Significance
Our ability to reduce stable small molecules, such as dinitrogen or carbon dioxide, is as vital as it is demanding and requires energetic electrons and a catalyst. In nature, these requirements are met by two-component enzymes: an electron-donating metallo-ATPase and the principal catalyst, a metalloprotein with a low-potential cofactor. Here, we present a two-component enzyme in which the catalyst houses a double-cubane type [Fe8S9]-cluster. Iron–sulfur clusters with so high nuclearity were so far only known from nitrogenase, an enzyme notorious for its capacity to reduce various small molecules. The enzyme not only shares structural features with nitrogenase, but is also able to reduce acetylene, indicating its potential employment for reductive reactions of our choice.
Keywords: Fe/S-cluster, nitrogenase, acetylene, ATPase, electron transfer
Abstract
Chemically demanding reductive conversions in biology, such as the reduction of dinitrogen to ammonia or the Birch-type reduction of aromatic compounds, depend on Fe/S-cluster–containing ATPases. These reductions are typically catalyzed by two-component systems, in which an Fe/S-cluster–containing ATPase energizes an electron to reduce a metal site on the acceptor protein that drives the reductive reaction. Here, we show a two-component system featuring a double-cubane [Fe8S9]-cluster [{Fe4S4(SCys)3}2(μ2-S)]. The double-cubane–cluster-containing enzyme is capable of reducing small molecules, such as acetylene (C2H2), azide (N3−), and hydrazine (N2H4). We thus present a class of metalloenzymes akin in fold, metal clusters, and reactivity to nitrogenases.
Biological redox reactions occur at a wide spectrum of reduction potentials. Some vital reactions take place at the perimeters of reduction potentials achievable in water, extending biological reactivity space to the limits set by the environment. Among these reactions are central processes, such as the oxidation of water to dioxygen or the reduction of dinitrogen to ammonia. These and related biochemical reactions are outstanding for two reasons: first they are coupled to energy-converting reactions in which light energy or the chemical energy stored in ATP are converted into reactive electrons/electron holes, and second they occur at complex metal clusters, together enabling valuable chemical transformations.
ATP-driven redox reactions are typically catalyzed by enzymes consisting of two components. The first component is an ATPase containing an Fe/S-cluster on which the electron to be transferred is resting. The second component, also a metalloenzyme, is the electron acceptor in need of a highly energetic electron because it contains one or more metal centers, which cannot be reduced at physiological reduction potentials. Three principal classes of electron-energizing ATPases are currently known: the nitrogenase-like proteins, the RACE (reductive activator of corrinoid enzymes)-type activators, and the benzoyl-CoA reductase/atypical dehydratase activators (1–3). The nitrogenase system represents the most extensively studied example for ATP-dependent electron transfer, not only because it was discovered first, but also because of the relevance of biological dinitrogen fixation and its parallels to the Haber–Bosch process. Additionally, nitrogenases catalyze several nonphysiological reductions, including the potentially very useful reduction and C-C coupling of CO to small hydrocarbons, opening the field to new applications (4–9). Moreover, homologs of nitrogenases act in different branches of tetrapyrroll biosynthesis, indicating that the same type of ATPase-coupled electron transfer may be used for very different chemical reactions (1, 10, 11). RACE-type activators reduce corrinoid-containing enzymes, which are difficult to reductively reactivate once they drop into the inactive Co(II) oxidation state (2, 12, 13). Finally, there are the ATPases driving electron transfers to reduce benzoyl-CoA or to trigger an atypical β,α-dehydration of 2-hydroxyacyl-CoA esters (3). These ATPases share with the first group the general architecture, with an [Fe4S4]-cluster placed in the dimer interface, likely triggering electron transfer by conformational changes.
We have characterized the two components of a widespread system of the third type and find that the electron-accepting component features a double-cubane [Fe8S9]-cluster. This [Fe8S9]-cluster, so far unknown to biology, catalyzes reductive reactions otherwise associated only with the complex iron–sulfur clusters of nitrogenases. Our results reveal several parallels between the double-cubane cluster-containing enzymes and nitrogenases and suggest that an unexplored biochemical reactivity space may be hidden among the diverse ATP-dependent two-component enzymes.
Results
Distribution of Double-Cubane Cluster Protein-Like Proteins.
