Significance
Assembly factors play key roles in the biogenesis of many multisubunit protein complexes regulating their stability, activity, or incorporation of essential cofactors. The bacterial assembly factor SdhE (also known as Sdh5 or SDHAF2 in mitochondria) promotes covalent attachment of flavin adenine dinucleotide (FAD) to SdhA and hence the assembly of functional succinate:quinone oxidoreductase (also known as complex II). Here, we present the crystal structure of Escherichia coli SdhE bound to its client protein SdhA. This structure provides unique insight into SdhA assembly, whereby SdhE constrains unassembled SdhA in an “open” conformation, promoting covalent attachment of FAD, but renders the holoprotein incapable of substrate catalysis. These data also provide a structural explanation for the loss-of-function mutation, Gly78Arg, in SDHAF2, which causes hereditary paraganglioma 2.
Keywords: SdhE, flavinylation, structure, SdhA, assembly
Abstract
Succinate:quinone oxidoreductase (SQR) functions in energy metabolism, coupling the tricarboxylic acid cycle and electron transport chain in bacteria and mitochondria. The biogenesis of flavinylated SdhA, the catalytic subunit of SQR, is assisted by a highly conserved assembly factor termed SdhE in bacteria via an unknown mechanism. By using X-ray crystallography, we have solved the structure of Escherichia coli SdhE in complex with SdhA to 2.15-Å resolution. Our structure shows that SdhE makes a direct interaction with the flavin adenine dinucleotide-linked residue His45 in SdhA and maintains the capping domain of SdhA in an “open” conformation. This displaces the catalytic residues of the succinate dehydrogenase active site by as much as 9.0 Å compared with SdhA in the assembled SQR complex. These data suggest that bacterial SdhE proteins, and their mitochondrial homologs, are assembly chaperones that constrain the conformation of SdhA to facilitate efficient flavinylation while regulating succinate dehydrogenase activity for productive biogenesis of SQR.
Succinate:quinone oxidoreductase (SQR) is a multisubunit membrane-associated enzyme found in the cytoplasm of bacteria and in the matrix of mitochondria (where it is commonly termed complex II). The enzyme is central to cellular metabolism and energy conversion, contributing to the tricarboxylic acid cycle and the electron transport chain. It catalyzes the oxidation of succinate to fumarate, which is coupled to electron transfer through flavin adenine dinucleotide (FAD) and three Fe–S clusters, resulting in the reduction of the electron carrier ubiquinone to ubiquinol. The overall architecture of the bacterial and mitochondrial complexes is highly conserved, with both enzymes forming a heterotetrameric protein complex (1, 2). The catalytic flavoprotein (SdhA in bacteria, Sdh1 in yeast, and SDHA in humans) contains a binding site for dicarboxylic acid and an N3-histidyl-8α-FAD linkage (1, 2). The covalent attachment of FAD to SdhA is essential for the oxidation of succinate by the enzyme (3). In the mature complex, SdhA connects to the inner membrane through interaction with the Fe–S protein SdhB, which makes direct contact to both integral membrane subunits, SdhC and SdhD. Ubiquinone binds at the interface of SdhB and the two membrane proteins (SdhC and SdhD), with the cofactors in SQR forming a direct path (∼40 Å long) for the transfer of electrons into the respiratory chain (2). Significantly, loss of complex II activity in humans is associated with Leigh syndrome and tumor syndromes including hereditary paraganglioma (PGL) (4–8).
The assembly of SQR is assisted by several assembly factors involved in cofactor biogenesis and regulation of assembly intermediates (9–12). The most widely conserved SQR assembly factor (10, 13, 14) is termed SdhE in bacteria [also known as Sdh5 in yeast and SDH assembly factor 2 (SDHAF2) or SDH5 in humans]. Initially characterized in Saccharomyces cerevisiae, Sdh5 is essential for the assembly of active complex II promoting the covalent attachment of FAD to Sdh1 (10). Likewise, bacterial SdhE is also required for SQR and fumarate reductase (FRD) activity promoting flavinylation of SdhA and FrdA (subunit A of the quinol:FRD), respectively (13, 15). The importance of this protein family, in normal cellular metabolism, is manifested by the identification of a mutation in human SDHAF2 (Gly78Arg), which is linked to an inherited neuroendocrine disorder, PGL2 (10). Currently, however, the role of SdhE in flavinylation remains poorly understood. To date, three different modes of action for SdhE/Sdh5 have been proposed, suggesting that SdhE facilitates the binding and delivery of FAD (13), acts as a chaperone for SdhA (10), or catalyzes the attachment of FAD (10). Moreover, the requirement for SdhE in SdhA biogenesis remains controversial, as recent studies have demonstrated that flavinylation of bacterial, archaeal, and mitochondrial SdhA can still occur in the absence of the assembly factor (16–19). In this study, we have determined the crystal structure of the Escherichia coli flavinylation factor SdhE in complex with its client protein SdhA to 2.15-Å resolution. This three dimensional structure of an SQR assembly intermediate provides valuable insights into the evolutionary conserved process of flavoprotein assembly.
Results and Discussion
Structure of SdhA in Complex with Its Assembly Factor SdhE.
