Significance
Protein–protein interactions are important in cellular signaling. However, they have presented a challenge for molecular dynamics (MD) simulations, due to increased system complexity and slowly evolving dynamics. In particular, binding of the medically important G-protein-coupled receptors (GPCRs) with intracellular signaling proteins has not been simulated with MD to date. In this study, we performed long-timescale enhanced MD simulations using the Gaussian accelerated MD (GaMD) method on the M2 muscarinic GPCR. The simulations revealed pathways and important low-energy intermediate states of the G-protein mimetic nanobody binding to the M2 receptor. Our simulation results are consistent with experimental data and provide important insights into the GPCR–nanobody binding mechanism. GaMD is thus applicable to enhanced simulations of protein–protein interactions.
Keywords: enhanced sampling, protein binding, pathways, biomolecular recognition, GPCR signaling
Abstract
Protein–protein binding is key in cellular signaling processes. Molecular dynamics (MD) simulations of protein–protein binding, however, are challenging due to limited timescales. In particular, binding of the medically important G-protein-coupled receptors (GPCRs) with intracellular signaling proteins has not been simulated with MD to date. Here, we report a successful simulation of the binding of a G-protein mimetic nanobody to the M2 muscarinic GPCR using the robust Gaussian accelerated MD (GaMD) method. Through long-timescale GaMD simulations over 4,500 ns, the nanobody was observed to bind the receptor intracellular G-protein-coupling site, with a minimum rmsd of 2.48 Å in the nanobody core domain compared with the X-ray structure. Binding of the nanobody allosterically closed the orthosteric ligand-binding pocket, being consistent with the recent experimental finding. In the absence of nanobody binding, the receptor orthosteric pocket sampled open and fully open conformations. The GaMD simulations revealed two low-energy intermediate states during nanobody binding to the M2 receptor. The flexible receptor intracellular loops contribute remarkable electrostatic, polar, and hydrophobic residue interactions in recognition and binding of the nanobody. These simulations provided important insights into the mechanism of GPCR–nanobody binding and demonstrated the applicability of GaMD in modeling dynamic protein–protein interactions.
G-protein-coupled receptors (GPCRs) represent the largest superfamily of membrane proteins that mediate cellular responses to hormones, neurotransmitters, and the senses of sight, olfaction, and taste. They have served as targets of about one-third of currently marketed drugs for treating a wide spectrum of diseases (1, 2). Muscarinic acetylcholine receptors are members of the class A GPCRs, comprising five subtypes, i.e., M1–M5 (3). The odd-numbered subtypes prefer to couple with the Gq/11 proteins, while the even-numbered subtypes predominantly couple with the Gi/o proteins. Particularly, the M2 receptor is widely distributed in mammalian tissues and is the only subtype found in the human heart. It plays a key role in modulating cardiac function. Activation of the M2 receptor mediated by the Gi/o-protein coupling typically results in a decrease in the heart rate and a reduction in heart contraction forces.
X-ray crystal structures have been determined for the M2 receptor in an inactive state bound by the antagonist 3-quinuclidinyl-benzilate (QNB) (4), as well as an active state bound by the agonist iperoxo (IXO) and G-protein mimetic nanobody Nb9-8 in the absence and presence of a positive allosteric modulator (PAM) LY2119620 (5). Relative to the inactive conformation, the active M2 receptor is characterized by opening of the orthosteric ligand-binding pocket and rearrangements of the transmembrane (TM) helices 5, 6, and 7. Binding of PAM LY2119620 in the receptor extracellular vestibule induces a marked conformational change in the Trp4227.35 side chain (5). The residue superscripts denote Ballesteros–Weinstein numbering of GPCRs (6).
In addition, X-ray structures have been obtained for several other GPCRs in the active state, including opsin coupled with the C-terminal peptide of the Gα subunit (7), the β2-adrenergic receptor (β2AR) and µ-opioid receptor (µ-OR) with G-protein mimetic nanobodies (8–10), the adenosine A2A receptor (A2AAR) with an engineered mini-Gs protein (11), and the β2AR with the Gs protein (12). Very recently, the cryo-EM structure was also reported for the calcitonin receptor (CTR; a class-B GPCR) coupled with the Gs protein (13). These high-resolution structures provided extremely valuable insights into the active state of GPCRs and the receptor–G-protein interactions in the bound conformation. However, the dynamic mechanism and pathways of intracellular protein binding to the GPCRs remain poorly understood (14).
Molecular dynamics (MD) is a widely used computational technique that probes biomolecular structural dynamics at an atomistic level (15). In particular, MD simulations have been applied on GPCRs and greatly helped in understanding the mechanisms of GPCR activation (16–18) and ligand-binding processes (19, 20), as reviewed earlier (21–24). Despite these remarkable advances, the conventional MD (cMD) simulations of GPCRs have been limited to typically tens of microseconds (22). This has greatly hindered the usage of cMD in studying biomolecular dynamics over longer timescales. The cMD simulation of the intracellular G-protein or mimetic nanobody binding to the GPCRs has not been accomplished so far.
