Significance
Mutations in the GRN gene cause frontotemporal dementia, a devastating neurological disease. The majority of these GRN mutations are nonsense and frameshift mutations. Here, we generated a knockin mouse model with a Grn mutation corresponding to the most prevalent human disease mutation, GRNR493X. We show that mice harboring this mutation phenocopy progranulin-deficient mice, and that the mutation triggers mRNA decay and, as a consequence, low production of Grn. However, the truncated mutant protein that would be produced from this allele is functional, suggesting inhibiting mRNA decay as a therapeutic approach for individuals with progranulin-deficient frontotemporal dementia caused by nonsense mutations.
Keywords: progranulin, frontotemporal dementia, neurodegeneration, nonsense-mediated mRNA decay, lysosome
Abstract
Frontotemporal dementia (FTD) is the most common neurodegenerative disorder in individuals under age 60 and has no treatment or cure. Because many cases of FTD result from GRN nonsense mutations, an animal model for this type of mutation is highly desirable for understanding pathogenesis and testing therapies. Here, we generated and characterized GrnR493X knockin mice, which model the most common human GRN mutation, a premature stop codon at arginine 493 (R493X). Homozygous GrnR493X mice have markedly reduced Grn mRNA levels, lack detectable progranulin protein, and phenocopy Grn knockout mice, with CNS microgliosis, cytoplasmic TDP-43 accumulation, reduced synaptic density, lipofuscinosis, hyperinflammatory macrophages, excessive grooming behavior, and reduced survival. Inhibition of nonsense-mediated mRNA decay (NMD) by genetic, pharmacological, or antisense oligonucleotide-based approaches showed that NMD contributes to the reduced mRNA levels in GrnR493X mice and cell lines and in fibroblasts from patients containing the GRNR493X mutation. Moreover, the expressed truncated R493X mutant protein was functional in several assays in progranulin-deficient cells. Together, these findings establish a murine model for in vivo testing of NMD inhibition or other therapies as potential approaches for treating progranulin deficiency caused by the R493X mutation.
Mutations in the progranulin gene (GRN) cause frontotemporal dementia (FTD) (1–3), accounting for 5–26% of disease cases in different populations (4–7). FTD is a devastating neurological disease (for reviews, see refs. 8–11), and currently no treatment or cure exists. Affected individuals display personality, behavior, and language abnormalities (12), but memory is typically spared. Pathologically, FTD is characterized by atrophy of the frontal and anterior temporal brain lobes, and postmortem analysis of brain tissue shows evidence of gliosis, swollen neurons, microvacuolation, and inclusion bodies that contain ubiquitin, tau, TAR DNA-binding protein 43 (TDP-43), or fused-in-sarcoma (8, 13). Complete progranulin deficiency in humans causes neural ceroid lipofuscinosis (NCL) (14), a neurodegenerative disease of multiple etiologies that is linked to lysosome deficiency.
How progranulin haploinsufficiency causes FTD is unclear, but likely involves progranulin’s roles in lysosomal (14), neurotrophic (15–17), and antiinflammatory functions (18–21). Recent studies in mice also indicate increases in complement activation and altered synaptic pruning may be important pathogenic factors (22). Progranulin localizes to lysosomes in cells (23, 24) and is expressed by many cell types, including neurons, microglia, macrophages, epithelial cells, and adipocytes. Progranulin is also secreted from cells and detectable in cerebrospinal fluid (25, 26) and blood (21, 26).
We and other laboratories generated mouse models of Grn gene deletions that yield total or heterozygous Grn knockouts (19, 21, 27, 28). Homozygous knockout models display microglial activation in the CNS (19, 22, 28–31) and recapitulate NCL with lysosomal defects reflected in altered lysosome morphology (14, 29), accumulation of lipofuscin (28–30), and increased expression of lysosomal genes (23). Heterozygous knockout mice exhibit limited phenotypes, including decreased sociability and altered social dominance (30, 32). Because these mouse models contain disrupted Grn alleles, they have limited utility in testing therapeutic approaches for progranulin-deficient FTD and NCL caused by nonsense mutations. Therefore, we sought to generate a mouse model that harbors a genetic mutation in a disease allele, and we targeted the mouse allele analogous to the most common human FTD mutation GRNR493X (3, 4, 33).
The majority (∼84%) of GRN mutations are nonsense and frameshift mutations that introduce premature termination codons (4). Among them, GRNR493X is the most prevalent nonsense mutation found in individuals with FTD (3, 4, 33). GRNR493X introduces a premature termination codon (PTC) and is predicted to encode for a C-terminally truncated protein lacking ∼17% of the protein, including one of progranulin’s 7.5 cysteine-rich granulin domains (34–36). Because of introduction of a PTC, the mutant mRNA transcribed from this allele could be a target of the nonsense-mediated mRNA decay pathway (NMD) (37).
Here, we show that mice harboring the GrnR493X mutation phenocopy Grn−/− mice and that the mutation results in reduced mRNA levels in part due to NMD. Furthermore, we test antisense oligonucleotides designed to inhibit NMD of the GrnR493X mRNA, and we characterize the truncated protein that would be produced from the GRNR493X allele, showing that it properly targets to lysosomes and has activity in several cell-based assays.
Results
Generation and Characterization of GrnR493X Mice.
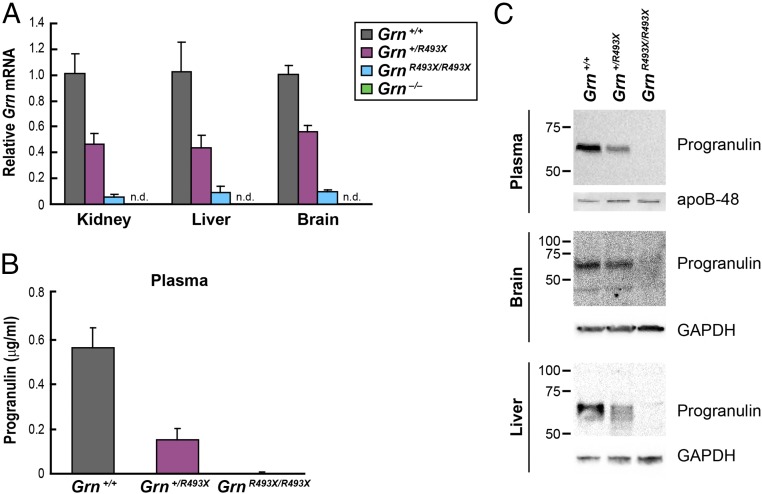
We used gene targeting in murine embryonic stem cells to generate mice harboring a Grn R504X nonsense mutation analogous to human R493X, the most common GRN mutation found in individuals with FTD (3, 4, 33) (Fig. S1B). Because these mice model the R493X human mutation, we refer to these mice as GrnR493X and the resulting truncated protein as progranulin R493X. Grn mRNA levels were ∼50% reduced in tissues of Grn+/R493X mice and >90% reduced in GrnR493X/R493X mice (Fig. 1A). Levels of the full-length progranulin protein (∼65 kDa) were ≥50% decreased in plasma and tissues of Grn+/R493X mice (Fig. 1 B and C). Although GrnR493X/R493X mice had detectable Grn mRNA (∼5–10% of wild-type levels), we did not detect the truncated progranulin R493X protein (∼54 kDa) in Western blots using an antibody that recognizes amino acids 198–214 and is able to detect the truncated progranulin R493X protein when this protein is overexpressed in cells (Fig. S2).
Fig. 1.
Homozygous GrnR493X targeted mice have markedly reduced Grn mRNA levels and lack progranulin protein. (A) Grn mRNA levels in tissues were determined by qPCR. (B) ELISA of progranulin in mouse plasma. Data are presented as mean ± SD. (C) Immunoblot analysis of progranulin in mouse tissues. apo, apolipoprotein; n.d., not detected.