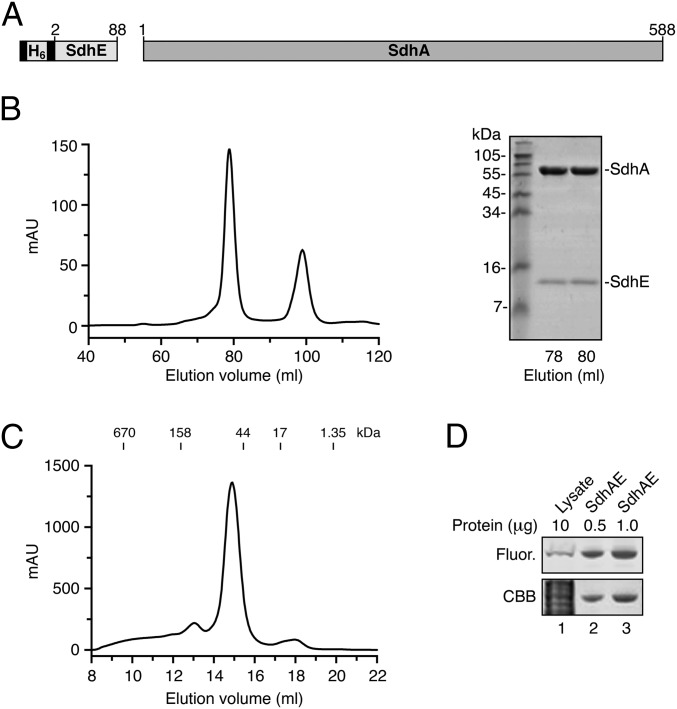
To obtain E. coli SdhA in complex with its assembly factor E. coli SdhE, we coexpressed untagged recombinant SdhA together with recombinant His6-tagged SdhE in E. coli (Fig. 1A). By using Ni2+-affinity chromatography, we isolated a mixture of free His6-SdhE and His6-SdhE in complex with untagged SdhA. A homogenous preparation of the SdhA/SdhE (SdhAE) complex was then purified by size-exclusion chromatography (Fig. 1B). The SdhAE complex eluted from an analytical size-exclusion column at a volume corresponding to the molecular weight of a heterodimer (Fig. 1C) and FAD was covalently bound (to SdhA) within the binary complex (10) (Fig. 1D). To elucidate the atomic structure of the SdhAE protein assembly, we crystallized the complex and determined its structure to 2.15-Å resolution by X-ray crystallography (Fig. 2A and Table S1).
Fig. 1.
Isolation of the binary protein complex containing SdhE, SdhA, and covalently bound FAD. (A) Schematic illustration showing His6-tagged SdhE and untagged SdhA constructs used in this study. The numbers reflect amino acid residues of the authentic portion of the protein constructs. (B) Elution profile (Left) showing separation of the SdhAE complex (first peak) from free SdhE (second peak) using size-exclusion chromatography. (Right) Confirmation of purified SdhAE complex by Coomassie Brilliant Blue (CBB)-stained SDS/PAGE. (C) Elution profile of purified SdhAE relative to indicated protein standards analyzed by size-exclusion chromatography. The molecular weight of the SdhAE complex is estimated to be 88.5 kDa. (D) Detection of FAD fluorescence (Fluor.) in protein samples (Upper) of E. coli lysate (lane 1) and purified SdhAE complex (lanes 2 and 3) following separation by SDS/PAGE. (Lower) The same gel following CBB staining. Only the region of the gel containing SdhA is shown.
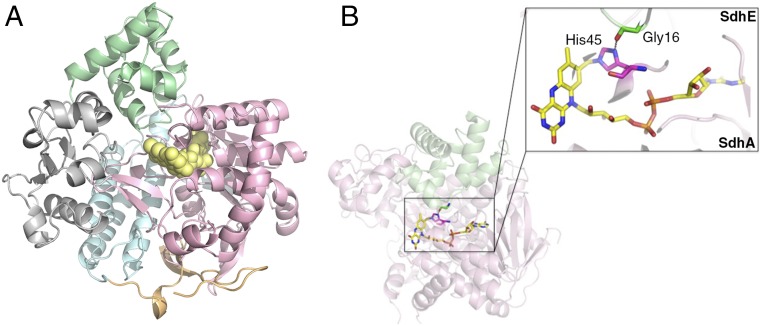
Fig. 2.
Structure of the SdhAE complex. (A) The SdhE protein (green) forms a 1:1 complex with SdhA. SdhE is positioned between the SdhA FAD-binding (pink) and capping (gray) domains. The helical and C-terminal domains of the SdhA protein are shown in pale blue and orange, respectively. FAD (yellow spheres) is covalently bound to the SdhA protein. (B) In the SdhAE complex, residue His45 (SdhA; pink) is covalently bonded to the FAD cofactor (yellow). The carbonyl group of SdhE residue Gly16 (green) participates in a hydrogen bonding interaction with SdhA His45.
