were obtained using the following equilibrium coupled binding model (SI Appendix, Fig. S4). For protein binding one light or heavy lipid,
| [3] |
or binding to multiple ligands,
| [4] |
where n is the number of bound light lipids, and KA,Ln represents the equilibrium association binding constant for the nth lipid binding to AmtB. A similar derivation is made for binding m number of heavy lipids. For heterogeneous lipid binding events, the binding to light lipid is first calculated using Eqs. 1 and 2 followed by binding to m number of heavy lipids,
| [5] |
The total protein ([P]total) in the system can be expressed as follows:
| [6] |
After substitution of Eqs. 1–3 into Eq. 4 and rearrangement, the mole fraction of P bound to n light lipids and/or m heavy lipids can be calculated,
| [7] |
| [8] |
The equilibrium association constants for each species PLnHm were determined by global fitting of the equilibrium coupled binding model to the experimental mole fraction (FPLnHm) data through minimization of the pseudoχ2 function,
| [9] |
where n and m are the number of light and heavy bound lipids, and d is the total number of data points.
The coupling factors (αx,y) for an x number of light and y number of heavy lipids bound were calculated using the calculated equilibrium association constants (KA),
| [10] |
AmtB Crystallization and Structure Determination.
AmtB in [50 mM Tris, pH 7.4 at room temperature, 130 mM sodium chloride, 10% (wt/vol) glycerol and 0.5% (wt/vol) C8E4] was concentrated to 12 mg/mL. Initial cocrystallization trials were carried out using Mosquito LCP (TTP Labtech) crystallization robot in hanging drop plates at 20 °C for 1 mg/mL to 12 mg/mL (8 μM to 95 µM) of AmtB combined with POPE and TFCDL in molar excess of 1:1.5:3, 1:3:3, and 1:3:1.5, respectively. The best diffracting crystals grew within 2 d to 3 d at 10 mg/mL (79 µM) AmtB mixed with 1:1.5:3 molar excess of POPE and TFCDL in crystallization solution 0.1 M Tris, pH 8.0, 0.3 M magnesium nitrate, 22% (wt/vol) PEG 8000 at a 1:1 sample to mother liquor ratio. No cryoprotection was needed, as determined by screening on an in-house source. Single crystals were mounted with CrystalCap HT Cryoloops (Hampton Research) before being flash frozen. Diffraction data were collected at the Advanced Photon Source (Argonne National Laboratory) on beamline 24-ID-E. The data were integrated, merged, and scaled using X-ray detector software (59) in space group H32 to a resolution of 2.45 Å. Initial phases were found using molecular replacement using the high-resolution, apo form of AmtB (PDB 1U7G) (49). Model refinement and building was performed using Phenix (60) and Crystallographic Object-Oriented Toolkit (61) programs, respectively. All structural figures were rendered in PyMOL (62). The atomic coordinates have been deposited into the PDB with the accession number 6B21.
Supplementary Material
Acknowledgments
We thank Dr. David Russell (Texas A&M University) for useful discussion. This work was supported by new faculty startup funds from the Institute of Biosciences & Technology, Texas A&M Health Science Center, and the Department of Chemistry, Texas A&M University, and support from National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) (DP2GM123486). Part of this work was conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS from NIH (P41 GM103403), and NIH Office of Research Infrastructure Programs High-End Instrumentation Grant (S10OD021527). This research used resources of the Advanced Photon Source under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6B21).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719813115/-/DCSupplemental.
References
- 1.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 2.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 3.Lee AG. Biological membranes: The importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Contreras FX, Ernst AM, Wieland F, Brügger B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb Perspect Biol. 2011;3:a004705. doi: 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanov M, Dowhan W, Vitrac H. Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta. 2014;1843:1475–1488. doi: 10.1016/j.bbamcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poveda JA, et al. Lipid modulation of ion channels through specific binding sites. Biochim Biophys Acta. 2014;1838:1560–1567. doi: 10.1016/j.bbamem.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saliba AE, Vonkova I, Gavin AC. The systematic analysis of protein-lipid interactions comes of age. Nat Rev Mol Cell Biol. 2015;16:753–761. doi: 10.1038/nrm4080. [DOI] [PubMed] [Google Scholar]
- 9.Landreh M, Marty MT, Gault J, Robinson CV. A sliding selectivity scale for lipid binding to membrane proteins. Curr Opin Struct Biol. 2016;39:54–60. doi: 10.1016/j.sbi.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Criado M, Eibl H, Barrantes FJ. Effects of lipids on acetylcholine receptor. Essential need of cholesterol for maintenance of agonist-induced state transitions in lipid vesicles. Biochemistry. 1982;21:3622–3629. doi: 10.1021/bi00258a015. [DOI] [PubMed] [Google Scholar]
- 11.Fong TM, McNamee MG. Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry. 1986;25:830–840. doi: 10.1021/bi00352a015. [DOI] [PubMed] [Google Scholar]
- 12.Baenziger JE, Hénault CM, Therien JP, Sun J. Nicotinic acetylcholine receptor-lipid interactions: Mechanistic insight and biological function. Biochim Biophys Acta. 2015;1848:1806–1817. doi: 10.1016/j.bbamem.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 14.Rohács T, et al. Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc Natl Acad Sci USA. 2003;100:745–750. doi: 10.1073/pnas.0236364100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara Y, Kubo Y. Regulation of the desensitization and ion selectivity of ATP-gated P2X2 channels by phosphoinositides. J Physiol. 2006;576:135–149. doi: 10.1113/jphysiol.2006.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng WW, D’Avanzo N, Doyle DA, Nichols CG. Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys J. 2011;100:620–628. doi: 10.1016/j.bpj.2010.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong X, Liu Y, Liu W, Liang X, Laganowsky A. Allosteric modulation of protein-protein interactions by individual lipid binding events. Nat Commun. 