Abstract
Environmental factors (e.g., malnutrition and physical inactivity) contribute largely to metabolic disorders including obesity, type 2 diabetes, cardiometabolic disease, and nonalcoholic fatty liver diseases. The abnormalities in metabolic activity and pathways have been increasingly associated with altered DNA methylation, histone modification, and noncoding RNAs, whereas lifestyle interventions targeting diet and physical activity can reverse the epigenetic and metabolic changes. Here we review recent evidence primarily from human studies that links DNA methylation reprogramming to metabolic derangements or improvements, with a focus on cross-tissue (e.g., the liver, skeletal muscle, pancreas, adipose tissue, and blood samples) epigenetic markers, mechanistic mediators of the epigenetic reprogramming, and the potential of using epigenetic traits to predict disease risk and intervention response. The challenges in epigenetic studies addressing the mechanisms of metabolic diseases and future directions are also discussed and prospected.
Keywords: epigenetic marker, DNA methylation, reprogramming, metabolic disorders, intervention
1. Introduction
Epigenetic changes induced by environmental factors have been increasingly associated with metabolic disorders including obesity, type 2 diabetes (T2D), and cardiovascular diseases (CVD) [1–4]. Importantly, the epigenetic differences may reflect metabolic health disparity in monozygotic twins who are considered genetically identical [5–10]. It is now recognized that genetic variants account poorly for the observed heritability of disease risk (less than 2% for obesity and 5–10% for T2D) [11–14], and the missing heritability is being revealed in epigenetic studies that target the impacts of prenatal and postnatal environment on the epigenome and metabolic disease risks [15, 16]. For instance, nutritional deficiency or excess (either prenatally or postnatally) leads to epigenetic reprogramming that is significantly correlated with increased obesity incidence [15–17]. Thus, understanding metabolic disorders from an epigenetic perspective may offer new strategies to prevent or treat these diseases.
Epigenetic mechanisms identified in metabolic disorders include DNA modification (e.g., methylation and hydroxymethylation), histone modification (e.g., methylation, acetylation, ubiquitylation, SUMOylation, citrullination and ADP-ribosylation), and altered expression of noncoding RNAs [17–19]. The epigenetic variants can stimulate or suppress gene expression depending on the individual mechanism, e.g., the type of modification and location it affects [17–19]. In general DNA methylation at gene promoters and enhancers tends to silence the gene, while DNA methylation in the gene body promotes gene expression [20, 21]. DNA wraps around a histone protein core (i.e., an octamer composed of two copies of H2A, H2B, H3, and H4) to form a nucleosome, and modifications of histone may increase the exposure of DNA to modification or transcription factors in the regulation of gene expression [19, 22]. Methylation of H3 histone may harbor a repressive mark (e.g., H3K27me3) or an active mark (e.g., H3K4me3) near genes with poised transcription [23]. Noncoding RNAs such as long noncoding RNAs (lncRNA) and small noncoding RNAs (e.g., microRNAs) represent the third epigenetic mechanism that can interact with chromatin or directly regulate gene expression in metabolic disorders [17–19]. The coordinated epigenetic changes not only control gene expression, but in some case may regulate DNA repair and replication [17–19, 22, 24].
The epidemics of obesity and its comorbidities (e.g., T2D and CVD) have been attributed largely to positive energy balance [4, 25–27]. As one of the major contributors, malnutrition can alter epigenetic profile in obese or diabetic patients, and the impact can be passed on to their offspring for generations [15–17]. It was shown that high fat diet caused unique variations in chromatin and epigenome trans-generationally, and low-birthweight subjects had lower DNA methylation plasticity [28–32]. Interestingly, undernutrition during pregnancy or lactation (e.g., famine) also imposes epigenetic influence that increases the risks of the offspring developing obesity and T2D [15–17]. By contrast, physical activity has been effective to prevent obesity and T2D by boosting energy expenditure or balance [33–38], and is associated with robust epigenetic reprogramming [4, 16, 17, 39–42]. In this article, we will summarize the evidence of epigenetic reprogramming in metabolic disorders, primarily from human studies of the plasticity of DNA methylation (the best-studied epigenetic mechanism). We also discuss the mechanistic mediators of epigenetic reprogramming and the potential of DNA methylation marker in disease risk and intervention assessment.
2. Epigenetic reprogramming in metabolic disorders
DNA methylation traits are discovered increasingly in the tissues that undergo metabolic alteration in obese and diabetic patients, including adipose tissue, liver, skeletal muscle, and pancreas (Table 1). The epigenetic reprogramming involves critical pathways or regulatory processes such as: (1) insulin signaling (e.g., IRS1, IRS2, SORBS2) [10, 43–45]; (2) insulin secretion (e.g., PPARGC1A, CCND2, CILP2, FHL2, CDKN1A, PDE7B, SEPT9 and EXOC3L2) [21, 46–48]; (3) adipocyte differentiation, transdifferentiation, and function (e.g., PPARG, PPARGC1A, PRDM16, LEP, ADIPOQ, HIF3A) [10, 49–55]; (4) mitochondrial function and redox regulation (e.g., PPARGC1A, TFAM, MT-ND6, CPT1A, TXNIP, SOD3) [46, 48, 49, 56–63]; (5) lipid and glucose homeostasis (e.g., SREBF1, ABCG1, CPT1A, SORBS2) [45, 48, 64]; (6) cytokine signaling and inflammation (e.g., SOCS3, ADIPOQ, ABCG1) [48, 53, 54, 64]; and (7) cell cycle, apoptosis and autophagy (e.g., DAPK3 and CDKN1A) [21, 65]. These pathways have been implicated in the pathogenesis of obesity and type 2 diabetes. For instance, adipocyte differentiation or expansion may contribute to increased adiposity and dysregulate cytokine secretion and signaling pathways, which is associated with chronic inflammation and insulin resistance [66, 67]. On the other hand, browning of white adipose tissue, where adipocyte transdifferentiation take places, may boost energy balance, prevent obesity and improve insulin sensitivity [68–71].
Table 1.