The crystal contains two almost identical SdhAE complexes in the asymmetric unit. The final SdhAE model includes residues 1–583 from the SdhA protein, apart from two regions: residues 52–67 and 107–137 in addition to residues 2–83 from the SdhE protein. The structure of SdhA is composed of four domains (20): an FAD-binding domain (residues 1–245 and 351–431), which includes a Rossmann-type fold and provides the binding site for FAD (Fig. 2A, pink); a capping domain composed of residues 245–351 (Fig. 2A, gray); a helical domain composed of residues 431–547 (Fig. 2A, blue); and a C-terminal domain composed of residues 547–583 (Fig. 2A, orange). FAD was well resolved in the electron density map and, consistent with the in-gel assay (Fig. 1D), was covalently attached to SdhA via a N3-histidyl-8α-FAD linkage (1.6 Å) between the Nε2 atom of His45 and the C8 atom of the isoalloxazine group (Fig. 2B). Although a dicarboxylate is required for the flavinylation reaction (21), the SdhAE structure presented here captures the assembly intermediate after flavinylation has occurred. Consistently, there was no evidence in the electron density for the presence of a dicarboxylate associated with SdhA. SdhE forms a single compact domain (Fig. 2A, green) composed of five α-helices and interfaces closely with SdhA, where it is wedged between the FAD and capping domains.
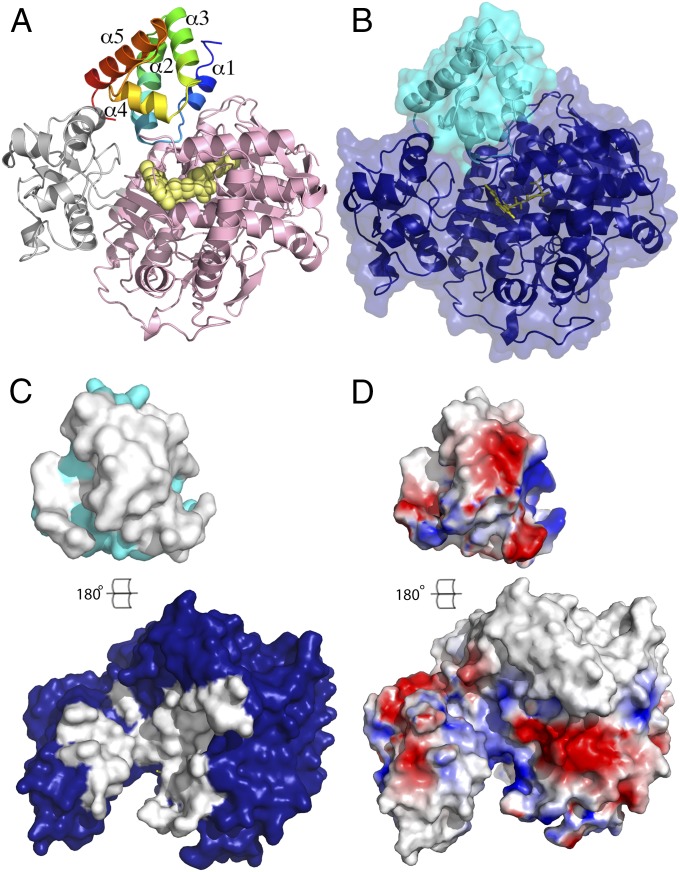
The SdhE protein forms an intimate complex with SdhA, with a buried surface area of 1,455 Å2 (Fig. 3 A–C). Three regions of SdhE make contact with the SdhA protein, namely residues 5–25 (which encompass helix α1, the N terminus of α2, and the loop that connects these two regions), residues 47–61 (which form helix α4), and residues 80–86 (which form the C terminus; Fig. 3A and Fig. S1). Overall, the interface is composed of electrostatic interactions, 18 hydrogen bonds, and hydrophobic contacts and exhibits a high degree of charge complementarity (Fig. 3D). Importantly, the interface defined by the SdhAE structure is consistent with a recent biochemical study that identified two SdhE residues (Arg8 and Met17), which could be photocrosslinked to SdhA or FrdA (18). Consistent with an important functional role for residues located within this interface, mutagenesis of Arg68 and Tyr71 in yeast Sdh5 (22) (equivalent to Arg8 and Trp11 in E. coli SdhE) prevented Sdh1 flavinylation. Similarly, the inherited mutation coding for Gly78Arg in human SDHAF2 (equivalent to Gly16 in E. coli SdhE, which is located in the loop between the α1 and α2 helices; Fig. S1) strongly destabilizes the SDHAF2 interaction with SDHA (10, 23). Collectively, these data suggest that the loop that connects the α1 and α2 helices of SdhE plays a crucial role in the stability of the complex. SdhA interacts with the SdhE protein in three main regions: residues 38–50, 138–143, and 210–217 of the FAD domain (which frame the FAD binding site); residues 247–251 of the upper surface of the capping domain; and residues 505–515 at the tip of the helical region of the FAD domain (nomenclature defined in ref. 18). Consistent with the SdhAE structure, FrdA residues 456–462, which are equivalent to the helical region in SdhA, were shown to photo–cross-link to Met17 of SdhE (18).
Fig. 3.