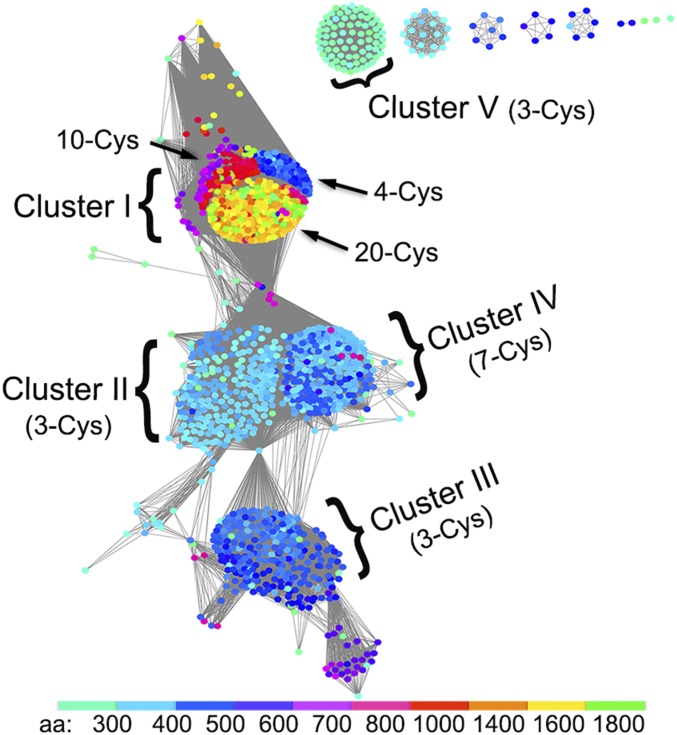
We analyzed InterPro entry IPR010327 (https://www.ebi.ac.uk/interpro/) that includes the protein families of benzoyl-CoA reductase B/C subunit (BcrB/C) and dehydratase α/β subunit (HadB/C). Sequences of this entry are widely distributed and found in the genome of various bacteria and archaea, including firmicutes, spirochaetes, actinobacteria, enterobacteria, deltaproteobacteria, and euryarchaeota. A sequence similarity network analysis of all 1,436 sequences reveals potential separation into five larger clusters (Fig. 1). Cluster I includes proteins of varying length (420 and 1,800 aa), where the longer proteins appear to be fusions between a dehydratase-type α/β unit and an ATP-dependent activator. Cluster I may be subdivided into three major subclusters, distinguished by length and number of conserved Cys-motifs (Fig. 1). Sequences in cluster II are ∼380 aa long and contain the α-subunits of benzoyl-CoA reductases (BcrC O87874) and the β-subunits of atypical dehydratases (HadC Q5U923), whose catalytic partner-subunits (BcrB O87875; HadB Q5U924), usually ∼410–430 aa, are clearly separated into cluster III. Both clusters II and III share a conserved 3-Cys motif, binding an [Fe4S4]-cluster with one open coordination site (14). Cluster IV contains proteins with 380–420 aa. Regardless of protein length, all sequences of cluster IV share a highly conserved 7-Cys motif (CX19CX18–19CX29CX166–170CX32–34CX32C) (Fig. S3B). A small isolated cluster (cluster V) features proteins with ∼330 aa sharing a strictly conserved 3-Cys motif.
Fig. 1.
Sequence similarity analysis. InterPro entry IPR010327 was analyzed using the EFI-EST server (36) with an alignment score of 20, visualized with Cytoscape (v3.3.0) (37) and colored by protein length (color code below). Distinct clusters (I−V) are indicated together with the number of conserved cysteine residues.
We investigated the properties of a 7-Cys-protein from cluster IV, encoded by CHY_0487 from Carboxydothermus hydrogenoformans Z-2901. In contrast to the 2-hydroxyacyl-CoA dehydratases and benzoyl-CoA reductases, which are encoded by two homologous genes forming α/β-dimers, no homologous protein is found near CHY_0487. Instead, CHY_0487 is located in an operon with only one other gene (CHY_0488), annotated to encode an ATP-dependent activator for CoA-specific enzymes, consistent with an ATP-dependent two-component system. In the following, the proteins encoded by CHY_0487 and CHY_0488 are called DCCPCh (double-cubane cluster protein from C. hydrogenoformans) and DCCP-RCh (DCCP-reductase). Both proteins were heterologously expressed, purified, and characterized.
Biochemical Properties.
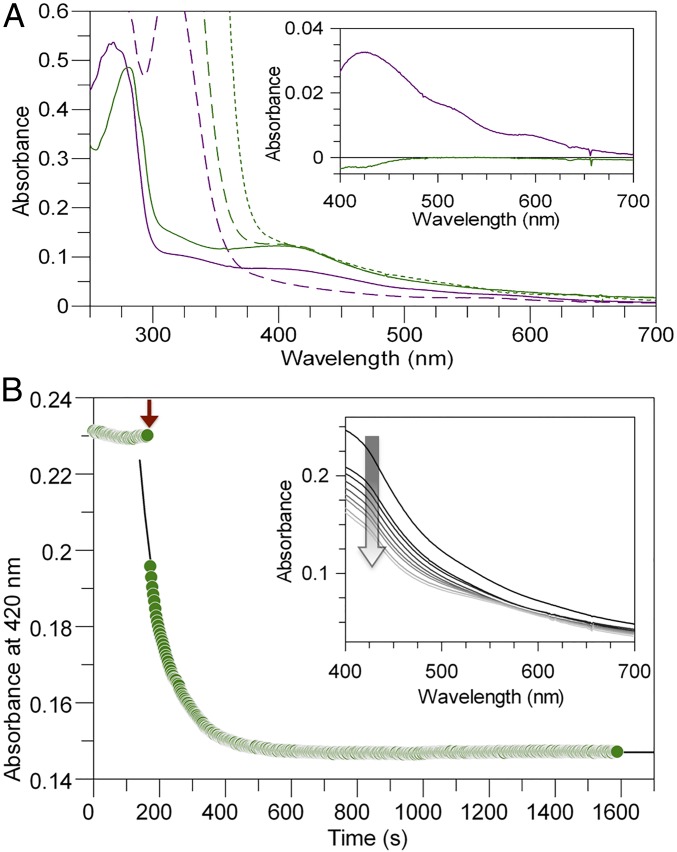
DCCPCh and DCCP-RCh were purified under strictly anoxic conditions by strep-tactin affinity chromatography. Both proteins show the brown color typically associated with Fe/S-proteins and, after addition of thionin, exhibit spectral features associated with oxidized [Fe4S4]-clusters, with a shoulder around 320 nm and a broad peak at 420 nm (Fig. 2A). We added an excess of reducing agents Na-dithionite (DT, E0′ ∼ −0.430 V at pH 7.0) and Ti(III)-citrate to investigate the reduced states. While the spectrum of DCCP-RCh shows the characteristic decrease in absorption between 400 and 700 nm, the spectrum of DCCPCh did not change, indicating a very low reduction potential for the Fe/S-cluster on DCCPCh.
Fig. 2.
