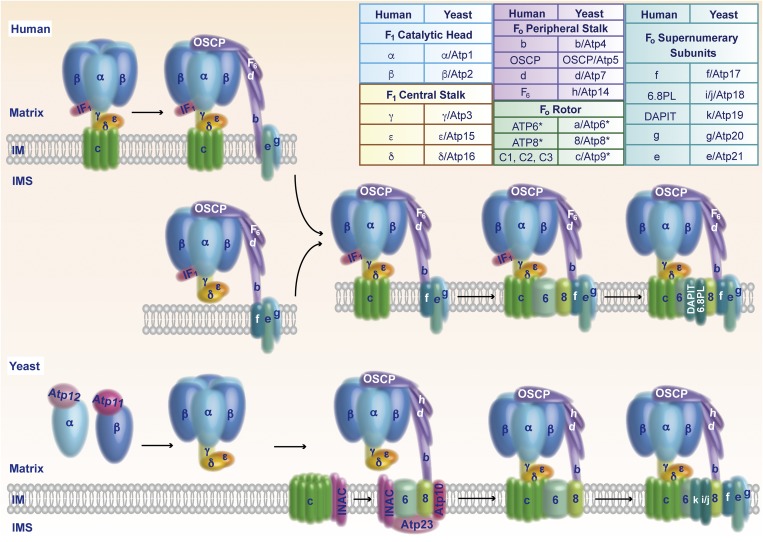
Mitochondria are known as the powerhouses of the cell. The F1Fo-ATP synthase of the mitochondrial inner membrane produces the bulk of cellular ATP. The respiratory chain complexes pump protons across the inner membrane into the intermembrane space and thereby generate a proton-motive force that drives the ATP synthase. In a fascinating molecular mechanism, the ATP synthase couples the synthesis of ATP to the transport of protons into the matrix (1–3). Formation of the ATP synthase depends on the association of 17 different structural subunits of dual genetic origin. Whereas a number of assembly factors and steps have been identified in the model organism baker’s yeast, little has been known about the assembly of the human ATP synthase. In PNAS, He et al. (4) report the identification of key assembly intermediates in the formation of the ATP synthase in human mitochondria. In a tour de force, they define the molecular composition of distinct vestigial ATP synthase complexes in cell lines lacking individual subunits of the enzyme and develop a model of how the membrane-bound domain of the ATP synthase is formed (Fig. 1).
Fig. 1.
Assembly pathways of the mitochondrial ATP synthase. (Upper) Assembly of human ATP synthase. The free F1 domain or an F1–c-ring intermediate binds to the peripheral stalk. The supernumerary subunits e, g, and f associate and promote the insertion of ATP6 and ATP8. Addition of 6.8PL and DAPIT stabilizes the inserted ATP6/ATP8, leading to formation of the proton-conducting channel between ATP6 and the c-ring. The inhibitory protein IF1 that blocks ATP hydrolysis of uncoupled ATP synthase is released. (Lower) Assembly of yeast ATP synthase. The chaperones Atp11 and Atp12 promote formation of the F1 domain. The INAC binds to the c-ring, as well as to an assembly intermediate that contains Atp6, Atp8, the peripheral stalk, the F1 domain, and the maturation factors Atp10 and Atp23. The INAC thus supports proper association of the c-ring with Atp6 to allow formation of the proton-conducting channel. Subsequently, the supernumerary subunits are added to promote dimerization of the enzyme. (Inset) Structural subunits of human and yeast ATP synthase. Asterisks mark mitochondrially encoded subunits. IM, inner mitochondrial membrane; IMS, intermembrane space; OSCP, oligomycin sensitivity-conferring protein.
The ATP synthase consists of the inner membrane-bound Fo region and the matrix-exposed F1 region. The catalytic head of the F1 region and the membrane-integrated rotor module of the Fo region are linked by two stalks, the F1 central stalk and the Fo peripheral stalk (Fig. 1). ATP6, together with the c-ring, forms the proton-conducting channel of the Fo region. The central c-ring is composed of eight or 10 identical subunits in human and yeast mitochondria, respectively. Transport of protons across the Fo rotor module results in rotation of the c-ring. The torque of the c-ring is transmitted to the catalytic head of the F1 region via the γ-subunit of the central stalk. Three heterodimers of subunits α and β form the reactive centers that undergo conformational changes upon rotation of subunit γ, leading to synthesis of ATP. The peripheral stalk prevents rotation of the catalytic head (1–3). Five so-called supernumerary subunits are part of the Fo region and contribute to the formation of dimers and oligomers of the ATP synthase (5–7). A recent high-resolution cryoelectron microscopic structure of the dimeric Fo region of yeast ATP synthase revealed that the subunits Atp6 and i/j form the contact sites between two ATP synthase monomers, supported by interaction between subunits e and k (8). In mitochondria, rows of ATP synthase dimers localize to the rims of the cristae membranes, which are invaginations of the inner membrane. The ATP synthase dimers bend the inner membrane and are crucial for forming the typical cristae shape (9, 10). The supernumerary subunits e and g, together with the N-terminal portion of the peripheral stalk subunit b, affect the curvature of the inner membrane (8, 9, 11). Thus, the ATP synthase not only synthesizes ATP but is also crucial for the architecture of the mitochondrial inner membrane.