Enhanced MD methods have proven useful in simulations of large biomolecules such as GPCRs (22, 24, 25). In particular, Gaussian accelerated MD (GaMD) is an enhanced sampling technique that works by applying a harmonic boost potential to smooth the potential energy surface, reduce the system energy barriers, and accelerate biomolecular structural dynamics by orders of magnitude (26–28). Moreover, because the boost potential follows Gaussian distribution, the original free-energy profiles of biomolecules can be recovered through cumulant expansion to the second order for characterizing biomolecular dynamics, such as ligand binding and graded activation of GPCRs (27, 29). GaMD does not require predefined collective variables (CVs), which is advantageous for studying protein folding (26, 27) and ligand binding (26–28).
In this study, we performed long-timescale GaMD simulations to investigate binding of the G-protein mimetic nanobody Nb9-8 and agonist IXO to the M2 receptor. Using X-ray structure of the agonist–M2 receptor–nanobody complex [Protein Data Bank (PDB) ID code 4MQS], we displaced the agonist IXO and nanobody Nb9-8 to be >20 Å away from the M2 receptor in the solvent. During GaMD simulations, although the agonist reached only the receptor extracellular vestibule, the nanobody was observed to bind the receptor intracellular G-protein-coupling site. GaMD simulations revealed important intermediate states during nanobody binding to the M2 receptor, despite the fact that the calculated free energies were not converged. Furthermore, allosteric coupling was identified between the nanobody binding and conformational changes in the orthosteric ligand-binding pocket.
Results
Five independent GaMD simulations lasting ∼4,500 ns were performed on the M2 receptor in the presence of the agonist IXO and nanobody Nb9-8. The average (∼16 kcal/mol) and SD (∼5 kcal/mol) of the boost potential were found to be closely similar among the GaMD simulations (Table 1). Although the agonist reached only the receptor extracellular vestibule in the GaMD simulations with minimum rmsd of 8–15 Å relative to the 4MQS X-ray conformation, the nanobody bound to the receptor G-protein-coupling site in one of the GaMD simulations (“Sim1” in Table 1), for which the minimum rmsd of the nanobody core domain was 2.48 Å compared with the X-ray conformation. During the other four GaMD simulations, the nanobody minimum rmsd ranged from 8.38 to 17.39 Å, suggesting that it did not bind to the G-protein-coupling site (Table 1).
Table 1.
Summary of GaMD simulations performed on the M2 receptor in the presence of the agonist IXO and G-protein mimetic nanobody Nb9-8
| ID | Length, ns | ∆V, kcal/mol | RMSDmin, Å | ||
| Avg | σ | Nb9-8 | IXO | ||
| Sim1 | 4,500 | 16.35 | 5.18 | 2.48 | 13.84 |
| Sim2 | 4,500 | 16.45 | 5.20 | 10.02 | 15.12 |
| Sim3 | 4,364 | 16.30 | 5.26 | 8.38 | 11.37 |
| Sim4 | 4,500 | 16.12 | 5.16 | 17.19 | 14.57 |
| Sim5 | 4,500 | 16.42 | 5.19 | 15.71 | 8.03 |
The average (Avg) and SD (σ) are calculated for the GaMD boost potential ∆V. Minimum rmsd (RMSDmin) was calculated for the Nb9-8 and IXO.
GaMD Simulation Revealed Nanobody Binding to the M2 Receptor.
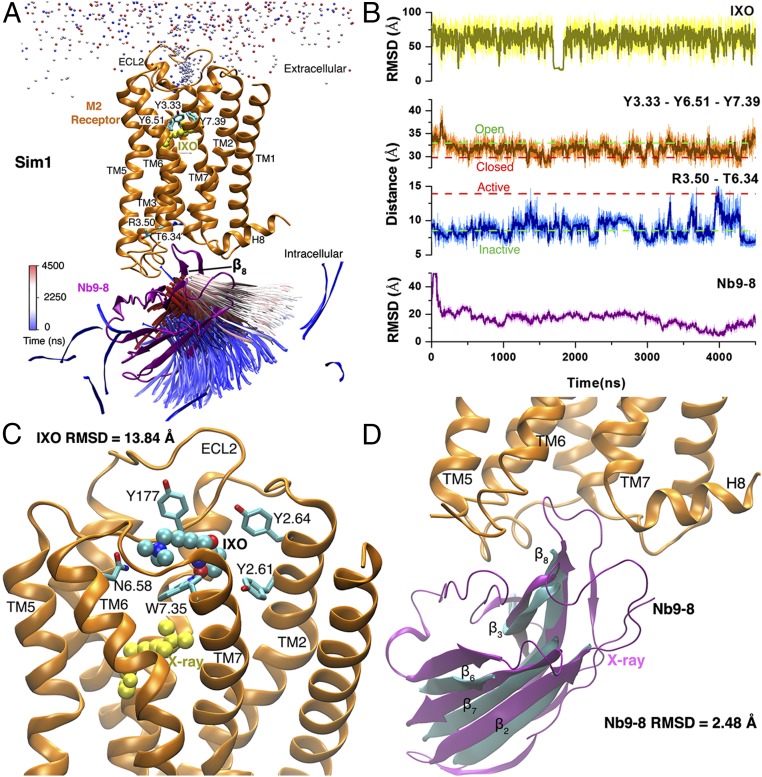
During the 4,500-ns Sim1 GaMD trajectory, starting from free diffusion in the solvent, the agonist IXO visited the extracellular vestibule of the M2 receptor and the nanobody Nb9-8 bound to the receptor intracellular G-protein-coupling site (Fig. 1A and Movie S1). The agonist bound to the receptor extracellular vestibule during ∼1,700–1,830 ns with <20 Å rmsd relative to the 4MQS X-ray conformation and visited the same site briefly several more times in the simulation (Fig. 1B). The nanobody approached the M2 receptor and formed initial contacts with the intracellular loop (ICL) 2. The TM6 cytoplasmic end became disordered in the C-terminal region of ICL3 and facilitated binding of the nanobody. The nanobody rmsd decreased to <20 Å after ∼560 ns. Then, the nanobody rearranged its conformation and fit into the receptor intracellular pocket. The nanobody interacted dynamically with the receptor flexible ICLs and the TM7–helix 8 (H8) hinge. The nanobody rmsd dropped <5 Å during ∼3,880−4,000 ns. Without the agonist at the orthosteric site, the nanobody deviated from the target G-protein-coupling site during ∼4,000–4,500 ns while maintaining interactions with the receptor intracellular domains (Movie S1).