SdhE forms an intimate contact with SdhA. (A) The SdhE protein is rainbow-colored (blue at the N terminus to red at the C terminus). Secondary structural elements are labeled (helices α1−α5). The capping domain of SdhA is colored gray and the remainder of SdhA is colored pink. The FAD cofactor is represented as yellow spheres. (B) Surface representation of the SdhAE complex with SdhE shown in cyan and SdhA in dark blue. The FAD cofactor is represented as yellow sticks. (C) “Open book” representation of the SdhAE complex from B (SdhE, cyan; SdhA, dark blue). The protein surfaces that are buried in the complex are highlighted in white. (D) The SdhAE complex as represented in C. The protein surfaces are colored according to the electrostatic potentials (red, negatively charged; blue, positively charged; white, uncharged). The protein–protein interface in the SdhAE complex shows charge complementarity.
Rotation of the SdhA Capping Domain Accompanies Formation of the SdhAE Complex.
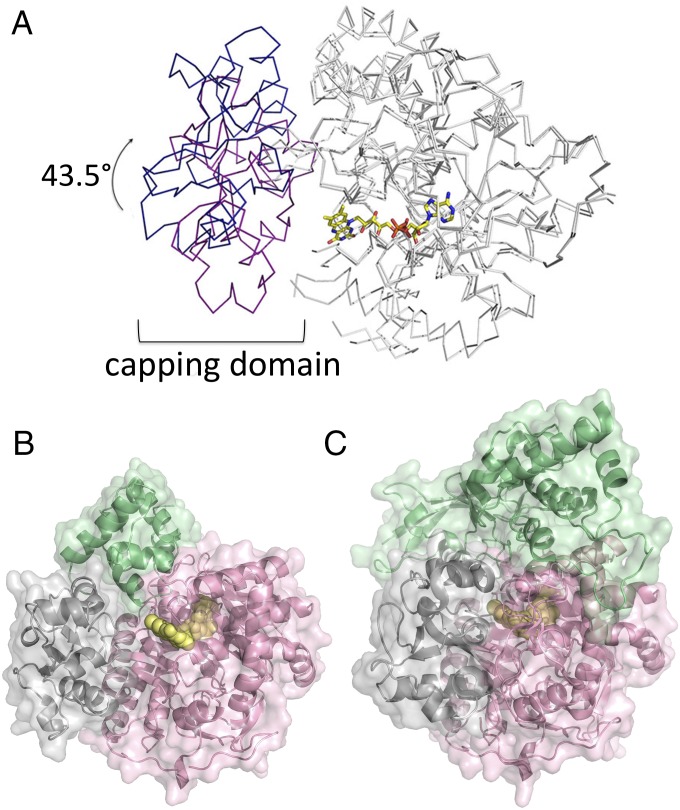
Superposition of SdhA from the SdhAE complex with SdhA from the intact SQR enzyme [Protein Data Bank (PDB) ID code 2WDQ; chain A (24)] reveals a number of significant differences between the two structures (rmsd of 4.14 Å for 536 common Cα positions). Although most of the structure of the FAD-binding domain is similar in both states, there are significant changes to three distinct regions of SdhA, specifically that residues 40–50 are displaced by as much as 7 Å, residues 88–105 are displaced by as much as 8 Å, and the capping domain (residues 246–350) is displaced by as much as 13 Å. In the SdhAE complex, the capping domain of SdhA is rotated away from the FAD-binding domain by 43.5° [as analyzed by DynDom (25)] relative to that observed for the intact SQR enzyme (Fig. 4A). The rotation of the capping domain, coupled with the apparent mobility of residues 52–67 and 107–137, exposes the isoalloxazine group of the bound FAD in the SdhAE complex to solvent (solvent-accessible surface area is 64 Å2; Fig. 4B), whereas the cofactor is completely buried in the intact SQR structure [PDB ID code 2WDQ (24); solvent accessible area is 5 Å2; Fig. 4C].
Fig. 4.
The position of the SdhA capping domain is markedly different in the SdhAE assembly relative to SQR. (A) Comparison of the orientation of the SdhA capping domain in the structure of SQR (PDB ID code 2WDQ; red) (24) with that in the SdhAE complex (blue). In SdhAE, the capping domain is observed in an orientation rotated 43.5° away from the FAD-binding domain. The FAD-binding, helical, and C-terminal domain structures for the SdhA protein within SdhAE and SQR complexes are shown in gray. The FAD cofactor is represented as yellow sticks. (B) Surface representation of the SdhAE complex (SdhE, green; SdhA capping domain, gray; FAD-binding, helical, and C-terminal domains, pink; FAD cofactor, yellow spheres). (C) Surface representation of the SdhAB structure within SQR (24) (PDB ID code 2WDQ; SdhB, green; SdhA colored as in B).
