Fig. 2.
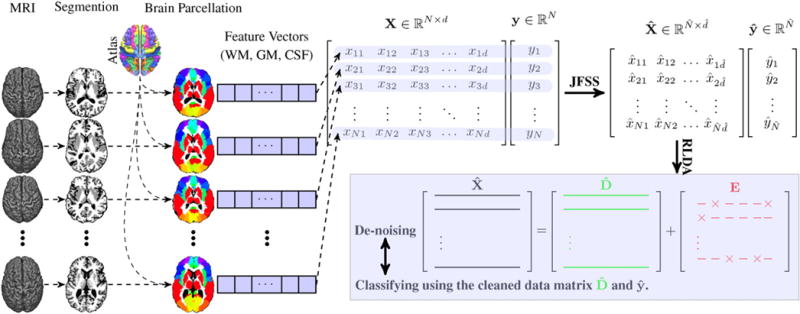
Overview of our proposed method. First, the MR images are processed and tissue-segmented. Then, the anatomical automatic labeling (AAL) atlas is non-linearly registered to each subject’s original MR image, and then the WM, GM and CSF volumes of each ROI are calculated as features. These features form X and the corresponding labels form Y. Through our proposed joint feature-sample selection (JFSS), we discard some uninformative features and samples, leading to and . Then, we train a robust classifier (i.e.,, Robust LDA), in which we jointly decompose into cleaned data and its noise component E, and classify the cleaned data.