2017;8:2203. doi: 10.1038/s41467-017-02397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebner M, Lucic I, Leonard TA, Yudushkin I. PI(3,4,5)P3 engagement restricts Akt activity to cellular membranes. Mol Cell. 2017;65:416–431.e416. doi: 10.1016/j.molcel.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh M, et al. Lipid-mediated regulation of embedded receptor kinases via parallel allosteric relays. Biophys J. 2017;112:643–654. doi: 10.1016/j.bpj.2016.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong L, Tombola F. Allostery: A lipid two-step. Nat Chem Biol. 2016;12:202–203. doi: 10.1038/nchembio.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swamy M, et al. A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity. 2016;44:1091–1101. doi: 10.1016/j.immuni.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Katta V, Chait BT. Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 24.Ganem B, Li YT, Henion JD. Detection of noncovalent receptor-ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 25.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Pacholarz KJ, Garlish RA, Taylor RJ, Barran PE. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem Soc Rev. 2012;41:4335–4355. doi: 10.1039/c2cs35035a. [DOI] [PubMed] [Google Scholar]
- 27.Marcoux J, Robinson CV. Twenty years of gas phase structural biology. Structure. 2013;21:1541–1550. doi: 10.1016/j.str.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Dyachenko A, Gruber R, Shimon L, Horovitz A, Sharon M. Allosteric mechanisms can be distinguished using structural mass spectrometry. Proc Natl Acad Sci USA. 2013;110:7235–7239. doi: 10.1073/pnas.1302395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cubrilovic D, et al. Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry. ACS Chem Biol. 2014;9:218–226. doi: 10.1021/cb4007002. [DOI] [PubMed] [Google Scholar]
- 30.Gülbakan B, Barylyuk K, Zenobi R. Determination of thermodynamic and kinetic properties of biomolecules by mass spectrometry. Curr Opin Biotechnol. 2015;31:65–72. doi: 10.1016/j.copbio.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Hilton GR, Benesch JL. Two decades of studying non-covalent biomolecular assemblies by means of electrospray ionization mass spectrometry. J R Soc Interface. 2012;9:801–816. doi: 10.1098/rsif.2011.0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng L, Kitova EN, Klassen JS. Dissociation kinetics of the streptavidin-biotin interaction measured using direct electrospray ionization mass spectrometry analysis. J Am Soc Mass Spectrom. 2013;24:49–56. doi: 10.1007/s13361-012-0533-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcoux J, et al. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc Natl Acad Sci USA. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson CV. From molecular chaperones to membrane motors: Through the lens of a mass spectrometrist. Biochem Soc Trans. 2017;45:251–260. doi: 10.1042/BST20160395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reading E, et al. The role of the detergent micelle in preserving the structure of membrane proteins in the gas phase. Angew Chem Int Ed Engl. 2015;54:4577–4581. doi: 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- 39.Rouse SL, Marcoux J, Robinson CV, Sansom MS. Dodecyl maltoside protects membrane proteins in vacuo. Biophys J. 2013;105:648–656. doi: 10.1016/j.bpj.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehmood S, et al. Charge reduction stabilizes intact membrane protein complexes for mass spectrometry. J Am Chem Soc. 2014;136:17010–17012. doi: 10.1021/ja510283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong X, et al. Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J Am Chem Soc. 2016;138:4346–4349. doi: 10.1021/jacs.6b01771. [DOI] [PubMed] [Google Scholar]
- 43.Allison TM, Landreh M, Benesch JLP, Robinson CV. Low charge and reduced mobility of membrane protein complexes has implications for calibration of collision cross section measurements. Anal Chem. 2016;88:5879–5884. doi: 10.1021/acs.analchem.6b00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liko I, Hopper JT, Allison TM, Benesch JL, Robinson CV. Negative ions enhance survival of membrane protein complexes. J Am Soc Mass Spectrom. 2016;27:1099–1104. doi: 10.1007/s13361-016-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Cong X, Liu W, Laganowsky A. Characterization of membrane protein-lipid interactions by mass spectrometry ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2017;28:579–586. doi: 10.1007/s13361-016-1555-1. [DOI] [PubMed] [Google Scholar]
- 46.Daneshfar R, Kitova EN, Klassen JS. Determination of protein-ligand association thermochemistry using variable-temperature nanoelectrospray mass spectrometry. J Am Chem Soc. 2004;126:4786–4787. doi: 10.1021/ja0316972. [DOI] [PubMed] [Google Scholar]
- 47.Sud M, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 49.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 50.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 51.Hulme EC, Trevethick MA. Ligand binding assays at equilibrium: Validation and interpretation. Br J Pharmacol. 2010;161:1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lever J, Krzywinski M, Altman N. Points of significance: Principal component analysis. Nat Methods. 2017;14:641–642. [Google Scholar]
- 53.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 54.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai CJ, Nussinov R. A unified view of “how allostery works.”. PLoS Comput Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Nussinov R. Allostery: An overview of its history, concepts, methods, and applications. PLoS Comput Biol. 2016;12:e1004966. doi: 10.1371/journal.pcbi.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raetz CR. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev. 1978;42:614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marty MT, et al. Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of COOT. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrödinger . The Pymol Molecular Graphics System. Version 1.8. Schrödinger; New York: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.