Summary
Large-scale phenotyping efforts have demonstrated that approximately 25-30% of mouse gene knockouts cause intra-uterine lethality. Analysis of these mutants has largely focussed on the embryo but not the placenta, despite the critical role of this extra-embryonic organ for developmental progression. Here, we screened 103 embryonic lethal and subviable mouse knockout lines from the Deciphering the Mechanisms of Developmental Disorders programme (https://dmdd.org.uk) for placental phenotypes. 68% of lines that are lethal at or after mid-gestation exhibited placental dys-morphologies. Early lethality (E9.5-E14.5) is almost always associated with severe placental malformations. Placental defects strongly correlate with abnormal brain, heart and vascular development. Analysis of mutant trophoblast stem cells and conditional knockouts suggests primary gene function in trophoblast for a significant number of factors that cause embryonic lethality when ablated. Our data highlight the hugely under-appreciated importance of placental defects in contributing to abnormal embryo development and suggest key molecular nodes governing placentation.
Keywords: mouse, embryo, placenta, phenotype, trophoblast, stem cells, development
Systematic identification of genes required for normal embryogenesis is essential if we are to successfully unravel the molecular framework underpinning embryo development. Such knowledge will help identify genetic causes of developmental abnormalities that manifest during pregnancy or at birth, and comprise a significant health burden. Large-scale phenotyping efforts have consistently found that 25-30% of mouse gene knockouts (KOs) result in non-viable offspring1–5. In almost all studies of developmentally critical genes, research has focussed on the impact of the mutation on the embryo. By comparison, little attention has been paid to possible effects of these mutations on extra-embryonic tissues, almost certainly resulting in under-representation of placental phenotypes in public databases. The Mouse Genome Informatics (MGI) database, for example, shows extra-embryonic defects in only 10% of embryonic lethal strains. Gaining a more accurate view of the actual frequency of placental abnormalities is critically important for our understanding of the contribution of this vital organ to the aetiology of developmental defects and congenital abnormalities6.
Several ground-breaking studies have highlighted the essential role of extra-embryonic tissues for normal development and long-term health. Placental insufficiency results in intrauterine growth retardation and, as a consequence, can cause fetal programming effects that predispose to later-onset disease7,8. Moreover, tetraploid complementation9 or conditional gene ablation experiments have identified embryonic lethal phenotypes where normal development can be entirely rescued solely by providing the embryo with a wild-type placenta10–15.
However, systematic efforts to discover genes required for normal placental development are still missing. The DMDD consortium16 is one of several ongoing programmes dedicated to identifying and characterising embryonic lethal genes in the mouse. In addition to detailed phenotypic assessment of structural abnormalities in mutant embryos, DMDD also investigates the impact of each mutation on placental development. Here we report the analysis of placental morphology for 103 such lines. Our results reveal a dramatically higher rate of placental phenotypes than had been previously appreciated, and a striking association of placental defects with specific abnormalities in the embryo itself. Our study identifies the placenta as a pivotal target organ for the effects of gene mutations contributing to developmental demise.
Results
Placental defects in embryonic lethals
We analysed 103 mouse KO lines that fail to produce mutant offspring at the expected Mendelian frequency at postnatal day (P)14, but yielded mutant embryos at either embryonic day (E)14.5 or E9.5. Lines for which mutant conceptuses could not be recovered at E9.5 were not included in this screen. Of the 103 lines analysed, 82 were classified as P14 lethal, since no mutant offspring were recovered at that stage. The remaining 21 lines were termed subviable, with mutant pups constituting 13% or less of all offspring obtained, a proportion significantly below the 25% expected from heterozygous crosses (Fig. 1a, Supplementary Table 1). Similar criteria were applied to further sub-categorise the P14 lethal group according to viability at E14.5 (Fig. 1a).
Figure 1. Placental defects are highly prevalent in gene mutants that affect embryonic viability.
(a) Summary of the 103 mouse lines screened. E: day of embryonic development; P: day of postnatal development. ‘Subviable’ identifies strains in which the proportion of mutant offspring is >0% but ≤13%. (b) Summary of non-viable mouse lines in which a placental phenotype has been annotated in Mouse Genome Informatics (MGI; http://www.informatics.jax.org) and in our DMDD programme. (c) Yolk sac appearance in wild-type (WT) and Dennd4c mutants. Images are representative of 3 independent mutants and >60 WT samples analysed. Sections were stained for E-Cadherin (green, demarcating the visceral endoderm) and Laminin (red, highlighting the basement membrane). Arrows point to the disconnected mesoderm and endoderm layers in mutants. (d) Breakdown of the proportion of placental phenotypes by stage of embryonic lethality. (e) Developmental progression of mutant embryos depending on presence or absence of a placental phenotype. TS: Theiler stage.
Placentas of all lines were subjected to histopathological analysis at E9.5, E14.5 or both (Supplementary Table 1). As expected, less than 1% of wild-type placentas showed an abnormal phenotype. By contrast, in mutant placentas we detected dys-morphologies in 56/82 (68%) of P14 lethal strains (Fig. 1b). Even when including the P14 subviable lines, the placental phenotype rate was still 59%, a far higher frequency than the ~10% annotated in MGI (Fig. 1b). All genes associated with placental abnormalities in mutants were expressed in the trophoblast lineage of this organ (Extended Data Fig. 1a), lending support to the notion that they contribute directly to placental growth or function.
We also assessed the conceptuses for yolk sac defects, an extra-embryonic structure that is especially important for nutrient provision during the first half gestation, before formation of the functional placenta at E9.5 (Supplementary Table 1). Since yolk sac was routinely used for genotyping it proved only possible to analyse this tissue in 66 lines. Amongst these, an abnormal yolk sac morphology was detected in 11% (7/66) of cases, compared to ~6% annotated amongst prenatal lethals in the MGI database. Strikingly, all 7 affected lines fell within the E9.5-E14.5 lethal group (7/24=29%; Fig. 1c). Thus, whilst yolk sac defects affecting its structure or hematopoietic function may contribute to the lethality of some of these early lethal strains, they occur at a much lower frequency than placental abnormalities.
When scoring the occurrence of placental defects as a function of developmental stage, we found that almost every line that died before E14.5 exhibited placental abnormalities (40/41; Fig. 1d; Extended Data Fig. 1b), compared to only 35% of lines that were viable beyond E14.5 (12/34) (Fig. 1d; Supplementary Table 1). These findings demonstrate that mutations resulting in embryonic lethality between E9.5-E14.5 are almost certainly associated with a defective placenta.
