Significance
Oxygenic phosphorylation entails the transfer of electrons from organic substrates to molecular oxygen by four large protein complexes in the mitochondrial inner membrane. Loss of electrons during this process can produce toxic side products and must be prevented. Three of the electron-transfer complexes form supercomplexes, which are thought to be instrumental in channeling electrons from the substrate to the acceptor. We used electron cryo-tomography and subtomogram averaging to determine the in situ structure and organization of the respiratory chain supercomplexes in three different eukaryotic lineages. We discovered that the mutual arrangement of the two largest components—complex I and complex III2—is essentially the same in all supercomplexes, indicating that this arrangement is important for electron transfer.
Keywords: respirasomes, respiratory chain supercomplexes, cryo-electron tomography, subtomogram averaging, mitochondria
Abstract
We used electron cryo-tomography and subtomogram averaging to investigate the structure of complex I and its supramolecular assemblies in the inner mitochondrial membrane of mammals, fungi, and plants. Tomographic volumes containing complex I were averaged at ∼4 nm resolution. Principal component analysis indicated that ∼60% of complex I formed a supercomplex with dimeric complex III, while ∼40% were not associated with other respiratory chain complexes. The mutual arrangement of complex I and III2 was essentially conserved in all supercomplexes investigated. In addition, up to two copies of monomeric complex IV were associated with the complex I1III2 assembly in bovine heart and the yeast Yarrowia lipolytica, but their positions varied. No complex IV was detected in the respiratory supercomplex of the plant Asparagus officinalis. Instead, an ∼4.5-nm globular protein density was observed on the matrix side of the complex I membrane arm, which we assign to γ-carbonic anhydrase. Our results demonstrate that respiratory chain supercomplexes in situ have a conserved core of complex I and III2, but otherwise their stoichiometry and structure varies. The conserved features of supercomplex assemblies indicate an important role in respiratory electron transfer.
Mitochondria are the main providers of ATP in nonphotosynthetic eukaryotes. The inner mitochondrial membrane contains five large protein complexes: NADH-dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome c reductase (complex III), cytochrome c oxidase (complex IV), and ATP synthase (sometimes referred to as complex V). The first four complexes form the respiratory chain, which transfers electrons from the electron donors NADH, FADH, and succinate to the final electron acceptor O2 (1, 2). In the course of electron transfer, complexes I, III, and IV pump protons from the matrix into the crista lumen, generating an electrochemical proton gradient. The ATP synthase uses the proton gradient to produce ATP by rotary catalysis (3–5).
In the respiratory chain, electrons pass from complex I and II to complex III via the hydrophobic electron carrier ubiquinol and from complex III to complex IV via cytochrome c, a 12-kDa soluble protein. The structures of individual respiratory chain complexes from mitochondria have been determined by X-ray crystallography (6–10) or electron cryo-microscopy (cryo-EM) (11–13). Complex I is an assembly of 44 different protein subunits in bovine or human mitochondria, 42 in the obligate aerobic Yarrowia lipolytica, and 49 in the plant Arabidopsis thaliana (13, 14). Together, the subunits form an L-shaped complex with a 20-nm membrane arm and a matrix arm that extends 12 nm from the membrane surface. The membrane arm carries out proton translocation, while the matrix arm is the site of electron transfer (15). Complex III is a 496-kDa dimeric multisubunit membrane protein that protrudes 8 nm into the matrix (6). The complex IV monomer has a mass of 204 kDa and extends by 4 nm from the membrane on both the lumenal and the matrix side (7). Bovine complex IV crystallizes as a dimer (7, 16, 17), but is likely to occur both as a monomer and a dimer in the membrane (18, 19).
Supramolecular assemblies, or supercomplexes, of respiratory chain complexes were first identified by blue-native gel electrophoresis (BN-PAGE) of detergent-solubilized inner mitochondrial membranes (20). Supercomplexes containing complexes I, III2, and IV are sometimes referred to as respirasomes. Negative-stain electron microscopy of samples extracted from gel bands yielded initial 3D maps at low resolution (21–23). The structures of supercomplexes I1III2 and I1III2IV isolated from bovine, porcine, and ovine mitochondria have been determined by single-particle electron cryo-EM at resolutions ranging from ∼30 Å to ∼4 Å (24–27) or by electron cryo-tomography (cryo-ET) at ∼30 Å resolution (28). In these structures, complex III2 borders the concave arc of the complex I membrane arm, and complex IV is located near complex III2 at the distal end of the complex I membrane arm.
Although the existence of supercomplexes in purified detergent extracts is now beyond doubt, the extent to which they exist in the mitochondrial membrane is under debate, as is the mutual arrangement of respiratory chain complexes in situ and its functional relevance. To address these questions, we applied cryo-ET and subtomogram averaging to mitochondrial membranes of three different eukaryotic lineages. We show that, in all three cases, complex I forms a conserved assembly with dimeric complex III. In mammals and fungi, up to two copies of monomeric complex IV are attached to the I1III2 core, but there appears to be no complex IV in the plant supercomplex. Principal component analysis indicated that a variety of supercomplexes can coexist in the same membrane. Our results demonstrate that supercomplexes are prevalent in the inner mitochondrial membrane, but their structure and composition varies between species and even within one species. Their conserved structural features signify an important role in respiratory electron transfer.