(A) UV-visible spectra. Solid lines show spectra of thionin-oxidized proteins [6 µM DCCPCh (green) and 28 µM DCCP-RCh (purple)]. Excess of DT and Ti(III)-citrate was added to oxidized samples, generating the dashed-line spectra (DCCP and DCCP-R) and dotted-line spectrum (DCCP only), respectively. (Inset) Difference spectra of oxidized to DT-added proteins. Absorption increased at ∼300–375 nm after excess addition of the reductants, because DT and Ti(III)-citrate absorb in this region. (B) ATP-dependent reduction. The assay mixture contained DCCPCh and prereduced DCCP-RCh in 50 mM Hepes-NaOH pH 8.0 with 2 mM DT. The red arrow indicates the start of DCCPCh reduction by addition of 2 mM Mg-ATP, recorded at 420 nm (green circle). The black solid-line shows a monoexponential fit to the time-dependent absorption. (Inset) Spectral changes over time. The arrow indicates decreasing absorbance at 420 nm.
The Fe/S-cluster on DCCPCh can be reduced by DT-reduced DCCP-RCh upon ATP-addition (Fig. 2B). When we add 2 mM Mg-ATP in the presence of prereduced DCCP-RCh, absorption of DCCPCh (1.6:1 = DCCP-RCh:DCCPCh) around 420 nm decreases. We approximated the absorbance change at 420 nm by a monoexponential function with an observed rate constant of 0.66 min−1 at 25 °C. When DCCP-RCh was supplied substoichiometrically (0.5:1 = DCCP-RCh:DCCPCh), we observed a similar amplitude, but substantially slower reduction, indicating the same extent of DCCPCh reduction at lower rate.
Next, we examined the ATP hydrolysis activity of DCCP-RCh. The ATPase activity of DCCP-RCh was determined at different conditions (Fig. S1A). The ATPase activity of oxidized DCCP-RCh is 30 ± 1.8 nmol/min/mgDCCP-R (kobs = 0.84 min−1) at 35 °C and was not changed when DCCP-RCh was reduced with DT. In contrast, when DCCPCh was present in the assay buffer, the ATPase activity increased almost threefold (83.2 ± 5.3 nmol/min/mgDCCP-R; kobs = 2.4 min−1, n = 3).
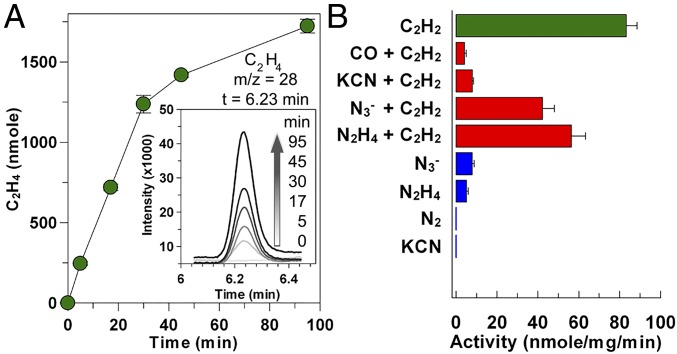
We tested the reductase activity of DCCPCh by following its ability to reduce acetylene. Acetylene reduction was strictly dependent on both the presence of Mg-ATP and reduced DCCP-RCh in the reaction, while in the absence of DCCPCh no acetylene reduction was observed. Reduction of acetylene reached 89.5 ± 5.9 nmol C2H4/min/mgDCCP (kobs = 4.32 ± 0.3 min−1, n = 6) with a molar DCCPCh:DCCP-RCh ratio of 1:4 at 35 °C in 50 mM Hepes-NaOH at pH 8.0 using 40% (vol/vol) acetylene in gas phase (Fig. 3A). High concentrations of KCl (100 mM) decreased acetylene reduction by ∼20% (Fig. S1B), similar to what has been observed for nitrogenase (15). Acetylene reduction was highest at 45 °C (Fig. S1C) and pH 8.0 (Fig. S1D). In the absence of an ATP-regeneration system, the rate of reduction dropped to approximately 10% of that at optimal conditions. Acetylene reduction is influenced by the molar ratio of DCCPCh to DCCP-RCh and upon going from a 1:4 to a 1:10 ratio, the rate doubled. Furthermore, the ATPase activity of reduced DCCP-RCh increased by 32% when, in addition to DCCPCh, acetylene was also present in the assay buffer (from 83.2 ± 5.3 to 117.8 ± 6 nmol/min/mgDCCP-R; kobs = 3.18 min−1) (Fig. S1A).
Fig. 3.
Reductive activities of DCCPCh. (A) Progress curve for reduction of acetylene (C2H2) to ethylene (C2H4). Time-dependent intensity profile derived from the mass spectra for ethylene. Area of intensity at each time was converted into amount of ethylene by a standard curve. (B) Reduction of acetylene (green bar) and its inhibition (red bars) were determined by quantifying ethylene production. The potential reduction of nitrogenous compounds (blue bars) was detected by NH3 production.
Acetylene reduction by DCCPCh was inhibited by several small molecules (Fig. 3B). The presence of 10 mM KCN decreased the activity to 10%, N3− to 50% and N2H4 to 60%. While neither the presence of 100% (vol/vol) N2 nor of 60% (vol/vol) H2 influence the acetylene reduction activity of DCCPCh, the presence of 60% (vol/vol) CO in the atmosphere decreased the activity to ∼8%. Inhibition by CO was reversible and activity was fully recovered when the reaction atmosphere was changed back to N2 and acetylene (Fig. S2A).
The reduction of small nitrogenous compounds was followed by detecting ammonia formation. Under our assay conditions, DCCPCh was not able to reduce N2 to ammonia when supplied with a pure N2 atmosphere, while time-dependent ammonia production was detected from N2H4 and N3− (Fig. 3B and Fig. S2B). DCCPCh produced 5.1 ± 0.8 nmol NH3/min/mgDCCP from hydrazine and 7.8 ± 0.9 nmol NH3/min/mgDCCP from azide at 35 °C (n = 4).
