A central goal of crop improvement is to breed varieties with broad-spectrum resistance (BSR) to pathogens, but most of the major resistance (R) genes identified to date confer race-specific resistance to their adapted pathogens. Although these R genes are effective for a specific pathogen, their durability in the field is typically short due to mutations in the pathogen population that overcome the resistance. An alternative strategy of incorporating multiple R genes against different pathogens into elite cultivars is time-consuming and technically challenging, and usually results in a yield penalty as the crop diverts energy to implementing disease resistance (1). Many pathogens infect rice (Oryza sativa), the staple food crop of over half of the world’s population. Among them, the fungal pathogen Magnaporthe oryzae and the bacterial pathogen Xanthomonas oryzae pv oryzae (Xoo) are two of the most destructive and can cause devastating yield losses in most rice-growing countries (2). Over the past two decades, many rice R genes have been identified, but none confers resistance to both pathogens. Although manipulating the expression of several defense-responsive genes, or genes in defense signaling pathways, has led to BSR (3, 4), few such genes have been successfully deployed in rice production for disease control. In PNAS, Zhou et al. (5) report the identification of the broad-spectrum resistance Kitaake-1 (Bsr-k1) gene, which negatively regulates BSR, and the bsr-k1 allele, which confers nonspecific BSR to both M. oryzae and Xoo without a yield penalty.
The resistant bsr-k1 mutant was identified from an ethylmethane sulfonate-treated mutant population of the japonica cultivar Kitaake that had been inoculated with seven Kitaake-compatible M. oryzae isolates in the field (5). In addition, the bsr-k1 mutant conferred enhanced resistance to 10 Kitaake-compatible Xoo isolates. Map-based cloning identified Bsr-k1, and further analysis showed that Bsr-k1 gene expression is not induced by M. oryzae infection and the BSR-K1 protein localizes in the cytoplasm.
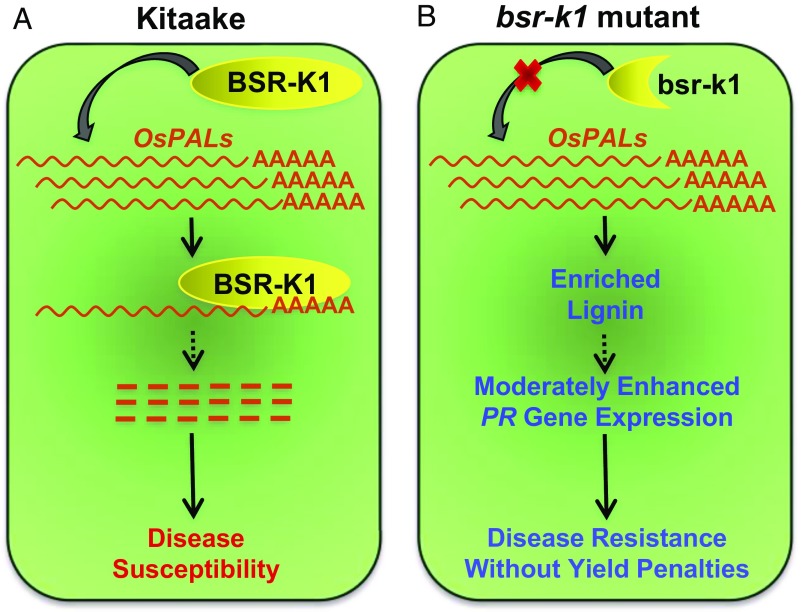
The disease resistance phenotype of the bsr-k1 mutant results from a mutation that causes early termination of a tetratricopeptide repeat (TPR)-containing protein (5). Some TPR-containing proteins are known to be involved in mRNA metabolism (6); therefore, Zhou et al. (5) performed in vitro RNA immunoprecipitation assays and found that the BSR-K1 protein binds to the mRNAs of the phenylalanine ammonia-lyase (PAL) genes and promotes mRNA turnover in rice (Fig. 1A). In contrast, the truncated bsr-k1 protein (1–276 aa) in the bsr-k1 mutant does not bind to OsPAL mRNAs (Fig. 1B). Consistent with the RNA binding results, overexpression of OsPAL1 in transgenic rice enhances resistance to M. oryzae and causes an increase in lignin content. Together, these results demonstrate that BSR-K1 negatively regulates BSR by modulating turnover of OsPAL mRNAs.