In line with the placenta being the essential nutrient-supplying organ from mid-gestation onwards, we also found that mutant E14.5 embryos in strains exhibiting a placental phenotype were shifted to a younger developmental stage compared to those in which placental development was normal (Fig. 1e; Extended Data Fig. 1c)17.
Categories of placental defects
In order to categorise the different types of defect, we examined the three main layers of the mature placenta: the labyrinth, which constitutes the main nutrient and gas exchange surface; the junctional zone consisting of spongiotrophoblast, glycogen cells and different giant cell subtypes; and the maternally derived decidua (Fig. 2; Extended Data Fig. 2a).
Figure 2. Summary of common placental defects and functional networks.
(a) Common phenotype criteria used to assess E9.5 mutant placentas (red = abnormality detected). TGC: trophoblast giant cell. (b) E14.5 placental phenotypes in mutant strains. SpT: spongiotrophoblast; Lab: labyrinth. (c) Top: Schematic representation of main structures of an E9.5 placenta. Below: In situ hybridisation for spongiotrophoblast marker Tpbpa and immunostaining against E-Cadherin (Cdh1) on WT and mutant placentas, as indicated. Large red arrows highlight Tpbpa-positive cells. Small red arrows in the Cdh1-stained WT placenta highlight nucleated blood cells in fetal blood vessels; arrowheads in the Pigl-/- placenta demarcate sites of chorionic ectoderm invagination but absence of blood vessels. (d) Top: Schematic representation of main structures of an E14.5 placenta. Below: Examples of histological analyses of E14.5 WT and mutant placentas: Tpbpa in situ hybridisation, red vertical line shows thickness of junctional zone. BSI-B4 isolectin staining demarcating the three main placental layers; red rectangle highlights the severely reduced complexity of labyrinth vascularisation in the Traf2 mutant. E-Cadherin (Cdh1) immunohistochemistry labelling syncytiotrophoblast; red arrowheads point to widened blood spaces, arrows to fibrotic areas. Images in (c) and (d) are representative of ≥3 independent mutants per line, see Methods. (e) Network created using esyN (www.esyn.org) of known interactors of L3mbtl2.
Haematoxylin & Eosin histology (Extended Data Fig. 2b, c) was complemented with three histological staining methods to accurately classify the cellular and tissue composition defects in abnormal placentas using a series of phenotype criteria (Fig. 2a, b). At E9.5, a frequently detected malformation affected the invagination of allantoic blood vessels into the chorionic ectoderm, a process critical for development of the labyrinth that will almost certainly result in developmental arrest (Fig. 2a, c; Extended Data Fig. 2b). At E14.5, by far the most prevalent abnormalities were defects in the growth and intricate organisation of the fetal and maternal blood conduits within the labyrinth layer (Fig. 2b, d; Extended Data Fig. 2c). Since these abnormalities diminish the surface area available for nutrient transport, they will compromise fetal growth and survival.
Collectively these histological characterisations of >300 mutant placentas provide a vast resource for the research community, with all data available at https://dmdd.org.uk.
Critical nodes in placental development
We next examined whether the identity of genes associated with placental defects suggested specific molecular pathways that may be pivotal for the formation or function of this organ. For genes affecting placental morphology at E9.5 in mutants, this network analysis highlighted several functional gene clusters centred around L3mbtl2, Bap1 and Arhgef7 (Fig. 2e; Extended Data Fig. 1d). Similarly, several factors identified in the E14.5 analysis formed specific molecular nodes, for example around Traf2, Nek9 and Rpgrip1l (Extended Data Fig. 1d). Although relatively few genes have been analysed for defects in extra-embryonic tissues in the literature, it is obvious that a large fraction of network components identified in our analyses have been associated with embryonic phenotypes. It therefore seems highly likely that mutants for many of these functionally connected genes will also exhibit placental abnormalities.
Embryo and placenta defects are linked
Since the DMDD programme scores both embryo and placental defects, it provides a unique opportunity to assess co-associations between specific phenotypes18,19. Importantly, DMDD phenotype calls are based on precise embryo sub-staging, therefore the analysis excludes any apparent phenotypes that simply reflect the developmental delay prevalent amongst embryos with placental defects. Nevertheless, mutant mouse lines exhibiting placental abnormalities were enriched for specific E14.5 embryo phenotypes that were distinct from those with normal placentas (Extended Data Fig. 3a).
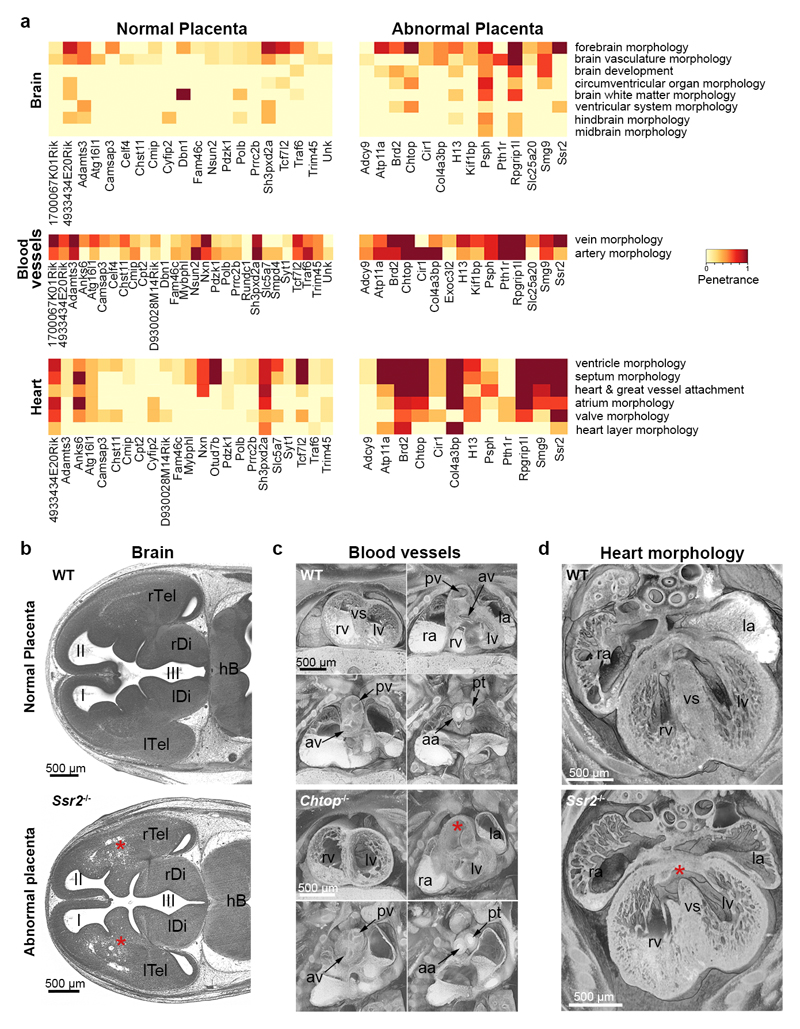
Embryo phenotype categories showing significant statistical correlation with placental defects included abnormalities in the heart, brain and vascular system (Fig. 3a; Extended Data Figs. 3b and 4a, b; Supplementary Table 2). In particular, this affected anomalies in forebrain development, heart chamber and septum morphology, subcutaneous edema, and overall artery or vein topology (Fig. 3b-d; Extended Data Fig. 4c, d). These phenotype co-associations suggest co-regulatory or inter-dependent mechanisms during the development of particular organ systems, notably between the placenta and morphogenesis of the brain, heart and vascular system.