The assembly of the mitochondrial ATP synthase is a complicated process that involves the coordinated association of mitochondrially and nuclear encoded subunits (12). In humans, the genes encoding the Fo subunits ATP6 and ATP8 are located in the mitochondrial genome. The other subunits are encoded in the nuclear genome, synthesized as precursor proteins on cytosolic ribosomes and imported into mitochondria. During assembly of the ATP synthase, the F1 domain seems to form independently of the membrane-integrated domain. In yeast, Atp11 and Atp12 bind to the newly imported subunits β and α, respectively. Loss of these assembly factors leads to aggregation of subunits α and β, and blocks formation of the ATP synthase (13). According to the current view, association of the central stalk leads to the release of Atp11 and Atp12 and the formation of the F1 domain (12).
A crucial step during assembly of the ATP synthase is the formation of the proton-conducting channel between ATP6 and the c-ring within the Fo rotor module. A proton-conducting channel that is not properly inserted into an ATP synthase would lead to a dissipation of the proton gradient across the inner membrane. He et al. (4) generated human cell lines with disruption of individual genes for the supernumerary subunits e, f, g, 6.8PL (6.8-kDa mitochondrial proteolipid), and diabetes-associated protein in insulin-sensitive tissues (DAPIT). They found that lack of individual supernumerary subunits leads to respiratory defects and loss of ATP synthase dimers or oligomers. They also determined the molecular composition of vestigial ATP synthases in various knockout cell lines that lacked single supernumerary subunits, peripheral stalk subunits, or the c-ring, as well as in ρ0 cells devoid of mitochondrial DNA (4, 14, 15). Based on their findings, they propose an elegant model of how the membrane domain of human ATP synthase is built (4) (Fig. 1, Upper). In one branch, an F1–c-ring intermediate associates with the peripheral stalk and the supernumerary subunits e and g (15, 16). In the other branch, the F1 domain first assembles with the peripheral stalk and supernumerary subunits e, g, and f. Both pathways merge in a key assembly intermediate that contains the F1 domain, the c-ring, the peripheral stalk, and the supernumerary subunits e, g, and f. In all these vestigial ATP synthase complexes, the inhibitory protein IF1 is enriched to prevent ATP hydrolysis by the uncoupled ATP synthase (4). The presence of the supernumerary subunits e, g, and f is crucial for the subsequent integration of the mitochondrially encoded subunits ATP6 and ATP8 that are stabilized by addition of 6.8PL. Thus, the proton-conducting channel between ATP6 and the c-ring is formed. At this stage, ATP synthesis is coupled to the proton-motive force and the inhibitory protein IF1 is released. Finally, DAPIT is added to the assembly line to promote dimerization and oligomerization of the ATP synthase.
In addition, He et al. (4) assigned 6.8PL and DAPIT as the likely functional orthologs of the yeast subunits i/j and k, respectively. Thus, the 17 main structural subunits of human and yeast ATP synthase are now directly comparable (Fig. 1, Inset).