Interestingly, in contrast to the SdhAE structure, in which the FAD binding site remains exposed in the presence of covalently bound FAD, the FAD binding pocket of most FAD-bound enzymes is enclosed. For example, the incorporation of FAD into l-aspartate oxidase triggers structural reorganization of the capping domain relative to the FAD-binding domain (26). In this case, the apo protein exists in an open conformation in which both domains are well separated. However, upon binding of FAD, the capping domain rotates 27° toward the FAD-binding domain, thereby closing the binding site. This type of “closed” conformation is commonly observed in these types of enzymes following FAD incorporation (27–29). Hence, our FAD-bound structure of SdhA in complex with SdhE is unique in presenting an FAD-bound, open arrangement of the FAD-binding and capping domains, and helps to reveal the role of SdhE in stabilizing this conformation.
Direct Contact Between SdhE and the Flavin Binding Site of SdhA Facilitates Covalent Flavinylation.
In contrast to the significant structural rearrangements in SdhA, which accompany formation of the SdhAE complex, the structure of SdhE is virtually unchanged. Superposition of the structure of SdhE alone (PDB ID code 1X6I) (30) with SdhE in the SdhAE complex reveals an rmsd of 0.39 Å (for 75 common core Cα atoms), with minor structural rearrangements localized to the C terminus. Interestingly, the SdhE loop, between helices α1 and α2, does not change conformation upon complex formation, despite its central location at the SdhAE interface. This part of the SdhE structure appears to be tailored for its interaction with SdhA and therefore may contribute directly to catalysis of SdhA flavinylation. Nevertheless, the autocatalytic flavinylation of several FAD-dependent enzymes has been well described in the literature. The mechanism of catalysis is proposed to involve five features (31): (i) abstraction of a proton from the FAD C8 methyl group (by bases near the residues that tether the flavin moiety); (ii) stabilization of the negative charge at the N1-C2=O2 locus of the isoalloxazine moiety; (iii) attack by the histidyl-imidazolyl group at C8α to form a covalent bond between the polypeptide chain and the reduced flavin; (iv) protonation of the negative charge at the N5 position of the reduced isoalloxazine ring system by a neighboring amide group; and (v) reoxidation of the reduced flavin by electron transfer to oxygen. As detailed in Table S2, the majority of the crucial interactions that support these steps are maintained not only in the final structures of bovine and E. coli enzymes but also in the SdhAE complex described here. Therefore, these data suggest that SdhE does not play a direct role in the catalysis of SdhA flavinylation, but rather supports a conformation of SdhA that facilitates autocatalysis.
Part of the interaction network between SdhA and SdhE includes a hydrogen bond between the carbonyl oxygen of Gly16 in SdhE and the Nδ1 atom of SdhA residue His45, which is covalently bound to the FAD cofactor (Fig. 2B). This glycine residue is highly conserved in SdhE/Sdh5 proteins across a broad range of species including bacteria, fungi, plants, and animals (Fig. S1B) (14, 32). Significantly, a point mutation at the equivalent Gly residue in human mitochondrial SDHAF2 (Gly78Arg) is associated with PGL2 disease, and this mutation dramatically impairs its interaction with human SDHA (10, 23). Consistent with an important role for this highly conserved Gly residue in the function of SDHAF2, tumor cells isolated from patients with PGL2 contain extremely low levels of flavinylated SDHA (10). Similarly, the equivalent mutation (Gly16Arg) in bacterial SdhE inhibits flavinylation of SdhA (32). Therefore, we propose that the hydrogen bond formed between SdhE residue Gly16 and SdhA residue His45 is crucial for efficient flavinylation of SdhA. This interaction ensures that the Nδ1 atom at SdhA residue His45 is protonated, promoting nucleophilic attack by the deprotonated Nε2 atom at FAD C8α to facilitate covalent bond formation between the polypeptide chain and the reduced flavin. This hydrogen bond may also lock the side chain of His45 in an orientation conducive to this reaction.
SdhE Stabilizes SdhA in a Nonactive Conformation During Assembly.
The SdhAE complex also reveals that flavinylation of SdhA does not trigger release of SdhE. Instead, SdhE is wedged between the FAD-binding and capping domains of SdhA, relocating its active site residues (24). Specifically, the side chains of SdhA residues His242, Thr254, Glu255, and Arg286 are displaced by distances of as much as 9.0 Å away from the dicarboxylate-binding site (in comparison with their positions in the SQR structure) (24), which is located between the isoalloxazine ring of FAD and the capping domain (2, 24) (Fig. 5). We propose that the significant positional displacement of these residues during SdhAE assembly prevents oxidation of succinate by the complex. This disruption of the active site is likely important in preventing electron “leakage” from free SdhA (via succinate dehydrogenase activity) and hence the subsequent production of damaging reactive oxygen species. So how does SdhA progress in the assembly pathway and associate with the iron–sulfur protein SdhB? Given that the surface area of the SdhA/SdhB interface in the SQR structure (2,400 Å2) (24) is significantly greater than the interface between SdhA and SdhE in the SdhAE complex (1,455 Å2), it is likely that the binding affinity of flavinylated SdhA is stronger for SdhB than it is for SdhE (33). Displacement of SdhE therefore likely occurs via competition with SdhB after the covalent attachment of the FAD, which allows “closure” of the capping and FAD-binding domains and repositioning of the catalytic residues, resulting in activation of the enzyme.
Fig. 5.