Figure 3. Phenotype co-associations between embryo and placenta.
(a) Enriched embryonic phenotype terms within the significantly co-associated categories of abnormal brain, blood vessel and heart morphology in mutant lines with abnormal placentas, compared to those with normal placentas (dark red: fully penetrant phenotype). For brevity, the description as “abnormal” has been removed from ontology terms. The most prevalent terms describing abnormalities observed in brain, blood vessel and heart development are shown. (b)-(d) HREM images showing embryonic phenotypes that correlate with the presence of placental defects. Upper row: normal morphology in stage-matched controls; bottom row: distinct developmental abnormalities in corresponding structures of mutants: (b) Abnormal forebrain morphology (asterisks) in Ssr2-/- embryo. (c) Double outlet right ventricle and bicuspid aortic valve in Chtop-/- embryo. Right ventricle (rv) with oblique outlet (asterisk). (d) Perimembraneous ventricular septal defect (asterisk) in Ssr2-/- embryo. I, II, III: 1st, 2nd and 3rd ventricle; aa: ascending aorta; av: aortic valve; hb: hindbrain; la, ra: left, right atrial appendix; lDi, rDi: left, right diencephalon; lTel, rTel: left, right telencephalon; lv, rv: left, right ventricle; pv: pulmonary valve; pt: pulmonary trunk; vs: ventricle septum. Defects shown in (b)-(d) are representative of ≥3 independent mutants.
Trophoblast-specific gene functions
Since the placenta comprises cell types of distinct lineage origins, a placental phenotype may be caused by trophoblast-intrinsic and/or extra-embryonic mesoderm-derived endothelial cell defects. To determine trophoblast-specific functions of genes identified as important for placental development, we used CRISPR-Cas9 mediated ablation in trophoblast stem cells (TSCs; Extended Data Fig. 5)20,21. We chose three genes for this analysis that caused lethality around E9.5-10.5 when ablated; the tumour suppressor BRCA1 associated protein 1 (Bap1), the Crumbs epithelial cell polarity complex family member 2 (Crb2), and the nucleotide binding protein-like factor (Nubpl) (Extended Data Fig. 6)22.
Nubpl-mutant TSCs exhibited a decreased stem cell potential, as evidenced by lower expression levels of Cdx2, Esrrb and Elf5, which may explain the dramatic size reduction of the trophoblast compartment in Nubpl-/- placentas. Moreover, severely impaired up-regulation of Gcm1 (an early marker of syncytializing trophoblast) and lower expression levels of Syna and, to a lesser extent, Synb showed that differentiation towards the syncytiotrophoblast lineage was inhibited in the absence of Nubpl (Fig. 4a; Extended Data Fig. 7a). We also detected a prominent phenotype in Bap1-deficient TSCs, as they displayed elevated expression of the key stem cell markers Cdx2 and Esrrb when grown under self-renewal conditions. When triggered to differentiate, Bap1-/- TSCs failed to up-regulate markers of syncytiotrophoblast, sinusoidal trophoblast giant cells and glycogen cells (Fig. 4b; Extended Data Fig. 7b). These TSC differentiation defects may well contribute to the labyrinth formation phenotype evident in both Nubpl and Bap1 mutants. By contrast, Crb2-null TSCs were indistinguishable from wild-type (empty vector) controls (Extended Data Fig. 7c).
Figure 4. Determining trophoblast-specific gene function.
(a) Analysis of Nubpl-/- TSCs grown in self-renewal conditions (“0d”) or upon differentiation for 3 and 6 days. Data are mean +/- S.E.M. *= p<0.05; **= p<0.01; ***= p<0.001 (ANOVA with Holm-Bonferroni’s post-hoc test). Specific defects are summarised in the schematic. (b) Equivalent analysis for Bap1-/- TSCs. EPC: ectoplacental cone; GlyT: glycogen cells; SpT: spongiotrophoblast; SynT: syncytiotrophoblast (layers I and II); TGC: trophoblast giant cells.
Lineage origins of placental defects
To gain further insights into trophoblast-intrinsic versus embryonic lineage-induced effects, we chose the same three genes that we studied in TSCs for conditional gene ablation in vivo. Thus, we used the Sox2-Cre transgene to remove their function in the embryo, while leaving expression intact in the trophoblast-derived cells of the placenta and the visceral yolk sac endoderm (Fig. 5a)23.
Figure 5. Dissecting lineage origins of placental phenotypes.
(a) Schematic representation of the genetic constitutions of embryo (E) and placenta (P) or trophoblast (T) achieved by conditional Sox2-Cre mediated knockout (KO), and corresponding E9.5 embryos of the Nubpl strain. Phenotypes are representative of ≥12 embryos per genotype. (b) Immunofluorescence staining of corresponding placentas for MCT4 (marker of SynT-II), E-Cadherin (Cdh1) and basement membrane component Laminin (Lam; demarcates fetal blood vessels). Nuclear counterstain with DAPI. Placental defects are representative of ≥3 independent mutants per genotype.