Other potential reactions were tested, but no turnover was detected under the conditions used (20 µM DCCPCh with 80 µM DCCP-RCh at 35 °C): neither was H2 formed from water, nor ethane from either acetylene (four-electron reduction) or ethylene (two-electron reduction). In addition acetaldehyde, a product from acetylene hydroxylation (16), was not detected.
Structure of DCCPCh.
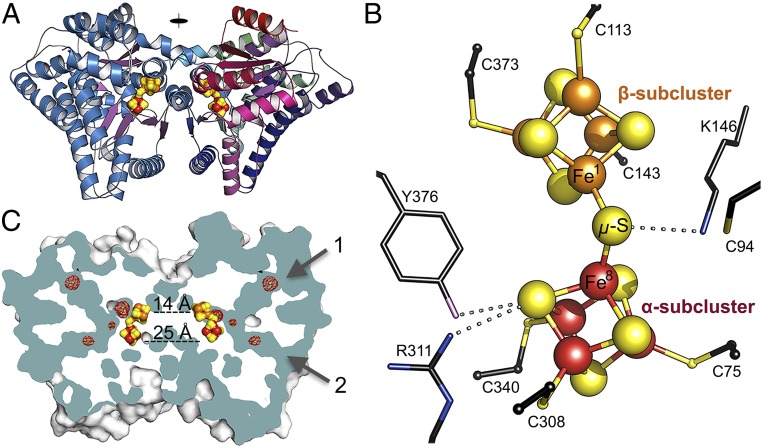
DCCPCh was crystallized under strictly anoxic conditions. The crystal structure was solved by single-wavelength anomalous diffraction phasing using the anomalous signals of the iron ions and refined to 1.63-Å resolution (Table S1). One subunit is found in the asymmetric unit of which a homodimeric assembly is created by twofold crystallographic symmetry (Fig. 4A), consistent with the homodimeric solution state (Fig. S8C). The subunits employ 13% of their overall accessible surface area (34,686 Å2) to form the dimer interface, consisting primarily of three helices H10-H19*, H19-H10* and H11-H11* (an asterisk denotes the symmetry mate) (see Fig. S3A for helix numbering). The subunit contains two domains: an N-terminal (1–193 aa) and a C-terminal (194–420 aa) domain, harboring an Fe/S-cluster between them (Fig. S3A). Each domain has a central β-sheet of four parallel β-strands surrounded on both sides by α-helices, resembling a Rossmann-fold. N- and C-terminal domains can be superimposed with an rmsd of 2.8 Å for Cα-atoms (Fig. S4A), with a sequence identity of 22.3% based on structure superimposition, indicating homology between both domains.
Fig. 4.
Crystal structure of DCCPCh. (A) Overall dimeric structure of DCCPCh at dmin = 1.63 Å. One subunit is depicted by blue α-helices and violet β-strands, while the other subunit is colored from green (N terminus) via blue to red (C terminus). Iron ions in the two subclusters are shown in different colors, red (α-subcluster) and orange (β-subcluster). Sulfur atoms are colored in yellow. (B) DCC. Residues within H-bonding distance (<3.5 Å; dotted lines) to the DCC are shown. N, O, and C atoms are colored blue, purple, and black, respectively. (C) Surface and channel representation. Arrows 1 and 2 indicate two channels (cyan) per subunit. Xenon atoms are indicated as green spheres with red mesh for their anomalous scattering at 5 σ. Nearest distances between the clusters are indicated by dashed lines.
DCCPCh contains an unusual iron–sulfur cluster. We used the anomalous scattering of iron and sulfur atoms for unambiguous identification of an [Fe8S9] cluster [{Fe4S4(SCys)3}2(μ2-S)] composed of two juxtaposed [Fe4S4]-clusters, termed a double -cubane cluster (DCC) for simplicity (Fig. 4B). Six of the eight iron ions are coordinated by Sγ atoms of conserved cysteine residues 75, 113, 143, 308, 340, and 373 (Fig. 4B and Fig. S5). The other two Fe ions (Fe1 and Fe8) are the nearest iron atoms between the two subclusters (Fe1-Fe8 distance of 3.74 Å) and share a μ2-ligand. Fe-ligand bond lengths as well as anomalous scattering (Fig. 4C) agree with a sulfide ion (μ2-S) as the bridging ligand, but a chloride ion can’t be ruled out. The two [Fe4S4]-subclusters are twisted relative to each other, likely to prevent intercluster repulsion (distance of 4.0 Å between S2 and S7) (Fig. S5). The two [Fe8S9]-clusters in the homodimer have shortest Fe-Fe distances of 14 Å (Fig. 4C). The α-subcluster is more deeply buried than the β-subcluster, with the latter being approximately 10 Å below the protein surface close to the twofold symmetry axis of the dimer. In the second coordination sphere of the DCC, we find Arg311 and Tyr376, which interact with S6 in the α-subcluster and a highly conserved Lys146 close to the bridging S9 (μ2-S) (Fig. 4B and Fig. S5).
Exchange of the conserved Cys94 to Ala increased acetylene reductase activity 3.5-fold, while exchanging Lys146 to Ala diminished the activity fivefold. Thus, both exchanges clearly point to the DCC as the place of acetylene reduction (Fig. 4B and Fig. S3B).
Two channels converge at the α-subcluster of the DCC (Fig. 4C). One channel is predominantly lined by hydrophobic residues (channel 1), while the other is filled with solvent molecules and surrounded by polar and charged side chains (channel 2). Both channels have diameters of less than ∼4 Å with a path length maximum of ∼20 Å. In Xe-pressurized DCCPCh crystals, the channels take up three of four Xe atoms (Fig. 4C).