The influence of bound SdhE on catalytic residues in SdhA. Superposition of the SdhA proteins from the SdhAE (gray) and SQR [PDB ID code 2WDQ (24); blue] structures. For clarity, the FAD cofactor from the SdhAE structure only is represented in yellow. Active site residues (labeled) are displaced by as much as 9.0 Å away from the substrate binding site in the SdhAE complex relative to SQR.
Mechanism of Action of SdhE in the Biogenesis of Holo-SdhA.
Recently, it was proposed that SdhE docks onto its target flavoprotein following closure of the capping domain over noncovalently bound FAD, and, as such, maintains the flavoprotein in the closed state to facilitate flavinylation (18, 19). In contrast to this recently proposed model, our structure clearly demonstrates that SdhE, in separating the FAD-binding and capping domains, holds holo-SdhA in an open conformation. Therefore, we propose an alternative pathway, at least for the biogenesis of holo-SdhA (Fig. 6). In the absence of SdhE, FAD binds noncovalently to the FAD binding domain of folded apo-SdhA. At this point, in the presence of dicarboxylate, spontaneous flavinylation may occur (Fig. 6, path I); however, under certain conditions and in certain cell types, this process is very inefficient (10, 13, 18), possibly because of nonproductive pathways of misfolding or unregulated assembly of apo-SdhA (Fig. 6, path III). In path II, SdhE and FAD bind to folded apo-SdhA sequentially (as illustrated in Fig. 6) or simultaneously. The docking of SdhE to SdhA not only maintains the capping domain of SdhA in an open conformation but also facilitates the formation of a critical hydrogen bond between the carbonyl oxygen of SdhE residue Gly16 and the Nδ1 atom of SdhA residue His45, which stabilizes SdhA in a state that is conducive to highly efficient autocatalytic flavinylation. The limited efficiency of flavinylation in the absence of SdhE (18) is most likely the result of the loss of exclusive protonation of the Nδ1 atom and tethering of the side chain of His45 in SdhA that forms the N3-histidyl-8α-FAD linkage. Following flavinylation of SdhA, SdhE remains bound to SdhA, thereby stabilizing holo-SdhA in an inactive conformation by displacement of catalytic residues (required for oxidation of succinate). This is likely important to prevent electron leakage and hence oxidative damage in the cell. Such a role may be particularly important under aerobic conditions in E. coli in which SQR is expressed (ref. 34 and references therein). Finally, we speculate that holo-SdhB, by virtue of a higher affinity for SdhA, displaces SdhE from SdhA, allowing the capping domain of SdhA to close over the FAD binding domain. Within this model, we cannot exclude the possibility that other factors act to passively or actively displace SdhE from SdhA before SdhB docking. Intriguingly, mitochondria contain an additional assembly factor, Sdh8 (also known as SDHAF4), which forms a stable complex with flavinylated SdhA and seemingly facilitates its assembly with SdhB (11). Nevertheless, a bacterial homolog (or functional equivalent) of Sdh8 has yet to be described. The assembly intermediate SdhAB subsequently associates with membrane-integrated SQR subunits to form the functional complex. In summary, SdhE has a dual function to promote efficient flavinylation of SdhA while simultaneously neutralizing its succinate dehydrogenase activity during assembly.
Fig. 6.
Proposed SdhA assembly pathway. Path I represents flavinylation of SdhA occurring in the absence of SdhE. In this case, the yield of flavinylated SdhA and hence active SQR is very low. Path II represents SdhE-assisted flavinylation of SdhA, the optimal route for efficient flavinylation and SQR activity. Path III also represents assembly of SdhA in the absence of SdhE. In this case, however, FAD is absent or remains associated with SdhA noncovalently. This leads to limited progression in the pathway or assembly of a nonfunctional SQR. This model is based largely on the structural (this study) and cellular studies of E. coli SdhA flavinylation and assembly under aerobic conditions. CAP, capping domain of SdhA; FBD, FAD binding domain of SdhA; E, SdhE; G16, Gly16 residue in SdhE; H45, His45 residue in SdhA. FAD is indicated in yellow.
Comparison with the E. coli FrdA–SdhE Complex.