DNA methylation reprogramming in metabolically critical tissues
| Tissue type* | Medical conditions | Genes that undergo DNA methylation reprogramming | Physiological or pathophysiological relevance | References |
|---|---|---|---|---|
|
| ||||
| Adipose tissue | Obesity | 5529 differentially methylated DNA sites for 2223 differentially expressed genes | 25 % genes linked to adipogenesis, 19 % to insulin signaling, 27 % to lipolysis | [143] |
| T2D | 15627 differentially methylated DNA sites covers 7046 genes including PPARG, KCNQ1, TCF7L2, and IRS1 | Insulin signaling, adipogenesis, and metabolism. | [10] | |
| Obesity | ADAMST4, ANGPT2/4, AOC3, AQP7, CETP, DOCK9, LIPE, SOD3, and TIMP4. | Adipogenesis/adiposity, redox, angiogenesis, glycaemia control, and lipolysis | [63] | |
| Obesity, insulin resistance | COL5A1, GAB1, IRS2, PFKFB3 and PTPRJ | Integrin cell surface interactions and insulin signaling | [43] | |
| Overweight, obesity, T2D, and aging | FHL2, NOX4, PLG, ELOVL2, KLF14, GLRA1, FTO, ITIH5, CCL18, MTCH2, IRS1, and SPP1, HIF3A | Glucose and fatty acid metabolism, mitochondrial function, oxidative stress, insulin signaling, chemokine signaling, adipocyte differentiation | [44] | |
| Obesity, T2D, and first-degree relatives of T2D | HIF3A | Glucose and amino acid metabolism, apoptosis, proteolysis, p53 and PPAR signaling, and adipocyte differentiation/adiposity | [50–52] | |
| High-T2D risk due to low birth weight | PPARGC1A | Mitochondrial biogenesis, energy expenditure and balance, browning of white adipose tissue | [49] | |
| Obesity | SORBS2 and ETV6 | Insulin mediated translocation of GLUT4 | [45] | |
| T2D | ABCG1, and SREBF1 | Lipogenesis, dyslipidemia, cytokine signaling, redox and insulin resistance | [48] | |
| Overweight, and obesity | CPT1A, ABCG1, LYS6GE, KDM2B, RALB, PRRL5, LGALS3BP, C7orf50, PBX1, EPB49 and BBS2 | Mitochondrial uptake of long-chain fatty acids and triglyceride metabolism; macrophage cholesterol and phospholipids transport, and lipid homeostasis; immune function and inflammatory pathways | [64] | |
| Severe obesity, diet induced obesity | LEP and ADIPOQ | Food intake, inflammation, insulin sensitivity, glucose and lipid homeostasis, energy balance | [53, 54] | |
|
| ||||
| Liver | Overweigh, obesity, and T2D | ABCG1, PHOSPHO1, SOCS3, SREBF1, and TXNIP | glucose and lipid homeostasis, redox, HDL-mediated increase in insulin secretion | [48] |
| T2D | 251 differentially methylated sites covering GRB10, ABCC3, MOGAT1, and PRDM16, and 29 genes showing differential gene expression besides DNA methylation, including PPP1R1A, RIPK4, H19, TICAM1, MYH10, and RAD50 | Inflammatory response, insulin sensitivity in liver, growth and metabolism, hepatic glycogen synthesis and glucose homoeostasis, fatty acid oxidation, energy homeostasis | [55] | |
| Hepatosteatosis, nonalcoholic steatohepatitis | DPP4 | Signal transduction, glucose and lipid homeostasis | [144] | |
| Obesity, nonalcoholic fatty Liver disease | DNA methylation ages | Mitochondrial function, insulin resistance, nonalcoholic fatty Liver disease activity score and liver cancer | [145] | |
| Obese and T2D | PRKCE, ABR, PDGFA, ARHGEF16, ADCY6, RPS6KA1, CTBP1, CCND1, WNT11, and ATF-motifs | Insulin signaling, hepatic glycolysis, de novo lipogenesis | [146] | |
| Nonalcoholic Fatty Liver Disease | TET, PPARGC1A, TFAM, and MT-ND6 | Mitochondrial biogenesis and function, oxidative stress, apoptosis, insulin resistance, lipid metabolism | [56–58]. | |
| Obesity, T2D, non-alcoholic steatohepatitis | LDHB | Glycolysis, cancer development and metastasis | [122] | |
|
| ||||
| Skeletal muscle | Obesity, T2D, or low birth weight | PPARGC1A and PDK4 | Mitochondrial function, age-associated metabolic dysfunction, inflammation, and dyslipidemia | [59–62] |
| T2D | DAPK3 | Cell proliferation, apoptosis, and autophagy, responding to insulin and glucose in a reciprocal manner | [65] | |
| Overweigh, obesity, and T2D | ABCG1, PHOSPHO1, SOCS3, SREBF1, and TXNIP | glucose and lipid homeostasis, redox, HDL-mediated increase in insulin secretion | [48] | |
|
| ||||
| Pancreas | T2D | PPARGC1A | Mitochondrial biogenesis and function, reduced in insulin secretion | [46] |
| Aging, genetic risk for T2D, pre-diabetes | CCND2, CILP2, FHL2, GNPNAT1, HLTF, KLF14, PBX4, SH2B3, SLC6A4, TCF7, and ZNF518B | Mitochondrial function, altered insulin secretion and glucose homeostasis | [47] | |
| T2D | Differential DNA methylation in 853 genes, 102 of which were differentially expressed in T2D islets: e.g., CDKN1A, PDE7B, SEPT9 and EXOC3L2 | Cell cycle, β-cell mass expansion β-cells and α-cells function, glucagon and insulin secretion, glucose homeostasis | [21] | |
| Overweigh, obesity, and T2D | ABCG1, SOCS3, SREBF1, and TXNIP | glucose and lipid homeostasis, redox, HDL-mediated increase in insulin secretion | [48] | |
| T2D | PDX1, TCF7L2, ADCY5, NR4A3, PARK2, PID1, SLC2A2, SOCS2, and more | β-cell proliferation, mitochondrial function, and insulin secretion | [147] | |
It also include the major cell type, such as adipocytes from adipose tissues, hepatocytes from liver, myotubes from skeletal muscle, and β-cells from pancreas.