Fig. 1.
Binding of agonist IXO and G-protein mimetic nanobody Nb9-8 to the M2 receptor was observed in one 4,500-ns GaMD simulation. (A) Trajectories of nitrogen in the trimethylamine group of IXO (beads) and the β8 strand of Nb9-8 (ribbons) colored by simulation time in a blue (0 ns)–white (2,250 ns)–red (4,500 ns) scale. The β8 strand of Nb9-8 moves into the X-ray conformation near the simulation end. X-ray conformations of the M2 receptor and Nb9-8 (PDB ID code 4MQS) are shown in orange and purple ribbons, respectively, and yellow spheres for IXO. Residues Tyr1043.33, Tyr4036.51, Tyr4267.39, Arg1213.50, and Thr3866.34 are represented by sticks. (B) The rmsds of the IXO and Nb9-8 relative to the X-ray structure, Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter and Arg1213.50−Thr3866.34 distance calculated from the simulation. Dashed lines indicate X-ray structural values of the M2 receptor (3UON, green; 4MQS, red). (C) Binding pose of IXO (spheres) in the receptor extracellular vestibule with 13.84-Å rmsd relative to the X-ray conformation (yellow spheres). Residues found within 5 Å of IXO are highlighted in sticks. (D) Binding pose of Nb9-8 (cyan) in the receptor intracellular pocket with 2.48-Å rmsd relative to the X-ray conformation (purple). The β2, β3, β6, β7, and β8 strands that represent the nanobody core domain were selected for calculating rmsd of Nb9-8 and heavy atoms for IXO.
Along with nanobody binding, conformational change occurred in both the orthosteric ligand-binding pocket and intracellular domains of the M2 receptor during the Sim1 GaMD trajectory (Fig. 1B). In the X-ray structures, the orthosteric pocket was “open” in the antagonist-bound receptor and “closed” in the agonist–nanobody-bound receptor, for which the triangle perimeter of the Tyr1043.33−Tyr4036.51−Tyr4267.39 “tyrosine lid” was 32.93 and 29.70 Å, respectively. Binding of the nanobody switched the orthosteric pocket from the open to closed conformation. Moreover, nanobody binding led to activation of the M2 receptor with an increase in the intracellular TM3−TM6 distance. When the nanobody diffused far away in the solution, the Arg1213.50−Thr3866.34 distance was ∼7 Å, as in the inactive X-ray structure of the M2 receptor. Upon nanobody binding to the intracellular pocket with rmsd <20 Å, it increased significantly and reached ∼13.9 Å (e.g., at ∼3,910 ns), as in the active receptor X-ray structure (Fig. 1B).
Fig. 1C shows the binding pose of the agonist IXO in the receptor extracellular vestibule. IXO exhibited 13.84-Å rmsd relative to the 4MQS X-ray conformation. Residues Tyr177ECL2, Tyr802.61, Tyr842.64, Asn4106.58, and Trp4227.35 formed polar and hydrophobic interactions with IXO. The extracellular vestibule has been shown to be an intermediate site for binding of orthosteric ligands (19, 27, 29), as well as the target site of allosteric modulators (5, 20). On the intracellular side, when the rmsd of the nanobody decreased to 2.48 Å, the β2, β3, β6, β7, and β8 strands of the nanobody core domain overlapped with the 4MQS X-ray structure at the receptor G-protein-coupling site (Fig. 1D). Therefore, the GaMD simulation successfully captured binding of the nanobody, accompanied by dynamic conformational changes in the M2 receptor.