Results
Complex I Is Visible in Tomographic Volumes.
Respiratory chain supercomplexes were visualized in situ by cryo-ET of mitochondrial membranes from bovine heart, the yeast Y. lipolytica, and the plant Asparagus officinalis. To improve contrast, matrix proteins of bovine and yeast mitochondria were removed by osmotic shock, while asparagus mitochondria underwent spontaneous disruption. Membrane fragments were resuspended in buffer before cryo-EM grid preparation.
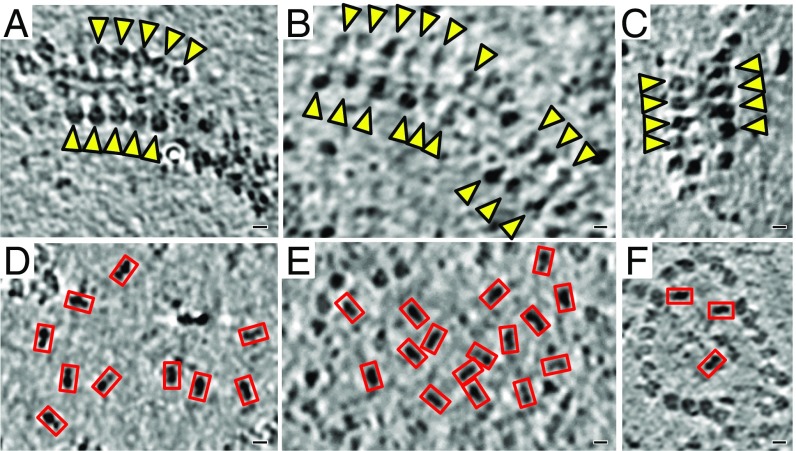
Lipid bilayer membranes were clearly visible in the tomographic volumes, and individual ATP synthase and complex I molecules in the inner membrane were easily identified by their shape (Fig. 1 and Fig. S1). When viewed parallel to the membrane, ATP synthase had the characteristic lollipop appearance of a 10-nm spherical head connected to the membrane by a thin 5-nm stalk (29–31), whereas the matrix arm of complex I had an angular, rhomboid profile (Fig. S1A) (31). Parallel slices at a distance of 10 nm above the membrane surface indicated a near-hexagonal shape for the ATP synthase F1 head and a bar-shaped density for the complex I matrix arm (Fig. S1B), as expected from the cross-sections and density envelopes of ATP synthase and complex I structures determined by subtomogram averaging or single-particle cryo-EM, Fourier-filtered to ∼25 Å resolution (Fig. S1 C–F). In tomographic slices at this level, only densities that were consistent with either the ATP synthase F1 head or the complex I matrix arm were observed. Mitochondrial ATP synthases form double rows in the inner membrane (Fig. 1 A–C) (30, 31), whereas the densities assigned to complex I are randomly distributed in flat membrane regions (Fig. 1 D–F). Complex I and ATP synthase can thus be distinguished unambiguously in electron cryo-tomograms of inner mitochondrial membranes.
Fig. 1.
Distribution of complex I and ATP synthase. Tomographic slices of mitochondrial membranes from bovine heart (A and D), Y. lipolytica (B and E), and A. officinalis (C and F) indicating the organization and orientation of ATP synthase (A–C, yellow arrowheads) and the complex I matrix arm (D–F, red rectangles). The random distribution and orientation of complex I in the membrane indicates that there are no respiratory strings (58). (Scale bar: 5 nm.)
Inner Mitochondrial Membranes Contain Respiratory Chain Supercomplexes of Different Structure and Stoichiometry.
To identify respiratory chain supercomplexes within the membrane, complex I densities and their surroundings were subjected to subtomogram averaging, followed by principle component analysis (PCA) (32). Tomographic volumes were aligned to the matrix arm of complex I (Fig. S2). The resulting average revealed good density for complex I and a less well-defined density at the position of complex III2 in single-particle averages (24–28). We performed PCA using a mask enveloping the expected position of complex III2 and classified the subvolumes. The majority of subvolumes contained density at the complex III position, of a shape typical for complex III2 at ∼40 Å resolution (Fig. S3 A–C). Some classes contained additional densities in contact with the complex I1III2 core. Further rounds of PCA were performed on subvolumes until no further additional densities were observed. The size and shape of the additional densities were consistent with the structure of monomeric complex IV at ∼40 Å resolution (Fig. S3 D–F). In conclusion, subtomogram averaging combined with PCA revealed several different types of respiratory chain supercomplexes in the inner membrane of mitochondria from the three organisms that we examined.
The in Situ Structure of the Bovine Respirasome Closely Resembles the Detergent-Solubilized Mammalian Supercomplex.