Nubpl null embryos associated with a heterozygous placenta were significantly more advanced in development than their complete KO counterparts at E9.5 and could still be recovered up to E11.5, a stage when the complete KO was already resorbed (Fig. 5a; Extended Data Fig. 8a). Histological examination of the E9.5 and E11.5 placentas showed that the trophoblast expansion, syncytiotrophoblast differentiation and labyrinth vascularisation defects were seemingly fully rescued in the conditional KOs (cKOs) (Fig. 5b; Extended Data Fig. 8b). This rescue was also suggested by the transcriptome-wide similarity between cKO and wild-type or heterozygous control placentas (Extended Data Fig. 9a). Thus, although the conditional Nubpl mutation is still lethal beyond E11.5 due to an essential role of this gene in the embryo proper, a functional trophoblast lineage rescues the placental phenotype and the resulting early mid-gestation embryonic lethality.
For Bap1, syncytiotrophoblast formation was partially restored in cKO placentas. Furthermore, global expression profiles of Bap1 cKO placentas were more similar to controls than to KOs (Extended Data Fig. 9a, b). However, placental vascularisation remained under-developed and the conceptuses still died at mid-gestation (Extended Data Fig. 9b). This indicates an essential additional function of Bap1 in the extra-embryonic mesoderm compartment that prevents placental labyrinth formation and also results in a yolk sac defect in KOs and cKOs (Extended Data Fig. 10a). Similarly, Crb2 null embryos could not be rescued by a genetically functional trophoblast lineage (Extended Data Fig. 10b), a result consistent with the lack of phenotype in mutant TSCs. Since the yolk sac phenotype also remained unchanged in cKOs, it can be concluded that the chorio-allantoic placentation defect is due to the critical role of Crb2 in mesoderm development24.
In summary, in vitro and in vivo analysis of 3 genes whose mutation causes mid-gestational lethality identified 2 factors (Nubpl and Bap1) with important roles in the proper expansion and differentiation capacity of trophoblast cells. One of these (Nubpl) is indeed causative of the embryonic lethal phenotype at E9.5.
Discussion
Systematic mouse KO phenotyping efforts undertaken to date have excluded the analysis of extra-embryonic tissues, most notably the placenta3–5,25. Ignoring placental defects as a major contributory factor to fetal demise has previously led to several prominent examples of misannotation of gene function, such as for the tumour suppressor Rb and the oncogene c-myc26–29. In both cases, subsequent studies revealed that restoring gene function to the trophoblast lineage could largely rescue the embryonic defects observed30,31. Here, we report the first systematic effort to assess the prevalence of placental abnormalities in P14 lethal or subviable mouse mutants that survive to at least mid-gestation.
We find a remarkably high percentage of placental abnormalities amongst these lines, with two-thirds of all P14 lethal strains exhibiting obvious defects. In particular, KOs resulting in mid-gestational lethality are almost certainly associated with an abnormal placenta, underpinning the notion that defects in placentation create a bottleneck for developmental progression past mid-gestation32. This frequency of placental defects illustrates the hugely under-estimated impact of gene mutations on extra-embryonic tissues. Given that approximately 25-30% of all mutations cause embryonic lethality, our data suggests that a placental phenotype has gone unnoticed and unreported in hundreds if not thousands of mutant strains.
Many of the genes associated with placental defects in our screen are part of specific functional hubs, such as the L3mbtl2 Polycomb group complex and the tumour necrosis factor-receptor associated factor (Traf2) network, which appear to be of major importance for placental development. Identification of such molecular nodes holds great promise as a way of gaining novel insights into the causes of placentation defects in humans. Consistent with this, at least three of the genes we assessed, TRAF2, PSPH and BAP1 (through its established interaction with ASXL3) have been implicated in the pathophysiology of human pregnancy disorders, many of which have their origin in defective placentation33–36.
A unique feature of our study is the integrated analysis of both embryo and placenta. This has revealed significant co-associations between the occurrence of a placental phenotype and particular defects within the embryo itself, notably affecting neurodevelopment, the heart, and the overall vascular system. A placenta-heart axis has been recognised before37–40, however, we can now identify highly specific pathologies such as a double outlet right ventricle and ventricular septal defects that strongly correlate with the presence of an abnormal placenta. Effects of placental insufficiency on brain development have also been reported31,41,42; our large-scale screen provides strong correlative evidence to support this developmental co-relationship. By contrast, a systematic impact of the placenta on vascular development, beyond overall hemodynamics43, has not previously been recognised. The significance of our findings may therefore extend not only through the immediate gestational period, but also into post-natal life and may help explain how placental insufficiency can have long-lasting consequences on cardiovascular disease risk, outweighing other behavioural factors44.
Taken together, in this study we demonstrate that placental malformations are far more common than previously thought in embryonic lethal mutations and co-occur specifically with heart, brain and vascular network defects. Our data highlight the importance of including extra-embryonic tissues in studies investigating the genetic basis of congenital abnormalities.
Methods
Mouse lines
The majority of mouse lines were generated using the EUCOMM/KOMP knockout first conditional-ready targeted ES cell resource (http://www.mousephenotype.org/about-ikmc/eucomm-program/eucomm-targeting-strategies; targeted trap “tm1a” allele and null “tm1b” allele). A few lines were generated by Crispr-Cas9 mediated gene deletion (“em1” allele). All lines were produced and maintained on a C57BL/6N genetic background at the Wellcome Trust Sanger Institute (http://www.mousephenotype.org/) as part of the DMDD project16. Use of all animals was in accordance with UK Home Office regulations, the UK Animals (Scientific Procedures) Act of 1986 and approved by the Wellcome Trust Sanger Institute’s Animal Welfare and Ethical Review Body. Gene KO lines were designated lethal if no homozygous mutants were present amongst a minimum of 28 pups at P14 and sub-viable if their proportion fell on or below 13% of total offspring from heterozygous intercrosses5. Corresponding cut-off criteria applied to the designation of sub-viability at E14.5. These “DMDD lines” were assessed at embryonic days E14.5 and/or E9.5, counting the day of the vaginal plug as E0.5. Embryos, placentas and yolk sacs were harvested; embryos were processed for HREM imaging19, placentas were fixed in 4% PFA and yolk sacs were used for genotyping.