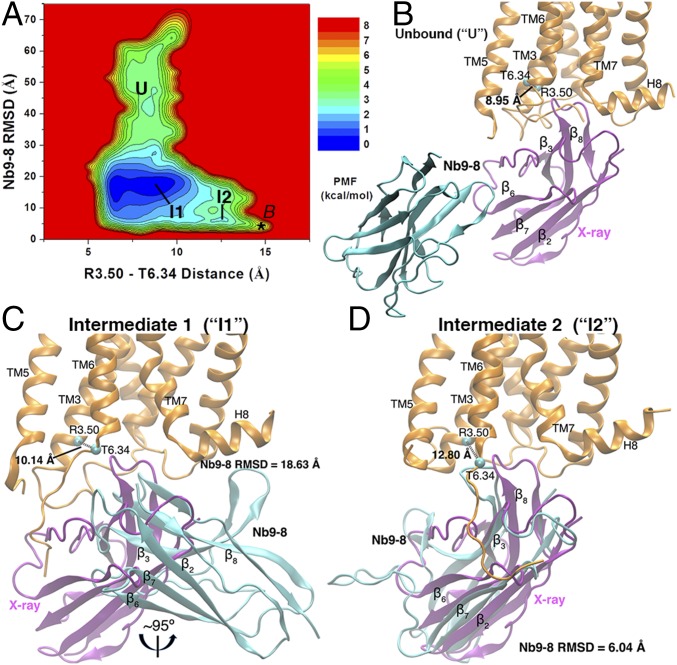
Nanobody Binding Pathways and Low-Energy States.
To characterize the nanobody binding pathways, we calculated a 2D potential of mean force (PMF) profile of the nanobody rmsd relative to the 4MQS X-ray conformation and the receptor Arg1213.50−Thr3866.34 distance using the Sim1 GaMD trajectory (Fig. 2A). Three low-energy conformational states were identified from the PMF profile, including unbound (U), intermediate 1 (I1), and intermediate 2 (I2). Relative to the zero-energy I1 state, free energies of the U and I2 states were 1.86 and 2.30 kcal/mol, respectively. The free energy barrier between the I1 and I2 states was ∼2.6 kcal/mol. The conformational transition from the unbound to the intermediate states exhibited a higher free energy barrier of ∼3.3 kcal/mol. The GaMD simulation sampled the bound (B) conformation as obtained in previous simulations of the 4MQS X-ray structure (29), although a low-energy well of the bound state did not appear in the PMF presented here. This was likely due to the absence of agonist binding at the orthosteric site and the boost potential applied to the system during GaMD simulation, which led to transient binding of the nanobody at the G-protein-coupling site.
Fig. 2.
(A) The 2D PMF calculated with the Arg1213.50−Thr3866.34 distance and rmsd of the Nb9-8 relative to the 4MQS X-ray conformation. Three low-energy conformational states are labeled as the Unbound (U), Intermediate 1 (I1), and Intermediate 2 (I2). The bound (B) state obtained from previous GaMD simulation of the 4MQS X-ray structure (29) is labeled a star. (B–D) Structural conformations of the M2 receptor and Nb9-8 in the U (B), I1 (C), and I2 (D) states, for which energy minima of the (Arg1213.50−Thr3866.34 distance, Nb9-8 rmsd) 2D PMF were found at (8.95 Å, 49.44 Å), (10.14 Å, 18.63 Å), and (12.80 Å, 6.04 Å), respectively. The reference X-ray conformations of the M2 receptor and nanobody are shown as orange and purple ribbons, respectively. The evolving nanobody is represented by cyan ribbons.
In the U state, the Arg1213.50−Thr3866.34 distance was 8.95 Å in the receptor (Fig. 2B). When the system visited the I1 state, the nanobody rotated around the membrane normal by ∼95° with 18.63-Å rmsd relative to the bound X-ray conformation, and the receptor Arg1213.50−Thr3866.34 distance increased to 10.14 Å (Fig. 2C). In the I2 state, the nanobody rmsd decreased further to 6.04 Å, and the receptor Arg1213.50−Thr3866.34 distance increased to 12.80 Å. The nanobody was aligned in a similar orientation as in the bound X-ray conformation (Fig. 2D).
Receptor ICLs Play a Key Role in Recognition and Binding of the Nanobody.
With low-energy states identified from the GaMD simulation, we examined residue interactions during nanobody binding to the M2 receptor (Fig. 3). Remarkably, the flexible receptor ICLs were able to dynamically rearrange their conformations to form favorable interactions with residues on the nanobody surface. They played a key role in the recognition and binding of the nanobody.
Fig. 3.
Residue interactions between the M2 receptor and the G-protein mimetic nanobody Nb9-8 in the low-energy conformational states. (A and B) The I1 conformation looking at the TM5–TM6–TM7 (A) and TM1–TM2–TM4 (B) helices. (C and D) The I2 conformation looking at the TM5–TM6–TM7 (C) and TM1–TM2–TM4 (D) helices. (E and F) The B conformation looking at the TM5–TM6–TM7 (E) and TM1–TM2–TM4 (F) helices. The M2 receptor is shown in orange ribbons, and residues found within 2.5 Å of the nanobody are represented by sticks. The nanobody is represented by surface and colored by the atomic charges in a blue (+0.5e)–white (0)–red (−0.5e) scale. Key residues in the nanobody that interact with the M2 receptor are labeled in purple.