DNA methylation controls gene activity in cell differentiation, embryogenesis, and development, and unique DNA methylation patterns may distinguish one type of cell (or tissue) from other types [72]. Intriguingly, DNA methylation reprogramming in PPARGC1A (the gene encoding PGC1α, a key regulator of mitochondrial biogenesis and function) has been identified in different tissues from obese and diabetic patients (Table 1) [46, 49, 56] [57, 59–62]. Thus, epigenetic regulation of mitochondria may represent a common mechanism in metabolic changes across tissues. Indeed, functional alterations of mitochondria have been observed across tissues in obesity, T2D, cardiometabolic diseases, and nonalcoholic fatty liver diseases [4, 73–77]. In the adipose tissue mitochondrial malfunction dysregulates adipocyte differentiation, trans-differentiation, cytokine secretion [68, 78, 79]. In the liver and skeletal muscle, mitochondrial impairment leads to incomplete fatty acid oxidation, ectopic fat accumulation, and insulin resistance [80, 81]. In pancreatic beta-cells, mitochondrial dysfunction dampens ATP-dependent insulin secretion [82]. Consistently, PPARGC1A shows increased methylation but reduced gene expression in diabetic islets [46], skeletal muscle from sedentary individuals [60], nonalcoholic fatty liver [56, 57], and adipose tissues from subjects with high T2D risk [49]. The downregulated PPARGC1A gene expression is associated with reduced mitochondrial content in the target tissues [57, 60]. Furthermore, PPARGC1A hypermethylation was found to cause gene downregulation and lower mitochondrial content, where DNA methyltransferase 3B (DNMT3B) is induced to promote PPARGC1A methylation [60].
In addition to mitochondrial alteration, obese and T2D patients also show impairment in glucose and lipid metabolism [83]. Recent studies identified DNA methylation changes in the genes of ATP-binding cassette (ABC) protein family, particularly in ABCG1 across tissues [48, 84]. The ABCG1 protein can remove excess cholesterol from peripheral tissues and transporting it to the liver [85], and ablation of ABCG1 in mice leads to sterol accumulation, impaired glucose tolerance and insulin secretion, and inflammation of pancreatic β-cells [86]. Cross-tissue dysregulation of DNA methylation was also identified in SREBF1, the gene encodes for sterol regulatory element-binding transcription factor 1 (or sterol regulatory element-binding protein 1 (SREBP1), a key regulator of lipid homeostasis [48, 84, 87, 88]. The altered DNA methylation in ABCG1 and SREBF1 genes is associated with downregulation of mRNA levels in the skeletal muscle and liver from T2D individuals compared with health controls [48].
Noteworthy is tissue-dependent epigenetic reprogramming, nevertheless, which may also play important roles in tissue and systemic metabolic changes. As an endocrine organ and energy reservoir, adipose tissues in obese and diabetic patients show altered DNA methylation in adipose PPARG [10], LEP and ADIPOQ [53, 54], and HIF3A [44, 50–52]. Functionally, PPARG encodes for adipogenic regulator PPARγ that underpins adipocyte differentiation and lipid droplet growth [89–91]. LEP encodes leptin that is anorexigenic and proinflammatory, and its action can be counteracted by ADIPOQ-encoded hormone adiponectin [92]. In obese individuals, leptin is elevated and adiponectin reduced, which contributes to chronic inflammation, insulin resistance, impaired glucose and lipid metabolism [53, 66, 92, 93]. Epigenetic studies suggested that DNA methylation in LEP was correlated negatively with body mass index (BMI), suggesting a trend of hypomethylation of LEP in obesity; however, DNA methylation in ADIPOQ were correlated positively with BMI, suggesting a trend of hypermethylation of ADIPOQ in obesity [54]. Indeed, hypermethylation in ADIPOQ suppresses adiponectin expression in obesity, where DNA methyltransferase 1 (DNMT1) is induced by proinflammatory cytokines tumour necrosis factor (TNFα) and interleukin (IL)-1β to stimulate ADIPOQ methylation [53]. By contrast, LEP DNA methylation level is reduced in obese individuals, which tends to upregulate leptin expression [54, 94]. The reciprocal regulation of DNA methylation and gene expression of LEP and ADIPOQ accounts, at least in part, for the chronic inflammation and metabolic changes in obesity.
Differing from other tissues, white adipose tissue undergo rapid expansion in obesity, which induces hypoxic stress, adipocyte dysfunction, and dysregulated adipokines secretion [66, 95–97]. Activation of hypoxia inducible factor HIF1α promotes macrophage recruitment to adipose tissues and increases adipocyte-derived pro-inflammatory cytokines [51, 52, 95]. However, the role of HIF3α is unsettled. In line with HIF3α counteracting HIF1α [98–100], HIF3A (HIF3α encoding gene) expression in adipose tissue is inversely associated with serum level of inflammation marker C-reactive protein [51]. By contrast, ablation of adipose HIF3α protects mice from weight gain, improving glucose tolerance and insulin sensitivity, indicative of a pro-obesity role of HIF3α [101]. The complexity of HIF3α role in obesity is further demonstrated by epigenetic studies that reveal positive associations between HIF3A DNA methylation and BMI, whereas HIF3A mRNA is associated negatively with BMI and positively with insulin sensitivity [44, 50, 52]. Although HIF3A DNA methylation was inversely correlated with HIF3A mRNA [50], the relationship was not that straightforward in other studies [51, 52]. The discrepancy may arise from heterogeneity of adipose tissues and fat depot specific difference in angiogenesis and cell population [51, 102, 103].
Methylated DNA can be oxidized into hydroxymethylated DNA, which is known as an intermediate for DNA demethylation [104]. However, recent studies have revealed roles of DNA hydroxymethylation in DNA repair [105], epigenomic remodeling of enhancers to activate transcription [106, 107], and lineage commitment [108]. Dynamic hydroxymethylation of DNA has been identified in different human tissues such as brain, heart, skeletal muscle, and spleen [109, 110]. Acute exercise may elicit robust hydroxymethylation of DNA, which is associated with DNA demethylation and induction of exercise-responsive gene (e.g., nuclear receptor subfamily 4 group A member 3) in skeletal muscle or myotubes [109]. In human adipose tissues, hydroxymethylation of DNA shows depot-dependent differences, with higher level of hydroxymethylated DNA in visceral fat than in subcutaneous fat [111]. Importantly, the DNA hydroxymethylation in human adipose tissue was correlated with clinical variables (e.g., lipid parameters) [111], which was also observed in human blood samples [112]. The responses to physiological and pathophysiological cues reported in these studies suggest that DNA hydroxymethylation may play a role in metabolic regulation.