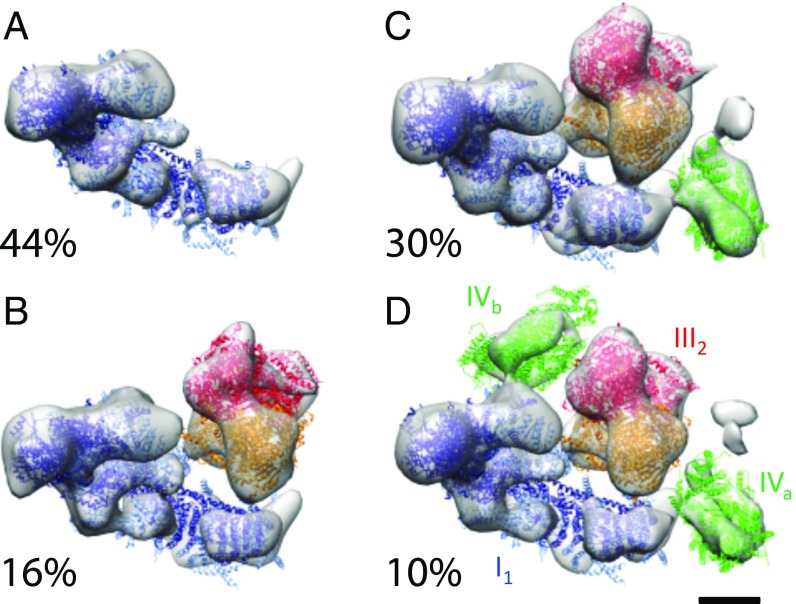
Four different assemblies of respiratory chain complexes were observed in the inner membrane of bovine heart mitochondria (Fig. 2). About 44% of the subvolumes contained a single copy of complex I (I1); 16% contained one complex I and one complex III dimer (I1III2); 30% contained one complex I, one complex III2, and one complex IV (I1III2IVa); and 10% contained one complex I, one complex III2, and two separate complex IV monomers (I1III2IVab). The I1III2IVa supercomplex is thus the most common respiratory chain supercomplex in the mammalian inner mitochondrial membrane under our conditions.
Fig. 2.
Respiratory chain supercomplexes in bovine heart mitochondria. Subvolumes centered on the matrix arm of complex I were aligned and averaged using a tight mask and then classified based on the presence of neighboring protein densities. Four classes were observed: (A) complex I alone (44%); (B) supercomplex I1III2IVa (30%); (C) supercomplex I1III2 (16%); and (D) supercomplex I1III2IVab (10%). A further possible protein density (gray above IVa) is observed in C and D but its identity is unknown. (Scale bar: 5 nm.) Blue, complex I [Protein Data Bank (PDB) ID code 4UQ8]; red/orange, complex III (PDB ID code 1BCC); green, complex IV (PDB ID code 1OCC).
The in situ structure of component complexes in the I1III2IVa supercomplex is very similar to the detergent-solubilized mammalian I1III2IV1 supercomplex (Fig. S4) determined by single-particle cryo-EM (24–28). The single-particle structures indicate some variation in the positions of complex III2 and complex IV1 relative to complex I. This is most notable for complex IV, which has been suggested to adopt a tight conformation in which complex IV is more closely appressed to complex III2, or a loose conformation, in which complex IV is up to 12 Å further removed from the I1III2 (25). Overlays with the single-particle maps suggest that the in situ assembly is most similar to the loose conformation (Fig. S4).
Yeast and Mammalian Supercomplexes Have the Same Stoichiometry but Different Structures.
Since the widely studied yeast Saccharomyces cerevisiae lacks a conventional complex I (33), we investigated the membranes of the obligate aerobic yeast Y. lipolytica. The structure of the Y. lipolytica complex I has been determined by X-ray crystallography (9, 13) and single-particle cryo-EM (11).
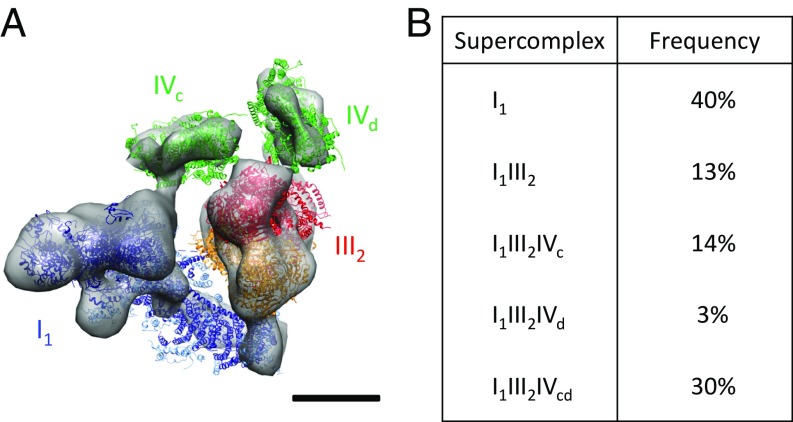
As with bovine mitochondria, several different arrangements of respiratory chain complexes were found in Y. lipolytica (Fig. 3). These included (i) complex I by itself (I1, 40%); (ii) complex I plus a complex III dimer (I1III2, 13%); (iii) complex I with complex III2 and one complex IV monomer (I1III2IVc, 14%); and (iv) complex I, complex III2, and two separate complex IV monomers (I1III2IVcd, 30%). The latter form is thus the most common supercomplex in Y. lipolytica. A fifth group containing complex I, complex III2, and the complex IVd monomer (I1III2IVd) was detected with low frequency (∼3%).
Fig. 3.