For conditional gene ablation in the embryo proper (“placental rescue”), lines were mated to Flp expressors to generate conditional “tm1c” alleles (http://www.mousephenotype.org/about-ikmc/eucomm-program/eucomm-targeting-strategies), and then crossed with Sox2-Cre transgenic mice23. Informative crosses were set up between females carrying at least one conditional allele at the locus of interest and heterozygous males that additionally carried the Sox2-Cre transgene. Embryos and placentas were collected as before; genotyping was performed on embryonic tail biopsies.
Histology
For histological analysis, at least 3 mutant and 3 wild-type placentas from at least 2 independent litters (with pairs of mutant and wild-type placentas recovered from the same litter if possible) were processed for routine paraffin histology and embedded side-by-side for each strain. Placentas of male and female conceptuses were analysed wherever possible. No other randomization is applicable for this study. In all cases, tissue appearance and cellular architecture of the placentas analysed confirmed they were in viable condition even if the associated embryo had been designated as dead or dying. Consecutive 7µm sections were produced, and alternate sections mounted. A series of sections per block was processed for haematoxylin and eosin (H&E) staining, using a standard protocol (https://dmdd.org.uk/placental-analysis-protocols/). Sections through the sagittal midline were chosen for imaging, indicated at E9.5 by the remnant of the uterine lumen and at E14.5 by the site of insertion of the umbilical cord. Slides were scanned on a Hamamatsu slide scanner and images deposited at https://dmdd.org.uk. Phenotypes of placentas were assessed for each strain, blinded for strain viability scores, and recorded by at least 2 independent investigators. In cases where all 3 mutant placentas exhibited a particular abnormality, that defect was scored as a phenotype. In cases where a defect was unambiguously detected only in 2 of the initial 3 placentas analysed, an additional 2-3 mutant placentas were added to confirm the call. Overall, a phenotype was scored when at least 67% of mutant placentas exhibited that particular abnormality. Criteria for assessing yolk sac morphology encompassed apposition of the visceral yolk sac endoderm and mesoderm layers, and the appearance of blood islands.
Immunostaining and in situ hybridisation
To gain a more precise view of the structural defects in mutant placentas, mutant placentas from all lines were stained for E-Cadherin (Cdh1) demarcating the labyrinthine syncytiotrophoblast (as well as parietal giant cells at E9.5) and with isolectin BSI-B4 outlining labyrinthine trophoblast and decidua. In situ hybridisation for Tpbpa was used to label the spongiotrophoblast and glycogen cells.
For immunostaining, sections were deparaffinised in xylene and processed through an ethanol series to PBS. Antigen retrieval was performed by boiling in 1mM EDTA pH7.2, 0.05% Tween-20 or in 10mM Na-citrate pH 6.0 buffer followed by blocking in PBS, 0.5% BSA, 0.1% Tween-20. Antibodies used were anti-Cdh1 (1:100 BD Biosciences 610181), anti-Laminin (1:100 Sigma L9393), anti-MCT4 (1:100 Merck Millipore AB3314P) and biotin-conjugated isolectin from Bandeiraea simplicifolia BSI-B4 (1:100 Sigma L2140). Primary antibodies were detected with appropriate fluorescence or horseradish peroxidase-conjugated secondary antibodies; BSI-B4 was detected with horseradish peroxidase-conjugated Streptavidin. Nuclei were counterstained with haematoxylin or 4,6-diamidino-2-phenylindole (DAPI). In situ hybridisation for Tpbpa was performed using a standard protocol45.
Phenotype Data analysis
All genes associated with a placental phenotype in mutant mouse lines were selected for interaction network analysis using esyN (http://www.esyn.org). Expression data for all genes assessed in mouse mutants was obtained by meta-analysis of published RNA-seq datasets22. For testing for co-associations between embryonic and placental defects, two separate analyses were performed. Firstly we examined the phenotypes of homozygous mutant embryos where the placentas have been scored for abnormalities (122 embryos), and secondly we analysed all homozygous mutant embryos scored for mutant phenotypes (241 embryos) according to placental abnormality observed within the line, which builds on our observation that placental abnormalities were fully penetrant in almost every line. The phenotypes scored in homozygous mutant embryos were summarised into broader phenotype categories within the Mammalian Phenotype Ontology by mapping the phenotype terms recorded onto the DMDD intermediate slim as described46.
Statistical analysis used Fisher’s exact test to assess for an association or increase in abnormality rate of the phenotypes when placentas were scored as abnormal. An orthogonal potential alpha-star filter was used prior to the statistical testing to reduce the multiple testing burden as recently described18. To assess the biological effect of placental abnormalities on the abnormality rate of mutant phenotypes we followed the procedure described in Karp et. al.(Ref. 18) of determining the difference in two binomial proportions and calculating the 95% confidence interval using Newcombe's recommended method 10 using the ci.pd function of the R Epi package. Significance was adjusted for the effects of multiple testing using the Benjamini-Hochberg procedure to control the false discovery rate at 5%.
Penetrance Analysis
The MP terms assigned during annotation of the embryos were summarised into the ontology slim categories, and the penetrance score for each slim terms observed for the line calculated as previously described46. The phenotype data was analysed using the DMDD intermediate slim terms, and lower hierarchy slims within ontology terms abnormal brain morphology, abnormal blood vessel morphology, and abnormal heart morphology (Supplementary Table 3).
Generation of mutant trophoblast stem cell (TSC) lines
The wild-type blastocyst-derived TS-Rs26 TSC line (a kind gift of the Rossant lab, Toronto, Canada and tested to be mycoplasma-free) was cultured as described previously20,21. Differentiation was induced by culturing in media lacking bFGF, Heparin and embryonic fibroblast-conditioned medium.
For generation of CRISPR/Cas9-mediated knockout TSCs, gRNAs that result in frameshift mutations were designed using the CRISPR.mit.edu design software and checked for high specificity by nucleotide blast searches. gRNA sequences were cloned into the Cas9.2A.EGFP plasmid (Plasmid #48138 Addgene) and sequence-verified. Empty vector Cas9.2A.EGFP and gene-specific gRNA + Cas9.2A.EGFP constructs were used to generate vector control TSCs and KO TSCs (Extended Data Fig. 6). Transfection was carried out with Lipofectamine 2000 (ThermoFisher Scientific 11668019) reagent according to the manufacturer’s protocol. KO clones were confirmed by genotyping using primers spanning the deleted exon, and by RT-qPCR with primers within, and downstream of, the deleted exon, as shown (Extended Data Fig. 6). Five or six independent KO clones were analysed for each gene mutation.