Fig. 1.
Model for BSR-K1–mediated disease resistance. (A) In the wild-type Kitaake plants, BSR-K1 proteins bind to the mRNA of OsPAL genes and promote their turnover, leading to disease susceptibility. (B) In the bsr-k1 mutant plants, the truncated bsr-k1 protein cannot bind to the OsPAL mRNAs. The accumulation of OsPAL transcripts can increase the levels of secondary compounds such as lignin, which may moderately enhance PR gene expression and increase disease resistance without a yield penalty.
PAL proteins are key enzymes in the phenylpropanoid pathway involved in the biosynthesis of lignin and flavonoids, and they contribute to disease resistance (7). In rice, there are nine members of the PAL gene family (8). Importantly, four of them (OsPAL1–4) are colocalized with the major resistance quantitative trait locus against Rhizoctonia solani and Xoo in the rice genome (8), and most OsPALs genes are induced by M. oryzae (9). The ospal4 mutant shows increases susceptibility to three rice pathogens (R. solani, Xoo, and M. oryzae) (8), and the OsPAL06 knockout mutant displays increased susceptibility to M. oryzae (9). These results demonstrate the importance of the OsPAL genes in BSR against diverse pathogens in rice. Similarly, Zhou et al. (5) found that the bsr-k1 mutant has higher transcript levels of OsPAL1-OsPAL7 than does Kitaake and that OsPAL1 overexpression lines have increased resistance to M. oryzae. However, unlike OsPAL4 and OsPAL06 (8, 9), suppression of OsPAL1 does not compromise resistance to M. oryzae in the bsr-k1 mutant or Kitaake background (5), probably due to function redundancy with other OsPAL genes.
Although dozens of BSR genes have been identified in crop plants (3), few of them have been used extensively in crop production due to the consequent yield penalty (1, 10). For example, the mlo gene of barley has an average yield penalty of 4.2% (11). Interestingly, the bsr-k1 mutation does not have clear adverse effects on major agronomic traits, compared with wild-type Kitaake, when growing in rice fields with or without high disease pressure. Furthermore, Zhou et al. (5) transferred the bsr-k1 allele into three elite rice cultivars that are popular restoring lines for hybrid rice in Southwest China. These bsr-k1–containing cultivars showed similar yields to those of their parental lines in three field tests at two locations.
Recently, the same team identified a natural allele of the transcription factor gene Bsr-d1 in rice that confers BSR to M. oryzae. A single-nucleotide mutation in the promoter of the bsr-d1 gene results in the binding of the repressive MYB transcription factor, leading to an inhibition of H2O2 degradation and enhanced resistance (12). The identification of the bsr-k1 allele provides another candidate gene for BSR breeding in rice.
TPR-containing proteins play central roles in the biogenesis of the photosynthetic apparatus and plant development (6). For example, the TPR-containing protein CGL71 participates in the assembly of photosystem I in Chlamydomonas reinhardtii (13). The chloroplast-localized TPR-containing proteins SG1 and WTG1 are required for chloroplast development in Arabidopsis (14, 15). TPR-containing proteins are also known to be involved in plant immunity. Suppressor of rps4-RLD (SRFR1), encoding a TPR-containing protein, modulates resistance to the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4 (16). SRFR1 negatively regulates the accumulation of multiple R proteins, including SNC1, RPS2, and RPS4. Interestingly, SRFR1 directly interacts with another TPR-containing protein, SGT1, which is required for the function of a number of R proteins (17). These results suggest that SRFR1 associates with SGT1 to suppress R gene-mediated disease resistance. Although BSR-K1 does not interact with the rice defensome complex, including OsSGT1, as shown in this study (5), it remains to be determined whether BSR-K1 associates with any R protein that recognizes factors from M. oryzae or Xoo. Another possibility is that BSR-K1 protein may act as a susceptibility locus targeted by M. oryzae and/or Xoo.