Respiratory chain supercomplexes of the yeast Y. lipolytica. (A) Subtomogram average of the I1III2IVcd supercomplex. Blue, complex I (PDB ID code 4WZ7); red/orange, complex III dimer (PDB ID code 1BCC); green, complex IV (PDB ID code 1OCC). (Scale bar: 5 nm.) (B) Composition and frequency of supercomplexes in the inner mitochondrial membrane.
The location of complex III2 relative to complex I was the same as in bovine mitochondria, but the position of the complex IV monomers varied. In particular, no complex IV was found in the position most commonly observed in the bovine heart supercomplexes. Instead, the complex IV monomers in Y. lipolytica occupy various positions around complex III2, including one adjacent to the complex I matrix arm (Fig. 3A). This position (IVc) coincides with the second complex-IV–binding site (IVb) in the bovine I1III2IVab supercomplex (Fig. 2D).
In the supercomplexes, complex IV can interact with complex I or complex III2. The fit of component complexes to the subtomogram average volumes shows which subunits interact in each case. Interactions with complex I always appeared to involve subunit 7A of complex IV and either ND5 or the 39-kDa subunit of complex I. The complex III2 interactions appeared to involve subunits cox7A and 8B of complex IV and regions of subunits QCR9, QCR8, or UCR1 of complex III2 (Fig. S5). Distances between component complexes were larger than would be expected for direct protein contacts (Cβ-Cβ of 10–20 Å), suggesting that some interactions are mediated by membrane lipid. None of the complex-IV–containing respirasomes contained a complex IV dimer as in the crystal structures of bovine complex IV (7, 16, 17).
Plant Respirasomes Do Not Contain Complex IV.
PCA classification of subvolumes from the inner mitochondrial membrane of the plant A. officinalis indicated that 42% contained a single copy of complex I, whereas the remaining 58% also contained a complex III2 dimer. Complex III2 occupied essentially the same position as in the bovine and Y. lipolytica supercomplexes but was rotated away from the complex I matrix arm by about 10°. No density resembling complex IV was observed in any of the plant subvolumes.
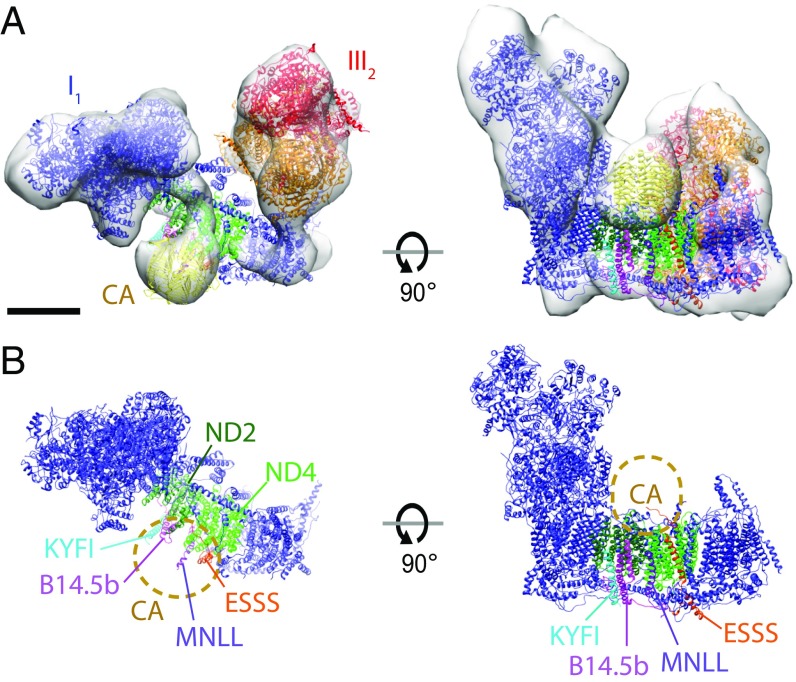
A conspicuous difference to the bovine and yeast respirasomes is a 4.5-nm, roughly spherical density on the matrix side of the plant complex I membrane arm. This density is at the interface of complex I subunits ND2/4 above the supernumerary subunits ESSS, MNLL, B14.5b, and KYFI (bovine heart nomenclature) (Fig. 4). Proteomic analysis suggests that plant complex I contains 49 subunits, of which 40 are homologous to the bovine enzyme (14). Of the nine plant-specific subunits, five are predicted to encode subunits of γ-carbonic anhydrase, an integral and vital part of plant complex I (34, 35). X-ray structures show that γ-carbonic anhydrases are homotrimers measuring roughly 4 × 4.5 nm (36). The atomic model of a γ-carbonic anhydrase fits the spherical 4.5-nm matrix density well. The other plant-specific complex I subunits include two 11 kDa proteins (P1 and P2), a 15–18 kDa translocase-like subunit, and a 68-kDa membrane-anchored l-galactono-1,4-lactone dehydrogenase (GLDH). These are either not associated with mature plant complex I, as is the case for GLDH, or are too small to account for the 4.5-nm density (14, 37). We conclude that the additional matrix density observed in the subtomogram averages of the plant complex I is γ-carbonic anhydrase.
Fig. 4.