3. Mechanistic mediators of epigenetic reprogramming
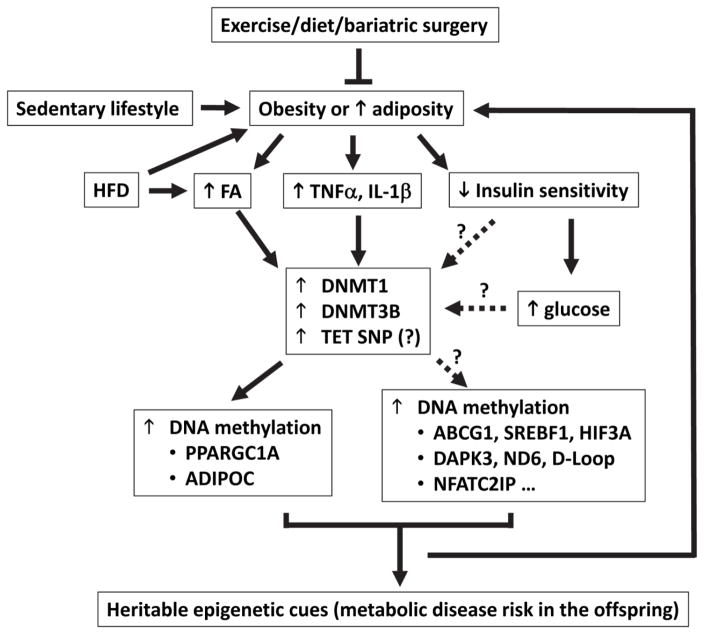
Cross-sectional studies have provided solid evidence linking obesity or BMI to epigenetic reprogramming and altered expression of related genes [24, 44, 113–115]. Until recently was a cause-effect relationship revealed between BMI and DNA methylation by emerging longitudinal studies that were coupled with genetic association analyses or Mendelian randomization (MR) analysis, i.e., BMI and adiposity per se causal to altered DNA methylation [113, 114]. Employing a weighted genetic risk score that combined effects of single-nucleotide polymorphism (SNP) on BMI, Wahl et al. suggested that the predicted effects of BMI genetic risk score on DNA methylation at certain loci (e.g., ABCG1) were significantly correlated with observed effects, and the effect of BMI on DNA methylation was further corroborated by longitudinal follow-up analyses of the participants [114]. In another longitudinal analysis, childhood BMI was correlated with HIF3A DNA methylation in adolescence but HIF3A DNA methylation in childhood was not associated with BMI in adolescence, suggesting that childhood BMI may precede or determine the altered DNA methylation in adolescence [113]. These findings suggest a causal role for adiposity or obesity in altered DNA methylation (Figure 1).
Figure 1.
The potential pathways that link the identified mediators to altered DNA methylation in metabolic disorders. Obesity due to sedentary lifestyle and energy overconsumption (e.g., HFD) is characterized by elevated fatty acids (FA), glucose, pro-inflammatory cytokines (TNFa, IL-1b), insulin resistance (or reduced insulin sensitivity), and resultant hyperglycemia (increased blood glucose). DNMT1 and DNMT3 were shown to account for DNA methylation reprogramming induced by FA, TNFa, and IL-1b in certain genes such as PPARGC1A and ADIPOC. However, it is largely unknown whether this represents a common mechanism for altered DNA methylation in other genes. In fact, TNFα induces DNMT3B in the regulation of skeletal muscle DNA methylation, whereas TNFα induces DNMT1 in adipose DNA methylation reprogramming, suggestive of a tissue-dependent selective induction of DNMTs by TNFα. Insulin and glucose alters DNA methylation of DAPK3 but not PPARGC1A in the muscle; whether and how DNMT1 or DNMT3 is involved and what accounts for the gene- or locus-specific DNA methylation reprogramming remains unknown. Genetic variant in TET2 is associated with altered PPARGC1A DNA methylation in fatty liver disease, and studies are need to examine whether TET2 SNP is related to dysregulated FA, pro-inflammatory cytokines, insulin signaling, and glucose. Longitudinal studies and MR analysis suggest that adiposity may cause DNA methylation reprogramming (e.g., in ABCG1 and HIF3A), while DNA methylation profiles (e.g., in PPARGC1A and NFATC2IP) can also predict adiposity. Maternal obesity confers epigenetic impacts on metabolic disease risk in the offspring, but maternal exercise may normalize DNA methylation (e.g., PPARGC1A hypermethylation) in the offspring. To this end, weigh loss intervention through exercise, diets or bariatric surgery have been shown to reverse the dysregulation of DNA methylation in subjects with metabolic disorders.
At molecular level, obesity may mediate DNA methylation reprogramming by dysregulating insulin signaling [24, 65, 115], glucose [65, 116], fatty acids or lipid [60, 117–121], and inflammatory cytokines [53, 60]. Insulin was reported to increase DNA methylation in the gene body of DAPK3 (a gene involved in cell proliferation, apoptosis, and autophagy), and in T2D patients who are insulin resistant, DAPK3 methylation was downregulated [65]. Consistently, DNA methylation profile in the liver from non-alcoholic fatty liver disease (NAFLD) patients is correlated with insulin action [57, 122]. Additional evidence shows that insulin resistance is associated with reprogramming of mitochondrial DNA methylation (e.g., ND6, and D-loop) in obese individuals as well as of PPARGC1A DNA methylation in NAFLD patients, concomitant with reduced mitochondrial DNA copy number [24, 57, 115]. In a reciprocal manner, however, an increased glucose dampens DNA methylation in DAPK3 gene body, which might constitute a feedback loop with insulin resistance in the control of the epigenome [65]. During pregnancy, fetal exposure to increased glucose (i.e., maternal hyperglycemia) leads to altered PPARGC1A DNA methylation in placenta samples and leptin level in cord blood [116]. Dysregulated leptin may arise from altered LEP promoter methylation in the adipose tissue in the offspring who had fetal exposure to hyperglycemia [123]. Currently, the molecular pathways by which insulin (or insulin resistance) and glucose induces DNA methylation reprogramming remains elusive, and it would be interesting to examine whether DNMT1 and DNMT3 are involved (Figure 1).