Interestingly, several maturation steps of yeast and human ATP synthase differ (Fig. 1). Yeast subunit c is mitochondrially encoded, whereas three nuclear genes code for identical c subunits in humans (only the precursors carry different mitochondrial targeting sequences). Yeast Atp6 and Atp8 associate first with a peripheral stalk-F1 intermediate, and the c-ring is added subsequently (17). In human mitochondria, ATP6 and ATP8 insert into an intermediate form of the ATP synthase that contains the c-ring, F1 domain, peripheral stalk, and supernumerary subunits (4). A number of assembly factors for formation of the membrane-bound Fo region have been identified in yeast. The two-domain protein Atp25 stimulates synthesis and assembly of the subunit c into the c-ring (18). The protease Atp23 processes membrane-inserted yeast Atp6, whereas human Atp6 is not proteolytically cleaved. In addition to its proteolytic activity, Atp23, along with the Atp10 chaperone, stabilizes and assembles yeast Atp6 (19, 20). The inner membrane assembly complex (INAC), composed of Ina17 and Ina22 (21), binds to two distinct assembly intermediates of the yeast ATP synthase: the newly assembled c-ring and an assembly intermediate composed of the F1 domain, the peripheral stalk, Atp6/Atp8, and the assembly factors Atp10 and Atp23 (17) (Fig. 1, Lower). The INAC prevents a premature interaction of the intermediates and supports the proper assembly of the c-ring with Atp6 to form the proton-conducting channel. Thus, although the overall composition of human and yeast ATP synthase are similar, the assembly pathways of the central proton-conducting channel differ.
Taken together, the analysis of vestigial ATP synthase complexes by He et al. (4, 14, 15) leads to a stepwise assembly model of the membrane domain of human ATP synthase. Future studies will address the molecular mechanisms and regulation of the distinct assembly steps. Important questions are if and which assembly factors promote the assembly of the human ATP synthase. Although humans and yeast belong to the same supergroup of eukaryotes, the opisthokonts, they use different assembly pathways for the proton-conducting channel. Studies addressing the assembly of the mitochondrial ATP synthase in different eukaryotic supergroups will likely reveal novel mechanisms for building this central cellular machinery.
Footnotes
The authors declare no conflict of interest.
See companion article on page 2988.
References
- 1.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 2.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1Fo-ATP synthases. Annu Rev Biochem. 2009;78:649–672. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 3.Junge W, Nelson N. ATP synthase. Annu Rev Biochem. 2015;84:631–657. doi: 10.1146/annurev-biochem-060614-034124. [DOI] [PubMed] [Google Scholar]
- 4.He J, et al. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc Natl Acad Sci USA. 2018;115:2988–2993. doi: 10.1073/pnas.1722086115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner K, et al. Mitochondrial F1Fo-ATP synthase: The small subunits e and g associate with monomeric complexes to trigger dimerization. J Mol Biol. 2009;392:855–861. doi: 10.1016/j.jmb.2009.07.059. [DOI] [PubMed] [Google Scholar]
- 7.Wagner K, Perschil I, Fichter CD, van der Laan M. Stepwise assembly of dimeric F1Fo-ATP synthase in mitochondria involves the small Fo-subunits k and i. Mol Biol Cell. 2010;21:1494–1504. doi: 10.1091/mbc.E09-12-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric Fo region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rühle T, Leister D. Assembly of F1Fo-ATP synthases. Biochim Biophys Acta. 2015;1847:849–860. doi: 10.1016/j.bbabio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Ackerman SH, Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci USA. 1990;87:4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, et al. Persistence of the mitochondrial permeability transition in the absence of subunit c of human ATP synthase. Proc Natl Acad Sci USA. 2017;114:3409–3414. doi: 10.1073/pnas.1702357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Carroll J, Ding S, Fearnley IM, Walker JE. Permeability transition in human mitochondria persists in the absence of peripheral stalk subunits of ATP synthase. Proc Natl Acad Sci USA. 2017;114:9086–9091. doi: 10.1073/pnas.1711201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujikawa M, Sugawara K, Tanabe T, Yoshida M. Assembly of human mitochondrial ATP synthase through two separate intermediates, F1-c-ring and b-e-g complex. FEBS Lett. 2015;589:2707–2712. doi: 10.1016/j.febslet.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Naumenko N, Morgenstern M, Rucktäschel R, Warscheid B, Rehling P. INA complex liaises the F1Fo-ATP synthase membrane motor modules. Nat Commun. 2017;8:1237. doi: 10.1038/s41467-017-01437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng X, Barros MH, Shulman T, Tzagoloff A. ATP25, a new nuclear gene of Saccharomyces cerevisiae required for expression and assembly of the Atp9p subunit of mitochondrial ATPase. Mol Biol Cell. 2008;19:1366–1377. doi: 10.1091/mbc.E07-08-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng X, Neupert W, Tzagoloff A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol Biol Cell. 2007;18:617–626. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 2011;30:920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lytovchenko O, et al. The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. EMBO J. 2014;33:1624–1638. doi: 10.15252/embj.201488076. [DOI] [PMC free article] [PubMed] [Google Scholar]