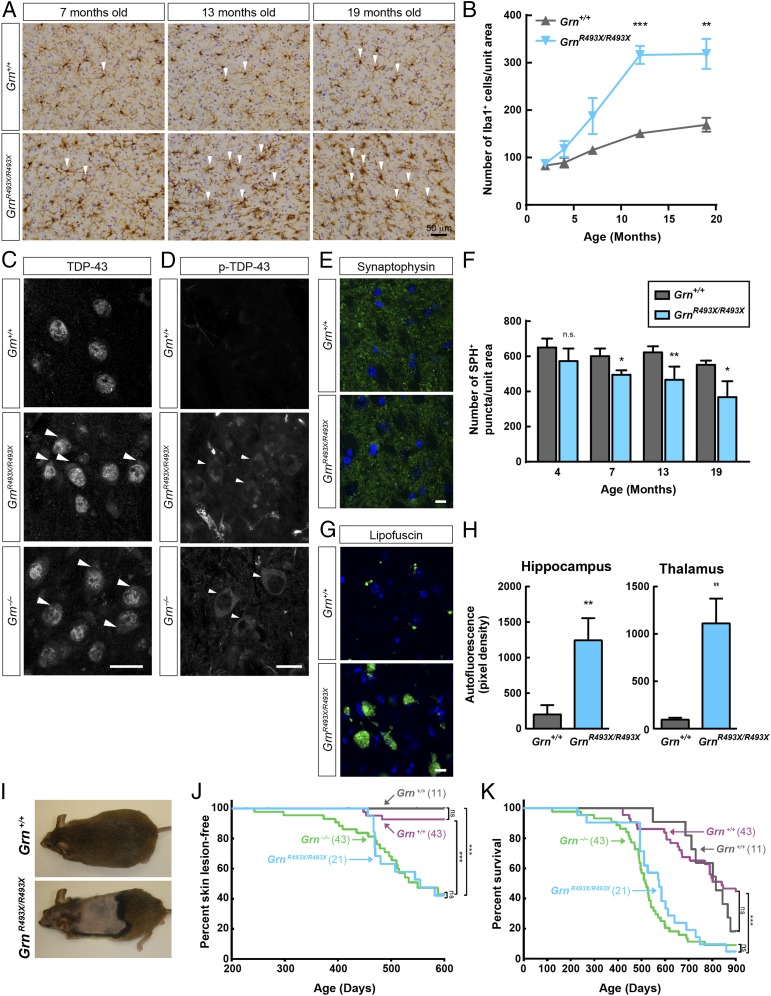
Progranulin knockout (Grn−/−) mice were generated by several laboratories, including ours (19, 21, 27, 28), and recapitulate some key features of FTD, such as neuroinflammation and behavioral changes. Given that the GrnR493X/R493X mice lack detectable progranulin protein, we expected that they would phenocopy Grn−/− mice. To assess this, we examined neuropathology, grooming behavior, and the inflammatory response in macrophages of GrnR493X/R493X mice. Consistent with progressive neuroinflammation and similar to Grn−/− mice (19, 22, 28–31, 38), GrnR493X/R493X mice exhibited age-dependent microgliosis, indicated by the increased Iba1+ staining of microglia in the thalamus (Fig. 2 A and B). Also similar to Grn−/− mice (31), GrnR493X/R493X mice have increased levels of total and phosphorylated TDP-43 in the cytoplasm of thalamic neurons (Fig. 2 C and D and Fig. S3). Like Grn−/− mice (22), GrnR493X/R493X mice exhibit an age-dependent reduction in synapse density, as reflected in the number of synaptophysin+ puncta in the thalamus (Fig. 2 E and F). Also similar to Grn−/− mice (28–30), GrnR493X/R493X mice also showed increased levels of lipofuscin in the brain (Fig. 2 G and H), which is characteristic of NCL models (39–42) and indicates lysosomal dysfunction.
Fig. 2.
Phenotype of GrnR493X/R493X mice recapitulates features of global knockout mice. (A and B) GrnR493X/R493X mice exhibit age-dependent microgliosis. (A) Immunostaining of Iba1 in the thalamus. Arrowheads indicate selected Iba1+ cells. (B) Quantification of Iba1+ microglial density in 12-mo-old mice, n = 3–4 mice per genotype. (C and D) Increased levels of total (C) and phosphorylated (D) TDP-43 in the cytoplasm of thalamic neurons of 12-mo-old GrnR493X/R493X and Grn−/− mice. Arrowheads indicate cytoplasmic accumulation of TDP-43. (Scale bars, 20 μm.) (E and F) GrnR493X/R493X mice exhibit age-dependent reduction of synaptic density. (E) Immunostaining of synaptophysin in the thalamus of 13-mo-old mice. (Scale bar, 10 μm.) (F) Quantification of synaptophysin density, n = 3–4 mice per genotype at each age. (G and H) GrnR493X/R493X mice have increased lipofuscin in the brain. (G) Images of autofluorescent lipofuscin (green channel) in the thalamus of 19-mo-old mice. (Scale bar, 10 μm.) (H) Quantification of autofluorescence. (I–K) GrnR493X/R493X mice have increased skin lesions resulting in decreased survival. (I) Skin lesions in 16-mo old mice. (I and J) Kaplan–Meier curves for skin lesion onset (J) and survival (K) in Grn+/+ (gray curves) and GrnR493X/R493X (blue curves) littermate mice. For comparison, curves for Grn+/+ (purple curves) and Grn−/− (green curves) littermate mice are also shown. Data are presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, as determined by Student’s t test (B, F, and H), Long-rank (Mantel–Cox) test (J and K). n.s., not significant; SPH, synaptophysin.
Grn−/− mice exhibit a number of behavioral changes, including abnormal social interactions, fear-avoidance, and increased grooming behavior (22, 28, 30–32, 38, 43). As a first step to test whether GrnR493X/R493X mice recapitulate the behavioral phenotype of Grn−/− mice, we focused on grooming behavior. In Grn−/− mice, increased grooming can lead to development of skin lesions, which often become infected and thereby reduce their survival (22). Compared with Grn+/+ littermates, GrnR493X/R493X mice displayed evidence of compulsive grooming (Fig. 2I), developed more skin lesions over time (Fig. 2J), and had 30% lower median survival [575 d for GrnR493X/R493X mice (blue curve in Fig. 2K) vs. 816 d for Grn+/+ mice (gray curve in Fig. 2K)]. For comparison, curves for Grn−/− mice (green curves in Fig. 2K) and Grn+/+ (purple curves in Fig. 2K) littermate mice are also shown. Of note, maximal lifespan for GrnR493X/R493X mice was similar to wild-type mice. Behavioral studies in the Grn deletion model required extensive analyses of large numbers of animals to detect differences (27, 28, 30, 31, 38). Therefore, complete behavioral analyses of GrnR493X/R493X mice have not yet been performed.
Macrophages and microglia isolated from Grn−/− mice exhibit a hyperinflammatory response when stimulated with proinflammatory agents (19, 21). In bone marrow–derived macrophages (BMDMs) isolated from GrnR493X/R493X mice, we similarly observed an exaggerated inflammatory response after treatment with lipopolysacharride (LPS) and IFN-γ (Fig. S4). Thus, GrnR493X/R493X mice appear to manifest the same phenotype as progranulin-knockout mice.
GrnR493X Mice Exhibit Reduced Grn mRNA in Part Due to Nonsense-Mediated Decay.
An advantage of GrnR493X mice over knockout animals is that they mimic a clinically relevant mutation that reduces mRNA levels, similar to what is found in patients with nonsense mutations (1–3, 44). Because the R493X mutation generates a premature stop codon, we tested whether GrnR493X mice yields a Grn mRNA that is degraded by the NMD pathway.