RT-qPCR expression analysis
Potential defects in TSC maintenance and differentiation capacity were investigated by analysing the expression levels and dynamics of trophoblast marker genes in mutant and control TSCs in stem cell conditions and following 3 and 6 days of differentiation. Total RNA was extracted using TRI reagent (Sigma T9424), DNase-treated and 1µg used for cDNA synthesis with RevertAid H-Minus reverse transcriptase (Thermo Scientific EP0451). Quantitative (q)PCR was performed using SYBR Green Jump Start Taq Ready Mix (Sigma S4438) and intron-spanning primer pairs (Supplementary Table 4)21 on a Bio-Rad CFX96 or CFX384 thermocycler. Normalised expression levels are displayed as mean relative to the vector control sample; error bars indicate standard error of the means (S.E.M.) of at least three replicates.
Transcriptomics analysis
Samples were lysed in Trizol with a 5 mm stainless steel bead (Qiagen) for 4 minutes at 20 Hz in a tissue lyser (Qiagen). After chloroform extraction for 30 minutes at room temperature, RNA was extracted from the aqueous phase using ethanol and a spin column (Qiagen RNeasy MinElute). After quantification (Qubit RNA BR) the sample was treated with DNAse enzyme (Qiagen) and purified over a spin column. Adapter indexed strand-specific RNA-seq libraries were generated from 1000 ng of total RNA following the dUTP method using the stranded mRNA LT sample kit (Illumina). Libraries were pooled and sequenced on Illumina HiSeq 2000 in 75bp paired-end mode. Sequence data were deposited in ENA under accession ERP023265. FASTQ files were aligned to the GRCm38.p5 reference genome using TopHat (v2.0.13, options: --library-type fr-firststrand). Counts for genes were produced using htseq-count (v0.6.1 options: --stranded=reverse) with the Ensembl v90 annotation as a reference. The data were assessed for technical quality (GC-content, insert size and gene body coverage) using QoRTs47 and poor quality samples removed. A variance stabilising transformation was applied to count data for each gene using the R package DESeq2's varianceStabilizingTransformation function48. Principal components analysis (PCA) was performed on the transformed count data for each gene using R's prcomp function.
Data availability
All placental phenotyping data are available at https://dmdd.org.uk. Sequence data were deposited in ENA under accession ERP023265. All primers sequences are provided.
Extended Data
Extended Data Figure 1. Potential trophoblast gene function in mutants with placental defect.
(a) Expression of trophoblast control genes and the 103 DMDD genes in trophoblast stem cells (TSCs), TSCs differentiated for 1 day (D) or 3D, and in E11.5 placentas. Log2-transformed expression values of RNA-seq data are displayed. Note that all genes associated with a placental phenotype in mutants (labelled in red font) are expressed in trophoblast. (b) Frequency of placental defects annotated in mid-gestational lethal mutants (MP: 0011098) as annotated in Mouse Genome Informatics, compared to the findings in DMDD where 40/41 E9.5-E14.5 lethals were found to exhibit placental abnormalities. (c) Left-hand side: Volume rendered 3D model of the surface of a wild-type (WT) embryo, staged as Theiler stage 23, and coronal section through the volume rendered model. Right-hand side: Equivalent images of a littermate E14.5 H13-/- embryo, staged as TS21. Note that the models are displayed in identical resolutions. Scale bar: 1mm. Images are representative of ≥5 embryos per genotype. (d) Network analysis using esyN (http://www.esyn.org) for all DMDD genes identified as causing a placental phenotype in mutants. BAP1 and ASXL3 are known interactors in humans. Red circles identify genes implicated in human trophoblast-based pathologies. The analysis reveals molecular nodes that appear to be of key importance for placental development.
Extended Data Figure 2. Identification of placental defects by H&E histology.
(a) Schematic representation of key stages and cell types in extra-embryonic development, complementing Fig. 2c, d. All: allantois; Ch: chorion; Epi: epiblast; EPC: ectoplacental cone; ExE: extra-embryonic ectoderm; PE: primitive endoderm; SynT-I, -II: syncytiotrophoblast layers I and II; TE: trophectoderm; VE: visceral endoderm. (b) Examples of E9.5 placental phenotypes. Dotted lines: boundary to maternal decidua; vertical bars: chorion trophoblast thickness; arrows in WT placenta: invagination sites of extra-embryonic mesoderm-derived blood vessels into chorionic trophoblast; arrowheads in Psph-/-: sites of chorion folding but missing blood vessels; arrowheads in Dpm1-/-: overabundant and enlarged trophoblast giant cells. (c) Examples of E14.5 placental phenotypes. Red arrows: abnormal maternal blood accumulations. Arrows in Traf2-/- and Col4a3bp-/- (incl. inset) placentas: fibrotic and/or necrotic areas; arrowheads in Chtop-/- and Pth1r-/- placentas: abnormal spongiotrophoblast inclusions. Representative mutant embryo images are also depicted. Images of mutant placentas in (b) and (c) are representative of ≥3 independent mutants per line, see Methods.
Extended Data Figure 3. Co-association analysis between embryo and placenta phenotypes.
(a) Mutant mouse lines were classified into those that exhibit a placental phenotype at E14.5 and those that do not. All embryos analysed by HREM imaging were tagged accordingly to either of these two groups. Enrichment of embryonic phenotype terms in mutant strains with normal or abnormal placentas is shown (dark red: fully penetrant phenotype). For brevity, the description as “abnormal” has been removed from ontology terms. (b) Significantly enriched embryonic phenotype terms in lines that exhibit an abnormal placenta (see also Supplementary Table 2) versus those with normal placenta. Following hypothesis testing using Fisher's exact test, adjusting for multiple testing using the Benjamini-Hochberg method, we estimated the magnitude of the abnormal placenta effect. This was determined by calculating independent binomial proportions for the two groups of embryos with normal (n=172) and abnormal (n=69) placenta. The percent difference between groups and the p-values are shown.
Extended Data Figure 4. Specific embryonic defects are significantly correlated with the occurrence of an abnormal placenta.