The plant respiratory chain supercomplex. (A) Subtomogram average (transparent gray) of the A. officinalis I1III2 supercomplex with fitted atomic models. Red/orange, complex III2 (PDB ID code 1BCC); blue, complex I (PDB ID code 4WZ7); yellow, γ-carbonic anhydrase (PDB ID code 1QRE). (B) Atomic model of complex I and γ-carbonic anhydrase (CA, yellow) and complex I subunits likely to interact with it. Subunits are labeled as in bovine complex I. (Scale bar: 5 nm.)
The in Situ Arrangement of Complexes I and III2 Is Conserved Across Phyla.
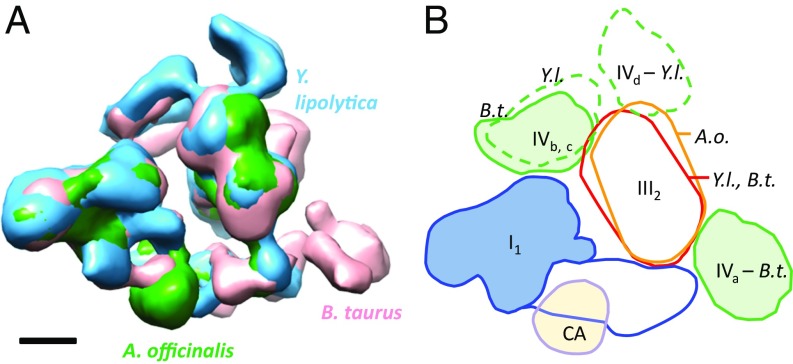
Aligning the various subtomogram averages to complex I indicated that the relative position of I and III2 in respiratory chain supercomplexes of mammals, fungi, and plants is essentially unchanged (Fig. 5). High-resolution single-particle maps of detergent-solubilized mammalian respirasomes implicate complex I subunits B14.7 and B22 (bovine nomenclature) and complex III subunits QCR9, QCR8, and UCR1 (24, 25, 38) in supercomplex I,III2 formation and stability. Interactions between these subunits or their fungal and plant equivalents are consistent with our subtomogram averages. When atomic models of the respiratory chain complexes are fitted to the maps, these subunits are next to one another (Fig. S6), indicating that the I,III2 interactions are conserved throughout the three eukaryotic lineages.
Fig. 5.
Comparison of supercomplexes from three eukaryotic lineages. (A) Subtomogram averages of bovine heart, Y. lipolytica, and A. officinalis supercomplexes were aligned on complex I volumes. (B) Schematic overlay of complex positions. The relative positions of complexes I and III2 in all three species are similar, but III2 is rotated clockwise by 12° in A. officinalis (A.o.) as seen from the matrix. Supercomplexes of A. officinalis contain no visible complex IV but a density consistent with γ-carbonic anhydrase (CA), a known component of plant complex I. Both bovine heart and Y. lipolytica supercomplexes contain up to two complex IV monomers. The position of IVc in Y. lipolytica coincides with the less frequently observed IVb position in bovine heart. Pink, Bos taurus (B.t.); blue, Y. lipolytica (Y.l.); green, A. officinalis (A.o.). (Scale bar: 5 nm.)
Discussion
Diversity of Respiratory Chain Supercomplexes.
Respiratory chain supercomplexes were first identified by nondenaturing BN-PAGE of mitochondrial membrane extracts (20). Using the mild detergents Triton X-100 and digitonin, Schägger and coworkers (20) observed several high-molecular-weight gel bands, which showed activity for complex I, complex III, and complex IV.
Cryo-ET and subtomogram averaging indicate several types of respiratory chain supercomplexes in the inner mitochondrial membranes of mammals, yeasts, and plants. The bovine subtomogram averages contain clear density for two copies of complex IV with occupancies of 40% for the primary IVa and 10% for the secondary IVb site, proving that supercomplexes I1III2IVa and I1III2IVab coexist in the inner membrane. BN-PAGE suggested that respirasomes of bovine heart mitochondria contain up to four complex IV monomers (20). In the gels, the most common supercomplexes were I,III2, I1III2IV1, and I1III2IV2, as in our subtomogram averages.
Single-particle cryo-EM structures of detergent-solubilized mammalian I1III2IV1 supercomplexes have been determined independently by four different groups (24, 25, 27, 28, 38) at medium-to-high resolution. All contain one copy of complex I and one complex III dimer. The relative orientations of complexes I and III2 in these structures are similar to those in our subtomogram averages (Fig. S4). The only exception is the recent 17-Å map of a respiratory chain supercomplex from digitonin-solubilized human mitochondria (39), which appears to contain two copies of complex I on either side of a central complex III dimer. Supercomplexes with more than one copy of complex I would be conspicuous in our tomographic volumes, but none were identified.
As in bovine mitochondria, supercomplex I1III2IV2 was the largest and most prevalent assembly in blue-native gels of Y. lipolytica mitochondria, whereas in plants the only apparent supercomplex was I1III2 (40, 41). Small amounts of complex IV have been observed by BN-PAGE, but only in freshly harvested potato tubers (42). All these biochemical results correlate closely with our subtomogram averages, indicating that BN-PAGE is an effective and reliable method for analyzing the supercomplex composition of mitochondrial membranes. The respiratory chain supercomplexes were found mostly in flat regions of the inner mitochondrial membrane (Fig. 1). They appeared to be randomly distributed, with the closest distances ranging from 15 nm up to more than 100 nm (Fig. S7).