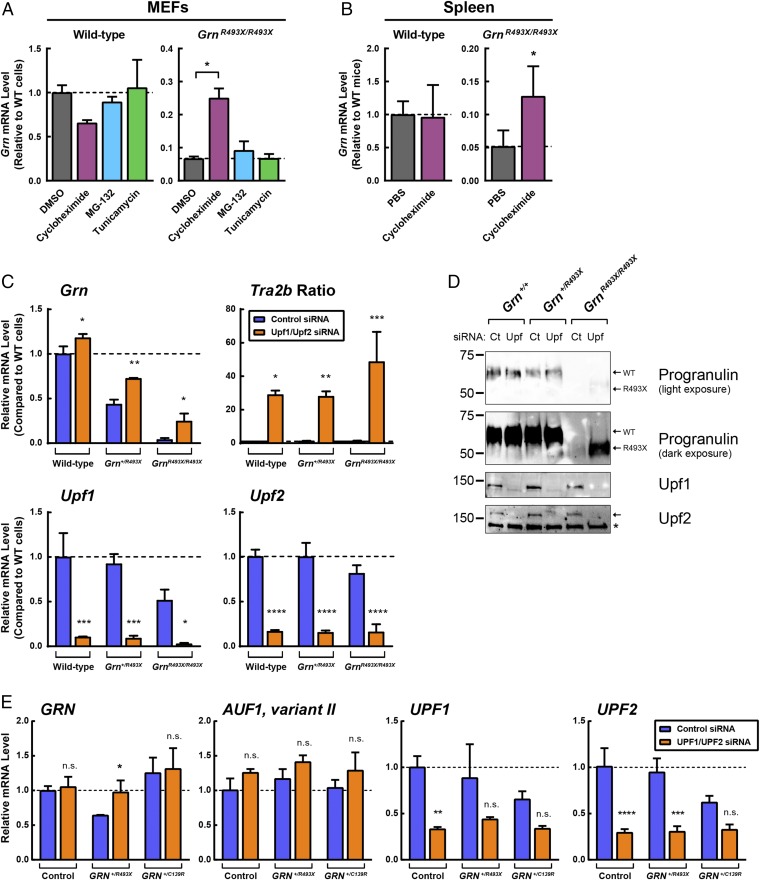
Because NMD requires a pioneer round of translation (45), cycloheximide treatment, which inhibits translation, also inhibits NMD (46). In two cell types isolated from GrnR493X/R493X mice, mouse embryonic fibroblasts (MEFs) and BMDMs, Grn mRNA was reduced to ∼6% of wild-type levels (Fig. 3A and Fig. S5). Cycloheximide treatment increased Grn mRNA ∼4-fold and ∼11-fold in GrnR493X/R493X MEFs and BMDMs, respectively. Comparable increases were not observed in wild-type cells. Similarly, in vivo treatment with cycloheximide via peritoneal injection increased Grn mRNA in the spleen of GrnR493X/R493X mice, but not in that of wild-type mice (Fig. 3B). Additionally, caffeine at high concentrations inhibits NMD (47–50). In GrnR493X/R493X BMDMs, we found caffeine increased mRNA levels of GrnR493X mRNA (Fig. S5). These data are consistent with the model that GrnR493X mRNA is degraded by the NMD pathway.
Fig. 3.
Blocking NMD increases Grn mRNA in GrnR493X mouse model and GRN+/R493X patient fibroblasts. (A) Wild-type and GrnR493X/R493X MEFs were treated for 5 h with 50 μM cycloheximide, 10 μM MG-132, or 10 μg/mL tunicamycin. Gene expression was determined by qPCR. (B) Wild-type and GrnR493X/R493X mice (n = 4–7 per group) were acutely injected with 20 mg/kg body weight cycloheximide dissolved in PBS. After 4 h, gene expression was determined by qPCR. (C and D) Upf1 and Upf2 were knocked down in wild-type, Grn+/R493X, and GrnR493X/R493X MEFs using DharmaFECT1. After 3 d, gene expression was determined by qPCR (C), and protein levels were determined by Western blot analyses (D). (E) UPF1 and UPF2 were knocked down in control and GRN+/R493X HDFs using DharmaFECT1. After 3 d, gene expression was determined by qPCR. All data are presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, as determined by one-way ANOVA followed by Tukey’s post hoc test (A), or two-way ANOVA followed by Bonferroni’s post hoc test (B, C, and E). Asterisk in D indicates a nonspecific band. n.s., not significant.
To block NMD more specifically, we utilized siRNA to knockdown Upf1 and Upf2, key components of the NMD machinery (51, 52) in GrnR493X MEFs. The effects of these knockdowns on GrnR493X mRNA levels were somewhat variable in our experiments, likely due to variable levels of Upf1 and Upf2 knockdown and consequently variable inhibition of NMD. In three independent experiments, we found a range of rescue of Grn mRNA levels (to 16–25% of wild-type levels) in homozygous GrnR493X cells, and the degree of Upf1 and Upf2 knockdown correlated with the increase in Grn mRNA levels. Fig. 3C shows an experiment in which Upf1/Upf2 siRNA increased Grn mRNA ∼sixfold (from 4% of wild-type levels to 25%) in GrnR493X/R493X MEFs but did not increase Grn mRNA to a similar extent in wild-type MEFs. In these experiments, we assessed NMD inhibition by measuring mRNA levels of variants of Tra2b, which are selectively degraded by NMD (53). The ratio of the NMD-sensitive isoform over the nonsensitive isoform of Tra2b was increased in Upf1/Upf2 knockdown cells, indicating that NMD was effectively inhibited. In parallel with the increased Grn mRNA levels in GrnR493X/R493X MEFs, Upf1/Upf2 siRNA also increased levels of the truncated progranulin protein as detected by Western blot analysis (Fig. 3D). Specifically, the lower molecular weight band, corresponding to the size of the progranulin R493X truncated mutant (Fig. S2), was increased.
In human dermal fibroblasts (HDFs) heterozygous for the GRNR493X mutation, siRNA knockdowns of UPF1 and UPF2 also increased GRN mRNA levels (from ∼60% to ∼95%), effectively normalizing levels to those found in control HDFs from a family member without the GRN mutation (Fig. 3E). In HDFs from a patient with a missense mutation (GRN+/C139R), which does not result in a premature stop codon, levels of GRN mRNA were not affected by UPF1 an UPF2 knockdowns. In these experiments, we assessed NMD inhibition by measuring mRNA levels of variant II of AUF1, which is selectively degraded by NMD (54). The mRNA levels of variant II of AUF1 trended higher with knockdowns of both UPF1 and UPF2. These results indicate that NMD contributes to the decreased mRNA levels in both GrnR493X mice and cells from individuals with FTD with GRN+/R493X mutations.
Antisense Oligonucleotides Designed to Block NMD Increase GrnR493X mRNA Levels.