In addition to insulin and glucose, blood lipids were shown to potentially alter DNA methylation in circulating cells according to a stepwise MR analysis, which suggests that triglyceride induced differential methylation at three CpGs, LDL at one CpG, and HDL at two CpGs [117]. Furthermore, the differential methylation was associated with altered expression of genes that regulate triglycerides (e.g., CPT1A and SREBF1), LDL (e.g., DHCR24) and HDL (e.g., ABCG1) [117]. Experimental evidence from THP-1 monocyte cell culture shows that treatment with fatty acids (arachidonic and oleic acid) induces drastic changes in DNA methylation in monocytes [118]. It was found that circulating level of arachidonic acid was positively associated with global and PDK4-specific DNA methylation in adult men, while it is inversely associated HDAC4 methylation [119]. Importantly, arachidonic acid-induced DNA methylation phenotype was similar to that induced by palmitic acid in metabolic disorders (e.g., atherosclerosis, diabetes, obesity) [118]. However, the diets rich in linoleic acid or palmitic acid were found to affect DNA methylation differently in human adipose tissue [121]. In human pancreatic islets, palmitate treatment leads to drastic epigenetic reprogramming, including global DNA methylation profile, DNA methylation at CpG island shelves and shores, 5′UTR, 3′UTR and gene body regions, and accounting for differential expression of 290 genes [120]. Together, the MR analyses and experimental evidence reveal a causal role for lipid or fatty acids in DNA methylation reprogramming.
The enzymes that regulate DNA methylation include DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) and ‘demethylating’ proteins, i.e., the ten-eleven translocation (TET) family of DNA dioxygenases (TET1, TET2 and TET3) [124, 125]. DNMT3A and DNMT3B catalyze de novo methylation of DNA, which is maintained by a DNMT1-mediated copying mechanism, or is removed by TET-mediated reductive reactions [125]. The pathophysiological role of theses enzymes was demonstrated by PPARGC1A methylation reprogramming in metabolic disorders. Genetic variation in TET2 has been associated with altered PPARGC1A-methylation and transcript level in the liver from patients with nonalcoholic fatty liver disease [56]. Further, DNMT3B promotes DNA methylation of PPARGC1A, which underlies fatty acid- or proinflammation cytokine (TNFα)-induced hypermethylation of PPARGC1A, transcript downregulation, and reduced mitochondrial density in human skeletal muscle cells [60]. Proinflammatory cytokines (e.g., TNFα and IL1-β) can also induce hypermethylation in ADIPQ via DNMT1, which downregulates adiponectin secretion from adipocytes and contributes to systemic metabolic changes in mice [53]. Given that obese and T2D subjects have chronic inflammation and elevated fatty acids and glucose in the circulation, DNMT1 and DNMT3B may account largely for the observed reprogramming in DNA methylation [53, 60, 120]. Indeed, skeletal muscle from T2D individuals showed hypermethylation in PPARGC1A and reduced PPARGC1A mRNA and mitochondrial DNA content compared with normal glucose-tolerant counterparts [60].
Overall, the mechanism of epigenetic reprogramming and its role in metabolic disorders (e.g., obesity, T2D, and NAFLD) remains largely undefined. However, DNMT1 and DNMT3 may serve as the key mediators. Figure 1 summarizes the potential connections among the identified mediators in the induction of DNA methylation reprogramming, with critical questions to be addressed: (1) what is the molecular pathway leading glucose and insulin to DNA methylation [65, 116]? It is interesting to note that insulin and glucose can alter DNA methylation in muscle DAPK3 [65] but not muscle PPARGC1A [60], and it is unknown what constitutes the gene- or locus-specific mechanism regulating muscle DNA methylation. (2) What are the receptors or molecular targets by which pro-inflammatory cytokines or fatty acids regulate DNMT1 and DNMT3 [53, 60]? TNFα induces DNMT3B but not DNMT1 or DNMT3A in skeletal muscle DNA methylation reprogramming [60]; by contrast, TNFα tends to induces DNMT1 other than DNMT3A or DNMT3B in adipose DNA methylation reprogramming [53]. It will be important to define the determinant(s) of tissue-dependent induction of DNMTs by TNFα. (3) Is DNA methylation reprogramming the cause or result of metabolic disorders? Although adiposity was shown to cause DNA methylation alteration, the other way around was also true because DNA methylation in certain locus may predict adiposity [114, 126]. For instance, the DNA methylation profiles of both NFATC2IP and PPARGC1A have been found to predict adiposity [114, 126]. Therefore, it is tempting to speculate that a feedback loop may exist between adiposity and DNA methylation reprogramming (Figure 1).
4. Epigenetic markers in disease risk and intervention assessment
The recognition of epigenetic reprogramming in metabolic diseases has stimulated the interest in identifying epigenetic markers for diagnosis and intervention assessment [24, 39, 115, 127, 128]. To this end, blood samples are being widely tested because it is more accessible than other tissues (Table 2). Cross-tissue examination of epigenetic changes in recent studies demonstrated considerable tissue equivalence between blood and other tissues, and suggested that epigenetic reprogramming in the blood might serve as a surrogate marker for metabolic disorders [44, 48, 52, 54.Bacos, 2016 #1023, 64, 114, 129–131].
Table 2.