While this manuscript was under review, the crystal structure of E. coli SdhE cross-linked to flavinylated FrdA (denoted here as the FrdA–SdhE complex) was reported to 2.6-Å resolution by Iverson and coworkers (35). The FrdA–SdhE structure shares a number of features with that of SdhAE reported here: (i) SdhE is located between the FAD and capping domains of the flavinylated protein in both structures, and makes a direct hydrogen bonding contact between Gly16 (in SdhE) and the histidyl FAD linkage in the respective flavoprotein; (ii) the SdhE binding site on the flavinylated protein overlaps with that of the FrdB/SdhB subunit within the final complex; and (iii) complex formation with SdhE accompanies a rotation of the appropriate capping domain. Importantly, comparison of the two structures also reveals a number of significant differences, which may reflect the relative stabilities of the protein complexes and/or the methods used to generate them. The SdhAE structure reported here represents a stable complex, which was recovered following size-exclusion chromatography. In contrast, the FrdA–SdhE structure required the generation of a FrdA–SdhE complex by chemical cross-linking, and, hence, as suggested by Iverson and coworkers, “reflects an imperfectly stabilized complex” (35). Interestingly, our comparison of the two structures reveals that the positioning of SdhE differs. In the FrdA–SdhE complex, SdhE is tilted toward the proposed position of the cross-link. The result is a smaller rotation of the FrdA capping domain (10.6–10.8°, dependent on whether malonate or acetate is present in the dicarboxylate binding site) in the FrdA–SdhE complex compared with the SdhAE complex (rotation of 43.5°). In addition, the size of the protein–protein interface within the FrdA–SdhE complex is smaller than that in the SdhAE structure (1,085 and 1,455 Å2, respectively). Significantly, the structure of the two FrdA–SdhE assemblies within the asymmetric unit are nonidentical, and the average B-factor for the SdhE protein is approximately twice that of the FrdA protein, indicating significant disorder. The authors also reported that the electron density for SdhE was “weaker” than for the FrdA subunit (35). In contrast, the crystal structure of both SdhAE complexes per asymmetric unit are almost identical, with consistent average B-factors across the SdhA and SdhE proteins and ordered electron density for the entire SdhE molecules (Table S1). Whether these differences are a result of the different experimental methods used to generate these complexes for structural analyses will await further investigation.
Materials and Methods
Protein Expression, Purification, and Characterization.
The SdhE and SdhA coding sequences were amplified by PCR by using E. coli (MC4100) genomic DNA and primer pairs (Bam_E, 5′-TGTAGCGGATCCGGACATTAACAACAAAGCCCG-3′; E_Hind3, 5′-CTATCGAAGCTTAAGCTTAGATTGCCACAGGACCA-3′; and Nde_A, 5′-ATGCGACATATGAAATTGCCAGTCAGAG-3′; Xho_A, 5′-TAGCACCTCGAGTTAGTAAGTACGAATCTTCGG-3′; restriction enzyme recognition sites are indicated in bold) and cloned into pETDuet-1. His6-SdhE and SdhA were coexpressed for 4 h at 37 °C in E. coli strain BL21(DE3) CodonPlus-RIL (Stratagene) transformed with sequence-verified pETDuet-1/His-sdhE/sdhA. Overexpressed His6-SdhE/SdhA was isolated from soluble lysate [in 20 mM Tris⋅HCl, pH 8.0, 200 mM KCl, 10% (vol/vol) glycerol, 1 mM EDTA, 0.05% (vol/vol) Triton X-100, 0.1 mg/mL lysozyme, 5 mM MgCl2, 10 µg/mL DNase I] by affinity chromatography by using Ni-NTA agarose (Qiagen) under standard conditions. Excess SdhE was separated from the SdhAE binary complex by using a Superdex 200 HiLoad 16/60 column (GE Healthcare) run at 0.5 mL/min in GF buffer (50 mM Tris⋅HCl, pH 8.0, 300 mM NaCl). The molecular weight of the purified SdhAE complex was estimated by size-exclusion chromatography by using a Superdex 200 Increase 10/300 GL column (GE Healthcare) calibrated by using a gel filtration protein standard mix (Bio-Rad). To analyze flavinylation, an in-gel fluorescence detection method was used (10).
Protein Crystallization and Data Collection.
SdhAE was crystallized at 20 °C by hanging-drop vapor diffusion with drops consisting of equal volumes (1 μL) of protein (6.3 mg/mL in 50 mM Tris⋅HCl, pH 8.0, 300 mM NaCl) and crystallization solution [0.1 M Hepes, pH 7.5, 0.2 M MgCl2.6H2O, 30% (wt/vol) PEG 3350, and 40 mM NaF]. Crystals were cryoprotected in reservoir solution with 30% (vol/vol) glycerol before flash-cooling in liquid nitrogen. Diffraction data were collected on an ADSC Quantum 315r detector at the Australian Synchrotron on beamline MX2 at 100 K and were processed with HKL2000 (36). Unit cell parameters and data collection statistics are presented in Table S1.
Structure Solution and Refinement.
The crystal structure of SdhAE was solved by molecular replacement by using PHASER (37) with search models from the crystal structures of the SdhA subunit from the E. coli SQR [chain A; PDB ID code 2WDQ (24)] and the E. coli “hypothetical protein” YgfY [now known to be SdhE; PDB ID code 1X6I (30)]. The resulting model was refined by iterative cycles of refinement with REFMAC (38) against anisotropically corrected diffraction data (39) interspersed with manual inspection and correction against calculated electron density maps by using COOT (40). The refinement of the model converged with residuals R = 0.199 and Rfree = 0.258 (Table S1). The structure was judged to have excellent geometry as determined by MOLPROBITY (41) (Table S1).
Supplementary Material
Acknowledgments
Aspects of this research were undertaken on the Macromolecular Crystallography beamline MX2 at the Australian Synchrotron (Victoria, Australia), and we thank the beamline staff for their enthusiastic and professional support. This work was funded by Australian Research Council Grants DP150104639 (to K.N.T. and D.A.D.), a La Trobe University Full Fee Research Scholarship (to A.S.H.), and a La Trobe University Postgraduate Research Scholarship (to A.S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6C12).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800195115/-/DCSupplemental.