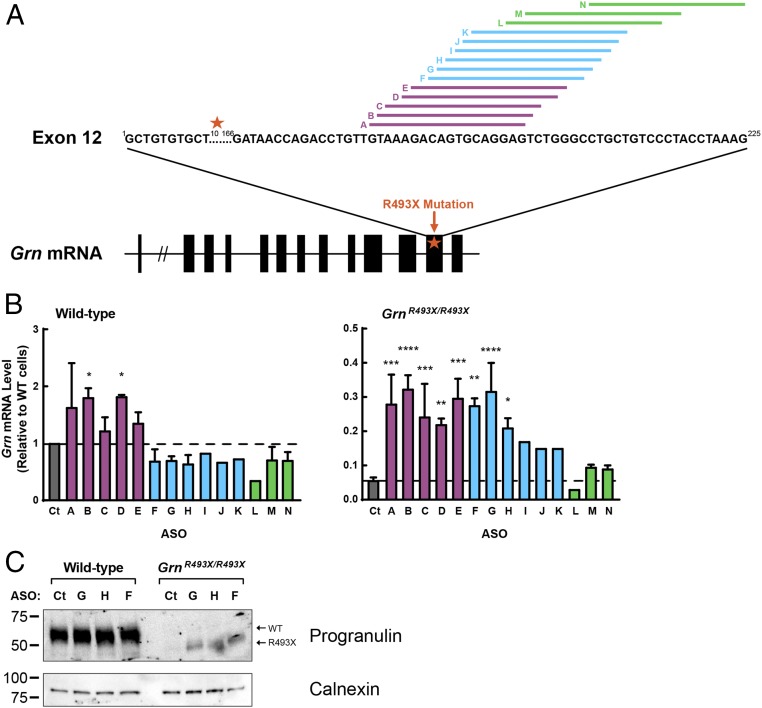
NMD can also be targeted by additional approaches, such as antisense oligonucleotides (ASOs). By targeting the 3′ end of exons that contain a premature termination codon, ASOs can prevent the deposition of the exon–exon junction complex and prevent the recruitment of NMD proteins, which otherwise would mark the mutant mRNA for degradation, thus stabilizing it. Such an approach would enable specific targeting of the Grn mRNA without affecting other mRNA substrates of NMD. Using this strategy, ASOs can effectively block degradation of a reporter mRNA construct (55). To test the potential of ASOs to increase endogenous Grn mRNA in the GrnR493X model, we designed a panel of 14 ASOs that target the region of the exon–exon junction complex at the 3′ end of exon 12 of the Grn mRNA (Fig. 4A and Table S1). As shown in Fig. 4B, a group of ASOs (F–K, blue bars in Fig. 4B) increased Grn mRNA in GrnR493X/R493X cells and not in wild-type cells, presumably by inhibiting NMD. Treatment with these ASOs also increased protein levels of R493X progranulin protein (Fig. 4C). Another group of ASOs (A–E, purple bars in Fig. 4B) unexpectedly increased Grn mRNA and protein in both wild-type and GrnR493X/R493X cells (Fig. 4B and Fig. S6). Because the wild-type Grn mRNA is largely unaffected by NMD inhibition (Fig. 3), these ASOs likely act predominantly through an NMD-independent mechanism. Finally, a group of ASOs (L–N, green bars in Fig. 4B), located closer to the 3′ end of exon 12, did not significantly increase Grn mRNA levels in either wild-type or GrnR493X/R493X cells. Of note, ASO N was designed to skip exon 12 and it led to production of a smaller protein with a size consistent with a progranulin protein lacking exon 12 in wild-type cells (Fig. S6).
Fig. 4.
ASOs designed to inhibit NMD of GrnR493X increase Grn mRNA in GrnR493X cells. (A) Schematic of ASO targets in exon 12 of the Grn mRNA. Black boxes, coding exons; star, R493X mutation. (B and C) MEFs were transfected with ASOs using Lipofectamine 2000. After 24 h, gene expression was determined by qPCR (B); after 48 h, protein levels in cell lysates were determined by immunoblot analysis (C). Data are presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, as determined by one-way ANOVA followed by Dunnett post hoc test. Ct, control.
The Mutant Protein Produced by the GrnR493X Allele Targets to Lysosomes and Is Functional in Cell-Based Assays.
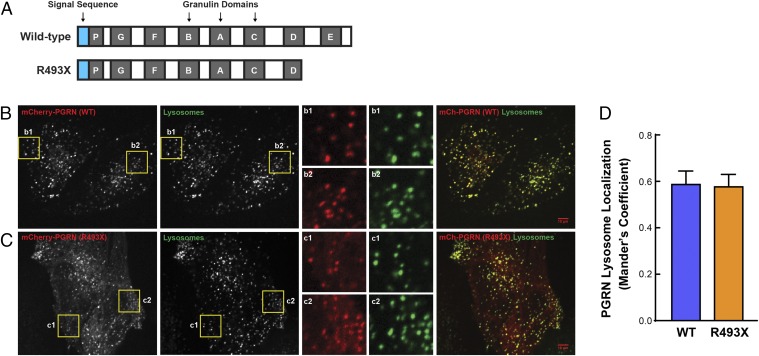
The Grn R493X mutation encodes for a truncated protein that lacks C-terminal 17% of the protein. Specifically, the R493X mutant lacks three amino acids of granulin D and the entire granulin E (Fig. 5A). Intracellular progranulin primarily localizes to lysosomes (23, 24). Because the C terminus of progranulin interacts with sortilin (56), which provides a pathway for progranulin targeting to lysosomes (57), we tested if the R493X mutant localizes to lysosomes by microscopy. In progranulin-deficient HeLa cells expressing low levels of N-terminally mCherry-tagged human progranulin, the wild-type protein and the R493X mutant both colocalized extensively with dextran-labeled lysosomes (Fig. 5 B–D). For the R493X mutant, we observed a larger pool of progranulin not localized to lysosomes, presumably due to partial impairment of lysosomal targeting. We also observed extensive colocalization of the R493X mutant and the lysosomal marker LAMP1 by immunofluorescence in progranulin-deficient MEFs expressing C-terminally FLAG-tagged mouse progranulin (Fig. S7). These results indicate that a large pool of the R493X mutant is able to target to lysosomes despite lacking the C-terminal 17%.
Fig. 5.
Progranulin R493X mutant is delivered to lysosomes. (A) Schematic representation of the wild-type progranulin and the R493X truncated mutant proteins. Blue boxes, signal sequence; gray boxes, granulin domains. (B and C) Images show the colocalization of mCherry-PGRN wild-type (B) and mCherry-PGRN R493X (C) with Alexa Fluor 647-dextran–labeled lysosomes in progranulin-deficient HeLa cells. Inset boxes (b1, b2, c1, and c2) obtained from the highlighted regions show the robust overlap of mCherry-PGRN puncta with lysosomes. (Scale bars, 10 μm.) (Magnification: B and C, Insets, 3×.) (D) Quantitative colocalization analysis shows similar lysosome localization for both wild-type and R493X forms of progranulin. Colocalization coefficient was quantified on a per cell basis, and 22–28 transfected cells were measured for each plasmid. Data are presented as mean ± SEM from four independent experiments. P = 0.897 (nonsignificant), as determined by unpaired t test.
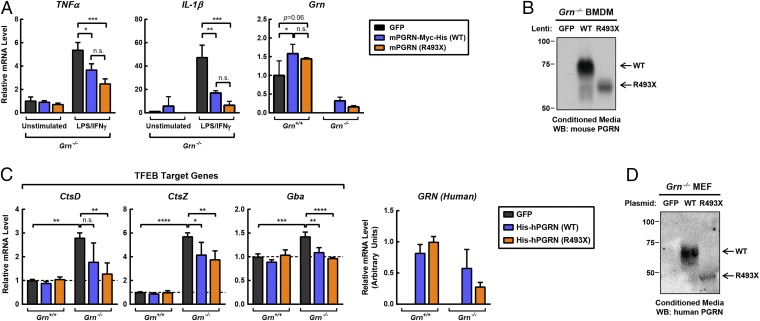
Progranulin can be cleaved by a number of proteases to yield individual granulins (18, 58–60), and it is currently unclear if the full-length progranulin or specific individual cleaved granulins are biologically active. Thus, we tested whether the progranulin R493X mutant protein is functional by overexpressing either wild-type progranulin or the R493X mutant progranulin in Grn−/− cells. We first expressed these proteins in progranulin-deficient macrophages, which exhibit a hyperinflammatory response (19). Overexpression of C-terminally tagged versions of wild-type mouse progranulin or the R493X mutant in Grn−/− macrophages by lentiviral transduction suppressed the inflammatory response triggered by LPS/IFN-γ treatment to similar extents (Fig. 6 A and B). The effects of the mutant protein appeared to be more potent than the wild-type protein inasmuch as inflammatory markers were suppressed more robustly even at lower expression levels.
Fig. 6.
Progranulin R493X mutant is functional in cell-based assays. (A) Overexpression of wild-type progranulin and the R493X mutant in Grn−/− BMDMs suppresses the proinflammatory response. Macrophages were infected with the indicated lentivirus for 24 h, and subsequently treated with 10 ng/mL LPS and 10 ng/mL IFN-γ for an additional 24 h. Gene expression was determined by qPCR. (B) Immunoblot analysis of progranulin in conditioned media from Grn−/− macrophages. (C) Overexpression of wild-type progranulin and the R493X mutant in Grn−/− MEFs restores normal expression of TFEB target genes. (D) Immunoblot analysis of progranulin in conditioned media from Grn−/− MEFs. All data are presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, as determined by two-way ANOVA followed by Bonferroni’s post hoc test. n.s., not significant.