A search for structurally similar proteins in the protein data bank using DALI (17) revealed the highest structural similarity of DCCPCh with the β-subunit followed by the α-subunit of 2-hydroxyisocaproyl-CoA dehydratase. DCCPCh also shows similarity to Ni,Fe-CO dehydrogenase and Mo,Fe-nitrogenase (Fig. S4 B–D). The arrangement of two Rossmann-fold domains of DCCPCh around the DCC is similar to the folds around the [Fe4S4]-clusters of 2-hydroxyisocaproyl-CoA dehydratase, the C-cluster of CODH and the P-cluster of Mo,Fe-nitrogenase. Superimpositions, although only calculated for the Cα-atoms also align the metal clusters.
As DCCPCh (CHY_0487) is annotated as a CoA-specific dehydratase family protein, we followed a titration of DCCPCh with CoA by isothermal titration calorimetry under strictly anoxic conditions, in which we could not observe a signal indicative of binding (Fig. S8A). This is consistent with the DCCPCh structure: although channel 1 of DCCPCh superimposes on the isocaproyl-CoA binding channel in the α-subunit of 2-hydroxyisocaproyl-CoA dehydratase (Fig. S6), it appears to be too narrow (diameter maximum of ∼4 Å) to accommodate a CoA molecule, indicating that the physiological substrate of DCCPCh is most probably not a CoA-ester.
Discussion
So far, a biological function for DCCPCh and DCCP-RCh has not been reported for any organism. DCCPCh shares the conserved 7 Cys-motif (CX19CX18–19CX29CX166–170CX32–34CX32C) with many enzymes (currently 499 in InterPro entry IPR010327), including YjiM from Escherichia coli. YjiM is encoded in an operon, which is up-regulated when E. coli switches from aerobic to anaerobic growth under control of the FNR (fumarate and nitrate reductase) transcriptional regulator (18). The other gene in the small operon encodes the putative ATP-dependent activator of YjiM, named YjiL, whose function has been linked to MdtM, a multidrug resistance transporter for efflux of antibiotics from E. coli (19). Their up-regulation under anaerobic conditions in E. coli may indicate a general function of DCCPs within the anaerobic metabolism.
All proteins joining DCCPCh in cluster IV (Fig. 1) share a common sequence motif of seven Cys residues, six of which are coordinating the [Fe8S9]-cluster and the seventh is present next to the cluster (Cys94 in Fig. 4B). As the sequence motif consists of all cluster coordinating Cys residues, it is very likely that all proteins will contain the same DCC. Therefore, we propose the name “double-cubane cluster proteins” (DCCPs).
Although double-cubane [Fe8S9] clusters have not been reported before for a biological system, they are well known to synthetic inorganic chemistry (20–23). The first example of a sulfide-bridged DCC was reported in 1989 by Stack, Carney, and Holm, who already speculated that these Fe/S-clusters may also occur in proteins (20). In the meantime, several variants of the DCC were reported, exploiting that the bridging μ2-S ligand is labile and replaceable by negatively charged small molecules (20, 21, 24, 25). The DCC of DCCPCh superimposes well on the inorganic DCCs (21–23) (Fig. S7), making inorganic DCCs valuable models for the active site of DCCPs.
Inorganic DCCs display a peculiar redox chemistry. Unless two [Fe4S4]-clusters are in very close proximity, they are only weakly coupled and show similar or identical midpoint reduction potentials (20). In inorganic DCCs, electron transfer is strongly coupled with two distinct reversible redox transitions at very low redox regime (< −1.0 V), which are separated by ∼190–220 mV (20, 22). Thus, DCCs react not as two individual [Fe4S4]-clusters, but due to strong coupling as a single entity.
In contrast to the inorganic DCCs, the two [Fe4S4]-subclusters of DCCPCh have different environments, likely modulating their reduction potentials. The β-subcluster rests in a hydrophobic pocket, while the α-subcluster is in hydrogen-bonding distance to H-donors in the protein matrix, which may make it easier to reduce than the β-subcluster (Fig. 4B and Fig. S5). As indicated by the UV-vis spectra (Fig. 2A), DCCPCh was resistant to reduction by sodium dithionite and Ti(III)-citrate, suggesting a low midpoint-potential for DCC reduction (< −600 mV). Thus, to reduce the DCC the ATP-driven electron transfer from DCCP-RCh is needed (Fig. 2B).
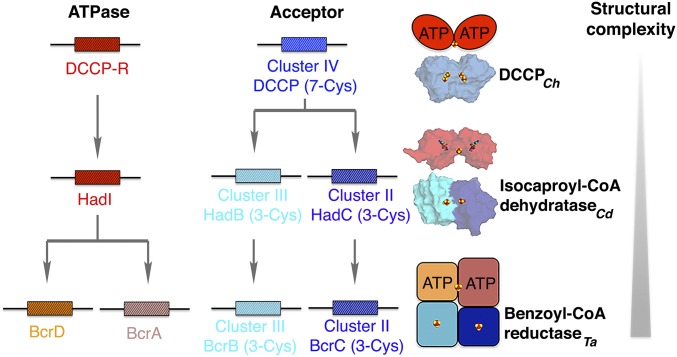
DCCPs are homologous to the ATP-dependent atypical dehydratases and benzoyl-CoA dehydratase (Fig. 1). Notably, DCCPs appear to be the most primitive members of this enzyme family. DCCPCh, as well as DCCP-RCh, are homodimers, whereas the atypical dehydratase isocaproyl-CoA dehydratase is a heterodimer of two homologous subunits, likely derived from a gene-duplication event (Fig. 5). Another gene-duplication, here of the gene encoding the ATPase, occurred in the evolution of the benzoyl-CoA reductases from Thauera aromatica and Azoarcus evansii, increasing the structural complexity (26) (Fig. 5).