DNA methylation reprogramming in blood samples
| Samples | Medical conditions | Genes that undergo DNA methylation reprogramming | Physiological or pathophysiological relevance | References |
|---|---|---|---|---|
|
| ||||
| Whole blood | Childhood obesity and cardiometabolic disease risk | PPARGC1A | Mitochondrial biogenesis and function, adiposity prediction | [126] |
| Overweight, T2D, and first-degree relatives of T2D | HIF3A | Glucose and amino acid metabolism, apoptosis, proteolysis, p53 and PPAR signaling, and adipocyte differentiation/adiposity | [50, 52] | |
| Overweight, Obesity | LEP and ADIPOQ | Food intake, inflammation, insulin sensitivity, glucose and lipid homeostasis, energy balance | [54, 148] | |
| Overweight, obesity, T2D, and aging | FHL2, NOX4, ELOVL2, FHL2, KLF14 and GLRA1 | Fatty acid metabolism, mitochondrial function, oxidative stress, chemokine signaling | [44] | |
| Overweigh, obesity, and T2D | ABCG1, PHOSPHO1, SOCS3, SREBF1, TXNIP | Nutrient sensing/metabolism, redox regulation, insulin secretion, mitochondrial function, oxidative stress, lipid homeostasis | [48, 84] | |
| Obesity | FYN, PIWIL4, and TAOK3 | Adipocyte proliferation, differentiation, and senescence, insulin signaling or resistance, inflammation, JNK-MAPK signaling, energy expenditure | [149] | |
| T2D | IGFBP-1 | Interaction with insulin-like growth factor, insulin resistance | [150] | |
| Overweight, obesity, siblings of breast cancer patients | ANGPT4, RORC, SOCS3, FSD2, XYLT1, ABCG1, STK39, ASB2 and CRHR2 | Inflammatory and cytokine signaling, angiogenesis, lipid metabolism, leptin resistance, cellular stress | [151] | |
| Overweight, obesity, Cardiometabolic Disease | BCG1, CPT1A, LGALS3BP, DHCR24, PHGDH, SARS, NOD2, CACNA2D3, HIF3A, SLC1A5, and SREBF1 | Energy homeostasis (glycolysis, lipogenesis, mitochondrial fatty acid oxidation), adipogenesis, immune response, amino acid synthesis, cardiac conduction | [152] | |
| Cardiometabolic disease | HIF3A, CPT1A and ABCG1 | Adipogenesis, response to hypoxia, macrophage cholesterol and phospholipids transport, glucose, insulin, lipid homeostasis | [64] | |
| T2D | TXNIP, ABCG1 and SAMD12 | Redox regulation, glucose metabolism, lipid homeostasis | [153, 154] | |
| T2D and Cardio-metabolic disease | CPT1A and ABCG1 | Mitochondrial fatty acid oxidation, macrophage cholesterol and phospholipids transport, and lipid homeostasis | [155] | |
| T2D | CPT1A, DQX1, SREBF1, SBNO2, PRR5L, LY6G6E, TXNIP | Mitochondrial uptake of fatty acids, triglyceride metabolism, cholesterol homeostasis, redox regulation. | [156] | |
| Obesity and pre-diabetes | MT-ND6 and D-loop | Mitochondrial replication and function | [24, 115, 133] | |
| Adiposity-related metabolic traits and T2D | ABCG1, LPIN1, HOXA5, LMNA, CPT1A, SOCS3, SREBF1, and PHGDH | Mitochondrial uptake of fatty acids, triglyceride metabolism, cholesterol homeostasis, Inflammatory and cytokine signaling | [114] | |
|
| ||||
| Peripheral blood leukocytes | Obesity | HIF3A | Responses to hypoxia, adipose differentiation, liver diseases | [142] |
| Obesity | CPT1A, SREBF1, ABCG1, ARID1B, TOP1 | Mitochondrial fatty acid oxidation, triglyceride metabolism, macrophage cholesterol and phospholipids transport | [157] | |
| Peripheral blood mononuclear cells | Obesity | SOCS3 | Regulation of cytokine signaling (insulin, leptin, growth hormone, IL-6, prolactin, and interferon), inflammation, energy balance | [158] |
| Obesity | ADAMTS2, FIP1L1, SAMD4A, | Procollagen processing, connective tissue disease | [43] | |
|
| ||||
| CD4+ T cells | Cardiometabolic disease | HIF3A, CPT1A and ABCG1 | Adipogenesis, response to hypoxia, macrophage cholesterol and phospholipids transport, glucose, insulin, lipid homeostasis | [64] |
| Cardiometabolic disease | CPT1A, PHGDH, CD38 | Mitochondrial uptake of fatty acids, serine biosynthesis, cellular growth, immunology | [159, 160] | |
In patients with metabolic disorders, altered DNA methylation profiles have been identified in whole blood and sub-type of blood cells (e.g., leukocytes, mononuclear cells, and CD4+ T cells) (Table 2). Of pathophysiological significance, the genes involved in lipid metabolism (e.g., CPT1A, SREBF1, and ABCG1) and inflammation (e.g., SOCS3) consistently show DNA methylation reprogramming (see the details and references in Table 2). These findings cast epigenetic light on the pathogenesis of metabolic diseases because such medical conditions (e.g., obesity, T2DM, and cardio-metabolic diseases) are characterized by dyslipidemia and chronic inflammation [53, 66, 67, 92, 93, 95–97]. In addition, the epigenetic changes appear to be systemic, as altered DNA methylation profiles in these genes (SREBF1, ABCG1, and SOCS3) were identified in adipose tissue [48, 63, 64], liver [48], skeletal muscle [48], and pancreatic islets [48]. In support of the link of mitochondrial dysfunction to metabolic disorders[74, 83, 132], DNA methylation reprogramming has been observed in mitochondrial genome (e.g., D-loop and ND6) or mitochondrially related genes (e.g., PPARGC1A) in blood samples from obese and prediabetic individuals [24, 115, 126].