Progranulin is enriched in lysosomes (23, 24) and likely functions in the lysosome. TFEB is a master regulator of lysosome biology and is activated with lysosomal dysfunction, and TFEB target gene expression is increased with progranulin deficiency (23). Consistent with progranulin having a lysosomal function, we found that levels of several TFEB target genes were increased in Grn−/− MEFs (Fig. S8). These TFEB target genes include cathepsins (CtsD, CtsZ), glucocerebrosidase (Gba), and hexosaminidase A (HexA), which encode enzymes that degrade proteins, glycolipid intermediates, and GM2 gangliosides, respectively. The increased expression of TFEB target genes was largely normalized by overexpression of either N-terminally tagged wild-type human progranulin or the R493X mutant in Grn−/− MEFs by electroporation (Fig. 6 C and D). Similar results were obtained with overexpression of C-terminally tagged mouse progranulin by lentiviral transduction (Fig. S9). It is worth noting that in these overexpression studies, it is currently unclear what levels of progranulin expression are required to restore function and how these levels relate to progranulin haploinsufficiency in human FTD and the mouse models.
These results indicate that the R493X mutant retains its activities related to the inflammatory response and lysosomal function.
Discussion
We report a mouse model of FTD (GrnR493X mice) that harbors a clinically relevant nonsense mutation at the residue corresponding to the GRN R493X mutation, the most common GRN mutation in individuals with FTD. This mutation triggers degradation of the mRNA through NMD in both the GrnR493X model and patient-derived fibroblasts, and ASOs designed to block NMD of the GrnR493X mRNA can increase progranulin mRNA and protein levels in GrnR493X cells. Moreover, the expressed truncated R493X mutant protein properly localizes to lysosomes and was functional in several assays in progranulin-deficient cells. Together, these findings establish a new murine model for in vivo testing of therapies for treating progranulin deficiency caused by the R493X mutation.
Consistent with the lack of detectable progranulin protein in GrnR493X/R493X mice, our characterization indicates that these mice phenocopy Grn−/− mice with CNS microgliosis, increased levels of total and phosphorylated TDP-43 in cytoplasm of thalamic neurons, reduced synaptic density, and lipofuscinosis, as well as hyperinflammatory macrophages, excessive grooming behavior, and reduced survival. We expect that GrnR493X mice will have additional behavioral changes that were observed in Grn−/− mice, including abnormal social interactions and fear-avoidance (22, 28, 30–32, 38, 43), and additional abnormalities in social dominance that were recently reported for heterozygous mice (30, 32). Taken together, GrnR493X/R493X mice are functional Grn knockout mice, with the additional feature of the Grn mRNA being subjected to NMD, thereby uniquely enabling testing of therapies aimed at increasing Grn mRNA levels in patients with nonsense mutations. Thus, heterozygous GrnR493X mice could serve as an excellent patient-oriented model, particularly for patients with this most common GRN mutation found in progranulin-deficient FTD.
Our data implicate NMD in the decreased Grn mRNA levels in GrnR493X mice. Inhibition of NMD in GrnR493X/R493X cell and mice, but not in wild-type cells and mice, with compounds known to inhibit NMD (i.e., cycloheximide and caffeine) increased Grn mRNA levels. Similarly, inhibition of NMD by siRNA knockdown of the key NMD proteins Upf1 and Upf2 increase Grn mRNA in GrnR493X/R493X cells. siRNA experiments in patient-derived fibroblasts indicate that GRN mRNA levels in GRN+/R493X cells are also reduced due to degradation of the mutant mRNA through NMD. Thus, in the GrnR493X model and in patient-derived cells, the mutant mRNA is subject to regulation by the same quality-control mechanism, which could be targeted therapeutically to restore normal progranulin levels as a potential treatment for progranulin-deficient FTD.
A number of chemicals have been reported to inhibit NMD, and the homozygous GrnR493X/R493X cells provide a cell-based system to test these inhibitors for efficacy. We tested three reported NMD inhibitors, including NMDI1 (61), NMDI9 (62), and NMDI14 (62), which have been reported to inhibit NMD in cells. However, in GrnR493X/R493X BMDMs, we did not observe increased mRNA levels of the NMD-sensitive isoforms of Tra2b and Hnrnpl or the mutant GrnR493X after treatment with these reported NMD inhibitors (Fig. S10). In contrast, the NMD-sensitive mRNAs were all increased in cells treated with cycloheximide and caffeine, which inhibit NMD (46–50). Nonetheless, our GrnR493X/R493X MEFs provide a cell line for testing NMD therapies against endogenous genes, as they exhibit NMD and the baseline level of mRNA expression is very low, making this an ideal cell line generally for screening for NMD inhibitors.
Our results in cell-based assays indicate that the progranulin R493X truncation mutant is likely to be functional. Specifically, when overexpressed, this mutant retains the ability to suppress expression of inflammatory markers in progranulin-deficient macrophages and the ability to restore normal TFEB target-gene expression in progranulin-deficient MEFs. This mutant lacks ∼17% of the protein at the C terminus, including three amino acids of granulin D and all of granulin E (Fig. 5A). By live-cell imaging and immunofluorescence microscopy, we found that the R493X mutant localizes in large part to lysosomes, similar to the wild-type protein, indicating that the C terminus is not essential for lysosomal localization of overexpressed progranulin. Two mechanisms have been reported for progranulin targeting to lysosomes: a sortilin-dependent mechanism (57) and a sortilin-independent mechanism that involves prosaposin, mannose 6-phosphate receptor, and low-density lipoprotein receptor-related protein 1 (24). The progranulin R493X mutant is not expected to interact with sortilin, as the last three C-terminal amino acids of progranulin are required for binding (56), and thus our finding suggests sortilin-independent mechanisms are sufficient for targeting overexpressed progranulin R493X to lysosomes in HeLa cells and MEFs. This is consistent with the recently proposed model that prosaposin and sortilin are independent and complementary mechanisms for targeting progranulin to lysosomes in fibroblasts (24) and that multiple individual granulin domains can support prosaposin interactions (63). Additionally, our finding that the progranulin R493X mutant is functional indicates that full-length progranulin is not required for function and suggests granulins may be the biologically active units. Further studies are necessary to directly test this model, which implies that other truncation mutants that contain at least one intact granulin domain are functional.
It is unclear why Grn mRNA levels in GrnR493X cells were not restored more completely to wild-type levels following NMD inhibition with pharmacological inhibitors, siRNA knockdown of key NMD proteins, and ASOs designed to inhibit NMD of the GrnR493X mRNA. In addition to the possibility of incomplete inhibition of NMD, feed-forward regulation of progranulin expression (64) may also contribute to this observation, as it would amplify changes in progranulin expression. Indeed, serum progranulin levels in patients with heterozygous GRN mutations are often reduced by 70–80% compared with controls (65) rather than the 50% expected by loss of one allele. Moreover, we cannot exclude possible contribution of additional RNA quality control pathways, such as microRNA (miRNA)-mediated mRNA decay (66), no-go decay (67), and nuclear RNA degradation pathways due to impaired RNA processing or export (68). In support of this, treatment with several ASOs increased mRNA levels in both wild-type and GrnR493X cells.