Fig. 5.
Architecture of DCCP homologs. Structural genes are indicated as boxes with distinct colors. Crystal structures of isocaproyl-CoA dehydratase (HadB and HadC; PDB ID code 3O3M) (14) and its activator (HadI; PDB ID code 4EHU) (42) are indicated. Benzoyl-CoA reductase is modeled as suggested in ref. 26. Fe/S-clusters are shown as spheres with iron in red and sulfur in yellow. AMPPNP bound in the dehydratase activator is shown as sticks. Cd: Clostridium difficile; Ta: Thauera aromatica.
DCCPCh not only shares similarities with the DCCP homologs, but has obvious parallels in structure and reactivity to nitrogenases, including activities so far only associated with nitrogenases (Table 1). A biological two-electron reduction of acetylene to ethylene was so far only known for nitrogenases, where it is also used as a marker for biological nitrogen fixation (27). Although DCCPCh reduces acetylene with lower rate than MoFe- and VFe-nitrogenase (Table 1), it is more active than FeFe-nitrogenase and NifEN, a maturation factor of nitrogenase, which is homologous to mature nitrogenase (Table 1) (28–30). In contrast to nitrogenases, nitrogenous compounds seem to be poor substrates for DCCPCh (Table 1).
Table 1.
Specific substrate-reducing activities of DCCPCh in comparison with other enzymes
| Substrate (product) | DCCPCh | Nitrogenases | NifEN† | DPOR | BCR | ||
| MoFe | VFe* | FeFe | |||||
| C2H2 (C2H4) | 169‡ | 2,144 | 1,007 | 28 | 52 | 0 | — |
| CN− (NH3) | 0 | 53 | — | — | 5 | — | — |
| N2 (NH3) | 0 | 1,112 | 822 | 38 | 0 | 0 | — |
| N3− (NH3) | 7.8 | 587 | — | — | 71 | 1.2 | 63 |
| N2H4 (NH3) | 5.1 | 1,145 | — | 0 | 1.07 | — | |
| H+ (H2)§ | 0 | 2,490 | 3,104 | 258 | 0 | — | — |
| Source | Present work | (30) | (28) | (29) | (30) | (43) | (44) |
Specific activity is shown in nmol product min−1 (milligram protein)−1. DPOR, dark-operative protochlorophyllide oxidoreductase; BCR, benzoyl-CoA reductase; NifEN, nitrogenase FeMo cofactor maturase/insertase.
Activity was measured at 45 °C.
Activity was measured in the presence of Fe-protein as a reductase.
H2 evolution activities were measured under Ar.
Activities were recalculated from nmol product min−1 (nmol protein)−1 to the specific activity using the molecular mass 135 kDa of VFe-nitrogenase (αβδ2) (28).
Acetylene reduction by nitrogenase was suggested to start by binding to the iron-molybdenum cofactor (FeMoCo). Thereby, acetylene may transiently act as a bridging ligand between two adjacent Fe-ions, ∼3.9 Å apart, before becoming reduced and protonated (31). In DCCPCh, Fe1-Fe8 are 3.74 Å apart, are facing the potential substrate channel, and are only bridged by a μ2-S ligand. In analogy to nitrogenase, we speculate that during catalysis the μ2-S ligand may dissociate allowing acetylene to bind to the vacant coordination site between Fe1-Fe8, initiating its activation and subsequent reduction.
But DCCPCh and nitrogenase do not only convert similar substrates, they are both reversibly inhibited by CO. CO inhibits MoFe-nitrogenase noncompetitively and binds between two iron atoms, replacing a bridging μ2-S ligand of the M cluster (32, 33). Interestingly, although CO strongly inhibits acetylene reduction of DCCPCh, activity is fully recovered after removal of CO. As inhibition by CO likely involves its binding to the [Fe8S9]-cluster, it is tempting to assume that nitrogenase and DCCPCh undergo similar inhibition reactions, namely the replacement of a labile μ2-S ligand. Notably, CO is slowly reduced by MoFe- and V-nitrogenase, forming small hydrocarbons by C-C bond formation (4–9). However, this reactivity is detected at scaled-up and optimized assays with high nitrogenase concentrations (6). Thus, although we did not observe conversion of CO so far, we currently can’t rule out a weak CO reduction activity of DCCPCh.
DCCPCh shares with nitrogenases not only the ATP-dependent ability to reduce acetylene and hydrazine, but also common structural motifs. Specifically, the arrangement of two Rossmann-fold domains with similar orientation, both contributing coordinating Cys residues for an [Fe8Sx]-cluster (DCC: x = 9; P-cluster: x = 7), indicates similar environments for both clusters (Fig. S4D). But there may be even an older link. The FeMoCo as well as the P-cluster of nitrogenase are thought to be formed from two closely spaced, paired [Fe4S4]-clusters, resembling DCCs (34). The P-cluster is produced on the nitrogenase scaffold by action of NifH from its precursor P*-cluster (35). Excitingly, in contrast to the mature P-cluster, the P*-cluster is catalytically active and catalyzes the reduction of acetylene and hydrazine, as well as the reduction of CO and CN to alkanes and alkenes (35). This led to the speculation that the P*-cluster is homologous to the active site of a primordial enzyme capable of reducing simple carbon compounds from which the FeMoCo, P*- and P-clusters in nitrogenase may have evolved. Is the more primitive DCCP family even more closely related to this elusive primordial enzyme?