In the I1, the TM6 cytoplasmic end became disordered as in the C-terminal region of the ICL3. Residues Arg3816.29 and Lys3836.31 formed salt-bridge interactions with Asp34 and Asp30 in the nanobody, respectively (Fig. 3A). Side chains of Arg3876.35 and Lys3846.32 interacted with Phe29 in the nanobody. Receptor residues Arg52ICL1 and Asn511.60 formed salt-bridge and H-bond interactions with Glu47 and Gln46 in the nanobody, respectively. Residue Lys134ICL2 formed polar and hydrophobic interactions with Leu104 and Ser107 in the nanobody (Fig. 3B). In the I2, receptor residues Arg3816.29 and Lys3836.31 in the disordered TM6 cytoplasmic end maintained electrostatic interactions with the nanobody. Side chains of Val3856.33 and Thr3866.34 formed hydrophobic interactions with Phe105 in the nanobody (Fig. 3C). The highly conserved Arg1213.50 in the receptor DRY motif formed a salt bridge with Asp108 in the nanobody. Residues Arg52ICL1 and Asn511.60 formed polar interactions with the nanobody N-terminal residues Gln3 and Gln1. His53ICL1 also contacted with Tyr110 in the nanobody. In the receptor ICL2, residues Lys127, Lys134, and Arg135 extended their side chains to form electrostatic interactions with the nanobody (Fig. 3D).
Finally, residue interactions at the interface of the receptor and nanobody were analyzed by using the 4MQS X-ray structure for comparison. Importantly, Arg1213.50 exhibited a cation-π interaction with Phe105 in the nanobody, which also formed hydrophobic interactions with side chains of the receptor residues Ile2095.61, Val3856.33, and Thr3886.36. Receptor residues Arg3816.29 and Lys3846.32 formed salt bridges with Asp58 and Asp34 in the nanobody, respectively. Asn4448.47 and Thr4468.49 in the TM7–H8 hinge formed H-bonds with Ser107 and Asp108 in the nanobody, respectively (Fig. 3E). In the receptor ICL1, His53 and Thr56 contacted with Tyr110, Asp103, and Ser107 in the nanobody. In the ICL2, Pro128, Val133, and Leu129 formed hydrophobic interactions with Ile62, Cys53, and Phe40 in the nanobody (Fig. 3F). These electrostatic, polar, and hydrophobic residue interactions are important for the M2 receptor–nanobody binding.
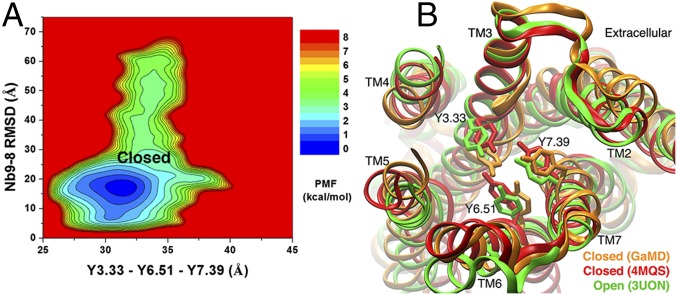
Allosteric Coupling Between Nanobody Binding and Conformational Change in the Ligand-Binding Pocket.
Next, we calculated another 2D PMF profile of the nanobody rmsd vs. the receptor Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter using the Sim1 GaMD trajectory (Fig. 4A). Analysis showed that when the nanobody bound to the receptor intracellular pocket, the tyrosine lid covering the orthosteric ligand-biding pocket predominantly adopted the closed conformation. The GaMD simulation-derived closed conformation of the M2 receptor was similar to the closed 4MQS X-ray structure (Fig. 4B). Both conformations exhibited significant inward movement of the side chains of Tyr1043.33, Tyr4036.51, and Tyr4267.39 compared with the open 3UON X-ray structure.
Fig. 4.
(A) The 2D PMF calculated by using the Sim1 GaMD trajectory regarding the Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter and rmsd of Nb9-8 relative to the 4MQS X-ray conformation. The low-energy conformational state is labeled as the closed. (B) Extracellular view of the GaMD simulation-derived closed conformational state (orange) with the closed (red) and open (green) X-ray conformations (PDB ID codes 4MQS and 3UON). The M2 receptor is represented by ribbons, and residues Tyr1043.33, Tyr4036.51, and Tyr4267.39 are highlighted in sticks.