Together, our findings suggest that agents that block NMD and thereby increase levels of the mutant GrnR493X mRNA will lead to production of a functional protein and potentially overcome progranulin haploinsufficiency associated with certain forms of FTD. Until now, testing such agents has not been technically feasible because cycloheximide blocks protein translation and other reported NMD inhibitors (NMDI1, NMDI9, and NMDI14) were not effective in our hands (Fig. S10). ASOs are a promising approach for specifically blocking NMD of the Grn mRNA, and they would enable the necessary long-term in vivo studies. Indeed, ASO-based approaches have been efficacious in mouse models of neurodegenerative diseases, including Alzheimer’s disease (69), amyotrophic lateral sclerosis (70), spinocerebellar ataxia type 2 (71), and Huntington’s disease (72). In humans, ASOs have also been efficacious in recent clinical trials for spinal muscular atrophy (73) and Duchenne muscular dystrophy (74). In the context of progranulin-deficient FTD, ASOs that increase expression of both the wild-type and the mutant alleles (such as ASOs A, B, and D) (Fig. 4 A and B and Fig. S6) in fibroblasts are especially promising. Future in vivo testing of these ASOs in heterozygous and homozygous GrnR493X mice with analysis of neuropathology and behavioral testing is warranted. While heterozygous GrnR493X mice are the model of choice for progranulin-deficient FTD, homozygous GrnR493X mice are ideal for in vivo testing of NMD inhibitors because they provide a very low basal Grn mRNA level to test for efficacy of potential therapies. With the existence of this model, such longer-term studies to examine glial pathology and neurodegenerative and behavioral phenotypes can now be performed.
Materials and Methods
Mice and Facilities.
Animal procedures were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco, and the Harvard Medical Area Standing Committee on Animals, and followed NIH guidelines. Mice were housed in a pathogen-free barrier facility with a 12-h light/12-h dark cycle and allowed food and water ad libitum. Grn−/− mice (21) were on the C57BL/6J background (backcrossed more than eight generations). To generate GrnR493X mice, a targeting vector for the Grn locus was created with loxP and FRT sites flanking the neomycin-resistance cassette (Fig. S1A). Neomycin-resistance and thymidine kinase (TK) genes were used for positive and negative selection, respectively. The vector was electroporated into RF8 (75) 129SV/Jae murine embryonic stem cells and positive clones were selected. The positive clones were injected into C57BL/6J blastocysts to create chimeras, which were then bred with C57BL/6J mice to confirm germline transmission of the GrnR493X allele. Disruption of the Grn allele was confirmed by Southern blotting of ScaI/XcmI-digested genomic DNA with a probe hybridizing downstream of sequences included in the targeting vector and by PCR (Fig. S1A). GrnR493X mice used in the experiments shown in Figs. 1 A and B, 2, and 3 A and B, and Figs. S1 C and D, S3, and S4 had a Grn allele that harbored the neomycin-resistance cassette. The Neomycin cassette was later deleted by crossing with actin-FLPe mice (76), and deletion of the Neomycin cassette did not alter Grn mRNA levels in the kidney, liver, and brain (Fig. S1E). GrnR493X mice used in the experiments shown in Fig. 2 and Fig. S3 were on a mixed-background consisting of 75% C57BL/6J and 25% 129Sv/Jae. In all other experiments, mice were back-crossed onto the C57BL/6J background (for eight generations). Genotyping was performed either by PCR using the following primers: S1 (5′-AAGATTCCTCGCTGGGACATG-3′), S2 (5′-CAGGACATAGCGTTGGCTACCC-3′), and AS (5′-GAATGCTGGTGTCAGAGGGCC-3′), or by real-time PCR (Transnetyx). GrnR493X are available through the Jackson Laboratory Repository (JAX stock number 029919). Analyses of skin lesion development and survival were carried out as described previously (22).
Immunohistochemistry.
Immunostaining of brain sections and quantification of Iba1+ cells and synaptophysin+ puncta was performed as described previously (22). For p-TDP43 staining, a Mouse on Mouse Immunodetection Kit (BMK-2202; Vector Laboratories) was used to reduce background. Images were acquired on a Nikon C2 confocal microscope with a 60× objective. For analysis of lipofuscinosis, DAPI-stained brain sections were imaged and total autofluorescence intensity per field in the green channel was quantified using ImageJ.
Cycloheximide Treatment in Mice.
Mice were treated with 20 mg/kg body weight cycloheximide (Sigma) dissolved in PBS, injected intraperitoneally. After 4 h, mice were killed and Grn mRNA levels measure progranulin expression in the spleen and liver by qPCR.
BMDMs.
BMDMs were isolated from femurs and tibias of age-matched Grn+/+, GrnR493X/R493X, and Grn−/− mice. Bone marrow was flushed with ice-cold PBS and pooled. Cells were centrifuged at 250 × g for 10 min at room temperature. Cell pellets were resuspended in 5 mL of cell culture medium [DMEM/F12 supplemented with 20% heat-inactivated FBS HI-FBS (HyClone), 20% (vol/vol) L929 conditioned medium, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1% GlutaMAX (Gibco)] for plating onto cell culture dishes.
After 7 d, macrophages were replated in either 6-well plates (106 cells per well) or 12-well plates (0.5 × 106 cells per well) in DMEM/F12 supplemented with 10% HI-FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1% GlutaMAX (Gibco). To determine response to proinflammatory stimuli, cells were treated with 100 ng/mL LPS and 100 ng/mL IFN-γ for 6 or 24 h before harvesting for RNA. Gene expression was determined by qPCR.
MEFs.
MEFs were isolated from embryos, cultured in DMEM supplemented with 10% HI-FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Grn−/− MEFs were immortalized by serial passaging; GrnR493X MEFs were not immortalized, and experiments were performed at or before passage 5. For siRNA experiments, knockdown was performed using 50 nM siRNA (Smartpools; Dharmacon) with DharmaFECT 1 transfection reagent. After 72 h, gene expression was determined by qPCR. For ASO experiments, 100 nM ASOs were transfected into cells using Lipofectamine 2000 (6 µL per well of six-well plates; ThermoFisher) in 1 mL Opti-MEM (ThermoFisher). After 4 h, cells were refed complete media. Gene expression was determined by qPCR at 24 h posttransfection, and protein levels were determined by immunoblot analysis at 48 h posttransfection. ASOs were uniformly modified with 2′-O-methoxyethyl nucleotides and phosphorothioate linkages. For overexpression experiments, DNA was electroporated into MEFs using the Neon system (ThermoFisher) using 10-μL tips with a single pulse at 1,350 V for 30 ms, and then plated into six-well plates containing DMEM supplemented with HI-FBS. After 24 h, gene expression was determined by qPCR.
HDFs.
This study was approved by the Institutional Review Board and Ethics Committees of the University of California, San Francisco, and written informed consent was obtained in all cases. Skin biopsies were collected and cut into small pieces (44), and HDFs were allowed to expand in culture dishes in DMEM supplemented with 10% HI-FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Control HDFs for the GRN+/R493X HDFs, isolated from a family member without the mutation, have been reported (77). siRNA knockdown was performed using 50 nM total siRNA with DharmaFECT 1 transfection reagent. After 3 d, gene expression was determined by qPCR.
HeLa Cells.
GRN−/− HeLa cells were generated by electroporation of a CRISPR gRNA-Cas9 ribonucleoprotein complex (RNP). CRISPR gRNA targeting the signal sequence of progranulin and tracer RNA duplex was prepared fresh by heating at 95 °C for 5 min and cooled down to room temperature. RNA duplex (150 pmol) was assembled with Cas9 protein (150 pmol) in Opti-MEM to form a CRISPR-Cas9 protein (RNP) complex. Freshly prepared RNP complex was electroporated into HeLa cells suspended in 100 µL of Nucleofector solution R (Lonza) using program A24 on a Nucleofector device (Amaxa Biosystems). Two days after electroporation, cells were plated at clonal density, expanded, and screened for GRN KO by immunoblotting (Fig. S11).
GRN−/− HeLa cells were grown in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. For transfection, cells were plated at 150,000 cells in 2 mL media per well in a six-well plate, and transfected with 1 µg mCherry-PGRN wild-type or 1 µg mCherry-PGRN R493X plasmids using 100 µL Opti-MEM and 3 µL Fugene 6 transfection reagent (Promega). mCherry-PGRN expression vectors were generated by transferring a previously reported mCherry-PGRN cassette (57) into the AAVS1_Puro_PGK1_3xFLAG_Twin_Strep vector (#68375; Addgene). After 24 h, 15,000 cells were plated on 35-mm MatTek dishes and incubated overnight with 10 µg/mL of Alexa Fluor 647-labeled dextran (10,000 MW; ThermoFisher Scientific). One hour before imaging, cells were rinsed twice with warm PBS and replaced with fresh media.