The potential of DNA methylation marker in identifying high-risk individuals and predicting metabolic disease incidence has been tested in several population studies. In a recent report, 187 DNA methylation markers in the blood were examined, 62 of which (including ABCG1 gene) were significantly associated with T2D incidence [114]. In addition, the DNA methylation risk score can predict incident T2D in two cohort studies, indicating a relative risk of 2.29 per 1 standard deviation change in methylation risk score (p = 4.2 × 10−52) in the LOLIPOP study, and 2.51 (p = 5.7 × 10−4) in the KORA study [114]. By examining the DNA methylation at specific loci in the LOLIPOP study, Chambers et al. found that the relative risk for future T2D incidence was 1·09 (p=1.3 × 10−17) per 1 percent increase in DNA methylation of ABCG1, 0.94 (p=4.2 × 10−11) of PHOSPHO1, 0.94 (p=1·4 × 10−9) of SOCS3, 1.07 (p=2·1 × 10−10) of SREBF1, and 0·92 (p=1·2 × 10−17) of TXNIP [84]. In the Botnia prospective study, blood DNA methylation at the ABCG1 locus also indicated an increased risk of future T2D, although smaller sample size and differences in study design did not allow the authors to replicate the associations with other loci as observed in the LOLIPOP study [48, 84]. Of note, the DNA methylation risk score predicted the risk of future T2D beyond traditional risk factors (e.g., BMI and waist–hip ratio), suggesting that DNA-methylation marker might be used to identify high-T2D-risk individuals who have normal adiposity or metabolically unfavourable adiposity [84, 114]. Indeed, it was shown that DNA methylation changes in the mitochondria may reflect early stage of prediabetes programming and have the potential to distinguish metabolically unhealthy obesity (MUO) from metabolically healthy obesity (MHO) [115, 133].
Another promising perspective of epigenetic marker resides in the potential to predict the response to interventions. In an 8-week obesity intervention using low calorie diet, the responders had lower DNA methylation levels (by 47%, p<0.05) in the promoter of LEP than the non-responders at baseline, suggesting that DNA methylation in LEP might predict the susceptibility to weight loss by dietary intervention [134]. The epigenetic predictors were also discovered in a 10-week lifestyle and nutritional educational weight loss program, where high and low responders among the overweight and obese adolescents showed differential methylation in 5 regions located in/near AQP9, DUSP22, HIPK3, TNNT1, and TNNI3 genes [135]. In fact, weight loss interventions through diets, exercise, and bariatric surgery can also induce robust reprogramming of DNA methylation [134, 136]; [41, 42, 59, 137–141]. The post-intervention methylation score was significantly associated with body fat mass loss, weight loss, and body mass index-standard deviation score [135]. Successful weight loss maintainers for up to 3 years after intervention have DNA methylation patterns that are similar to normal weight individuals but different from obese counterparts [128].
As the well-recognized epigenetic marker across tissues (Table 1, Table 2), PPARGC1A methylation is highly responsive to exercise and bariatric surgery [59, 138]. In individuals with a sedentary lifestyle, acute exercise can induce a marked hypomethylation in the promoter of PPARGC1A, which is concomitant with an exercise dose-dependent upregulation of PPARGC1A transcript [138]. A beneficial effect was also documented for maternal exercise, which reverses high fat diet (HFD)-induced PPARGC1A hypermethylation and downregulation of PPARGC1A transcript in the offspring [61]. In addition, dysregulated DNA methylation in PPARGC1A promoter in obese patients can be normalized 6 months after Roux-en-Y gastric bypass (RYGB) surgery, to a similar level observed in healthy individuals [59]. Consistently, dysregulated PPARGC1A gene expression was restored to the level of healthy controls [59]. Thus, PPARGC1A methylation is a sensitive epigenetic marker in response to weight loss interventions.
5. Conclusions
DNA methylation reprogramming is generally associated with transcript expression in metabolic disorders, which has revealed an epigenetic link to insulin secretion and signaling, glucose and lipid metabolism, adipocyte differentiation and trans-differentiation, mitochondrial function, inflammation, cell death and autophagy. Although certain epigenetic traits show tissue dependence (e.g., DNA methylation changes in adipocytokine genes LEP and ADIPOQ), mounting evidence has established tissue equivalence for epigenetic markers that regulate mitochondrial homeostasis (e.g., PPARGC1A) among adipose tissue, liver, skeletal muscle, pancreas and the blood samples (Tables 1 and 2). Tissue equivalence is also being revealed for epigenetic changes in genes that mediate lipid metabolism (e.g., ABCG1 and SREBF1) [48], hypoxia response (HIF3A) during obesity [50–52, 64, 142], and mitochondrial function (e.g., ND6) [58, 115] between blood and other tissues. As such, the epigenetic changes (e.g., ABCG1 and SREBF1) in the blood have been tested and shown promising potential as surrogate markers for metabolic diseases [48, 84]. In addition, the epigenetic traits (e.g., DNA methylation in PPARGC1A) respond robustly to lifestyle and surgical interventions [59, 61, 138], underscoring the potential of using epigenetic markers to assess disease risk and intervention efficacy [128, 134, 135].
The mechanism underlying epigenetic changes in metabolic disorders remains largely undefined. The literature suggests that DNA methylation reprogramming can be induced by insulin, glucose, fatty acids or lipids, and inflammatory cytokines [53, 60, 65, 116–120]. In particular, DNMT1 and DNMT3 mediate fatty acid- or inflammatory cytokine-induced DNA methylation changes [53, 60]. Further studies are warranted to dissect the pathways by which DNMT1 and DNMT3 are induced, and whether it is the mechanism shared by insulin- and glucose-induced epigenetic reprogramming. As TET proteins are responsible for DNA demethylation, it would be of interest to examine whether and how TET proteins are involved in the epigenetic reprogramming in metabolic disorders. Regardless of the well-established association between epigenetic changes and metabolic disorders, the evidence examining a cause-effect relationship has just emerged [53, 60, 114, 126]. Therefore, large-scale longitudinal studies of human subjects and functional characterization of epigenetic changes through post-developmental modulation in cell or animal models are critical to address these questions.
Highlights.
Abnormalities in metabolic activity and pathways are associated with DNA methylation reprogramming.
Metabolic disorders have shown cross-tissue epigenetic changes among the liver, skeletal muscle, pancreas, adipose tissue, and blood samples.
Adiposity, fatty acids or lipids, pro-inflammatory cytokines (e.g., TNFα and IL-1β), insulin, and glucose may induce DNA methylation reprogramming, with DNMT1 and DNMT3 as the key mediators in certain cases.
DNA methylation changes (e.g., PPARGC1A and ADIPOQ) may serve as sensitive epigenetic markers to predict metabolic disease risk and intervention responses.