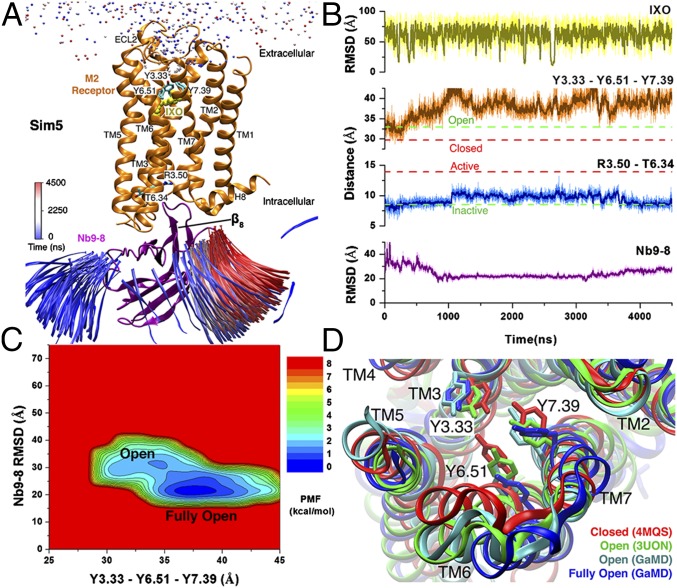
In the “Sim5” 4,500-ns GaMD trajectory, the agonist visited the receptor extracellular vestibule with a minimum rmsd of 8.03 Å, and the nanobody did not bind to the receptor G-protein-coupling site with rmsd >20 Å relative to the 4MQS X-ray conformation (Fig. 5 A and B). In the absence of nanobody binding, the receptor remained inactive as characterized by the ∼8- to 10-Å distance between Arg1213.50 and Thr3866.34. The orthosteric pocket adopted the open to “fully open” conformations, for which the triangle perimeter of the tyrosine lid was ∼33 and ∼35−40 Å, respectively (Fig. 5B). These two conformations were confirmed as low-energy states in the 2D PMF profile of the receptor Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter and nanobody rmsd, as shown in Fig. 5C. The simulation-derived open conformation was similar to the 3UON antagonist-bound X-ray structure. The newly discovered fully open conformation showed significant outward movement of the three tyrosine side chains relative to the open conformation and even more compared with the closed 4MQS X-ray structure (Fig. 5D).
Fig. 5.
One 4,500-ns GaMD simulation of the M2 receptor (Sim5 in Table 1), during which IXO and Nb9-8 bind to the receptor with the smallest rmsds of 8.03 and 15.71 Å, respectively, relative to the 4MQS X-ray conformation. (A) Trajectories of nitrogen in the trimethylamine group of IXO (beads) and the β8 strand of Nb9-8 (ribbons) colored by the simulation time in a blue (0 ns)–white (2,250 ns)–red (4,500 ns) scale. The structural representations are similar to Fig. 1A. (B) The rmsds of the IXO and Nb9-8 relative to the 4MQS X-ray conformations, Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter, and Arg1213.50−Thr3866.34 distance calculated from the simulation. Dashed lines indicate X-ray structural values of the M2 receptor (3UON, green; 4MQS, red). (C) The 2D PMF calculated with the Tyr1043.33−Tyr4036.51−Tyr4267.39 triangle perimeter and rmsd of the Nb9-8 relative to the 4MQS X-ray conformation. Two low-energy conformational states are labeled as open and fully open. (D) Extracellular view of the GaMD simulation-derived open (cyan) and fully open (blue) conformational states compared with the closed (red) and open (green) X-ray conformational states (PDB ID codes 4MQS and 3UON). The M2 receptor is represented by ribbons, and residues Tyr1043.33, Tyr4036.51, and Tyr4267.39 are highlighted in sticks.
In addition to Sim1 and Sim5, detailed analysis was further performed on the other three GaMD trajectories (Figs. S1–S3). Particularly in the “Sim2” trajectory, the nanobody diffused into the receptor intracellular pocket with rmsd decreased to ∼15 Å, but was not able to bind further to the target G-protein-coupling site as in the 4MQS X-ray conformation (Fig. S1). PMF calculations showed that the M2 receptor sampled the inactive state with ∼5- to 8-Å distance between Arg1213.50 and Thr3866.34 (Fig. S1C) and the closed to open conformations in the orthosteric pocket with ∼30- to 35-Å triangle perimeter of the tyrosine lid (Fig. S1D). Similar results were obtained in the “Sim3” GaMD trajectory (Fig. S2). During the “Sim4” GaMD trajectory, the nanobody did not diffuse into the intracellular pocket of the M2 receptor with >20-Å rmsd compared with the 4MQS X-ray structure (Fig. S3). The M2 receptor adopted mostly the inactive state (Fig. S3 B and C) and open to fully open conformations in the orthosteric pocket (Fig. S3D).
Taking the five GaMD simulations together, we observed allosteric coupling between intracellular nanobody binding and conformational change in the ligand-binding pocket of the M2 receptor. In the absence of nanobody binding in the intracellular pocket, the receptor ligand-binding pocket adopted mostly the open to fully open conformations as observed in the Sim4 and Sim5 GaMD trajectories. In contrast, binding of the nanobody led to conformational transition of the ligand-binding pocket to the closed state as shown in the Sim1 trajectory.
Discussion
In this study, we have performed, to our knowledge, the longest GaMD enhanced simulations to date (4,500 ns), which have successfully captured binding of the G-protein mimetic nanobody to the M2 muscarinic GPCR. Because only one binding event has been observed through the simulations, it is important to note that the presented free energy profiles are not converged. Each of the GaMD simulations likely sampled only a subset of the large conformational space. Future developments in sampling methodology and computing power will be needed to achieve converged simulations of such complex systems. Nevertheless, the GaMD simulations have allowed us to identify the binding pathways and important intermediate states of the nanobody. This work has extended the application of GaMD to protein–protein interactions (PPIs). PPIs are important in cellular signaling. However, they have presented a challenge for cMD simulations, due to increased system complexity and slowly evolving dynamics. Furthermore, it is difficult to define a priori CVs to simulate PPIs, which often involve large-scale translations and rotations of the binding partners, as well as complicated conformational changes inside each protein. In this regard, GaMD that does not require predefined CVs is well suited for unconstrained enhanced simulations of PPIs.