Independent of their early evolution, the potential catalytic space of DCCPs makes them very attractive for further developments and applications, and given the wide spread and variety of DCCPs (Fig. 1), an abundance of reductive reactivities, analogous to those of the nitrogenase family, waits to be uncovered.
Materials and Methods
Detailed materials and methods are provided in SI Materials and Methods.
Cloning, Expression, and Purification.
Genes CHY_0487 and CHY_0488 for DCCPCh and DCCP-RCh, respectively, were amplified from the genomic DNA of C. hydrogenoformans Z-2901 by PCR and cloned into a vector containing a strep-tag. DCCPCh and DCCP-RCh were expressed anaerobically in E. coli Rosetta(DE3). The proteins were purified to homogeneity by affinity chromatography.
Network Analysis.
InterPro entry IPR010327 was analyzed using enzyme function initiative-enzyme similarity tool (EFI-EST) server (36) with an alignment score of 20. Protein networks were visualized using Cytoscape (v3.3.0) (37).
ATPase Activity of DCCP-RCh.
The malachite green assay (38) was used to determine the amount of inorganic phosphate (Pi) released during hydrolysis of ATP catalyzed by DCCP-RCh.
ATP-Dependent Reduction of DCCPCh and DCCP-RCh.
ATP-dependent reduction of DCCPCh by DCCP-RCh was initiated by adding Mg-ATP and monitoring the decrease of absorption at 420 nm. The observed reduction rate constant was determined by fitting the time-dependent absorption to a single exponential equation.
Acetylene Reduction Activity.
Acetylene reduction was initiated in a calibrated 7-mL reaction tube sealed with a butyl septum and a screw cap in a shaking water-bath. The reaction solution with a volume of 1 mL contains a 1:4 molar ratio of DCCPCh to DCCP-RCh with DT as electron donor and an ATP regeneration mix of creatine phosphokinase and phosphocreatine. The gas phase contains 40% (vol/vol) acetylene in N2 atmosphere. Acetylene reduction was initiated by adding Mg-ATP. The product of the two-electron reduction of acetylene, ethylene, was detected by a GC/MS-QP2010 ultra equipped with a Carboxene-1010 PLOT column (Shimadzu Europa) using helium as carrier gas.
Ammonia Detection.
Ammonia was quantified using a fluorescence method described previously (39) with some modifications (40).
X-Ray Crystallography.
Crystals of DCCPCh were obtained by hanging drop vapor diffusion under anoxic condition. Diffraction data were collected at the beamline 14.1 (BESSY, Berlin, Germany) (41). The structure of DCCPCh was solved by SAD phasing using anomalous diffraction data collected at the Fe edge (1.74 Å) and was refined to dmin = 1.63 Å.
Supplementary Material
Acknowledgments
We thank Rainer Dietrich and Sabine Niklisch for excellent technical support, and Dr. Kathryn A. Pérez for advice on ammonia detection. The authors acknowledge funding through the German excellence initiative (EXC 314 – “Unifying concepts in Catalysis – UniCat”).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6ENO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720489115/-/DCSupplemental.
References
- 1.Hu Y, Ribbe MW. Nitrogenase and homologs. J Biol Inorg Chem. 2015;20:435–445. doi: 10.1007/s00775-014-1225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennig SE, Jeoung JH, Goetzl S, Dobbek H. Redox-dependent complex formation by an ATP-dependent activator of the corrinoid/iron-sulfur protein. Proc Natl Acad Sci USA. 2012;109:5235–5240. doi: 10.1073/pnas.1117126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckel W, Hetzel M, Kim J. ATP-driven electron transfer in enzymatic radical reactions. Curr Opin Chem Biol. 2004;8:462–467. doi: 10.1016/j.cbpa.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, Ribbe MW. Nitrogenases—A tale of carbon atom(s) Angew Chem Int Ed Engl. 2016;55:8216–8226. doi: 10.1002/anie.201600010. [DOI] [PubMed] [Google Scholar]
- 5.Lee CC, Hu Y, Ribbe MW. Catalytic reduction of CN-, CO, and CO2 by nitrogenase cofactors in lanthanide-driven reactions. Angew Chem Int Ed Engl. 2015;54:1219–1222. doi: 10.1002/anie.201410412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Lee CC, Ribbe MW. Extending the carbon chain: Hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science. 2011;333:753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seefeldt LC, Yang Z-Y, Duval S, Dean DR. Nitrogenase reduction of carbon-containing compounds. Biochim Biophys Acta. 2013;1827:1102–1111. doi: 10.1016/j.bbabio.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CC, Hu Y, Ribbe MW. Vanadium nitrogenase reduces CO. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Lee CC, Ribbe MW. Vanadium nitrogenase: A two-hit wonder? Dalton Trans. 2012;41:1118–1127. doi: 10.1039/c1dt11535a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng K, Ngo PD, Owens VL, Yang X-P, Mansoorabadi SO. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science. 2016;354:339–342. doi: 10.1126/science.aag2947. [DOI] [PubMed] [Google Scholar]
- 11.Moore SJ, et al. Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature. 2017;543:78–82. doi: 10.1038/nature21427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson T, Soares JA, Lienard T, Gottschalk G, Krzycki JA. RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea. J Biol Chem. 2009;284:2285–2295. doi: 10.1074/jbc.M807392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilhabel A, et al. The ether-cleaving methyltransferase system of the strict anaerobe Acetobacterium dehalogenans: Analysis and expression of the encoding genes. J Bacteriol. 2009;191:588–599; erratum in J Bacteriol 2009 191:1994. doi: 10.1128/JB.01104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knauer SH, Buckel W, Dobbek H. Structural basis for reductive radical formation and electron recycling in (R)-2-hydroxyisocaproyl-CoA dehydratase. J Am Chem Soc. 2011;133:4342–4347. doi: 10.1021/ja1076537. [DOI] [PubMed] [Google Scholar]