Real-Time qPCR.
Total RNA was isolated from cultured cells or 100 mg of mouse liver, kidney, or brain using the RNeasy Mini kit (Qiagen) with on-column DNase digestion. RNA was reverse-transcribed to obtain cDNA using the iScript cDNA synthesis kit (Bio-Rad), and qPCR was performed using Power SYBR Green Master Mix (ThermoFisher) with a Bio-Rad CFX96 Real-Time System. Primers for each gene (Table S2) were validated by analysis of template titration and dissociation curves. Results for qPCR were normalized to the housekeeping gene Cyclo and evaluated by comparative CT method.
ELISA and Immunoblot Analysis.
An in-house sandwich ELISA for mouse progranulin was performed as described previously (21, 43). Tissues were homogenized in RIPA buffer containing protease inhibitors (Roche). Sample buffer was added to the lysates, and the samples were heated at 95 °C for 10 min before separating on SDS/PAGE gels. Proteins were transferred to nitrocellulose membranes using the iBlot system (ThermoFisher). After blocking and antibody incubations, membranes were incubated with SuperSignal West Pico or Femto enhanced chemiluminescent HRP substrate (ThermoFisher) and visualized using a Chemi-Doc system (Bio-Rad). HeLa cell lysates were prepared by lysing cells in ice-cold lysis buffer containing 1% Triton X-100 and protease and phosphatase inhibitors (Roche Diagnostics), followed by centrifugation at 20,817 × g for 5 min. The resultant supernatant was used to determine the protein concentration of the sample by Bradford assay reagent. Protein sample mixed with sample buffer containing 50 mM Tris (2-carboxyethyl) phosphine (TCEP) was heated at 70 °C for 15 min before resolving on a 4–20% Tris-Glycine Mini-PROTEAN precast polyacrylamide gel (Bio-Rad) (78), and transferred onto nitrocellulose membrane. The membrane blot was blocked in 5% milk, incubated in primary antibody dissolved in 5% BSA, followed by incubation with HRP-conjugated secondary antibody prepared in 5% milk. Next, 5% milk or 5% BSA were prepared in Tris buffered saline (TBS) containing 0.1% Tween 20. The membrane was developed with SuperSignal West Pico or Femto chemiluminescent substrate (ThermoFisher Scientific) and signals were detected using the VersaDoc imaging system (Bio-Rad).
Antibodies.
Primary antibodies used for immunoblot analysis include an anti-mouse progranulin polyclonal antibody that recognizes an epitope between amino acids 198 and 214 (79), an anti-human progranulin polyclonal antibody (NBP1-87324; Novus Biologicals) that recognizes an epitope in amino acids 140–285, an anti-mouse apoB polyclonal antibody (sc-11795; Santa Cruz), an anti-GAPDH monoclonal antibody (MMS-580S; Covance), an anti-UPF1 polyclonal antibody (A301-902A; Bethyl Laboratories), and an anti-UPF2 polyclonal antibody (A303-929A; Bethyl Laboratories), an anti-human progranulin antibody (HPA008763; Sigma) for Fig. S11, and an anti-Vinculin antibody (V4505; Sigma). HRP-conjugated goat anti-rabbit IgG and goat anti-mouse IgG secondary antibodies were from Santa Cruz. Primary antibodies used for immunofluorescence were an anti-FLAG monoclonal antibody (M2; Sigma) and anti-LAMP1 rat monoclonal antibody (clone 1D4B-c; Developmental Studies Hybridoma Bank). Secondary antibodies used were Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-rat IgG (ThermoFisher). Primary antibodies used for immunohistochemistry include an anti-Iba1 polyclonal antibody (019-19741; Wako), an anti-TDP-43 polyclonal antibody that recognizes the N terminus (10782-2-AP; Protein Tech), an anti–p-TDP43 monoclonal antibody (TIP-PTD-M01; Cosmo Bio), an anti-TuJ1 monoclonal antibody (MMS-435P; Covance), and an antisynaptophysin monoclonal antibody (S5768; Sigma).
Microscopy.
For HeLa cells, images were acquired for mCherry-PGRN with a 561-nm laser line and for dextran with a 640-nm laser line, using a 60× 1.49 NA objective on a spinning disk confocal microscope [UltraVIEW VoX system (PerkinElmer) including a Nikon Ti-E Eclipse inverted microscope and a Yokogawa CSU-X1 spinning disk confocal scan head]. The microscope was driven by Volocity software (PerkinElmer) and images (1,000 × 1,000 pixels) were acquired at 2 × 2 binning with a 14-bit EMCCD camera (C9100-50; Hamamatsu Photonics). For immunohistochemistry with MEFs, cells grown on glass-bottom 96-well plates were fixed in 4% (wt/vol) paraformaldehyde, permeabilized with 0.1% saponin, immunostained, and imaged on a spinning disk confocal (491 CSU-X1; Yokogawa) set up on a Nikon 492 Eclipse Ti inverted microscope with a 60× Plan Apo λ 1.4 NA objective (Nikon) and an iXon Ultra 897 EMCCD camera (Andor). Buffers used for blocking and antibody incubations included 5% normal goat serum and 0.05% saponin.
Quantitative Colocalization Analysis.
Images were quantified for the colocalization of mCherry-PGRN (wild-type) or mCherry-PGRN (R493X) with dextran-labeled lysosomes using Cell Profiler (80). The Mander’s colocalization coefficient was measured using the “measure correlation function” in Cell Profiler by setting the Otsu automatic intensity thresholding for puncta of size >0.48 µm.
Statistical Analysis.
Data are presented as mean ± SD (except, data in Fig. 5D are mean ± SEM) and were analyzed with GraphPad Prism software (GraphPad) using the statistical tests described in the figure legends. P values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Florence Mahuteau-Betzer (Institut Curie) for kindly providing NMDI1; Sarah Coughlin for early contributions to the project; Laura Mitic (University of California, San Francisco) and members of the R.V.F. and T.C.W. laboratory for helpful discussions; Lawrence Gardner (New York University) for advice; and Gary Howard for editorial assistance. This work is supported by National Institutes of Health Grants AG023501 (to R.V.F.), GM105718 (to S.M.F.), AG047270 (to S.M.F.), and AG047339 (to A.D.N.); VA Merit Award BX002978 (to E.J.H.); a Bluefield Project postdoctoral fellowship (to S.D.); the Consortium for Frontotemporal Dementia Research (R.V.F., T.C.W., E.J.H., S.M.F., and B.L.M.); and the Gladstone Institutes. T.C.W. is an investigator of the Howard Hughes Medical Institute. The Gladstone Institutes received support from National Center for Research Resources Grant RR18928.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722344115/-/DCSupplemental.
References
- 1.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 2.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 3.Gass J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 4.Cruts M. 2012 The AD & FTLD mutation database. Available at www.molgen.vib-ua.be/FTDMutations. Accessed November 8, 2016.