In the GaMD simulation, the G-protein mimetic nanobody diffused spontaneously from the bulk solvent to the intracellular target site of the M2 receptor. Although a low-energy well did not appear in the calculated PMF for the bound state, the GaMD simulation sampled this conformation as in previous simulations of the 4MQS X-ray structure (29). Unstable binding of the nanobody could result from the absence of agonist binding at the orthosteric site. In addition, the GaMD boost potential that was applied to both the dihedral and total potential energetic terms could contribute to transient binding of the nanobody. It is subject to future studies using different GaMD parameters for various acceleration levels (28) to investigate the PPIs. Nevertheless, in the simulation-derived bound conformation, the nanobody core domain showed a minimum rmsd of 2.48 Å compared with the 4MQS X-ray structure (5).
Furthermore, the GaMD simulations revealed two important low-energy intermediate states, I1 and I2, during nanobody binding to the M2 receptor. Close examination of residue interactions at the interface revealed that the flexible ICLs played a key role in the recognition and binding of the G-protein mimetic nanobody. Notably, residues Val3856.33, Thr3866.34, Ile3896.37, and Leu3906.38 in ICL3 of the M2 receptor were suggested to be important for G-protein coupling in previous mutagenesis experiments (30, 31). The ICLs in the β2AR and A2AAR were also shown to form extensive interactions with the Gs and mini-Gs proteins in the X-ray structures (11, 12). Conformational flexibility of the ICL2 and ICL3 were critical for G-protein coupling of several GPCRs (14). Our GaMD simulations were in line with these findings and have provided a detailed view of the dynamic process at an atomistic level.
In the receptor orthosteric ligand-binding pocket, a fully open conformation of the tyrosine lid was discovered from the GaMD simulations, in addition to the closed and open conformations as determined in previous X-ray structures of the QNB-bound (4) and IXO-nanobody-bound (5) M2 receptor. The fully open orthosteric pocket may be able to accommodate larger ligands, especially the antagonists with nearly two times the molecular weight of QNB (32). This can be potentially validated through future experimental structural studies of the M2 receptor with the larger ligands.
Allosteric coupling between the nanobody binding and conformational change in the orthosteric ligand-binding pocket of the M2 receptor took place in the GaMD simulations. In the absence of nanobody binding, the orthosteric pocket sampled open to fully open conformations. In contrast, the orthosteric pocket became closed upon nanobody binding to the receptor G-protein-coupling site. This is consistent with the recent experimental finding that nanobody binding allosterically closed the ligand-binding pocket of the β2AR (33).
In summary, long-timescale GaMD simulations have captured nanobody binding to the M2 receptor. While engineered G proteins such as the mini-Gs (11) have a similar size as the nanobody, the entire heterotrimeric G proteins are significantly larger than the nanobody. It may be more challenging to simulate binding of the G proteins to the GPCRs. On the other hand, the GaMD method can be further improved for more powerful enhanced sampling. Particularly, the GaMD simulations herein applied boost potential to the system dihedrals and total potential energy. Since nonbonded interactions are mainly involved in protein–protein binding, adding boost potential based on the “essential” nonbonded potential energy may be more effective for the GaMD simulations (28, 34).
The GaMD simulations have provided important insights into the dynamic mechanism of GPCR–nanobody binding. The work also paves the way for enhanced simulations of, among many PPIs, the binding of G proteins or engineered G proteins (e.g., mini-G) to the GPCRs. This will be facilitated by continued advances in the computing power and enhanced simulation methodology. A growing number of GPCR protein complex structures, including the β2AR-Gs (12), A2AAR-mini Gs (11), and CTR-Gs (13), can be used to validate the GaMD simulations. In turn, GaMD simulations will be valuable for us to determine mechanisms of PPIs and understand the structural dynamics and function of the GPCR protein complexes.
Methods
GaMD simulations were performed on the M2 receptor in the presence of the G-protein mimetic nanobody Nb9-8 and agonist IXO. Energy minimization, thermalization, and 100-ns cMD equilibration were first performed by using NAMD2.10 (35). By using the NAMD output structure, along with the system topology and CHARMM36 (36) force field files, ParmEd was used to convert the simulation files into the AMBER format (37). The GaMD module implemented in the graphics processing unit (GPU) version of AMBER14 (26, 37) was then applied to perform GaMD simulations, which included 10-ns short cMD simulation used to collect potential statistics for calculating the GaMD acceleration parameters, 50-ns equilibration after adding the boost potential, and, finally, multiple independent GaMD production runs with randomized initial atomic velocities. The simulation frames were saved every 0.1 ps for analysis. The five GaMD production simulations lasting ∼4,500 ns are listed in Table 1. Details of the GaMD method, energetic reweighting, system setup, simulation protocol, and analysis are provided in SI Text.