Structural Variability of Respirasomes.
Comparison of cryo-EM structures of the mammalian supercomplex I1III2IV1 at subnanometer resolution has indicated some degree of variability in the relative positions of the three complexes in the detergent-extracted supercomplex. Complex III2 can be twisted by 25° relative to complex I (24), and complex IV can form either a tight or loose association with complex III2 (25). The more loosely assembled complexes were suggested to be artifacts of purification, as they were not abundant or their abundance increased with time, suggesting gradual dissociation in detergent (24, 25). However, the loose conformation most closely resembles that of respirasomes in the membrane. The slightly tighter packing of component complexes in isolated supercomplexes may reflect the removal of lipids by detergent upon isolation.
Subunit Contacts.
Detailed comparisons of subtomogram average volumes to single-particle cryo-EM structures reveal which subunits establish contacts between component complexes. Interactions involve subunits B14.7 and B22 of complex I and QCR8 of complex III2. Homologs for all three subunits are found in mammals, fungi, and plants (14), consistent with our observation that the I,III2 interaction is conserved in the three eukaryotic lineages.
Recent proteomic and mutational analysis suggested that cox7A2L (also known as the supercomplex assembly factor 1, or SCAF1) is essential for the interaction of complexes III2 and IV in mammals (18). Homologs of cox7A2L are present in yeasts, but a Blast search (43) did not identify a plant homolog. The absence of cox7A2L in plants would explain why the A. officinalis supercomplex does not contain complex IV.
Osmotic shock, as applied to the bovine heart and Y. lipolytica membranes that we examined, is unlikely to affect subunit contacts in the supercomplexes, as these interactions are mediated primarily by hydrophobic protein surfaces within the lipid bilayer, or by lipids. Neither would be particularly sensitive to changes in ionic strength or the concentration of osmolytes, which would act primarily on the membrane-exposed, hydrophilic parts of the complex. A well-documented example is the interaction between ATP synthase dimers in the inner mitochondrial membrane. We and others have shown that rows of ATP synthase dimers are ubiquitous in the cristae of intact mitochondria of Podospora anserina (31, 44) and Polytomella sp. (45). ATP synthase dimer rows are conspicuous in inner membrane fragments of mitochondria from more than 10 species that we investigated (30, 31, 46, 47). The dimer rows persist, irrespective of whether the inner membrane is fragmented by osmotic shock (Fig. 1 A and B) or under isotonic conditions (Fig. 1C). Although the rows tend to be shorter (because in intact mitochondria they are longer than the membrane fragments), their structure appears to be unchanged.
The interaction between ATP synthase dimers in the dimer rows is less strong and specific than the interaction between the components of respiratory chain supercomplexes. This is indicated by (i) the variable interdimer distance (30, 31), which suggests weak interactions, and (ii) the defined structure of supercomplexes, which can be isolated by detergent solubilization, whereas this is not the case for ATP synthase dimer rows. If the weak interactions between ATP synthase dimers persist, osmotic shock will not affect the stronger interactions in the supercomplexes.
Our present study shows that the interface between complex I and III2 in the supercomplexes from bovine heart, Yarrowia, and plants is essentially the same, whether or not the mitochondria were osmotically shocked (Fig. 5). This supports our conclusion that osmotic shock has no effect on the supercomplex structure.
Complex IV Dimers.
All X-ray structures of bovine complex IV (7, 16, 17) show a dimer, whereas there are no complex IV dimers in any of our supercomplex volumes. Apparently, dimer formation depends on the presence of subunits cox6A2 and cox7A1 in complex IV. If cox6A1 and cox7A2L (or SCAF1) are present instead, this results in the formation of complex-IV–containing supercomplexes (18). As cox6A1 is an isoform of cox6A2, and cox7A1 an isoform of cox7A2, complex IV would contain only one of each. The isoforms thus determine whether complex IV assembles into dimers or supercomplexes. Most likely complex IV dimers and monomers that are not part of a supercomplex coexist in the membrane, but both would be too small to be easily detected in our tomographic volumes.
Role in Electron Transfer.
The evidence in support of mitochondrial respiratory chain supercomplexes in detergent solution is unambiguous, and we now show that the supercomplexes are ubiquitous in the mitochondrial inner membrane. However, their functional significance has not been clear. For many years, a role in quinol substrate channeling has been discussed. Substrate channeling implies a defined conduit for the hydrophobic quinol electron carrier from complex I, where it is reduced, to complex III, where it is oxidized. Neither the single-particle maps nor the subtomogram averages indicate a protein-defined conduit between complex I and III2. However, there appears to be no need for such a conduit, provided that the quinol-binding sites are close enough. Ubiquinol reduced by complex I can diffuse freely within the lipid bilayer, but the close proximity and mutual orientation of the binding sites would ensure that it is trapped by complex III, as soon as a binding site becomes available. The observed functional asymmetry of isolated bovine respirasomes (24) suggests that, of the two monomers in the complex III dimer, only one is active. In the single-particle structure (24), the active monomer faces the complex I quinol site, enhancing the efficiency of electron transfer in the supercomplex.