- 5.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gijselinck I, Van Broeckhoven C, Cruts M. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: An update. Hum Mutat. 2008;29:1373–1386. doi: 10.1002/humu.20785. [DOI] [PubMed] [Google Scholar]
- 7.Yu CE, et al. The spectrum of mutations in progranulin: A collaborative study screening 545 cases of neurodegeneration. Arch Neurol. 2010;67:161–170. doi: 10.1001/archneurol.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386:1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37:388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Neumann M, Tolnay M, Mackenzie IR. The molecular basis of frontotemporal dementia. Expert Rev Mol Med. 2009;11:e23. doi: 10.1017/S1462399409001136. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen AD, Nguyen TA, Martens LH, Mitic LL, Farese RV., Jr Progranulin: At the interface of neurodegenerative and metabolic diseases. Trends Endocrinol Metab. 2013;24:597–606. doi: 10.1016/j.tem.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruts M, Van Broeckhoven C. Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet. 2008;24:186–194. doi: 10.1016/j.tig.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Neary D, Snowden J, Mann D. Frontotemporal dementia. Lancet Neurol. 2005;4:771–780. doi: 10.1016/S1474-4422(05)70223-4. [DOI] [PubMed] [Google Scholar]
- 14.Smith KR, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Damme P, et al. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol. 2008;181:37–41. doi: 10.1083/jcb.200712039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan CL, et al. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 2009;10:130. doi: 10.1186/1471-2202-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gass J, et al. Progranulin regulates neuronal outgrowth independent of sortilin. Mol Neurodegener. 2012;7:33. doi: 10.1186/1750-1326-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, et al. Conversion of proepithelin to epithelins: Roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–878. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 19.Yin F, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207:117–128. doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens LH, et al. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest. 2012;122:3955–3959. doi: 10.1172/JCI63113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lui H, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka Y, Matsuwaki T, Yamanouchi K, Nishihara M. Increased lysosomal biogenesis in activated microglia and exacerbated neuronal damage after traumatic brain injury in progranulin-deficient mice. Neuroscience. 2013;250:8–19. doi: 10.1016/j.neuroscience.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210:991–1002. doi: 10.1083/jcb.201502029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Riz M, et al. Cerebrospinal fluid progranulin levels in patients with different multiple sclerosis subtypes. Neurosci Lett. 2010;469:234–236. doi: 10.1016/j.neulet.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feneberg E, et al. Progranulin as a candidate biomarker for therapeutic trial in patients with ALS and FTLD. J Neural Transm (Vienna) 2016;123:289–296. doi: 10.1007/s00702-015-1486-1. [DOI] [PubMed] [Google Scholar]
- 27.Kayasuga Y, et al. Alteration of behavioural phenotype in mice by targeted disruption of the progranulin gene. Behav Brain Res. 2007;185:110–118. doi: 10.1016/j.bbr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 28.Petkau TL, et al. Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis. 2012;45:711–722. doi: 10.1016/j.nbd.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed Z, et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311–324, and erratum (2010) 177:2146. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filiano AJ, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J Neurosci. 2013;33:5352–5361. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin F, et al. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: A mouse model of frontotemporal dementia. FASEB J. 2010;24:4639–4647. doi: 10.1096/fj.10-161471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arrant AE, Filiano AJ, Warmus BA, Hall AM, Roberson ED. Progranulin haploinsufficiency causes biphasic social dominance abnormalities in the tube test. Genes Brain Behav. 2016;15:588–603. doi: 10.1111/gbb.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen-Plotkin AS, et al. Genetic and clinical features of progranulin-associated frontotemporal lobar degeneration. Arch Neurol. 2011;68:488–497. doi: 10.1001/archneurol.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrabal R, Chen Z, James S, Bennett HP, Ni F. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat Struct Biol. 1996;3:747–752. doi: 10.1038/nsb0996-747. [DOI] [PubMed] [Google Scholar]
- 35.Tolkatchev D, et al. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 2008;17:711–724. doi: 10.1110/ps.073295308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palfree RG, Bennett HP, Bateman A. The evolution of the secreted regulatory protein progranulin. PLoS One. 2015;10:e0133749. doi: 10.1371/journal.pone.0133749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 38.Ghoshal N, Dearborn JT, Wozniak DF, Cairns NJ. Core features of frontotemporal dementia recapitulated in progranulin knockout mice. Neurobiol Dis. 2012;45:395–408. doi: 10.1016/j.nbd.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koike M, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultheis PJ, et al. Atp13a2-deficient mice exhibit neuronal ceroid lipofuscinosis, limited α-synuclein accumulation and age-dependent sensorimotor deficits. Hum Mol Genet. 2013;22:2067–2082. doi: 10.1093/hmg/ddt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thelen M, et al. Disruption of the autophagy-lysosome pathway is involved in neuropathology of the nclf mouse model of neuronal ceroid lipofuscinosis. PLoS One. 2012;7:e35493, and erratum (2012) 7, 10.1371/annotation/a4b06d46-8eb9-4d15-a15a-41bf4b5ccb8b. doi: 10.1371/journal.pone.0035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traina G, et al. Lipofuscin accumulation and gene expression in different tissues of mnd mice. Mol Neurobiol. 2012;45:247–257. doi: 10.1007/s12035-012-8248-y. [DOI] [PubMed] [Google Scholar]
- 43.Minami SS, et al. Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat Med. 2014;20:1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almeida S, et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2:789–798. doi: 10.1016/j.celrep.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: Features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter MS, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usuki F, et al. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich’s disease. Ann Neurol. 2004;55:740–744. doi: 10.1002/ana.20107. [DOI] [PubMed] [Google Scholar]
- 49.Keeling KM, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nickless A, et al. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat Med. 2014;20:961–966. doi: 10.1038/nm.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Vuong JK, Zhang M, Stork C, Zheng S. Inhibition of nonsense-mediated RNA decay by ER stress. RNA. 2017;23:378–394. doi: 10.1261/rna.058040.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banihashemi L, Wilson GM, Das N, Brewer G. Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay. Mol Cell Biol. 2006;26:8743–8754. doi: 10.1128/MCB.02251-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nomakuchi TT, Rigo F, Aznarez I, Krainer AR. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol. 2016;34:164–166. doi: 10.1038/nbt.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Y, Brady OA, Meng PS, Mao Y, Hu F. C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS One. 2011;6:e21023. doi: 10.1371/journal.pone.0021023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu F, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai XH, et al. ADAMTS-7, a direct target of PTHrP, adversely regulates endochondral bone growth by associating with and inactivating GEP growth factor. Mol Cell Biol. 2009;29:4201–4219. doi: 10.1128/MCB.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kessenbrock K, et al. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J Clin Invest. 2008;118:2438–2447. doi: 10.1172/JCI34694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suh HS, Choi N, Tarassishin L, Lee SC. Regulation of progranulin expression in human microglia and proteolysis of progranulin by matrix metalloproteinase-12 (MMP-12) PLoS One. 2012;7:e35115. doi: 10.1371/journal.pone.0035115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand S, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin L, et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou X, Sullivan PM, Sun L, Hu F. The interaction between progranulin and prosaposin is mediated by granulins and the linker region between saposin B and C. J Neurochem. 2017;143:236–243. doi: 10.1111/jnc.14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neill T, et al. EphA2 is a functional receptor for the growth factor progranulin. J Cell Biol. 2016;215:687–703. doi: 10.1083/jcb.201603079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finch N, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132:583–591. doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 67.Graille M, Séraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 68.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 69.DeVos SL, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017;9:eaag0481. doi: 10.1126/scitranslmed.aag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becker LA, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scoles DR, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544:362–366. doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci. 2017;20:497–499. doi: 10.1038/nn.4508. [DOI] [PubMed] [Google Scholar]
- 74.Niks EH, Aartsma-Rus A. Exon skipping: A first in class strategy for Duchenne muscular dystrophy. Expert Opin Biol Ther. 2017;17:225–236. doi: 10.1080/14712598.2017.1271872. [DOI] [PubMed] [Google Scholar]
- 75.Meiner VL, et al. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: Evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc Natl Acad Sci USA. 1996;93:14041–14046. doi: 10.1073/pnas.93.24.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 77.Cenik B, et al. Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: Rational therapeutic approach to frontotemporal dementia. J Biol Chem. 2011;286:16101–16108. doi: 10.1074/jbc.M110.193433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holler CJ, Taylor G, Deng Q, Kukar T. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro. 2017;4:ENEURO.0100-17.2017. doi: 10.1523/ENEURO.0100-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholson AM, et al. Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun. 2016;7:11992. doi: 10.1038/ncomms11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.