This intriguing study establishes a link between a TPR-containing protein and PAL mRNA turnover in the context of plant BSR (Fig. 1); it also suggests a new strategy to breed durably resistant plants. However, the results also suggest several important areas of future investigation. First, the mechanism by which the BSR-K1 protein preferentially binds to different OsPAL mRNAs and promotes their turnover during pathogen infection is not known. Second, the relationship between BSR-K1 and R proteins, the OsRac1-mediated defensome, and other defense signaling components needs to be elucidated. Third, the location in the cytoplasm where BSR-K1 binds to OsPAL mRNAs warrants study. Fourth, it is unclear whether, and how, the higher level of lignin content directly induces the pathogenesis-related (PR) gene expression in the bsr-k1 mutant. Finally, an important question is whether moderate overexpression of OsPAL1, OsPAL2, OsPAL3, and OsPAL4 in rice leads to BSR without a yield penalty. Addressing these questions will result in deeper mechanistic understanding of BSR-K1–mediated BSR, and ultimately result in better strategies for breeding durable resistance in crops.
Acknowledgments
Our research is supported by the National Natural Science Foundation of China (Grant 31571944), the National Key Research and Development Program of China (Grant 2016YFD0100600), and the Young Elite Scientist Sponsorship of China Association for Science and Technology (Grant 2015QNRC001).
Footnotes
The authors declare no conflict of interest.
See companion article on page 3174.
References
- 1.Ning Y, Liu W, Wang GL. Balancing immunity and yield in crop plants. Trends Plant Sci. 2017;22:1069–1079. doi: 10.1016/j.tplants.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Liu W, Liu J, Triplett L, Leach JE, Wang GL. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- 3.Ke Y, Deng H, Wang S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017;90:738–748. doi: 10.1111/tpj.13438. [DOI] [PubMed] [Google Scholar]
- 4.Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc Natl Acad Sci USA. 2018;115:3174–3179. doi: 10.1073/pnas.1705927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohne AV, Schwenkert S, Grimm B, Nickelsen J. Roles of tetratricopeptide repeat proteins in biogenesis of the photosynthetic apparatus. Int Rev Cell Mol Biol. 2016;324:187–227. doi: 10.1016/bs.ircmb.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Dixon RA, et al. The phenylpropanoid pathway and plant defence-A genomics perspective. Mol Plant Pathol. 2002;3:371–390. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]
- 8.Tonnessen BW, et al. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol. 2015;87:273–286. doi: 10.1007/s11103-014-0275-9. [DOI] [PubMed] [Google Scholar]
- 9.Duan L, Liu H, Li X, Xiao J, Wang S. Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant. 2014;152:486–500. doi: 10.1111/ppl.12192. [DOI] [PubMed] [Google Scholar]
- 10.Brown JK. Yield penalties of disease resistance in crops. Curr Opin Plant Biol. 2002;5:339–344. doi: 10.1016/s1369-5266(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 11.Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. [Google Scholar]
- 12.Li W, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126.e15. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Heinnickel M, et al. Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci USA. 2016;113:2774–2779. doi: 10.1073/pnas.1524040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, et al. The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in Arabidopsis. J Exp Bot. 2014;65:1111–1123. doi: 10.1093/jxb/ert463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma F, et al. A novel tetratricopeptide repeat protein, WHITE TO GREEN1, is required for early chloroplast development and affects RNA editing in chloroplasts. J Exp Bot. 2017;68:5829–5843. doi: 10.1093/jxb/erx383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon SI, Kim SH, Bhattacharjee S, Noh JJ, Gassmann W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009;57:109–119. doi: 10.1111/j.1365-313X.2008.03669.x. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010;6:e1001111. doi: 10.1371/journal.ppat.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]