The fact that the mutual arrangement of complexes I and III2 is essentially conserved in organisms as diverse as yeasts, mammals, and plants argues strongly for a role in the conduction of reduced quinol. Thus, the main function of supercomplexes in the mitochondrial inner membrane may be to avoid the loss of reducing power by maintaining the close proximity and favorable orientation of the key respiratory chain components. This would ensure that ubiquinol diffusion does not become rate-limiting in the crowded mitochondrial inner membrane (48, 49).
Methods
Isolation of Mitochondria.
Bovine heart mitochondria were isolated in 0.25 M sucrose and 0.1 M Tris⋅HCl, pH 7.4 (50, 51). Yeast mitochondria were isolated from Y. lipolytica strain E129 in 0.6 M sorbitol and 0.1 M Tris⋅HCl, pH 7.4 (31). White asparagus shoots were purchased at a local supermarket and ground in a Waring blender in 400 mM mannitol, 25 mM Mops–KOH, pH 7.8, 10 mM EDTA, 10 mM DTT, 1% (wt/vol) PVP-40, and 0.01% (wt/vol) BSA (fatty acid-free), with added protease inhibitors. Mitochondria were isolated, fragmented, and washed in 400 mM mannitol, 10 mM Mops–KOH, pH 7.4, and 1 mM EDTA essentially as described for potato tubers (31), except that a self-forming Percoll density gradient (28%) was used instead of a step gradient.
Sample Preparation for Cryo-ET.
For bovine heart and Yarrowia, 20 µL of isolated mitochondria at a concentration of 4–5 mg/mL was diluted with 25 vol of osmotic shock buffer (10 mM Tris, pH 7.8). Asparagus mitochondria were diluted with 25 vol of SEM buffer (250 mM sucrose, 10 mM Mops–KOH, pH 7.2, 1 mM EDTA). Membrane fragments were pelleted by centrifugation (13,000 × g at 4 °C for 20 min) and resuspended in 15 µL of buffer. The suspension was mixed 1:1 with Fiducial markers (6- or 10-nm gold particles conjugated to protein A; Aurion) before being applied to glow-discharged quantifoil grids (R2/2) and plunge-frozen in liquid ethane using a home-made guillotine (52).
Data Collection.
Tomograms were collected as described (52) using a Krios transmission electron microscope (FEI) operating at 300 kV in microprobe mode. Single-axis tilt series (±60°, 1.5° intervals, start angle of 24°) were recorded on a K2 summit detector with postcolumn energy filter (Gatan Quantum 976) at a slit width of 20 eV. Nominal magnification in EFTEM mode was 33,000 (0.43 nm/px) for Y. lipolytica and 44,000 (0.33 nm/px) for bovine and asparagus membranes. Exposure times were adjusted automatically during tilt series acquisition to maintain a constant dose of 8 e−/px. The total dose per tilt series was kept below 80 e−/Å2. All tilt series were collected at 4- to 6-µm defocus with the automatic data collection program Latitude (Gatan).
Tomogram Reconstruction and Subtomogram Averaging.
Tilt series were aligned and tomograms were reconstructed using IMOD (53). Contrast was enhanced by nonlinear anisotropic diffusion (54). Subvolumes containing complex I were extracted from the unfiltered tomogram and aligned in PEET (55) using predefined angles calculated from the vector describing the position of the peripheral arm in relation to the membrane. Initial alignment was restricted to ±180° about the axis of the peripheral arm, using a single subvolume as initial reference. Subsequent alignments were performed with the degree of rotation relative to the peripheral arm restricted to ±20°. The average from each round of refinement was used as the reference for the next round. All references were masked to contain only one complex I peripheral arm density. For the bovine supercomplex, 324 subvolumes were averaged: 316 for Yarrowia, and 168 for Asparagus. Principal component analysis (32) was performed on all volumes using a mask around the expected position of complex III2 or complex IV. Resolution was estimated by Fourier shell correlation on two half-sets after alignment. Subtomogram averages were filtered to reduce noise with the median filter (IMOD, ref. 53) or the Fermi Fourier filter [SPIDER (56)] (Fig. S8). All molecular fits and figures were produced in Chimera (57).
Supplementary Material
Acknowledgments
We thank Deryck Mills for maintenance and running the electron microscope facility and Özkan Yildiz and Juan Francisco Castillo Hernandez for computer support. This work was supported by the Max Planck Society and the Cluster of Excellence Frankfurt “Macromolecular Complexes” funded by the Deutsche Forschungsgemeinschaft (K.M.D. and W.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720702115/-/DCSupplemental.