References
- 1.Sun F, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 3.Robinson KM, Rothery RA, Weiner JH, Lemire BD. The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur J Biochem. 1994;222:983–990. doi: 10.1111/j.1432-1033.1994.tb18949.x. [DOI] [PubMed] [Google Scholar]
- 4.Astuti D, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeron T, et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 7.Burnichon N, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 9.Ghezzi D, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 10.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Vranken JG, et al. SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014;20:241–252. doi: 10.1016/j.cmet.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na U, et al. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014;20:253–266. doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC. SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem. 2012;287:18418–18428. doi: 10.1074/jbc.M111.293803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Taylor NL, Ströher E, Fenske R, Millar AH. Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 2013;73:429–441. doi: 10.1111/tpj.12041. [DOI] [PubMed] [Google Scholar]
- 15.McNeil MB, et al. The succinate dehydrogenase assembly factor, SdhE, is required for the flavinylation and activation of fumarate reductase in bacteria. FEBS Lett. 2014;588:414–421. doi: 10.1016/j.febslet.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Bezawork-Geleta A, Dong L, Rohlena J, Neuzil J. The assembly factor SDHAF2 is dispensable for flavination of the catalytic subunit of mitochondrial complex II in breast cancer cells. J Biol Chem. 2016;291:21414–21420. doi: 10.1074/jbc.C116.755017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kounosu A. Analysis of covalent flavinylation using thermostable succinate dehydrogenase from Thermus thermophilus and Sulfolobus tokodaii lacking SdhE homologs. FEBS Lett. 2014;588:1058–1063. doi: 10.1016/j.febslet.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Maklashina E, et al. Binding of the covalent flavin assembly factor to the flavoprotein subunit of complex II. J Biol Chem. 2016;291:2904–2916. doi: 10.1074/jbc.M115.690396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starbird CA, et al. Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli complex II homolog quinol:fumarate reductase. J Biol Chem. 2017;292:12921–12933. doi: 10.1074/jbc.M117.795120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lancaster CR, Kröger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 21.Brandsch R, Bichler V. Covalent cofactor binding to flavoenzymes requires specific effectors. Eur J Biochem. 1989;182:125–128. doi: 10.1111/j.1432-1033.1989.tb14808.x. [DOI] [PubMed] [Google Scholar]
- 22.Eletsky A, et al. Solution NMR structure of yeast succinate dehydrogenase flavinylation factor Sdh5 reveals a putative Sdh1 binding site. Biochemistry. 2012;51:8475–8477. doi: 10.1021/bi301171u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bezawork-Geleta A, Saiyed T, Dougan DA, Truscott KN. Mitochondrial matrix proteostasis is linked to hereditary paraganglioma: LON-mediated turnover of the human flavinylation factor SDH5 is regulated by its interaction with SDHA. FASEB J. 2014;28:1794–1804. doi: 10.1096/fj.13-242420. [DOI] [PubMed] [Google Scholar]
- 24.Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem. 2009;284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 26.Bossi RT, Negri A, Tedeschi G, Mattevi A. Structure of FAD-bound L-aspartate oxidase: Insight into substrate specificity and catalysis. Biochemistry. 2002;41:3018–3024. doi: 10.1021/bi015939r. [DOI] [PubMed] [Google Scholar]
- 27.Stoltz M, Brandsch R. The conformational change induced by FAD in covalently flavinylated 6-hydroxy-D-nicotine oxidase does not require (8alpha)FAD-(N3)histidyl bond formation. J Biochem. 1998;123:445–449. doi: 10.1093/oxfordjournals.jbchem.a021957. [DOI] [PubMed] [Google Scholar]
- 28.Koetter JW, Schulz GE. Crystal structure of 6-hydroxy-D-nicotine oxidase from Arthrobacter nicotinovorans. J Mol Biol. 2005;352:418–428. doi: 10.1016/j.jmb.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 29.Fraaije MW, van Den Heuvel RH, van Berkel WJ, Mattevi A. Structural analysis of flavinylation in vanillyl-alcohol oxidase. J Biol Chem. 2000;275:38654–38658. doi: 10.1074/jbc.M004753200. [DOI] [PubMed] [Google Scholar]
- 30.Lim K, et al. Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation. Proteins. 2005;58:759–763. doi: 10.1002/prot.20337. [DOI] [PubMed] [Google Scholar]
- 31.Heuts DP, Scrutton NS, McIntire WS, Fraaije MW. What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 2009;276:3405–3427. doi: 10.1111/j.1742-4658.2009.07053.x. [DOI] [PubMed] [Google Scholar]
- 32.McNeil MB, Fineran PC. The conserved RGxxE motif of the bacterial FAD assembly factor SdhE is required for succinate dehydrogenase flavinylation and activity. Biochemistry. 2013;52:7628–7640. doi: 10.1021/bi401006a. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Sawyer N, Regan L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 35.Sharma P, Maklashina E, Cecchini G, Iverson TM. Crystal structure of an assembly intermediate of respiratory complex II. Nat Commun. 2018;9:274. doi: 10.1038/s41467-017-02713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.