Supplementary Material
Acknowledgments
Computing time was provided on the Gordon and Comet supercomputers through the Extreme Science and Engineering Discovery Environment Award TG-MCA93S013 and the Edison and Cori supercomputers through the National Energy Research Scientific Computing Center project M2874, as well as the GPU clusters at San Diego Supercomputer Center. This work was supported by NSF Grant MCB1020765, NIH Grant GM31749, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, and American Heart Association Award 17SDG33370094. Y.M. was supported by startup funding in the College of Liberal Arts and Sciences at the University of Kansas.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800756115/-/DCSupplemental.
References
- 1.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: New agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruse AC, et al. Muscarinic acetylcholine receptors: Novel opportunities for drug development. Nat Rev Drug Discov. 2014;13:549–560. doi: 10.1038/nrd4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spalding TA, Burstein ES. Constitutive activity of muscarinic acetylcholine receptors. J Recept Signal Transduct Res. 2006;26:61–85. doi: 10.1080/10799890600567349. [DOI] [PubMed] [Google Scholar]
- 4.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In: Stuart CS, editor. Methods in Neurosciences. Vol 25. Academic; New York: 1995. pp. 366–428. [Google Scholar]
- 7.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 8.Ring AM, et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen SGF, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, et al. Structural insights into µ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter B, Nehmé R, Warne T, Leslie AGW, Tate CG. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen SGF, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang YL, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira IS. Structural features of the G-protein/GPCR interactions. Biochim Biophys Acta. 2014;1840:16–33. doi: 10.1016/j.bbagen.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 16.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan J, Khelashvili G, Mondal S, Mehler EL, Weinstein H. Ligand-dependent conformations and dynamics of the serotonin 5-HT(2A) receptor determine its activation and membrane-driven oligomerization properties. PLoS Comput Biol. 2012;8:e1002473. doi: 10.1371/journal.pcbi.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chem. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dror RO, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci USA. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dror RO, et al. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 21.Grossfield A. Recent progress in the study of G protein-coupled receptors with molecular dynamics computer simulations. Biochim Biophys Acta. 2011;1808:1868–1878. doi: 10.1016/j.bbamem.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Johnston JM, Filizola M. Showcasing modern molecular dynamics simulations of membrane proteins through G protein-coupled receptors. Curr Opin Struct Biol. 2011;21:552–558. doi: 10.1016/j.sbi.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanni S, Rothlisberger U. A closer look into G protein coupled receptor activation: X-ray crystallography and long-scale molecular dynamics simulations. Curr Med Chem. 2012;19:1135–1145. doi: 10.2174/092986712799320493. [DOI] [PubMed] [Google Scholar]
- 24.Miao Y, McCammon JA. G-protein coupled receptors: Advances in simulation and drug discovery. Curr Opin Struct Biol. 2016;41:83–89. doi: 10.1016/j.sbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YH, Lin JH. Can ligands of different functional types induce distinct dynamics in G protein-coupled receptors? Curr Top Med Chem. 2017;17:2370–2380. doi: 10.2174/1568026617666170328100359. [DOI] [PubMed] [Google Scholar]
- 26.Miao Y, Feher VA, McCammon JA. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. J Chem Theory Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang YT, Miao Y, Wang Y, McCammon JA. Gaussian accelerated molecular dynamics in NAMD. J Chem Theory Comput. 2017;13:9–19. doi: 10.1021/acs.jctc.6b00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y, McCammon JA. Gaussian accelerated molecular dynamics: Theory, implementation and applications. Annu Rep Comput Chem. 2017;13:231–278. doi: 10.1016/bs.arcc.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao Y, McCammon JA. Graded activation and free energy landscapes of a muscarinic G-protein-coupled receptor. Proc Natl Acad Sci USA. 2016;113:12162–12167. doi: 10.1073/pnas.1614538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Conklin BR, Blin N, Yun J, Wess J. Identification of a receptor/G-protein contact site critical for signaling specificity and G-protein activation. Proc Natl Acad Sci USA. 1995;92:11642–11646. doi: 10.1073/pnas.92.25.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostenis E, Conklin BR, Wess J. Molecular basis of receptor/G protein coupling selectivity studied by coexpression of wild type and mutant m2 muscarinic receptors with mutant G alpha(q) subunits. Biochemistry. 1997;36:1487–1495. doi: 10.1021/bi962554d. [DOI] [PubMed] [Google Scholar]
- 32.Gatica EA, Cavasotto CN. Ligand and decoy sets for docking to G protein-coupled receptors. J Chem Inf Model. 2012;52:1–6. doi: 10.1021/ci200412p. [DOI] [PubMed] [Google Scholar]
- 33.DeVree BT, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–186. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao Y, McCammon JA. Unconstrained enhanced sampling for free energy calculations of biomolecules: A review. Mol Simul. 2016;42:1046–1055. doi: 10.1080/08927022.2015.1121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanommeslaeghe K, MacKerell AD., Jr CHARMM additive and polarizable force fields for biophysics and computer-aided drug design. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagen.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case D, et al. 2014. Amber 14 (University of California, San Francisco)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.