(a) Further, detailed co-association statistics between the occurrence of a placental phenotype and specific abnormalities in the embryo proper in DMDD lines. As before, mutant mouse lines were classified into those that exhibit a placental phenotype at E14.5 and those that do not. All embryos analysed by HREM imaging were tagged accordingly to either of these two groups. Significant differences in the frequency of specific embryonic defects was determined between these two groups, and scored for the size of the effect and for its significance. Following hypothesis testing using Fisher's exact test, adjusting for multiple testing using the Benjamini-Hochberg method, we estimated the magnitude of the abnormal placenta effect. This was determined by calculating independent binomial proportions for the two groups of embryos with normal (n=172) and abnormal (n=69) placenta. The figure shows the differences in the estimated abnormality rates of the two embryo groups, and the extent of the bars represent the 95% Newcombe confidence interval (see Methods). “TRUE” means that these associations are significant, “FALSE” that they fall below the significance threshold. Please note that some terms, such as eye development and growth/size/body region are likely a consequence of developmental retardation. However, the highlighted terms such as heart, brain and vascular system morphology are definitely based on abnormalities that are not merely due to developmental delay. (b) Same analysis as in (a) but only including those specific embryos whose placenta was analysed histologically (as opposed to all embryos per strain; n=81 and n=41 embryos having normal and abnormal placenta, respectively). Please note that the important and meaningful terms hold up to significance irrespectively. (c) HREM image of an example of a massive subcutaneous edema (asterisk) covering the entire back of a Psph-/- embryo. Volume rendered 3D model. Axial section through the level of the heart is shown as inlay. Note also the delay in developmental progress. (d) Muscular ventricular septal defect (arrowhead) in an Atp11a-/- embryo. Coronal section through volume rendered 3D model. Axial HREM-image is shown as inlay. la: left atrial appendix; lv: left ventricle; pt: pulmonary trunk; ra: right atrial appendix; rv: right ventricle; vs: ventricular septum. Embryo defects shown in (c) and (d) are representative of ≥3 independent mutants.
Extended Data Figure 5. Major routes of Trophoblast Stem Cell differentiation.
Diagram of the main differentiation routes of trophoblast stem cells (TSCs), including representative cell type-specific marker genes. EPC: ectoplacental cone; GlyT: glycogen cells; SpT: spongiotrophoblast; SynT: syncytiotrophoblast (layers I and II); TGC: trophoblast giant cells.
Extended Data Figure 6. Selection of genes for in-depth analysis of trophoblast contribution to embryonic lethality.
(a) E9.5 phenotypes of mutant placentas of the three genes (Nubpl, Bap1, Crb2) chosen for ablation in TSCs, as well as for placental rescue analysis in vivo (Fig. 5, Extended Data Figs. 8-10). Black arrows (WT placenta): fetal blood vessels penetrating into the chorionic ectoderm. Vertical bars: unpatterned appearance of chorion. Orange arrows: empty or fibrotic maternal blood spaces. Images are representative of ≥3 mutants per line. (b) Details of CRISPR design and TSC clone screening strategy for the three selected genes Nubpl, Bap1 and Crb2. All targeted exons were first confirmed to be trophoblast-expressed. RT-qPCR (performed in technical triplicate per clone) and genomic genotyping PCR analysis (performed in duplicate per sample, with results independently confirmed by RT-qPCR data) were performed on individual, single-cell expanded TSC clones to confirm homozygous knockout (KO). Of note, even though splicing may occur across the deleted exon, all CRISPR-Cas9 deletions were designed to result in a premature stop codon. RT-qPCR data are mean +/- S.E.M. of n=3 technical replicates.
Extended Data Figure 7. Analysis of mutant TSCs for defects in TSC maintenance and differentiation.
(a) Nubpl-/- TSC clones assessed for additional trophoblast marker genes by RT-qPCR. (b) Additional marker gene analysis on Bap1-mutant TSCs. (c) Analysis of Crb2-/- TSC clones for a phenotype in stem cell maintenance (“0d”) or during differentiation (“3d”, “6d”). No significant difference in cell morphology, growth behaviour and gene expression pattern was observed compared to wild-type (WT) vector control clones. Data are mean +/- S.E.M. *= p<0.05; **= p<0.01 (ANOVA with Holm-Bonferroni’s post-hoc test).
Extended Data Figure 8. Placental rescue of Sox2-Cre mediated conditional knockout (cKO) of Nubpl.
(a) Additional images of Nubpl-mutant embryos showing that a wild-type trophoblast compartment significantly rescues the developmental retardation phenotype and embryonic defects observed in the full KO at E9.5. At E11.5, Nubpl-/- embryos can still be recovered while complete KO embryos are not retrievable any more. Images are representative of ≥10 independent embryos with the corresponding genotype. (b) Histological analysis of the corresponding placentas at E11.5 shows a complete rescue of the placental defect in cKOs with a genetically functional trophoblast lineage. Sections were stained for MCT4 (SynT-II marker), E-Cadherin (Cdh1, global SynT marker) and Laminin (Lam, blood vessel basement membrane marker).Images are representative of 3 placentas per genotype.
Extended Data Figure 9. Transcriptomic analysis of placentas from rescue experiments and developmental performance of Bap1 cKOs.
(a) Principal component analysis of global transcriptomes of E9.5 placentas with the indicated genotype. “Res” refers to placentas from Sox2-Cre mediated conditional KOs in which the trophoblast lineage remains functional, whereas the embryo is ablated for the gene-of-interest (E:KO; T: HET). (b) Top row: E9.5 embryo photos of the depicted genotypes for the Bap1 strain. The embryonic lethality of the complete Bap1 KO cannot be rescued by a functional trophoblast compartment. Images are representative of ≥12 independent embryos per genotype. Bottom row: Histological analysis of the corresponding placentas, stained as in Fig. 5b and Extended Data Fig. 8b. Arrows point to partially rescued syncytiotrophoblast loops and some vascular invaginations into the chorionic ectoderm. Yet the vascularisation of the forming labyrinth layer remains under-developed compared to controls. Images are representative of 3 placentas per genotype.
Extended Data Figure 10. Analysis of yolk sac morphology in Nubpl, Bap1 and Crb2 mutants and developmental performance of Crb2 cKOs.
(a) Immunofluorescence staining of yolk sacs for E-Cadherin (Cdh1, green) and Laminin (Lam, red) demarcating the visceral endoderm (VE) and basement membrane of the yolk sac mesoderm (YSM), respectively. Bl: Blood cells. Bap1 and Crb2 mutants show a defect characterised by the lack of attachment of the two visceral yolk sac layers (arrows). This defect cannot be rescued by Sox2-Cre mediated cKO, indicating that its cause resides in the extra-embryonic mesoderm lineage. (b) Developmental performance of Crb2 KO and cKO embryos and analysis of placental morphology, equivalent to Extended Data Fig. 8b. No rescue of embryonic lethality or placental defects is observed in the cKOs (E: KO; T: HET). Images are representative of ≥3 independent conceptuses per genotype.