References
- 1.Hatefi Y, Haavik AG, Fowler LR, Griffiths DE. Studies on the electron transfer system. XLII. Reconstitution of the electron transfer system. J Biol Chem. 1962;237:2661–2669. [PubMed] [Google Scholar]
- 2.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. IV. The respiratory chain. J Biol Chem. 1955;217:429–438. [PubMed] [Google Scholar]
- 3.Boyer PD. The ATP synthase: A splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T, Ueno H, Mitome N, Suzuki J, Yoshida M. F(0) of ATP synthase is a rotary proton channel. Obligatory coupling of proton translocation with rotation of c-subunit ring. J Biol Chem. 2002;277:13281–13285. doi: 10.1074/jbc.M111210200. [DOI] [PubMed] [Google Scholar]
- 5.Klusch N, Murphy BJ, Mills DJ, Yildiz Ö, Kühlbrandt W. Structural basis of proton translocation and force generation in mitochondrial ATP synthase. Elife. 2017;6:e33274. doi: 10.7554/eLife.33274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwata S, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 7.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 8.Sun F, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 11.D’Imprima E, et al. Cryo-EM structure of respiratory complex I reveals a link to mitochondrial sulfur metabolism. Biochim Biophys Acta. 2016;1857:1935–1942. doi: 10.1016/j.bbabio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirth C, Brandt U, Hunte C, Zickermann V. Structure and function of mitochondrial complex I. Biochim Biophys Acta. 2016;1857:902–914. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Senkler J, Senkler M, Braun HP. Structure and function of complex I in animals and plants: A comparative view. Physiol Plant. 2017;161:6–15. doi: 10.1111/ppl.12561. [DOI] [PubMed] [Google Scholar]
- 15.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaila VR, et al. A combined quantum chemical and crystallographic study on the oxidized binuclear center of cytochrome c oxidase. Biochim Biophys Acta. 2011;1807:769–778. doi: 10.1016/j.bbabio.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 17.Luo F, et al. Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent. Acta Crystallogr F Struct Biol Commun. 2017;73:416–422. doi: 10.1107/S2053230X17008834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogliati S, et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- 19.Hakvoort TB, et al. Separation, stability and kinetics of monomeric and dimeric bovine heart cytochrome c oxidase. Biochim Biophys Acta. 1987;894:347–354. doi: 10.1016/0005-2728(87)90112-5. [DOI] [PubMed] [Google Scholar]
- 20.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schäfer E, et al. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 22.Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 23.Bultema JB, Braun HP, Boekema EJ, Kouril R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Sousa JS, Mills DJ, Vonck J, Kühlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. Elife. 2016;5:e21290. doi: 10.7554/eLife.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 26.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 27.Althoff T, Mills DJ, Popot JL, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci USA. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagawa Y, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase. J Biol Chem. 1966;241:2467–2474. [PubMed] [Google Scholar]
- 30.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcet-Houben M, Marceddu G, Gabaldón T. Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol Biol. 2009;9:295. doi: 10.1186/1471-2148-9-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Córdoba JP, et al. The CA domain of the respiratory complex I is required for normal embryogenesis in Arabidopsis thaliana. J Exp Bot. 2016;67:1589–1603. doi: 10.1093/jxb/erv556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fromm S, Braun HP, Peterhansel C. Mitochondrial gamma carbonic anhydrases are required for complex I assembly and plant reproductive development. New Phytol. 2016;211:194–207. doi: 10.1111/nph.13886. [DOI] [PubMed] [Google Scholar]
- 36.Iverson TM, Alber BE, Kisker C, Ferry JG, Rees DC. A closer look at the active site of gamma-class carbonic anhydrases: High-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry. 2000;39:9222–9231. doi: 10.1021/bi000204s. [DOI] [PubMed] [Google Scholar]
- 37.Schimmeyer J, Bock R, Meyer EH. L-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol Biol. 2016;90:117–126. doi: 10.1007/s11103-015-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609.e10. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Guo R, Zong S, Wu M, Gu J, Yang M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257.e12. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 40.Nübel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9:2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- 41.Eubel H, Heinemeyer J, Sunderhaus S, Braun HP. Respiratory chain supercomplexes in plant mitochondria. Plant Physiol Biochem. 2004;42:937–942. doi: 10.1016/j.plaphy.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 42.Eubel H, Heinemeyer J, Braun HP. Identification and characterization of respirasomes in potato mitochondria. Plant Physiol. 2004;134:1450–1459. doi: 10.1104/pp.103.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson M, et al. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci USA. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudkina NV, Oostergetel GT, Lewejohann D, Braun HP, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography. Biochim Biophys Acta. 2010;1797:272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Mühleip AW, et al. Helical arrays of U-shaped ATP synthase dimers form tubular cristae in ciliate mitochondria. Proc Natl Acad Sci USA. 2016;113:8442–8447. doi: 10.1073/pnas.1525430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mühleip AW, Dewar CE, Schnaufer A, Kühlbrandt W, Davies KM. In situ structure of trypanosomal ATP synthase dimer reveals a unique arrangement of catalytic subunits. Proc Natl Acad Sci USA. 2017;114:992–997. doi: 10.1073/pnas.1612386114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Fabuel I, et al. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc Natl Acad Sci USA. 2016;113:13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hackenbrock CR. Energy-linked ultrastructural transformations in isolated liver mitochondria and mitoplasts. Preservation of configurations by freeze-cleaving compared to chemical fixation. J Cell Biol. 1972;53:450–465. doi: 10.1083/jcb.53.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crane FL, Glenn JL, Green DE. Studies on the electron transfer system. IV. The electron transfer particle. Biochim Biophys Acta. 1956;22:475–487. doi: 10.1016/0006-3002(56)90058-0. [DOI] [PubMed] [Google Scholar]
- 52.Davies KM, et al. Visualization of ATP synthase dimers in mitochondria by electron cryo-tomography. J Vis Exp. 2014:51228. doi: 10.3791/51228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 54.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 55.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 56.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Wittig I, Carrozzo R, Santorelli FM, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.