- 15.Deits TL, Howard JB. Effect of salts on Azotobacter vinelandii nitrogenase activities. Inhibition of iron chelation and substrate reduction. J Biol Chem. 1990;265:3859–3867. [PubMed] [Google Scholar]
- 16.Kroneck PMH. Acetylene hydratase: A non-redox enzyme with tungsten and iron-sulfur centers at the active site. J Biol Inorg Chem. 2016;21:29–38. doi: 10.1007/s00775-015-1330-y. [DOI] [PubMed] [Google Scholar]
- 17.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krizsan A, Knappe D, Hoffmann R. Influence of the yjiL-mdtM gene cluster on the antibacterial activity of proline-rich antimicrobial peptides overcoming Escherichia coli resistance induced by the missing SbmA transporter system. Antimicrob Agents Chemother. 2015;59:5992–5998. doi: 10.1128/AAC.01307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stack T, Carney MJ, Holm RH. Formation of bridged [4Fe-4S]2+ double cubanes by site-specific reactions: Electron-transfer coupling across sulfur-containing bridges of variable length. J Am Chem Soc. 1989;111:1670–1676. [Google Scholar]
- 21.Challen PR, Koo SM, Dunham WR. New μ2-S2--coupled, singly bridged double cubane with the [(Fe4S4Cl3)2S]4- core. The stepwise synthesis and structural characterization of (n-Bu4N)2(Ph4P)2. J Am Chem Soc. 1990;112:2455–2456. [Google Scholar]
- 22.Terada T, et al. Tridentate thiolate ligands: Application to the synthesis of the site-differentiated [4Fe-4S] cluster having a hydrosulfide ligand at the unique iron center. Chem Asian J. 2012;7:920–929. doi: 10.1002/asia.201200039. [DOI] [PubMed] [Google Scholar]
- 23.Gerlach DL, Coucouvanis D, Kampf J, Lehnert N. Isolation and characterization of single and sulfide-bridged double [4Fe-4S] cubane clusters with 4-pyridinethiolato ligands. Eur J Inorg Chem. 2013;2013:5253–5264. [Google Scholar]
- 24.Venkateswara Rao P, Holm RH. Synthetic analogues of the active sites of iron-sulfur proteins. Chem Rev. 2004;104:527–559. doi: 10.1021/cr020615+. [DOI] [PubMed] [Google Scholar]
- 25.Lee SC, Lo W, Holm RH. Developments in the biomimetic chemistry of cubane-type and higher nuclearity iron-sulfur clusters. Chem Rev. 2014;114:3579–3600. doi: 10.1021/cr4004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckel W, Kung JW, Boll M. The benzoyl-coenzyme A reductase and 2-hydroxyacylcoenzyme A dehydratase radical enzyme family. ChemBioChem. 2014;15:2188–2194. doi: 10.1002/cbic.201402270. [DOI] [PubMed] [Google Scholar]
- 27.Somasegaran P, Hoben H. Handbook for Rhizobia: Methods in Legume-Rhizobium Ttechnology. Springer; New York: 1994. [Google Scholar]
- 28.Lee CC, Hu Y, Ribbe MW. Unique features of the nitrogenase VFe protein from Azotobacter vinelandii. Proc Natl Acad Sci USA. 2009;106:9209–9214. doi: 10.1073/pnas.0904408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chisnell JR, Premakumar R, Bishop PE. Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. J Bacteriol. 1988;170:27–33. doi: 10.1128/jb.170.1.27-33.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, et al. Catalytic activities of NifEN: Implications for nitrogenase evolution and mechanism. Proc Natl Acad Sci USA. 2009;106:16962–16966. doi: 10.1073/pnas.0907872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H-I, et al. Characterization of an intermediate in the reduction of acetylene by the nitrogenase α-Gln 195MoFe protein by Q-band EPR and 13C, 1H ENDOR. J Am Chem Soc. 2000;122:5582–5587. [Google Scholar]
- 32.Cameron LM, Hales BJ. Unusual effect of CO on C2H2 reduction by V nitrogenase from Azotobacter vinelandii. J Am Chem Soc. 1996;118:279–280. [Google Scholar]
- 33.Spatzal T, Perez KA, Einsle O, Howard JB, Rees DC. Ligand binding to the FeMo-cofactor: Structures of CO-bound and reactivated nitrogenase. Science. 2014;345:1620–1623. doi: 10.1126/science.1256679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribbe MW, Hu Y, Hodgson KO, Hedman B. Biosynthesis of nitrogenase metalloclusters. Chem Rev. 2014;114:4063–4080. doi: 10.1021/cr400463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CC, Hu Y, Ribbe MW. ATP-independent substrate reduction by nitrogenase P-cluster variant. Proc Natl Acad Sci USA. 2012;109:6922–6926. doi: 10.1073/pnas.1202429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerlt JA, et al. Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 39.Corbin JL. Liquid chromatographic-fluorescence determination of ammonia from nitrogenase reactions: A 2-min assay. Appl Environ Microbiol. 1984;47:1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spatzal T, Perez KA, Howard JB, Rees DC. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor. eLife. 2015;4:e11620. doi: 10.7554/eLife.11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller U, et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur Phys J Plus. 2015;130:141. [Google Scholar]
- 42.Knauer SH, Buckel W, Dobbek H. On the ATP-dependent activation of the radical enzyme (R)-2-hydroxyisocaproyl-CoA dehydratase. Biochemistry. 2012;51:6609–6622. doi: 10.1021/bi300571z. [DOI] [PubMed] [Google Scholar]
- 43.Moser J, et al. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc Natl Acad Sci USA. 2013;110:2094–2098. doi: 10.1073/pnas.1218303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.