Supplementary Material
Acknowledgements
We would like to thank Dr Natasha Karp for expert advice on statistical analyses, Ian Sealy for help with PCA analyses, the Flow Cytometry Facility at the Babraham Institute, as well as all contributors to the DMDD programme. This work was supported by Wellcome Trust Strategic Award WT100160MA.
Footnotes
Author contributions
VPG, EF, AM, AS and MH performed the core experiments including histological analyses and TSC work; RW performed statistical co-association analyses and DMDD webpage data handling; CIM, CT, JKW, ET, EJR, DG, ES, HWJ, AG performed all mouse colony management, breeding, sample collection and genotyping work; NS, NW, JC, EMBN performed transcriptomics analyses; SG, WW, TM performed HREM imaging and analyses, JCS, EJR, DJA, TM and MH designed the study, interpreted results and wrote the manuscript.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Ayadi A, et al. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome. 2012;23:600–610. doi: 10.1007/s00335-012-9418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Angelis MH, et al. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet. 2015;47:969–978. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White JK, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams D, et al. Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis Models Mech. 2013;6:571–579. doi: 10.1242/dmm.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 7.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 8.Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. BMJ. 1990;301:259–262. doi: 10.1136/bmj.301.6746.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossant J. Development of the extraembryonic lineages. Semin Dev Biol. 1995;6:237–247. [Google Scholar]
- 10.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 11.Luo J, et al. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto H, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 14.Shi W, et al. Choroideremia gene product affects trophoblast development and vascularization in mouse extra-embryonic tissues. Dev Biol. 2004;272:53–65. doi: 10.1016/j.ydbio.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber M, et al. Placental vascularisation requires the AP-1 component Fra1. Development. 2000;127:4937–4948. doi: 10.1242/dev.127.22.4937. [DOI] [PubMed] [Google Scholar]
- 16.Mohun T, et al. Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice. Disease Models and Mechanisms. 2013;6:562–566. doi: 10.1242/dmm.011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer SH, et al. A staging system for correct phenotype interpretation of mouse embryos harvested on embryonic day 14 (E14.5) J Anat. 2017;230:710–719. doi: 10.1111/joa.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karp NA, Heller R, Yaacoby S, White JK, Benjamini Y. Improving the Identification of Phenotypic Abnormalities and Sexual Dimorphism in Mice When Studying Rare Event Categorical Characteristics. Genetics. 2017;205:491–501. doi: 10.1534/genetics.116.195388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weninger WJ, et al. Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis Model Mech. 2014;7:1143–1152. doi: 10.1242/dmm.016337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 21.Murray A, Sienerth AR, Hemberger M. Plet1 is an epigenetically regulated cell surface protein that provides essential cues to direct trophoblast stem cell differentiation. Sci Rep. 2016;6:25112. doi: 10.1038/srep25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latos PA, et al. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 2015;29:2435–2448. doi: 10.1101/gad.268821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 24.Xiao Z, et al. Deficiency in Crumbs homolog 2 (Crb2) affects gastrulation and results in embryonic lethality in mice. Dev Dyn. 2011;240:2646–2656. doi: 10.1002/dvdy.22778. [DOI] [PubMed] [Google Scholar]
- 25.Bradley A, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee EY, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 27.Clarke AR, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 28.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 29.Trumpp A, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 30.Dubois NC, et al. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- 31.Wu L, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 32.Copp AJ. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 33.Fu J, Zhao L, Wang L, Zhu X. Expression of markers of endoplasmic reticulum stress-induced apoptosis in the placenta of women with early and late onset severe pre-eclampsia. Taiwan J Obstet Gynecol. 2015;54:19–23. doi: 10.1016/j.tjog.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Haider S, Knofler M. Human tumour necrosis factor: physiological and pathological roles in placenta and endometrium. Placenta. 2009;30:111–123. doi: 10.1016/j.placenta.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acuna-Hidalgo R, et al. Neu-Laxova syndrome is a heterogeneous metabolic disorder caused by defects in enzymes of the L-serine biosynthesis pathway. Am J Hum Genet. 2014;95:285–293. doi: 10.1016/j.ajhg.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava A, et al. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome. Hum Mol Genet. 2016;25:597–608. doi: 10.1093/hmg/ddv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 38.Adams RH, et al. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- 39.Raffel GD, et al. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol Cell Biol. 2009;29:333–341. doi: 10.1128/MCB.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama EO, et al. Extraembryonic but not embryonic SUMO-specific protease 2 is required for heart development. Sci Rep. 2016;6:20999. doi: 10.1038/srep20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 42.Adelman DM, Gertsenstein M, Nagy A, Simon MC, Maltepe E. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 2000;14:3191–3203. doi: 10.1101/gad.853700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linask KK, Han M, Bravo-Valenzuela NJ. Changes in vitelline and utero-placental hemodynamics: implications for cardiovascular development. Front Physiol. 2014;5:390. doi: 10.3389/fphys.2014.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthiesen NB, et al. Congenital Heart Defects and Indices of Placental and Fetal Growth in a Nationwide Study of 924 422 Liveborn Infants. Circulation. 2016;134:1546–1556. doi: 10.1161/CIRCULATIONAHA.116.021793. [DOI] [PubMed] [Google Scholar]
- 45.Hemberger M, Nozaki T, Masutani M, Cross JC. Differential expression of angiogenic and vasodilatory factors by invasive trophoblast giant cells depending on depth of invasion. Dev Dyn. 2003;227:185–191. doi: 10.1002/dvdy.10291. [DOI] [PubMed] [Google Scholar]
- 46.Wilson R, McGuire C, Mohun T, Project D. Deciphering the mechanisms of developmental disorders: phenotype analysis of embryos from mutant mouse lines. Nucleic Acids Res. 2016;44:D855–861. doi: 10.1093/nar/gkv1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartley SW, Mullikin JC. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 2015;16:224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All placental phenotyping data are available at https://dmdd.org.uk. Sequence data were deposited in ENA under accession ERP023265. All primers sequences are provided.