Acknowledgments
This work was funded in part by USDA National Institute of Food and Agriculture Hatch Project 1007334 (Zhiyong Cheng) and NIH Grant 5R18DK091811-02 (Fabio A. Almeida).
Abbreviations
- ABCG1
ATP-binding cassette sub-family G member 1
- ADIPOQ
adiponectin encoding gene
- CPT1A
Carnitine palmitoyltransferase 1 A
- CCND2
Cyclin D2
- CDKN1A
cyclin-dependent kinase inhibitor 1
- CILP2
Cartilage Intermediate Layer Protein 2
- CVD
cardiovascular diseases
- DAPK3
Death-associated protein kinase 3
- D-loop
displacement loop
- DNMT1
DNA methyltransferase 1
- DNMT3B
DNA methyltransferase 3B
- EXOC3L2
exocyst complex component 3 like 2
- FHL2
four and a half LIM Domains 2
- HFD
high fat diet
- HIF1α
hypoxia inducible factor 1α
- HIF3a
hypoxia inducible factor 3α
- IL1β
interleukin (IL)-1β
- lncRNA
long noncoding RNAs
- IRS1
insulin receptor substrate 1
- IRS2
insulin receptor substrate 2
- LEP
Leptin encoding gene
- NAFLD
non-alcoholic fatty liver disease
- ND6
NADH:ubiquinone oxidoreductase core subunit 6
- PDE7B
phosphodiesterase 7B
- PPARG
peroxisome proliferator-activated receptor gamma
- PPARGC1A
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PRDM16
PR domain containing 16
- RYGB
Roux-en-Y gastric bypass
- SEPT9
septin 9
- SNP
single-nucleotide polymorphism
- SOCS3
suppressor of cytokine signaling 3
- SOD3
superoxide dismutase 3
- SORBS2
sorbin and SH3 domain containing 2
- SREBP1
sterol regulatory element-binding protein 1, also known as SREBF1
- SREBF1
Sterol regulatory element-binding transcription factor 1, also known as SREBP1
- T2D
type 2 diabetes
- TET
ten-eleven translocation family of DNA dioxygenases
- TNFα
tumour necrosis factor α
- TXNIP
Thioredoxin-interacting Protein.
Footnotes
Disclosure statements
The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karlsson O, Baccarelli AA. Environmental Health and Long Non-coding RNAs. Current environmental health reports. 2016;3:178–87. doi: 10.1007/s40572-016-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keating ST, El-Osta A. Epigenetics and metabolism. Circ Res. 2015;116:715–36. doi: 10.1161/CIRCRESAHA.116.303936. [DOI] [PubMed] [Google Scholar]
- 3.Delgado-Cruzata L, Zhang W, McDonald JA, Tsai WY, Valdovinos C, Falci L, et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J Nutr. 2015;145:783–90. doi: 10.3945/jn.114.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Z, Almeida FA. Mitochondrial alteration in type 2 diabetes and obesity: an epigenetic link. Cell Cycle. 2014;13:890–7. doi: 10.4161/cc.28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietilainen KH, Ismail K, Jarvinen E, Heinonen S, Tummers M, Bollepalli S, et al. DNA methylation and gene expression patterns in adipose tissue differ significantly within young adult monozygotic BMI-discordant twin pairs. Int J Obes (Lond) 2016;40:654–61. doi: 10.1038/ijo.2015.221. [DOI] [PubMed] [Google Scholar]
- 6.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PloS one. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan W, Xia Y, Bell CG, Yet I, Ferreira T, Ward KJ, et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nature communications. 2014;5:5719. doi: 10.1038/ncomms6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes. 2012;61:542–6. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ollikainen M, Ismail K, Gervin K, Kyllonen A, Hakkarainen A, Lundbom J, et al. Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. Clinical epigenetics. 2015;7:39. doi: 10.1186/s13148-015-0073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–76. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 11.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basile KJ, Johnson ME, Xia Q, Grant SF. Genetic susceptibility to type 2 diabetes and obesity: follow-up of findings from genome-wide association studies. International journal of endocrinology. 2014;2014:769671. doi: 10.1155/2014/769671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23:5492–504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwenk RW, Vogel H, Schurmann A. Genetic and epigenetic control of metabolic health. Molecular metabolism. 2013;2:337–47. doi: 10.1016/j.molmet.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS. Epigenetics and human obesity. Int J Obes (Lond) 2015;39:85–97. doi: 10.1038/ijo.2014.34. [DOI] [PubMed] [Google Scholar]
- 16.van Dijk SJ, Tellam RL, Morrison JL, Muhlhausler BS, Molloy PL. Recent developments on the role of epigenetics in obesity and metabolic disease. Clinical epigenetics. 2015;7:66. doi: 10.1186/s13148-015-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barres R, Zierath JR. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat Rev Endocrinol. 2016;12:441–51. doi: 10.1038/nrendo.2016.87. [DOI] [PubMed] [Google Scholar]
- 18.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes (Lond) 2016;40:88–101. doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schones DE, Leung A, Natarajan R. Chromatin Modifications Associated With Diabetes and Obesity. Arterioscler Thromb Vasc Biol. 2015;35:1557–61. doi: 10.1161/ATVBAHA.115.305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 21.Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS genetics. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 24.Zheng LD, Linarelli LE, Liu L, Wall SS, Greenawald MH, Seidel RW, et al. Insulin resistance is associated with epigenetic and genetic regulation of mitochondrial DNA in obese humans. Clinical epigenetics. 2015;7:60. doi: 10.1186/s13148-015-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Zheng LD, Donnelly SR, Emont MP, Wu J, Cheng Z. Isolation of Mouse Stromal Vascular Cells for Monolayer Culture. Methods Mol Biol. 2017;1566:9–16. doi: 10.1007/978-1-4939-6820-6_2. [DOI] [PubMed] [Google Scholar]
- 26.Hall KD, Heymsfield SB, Kemnitz JW, Klein S, Schoeller DA, Speakman JR. Energy balance and its components: implications for body weight regulation. Am J Clin Nutr. 2012;95:989–94. doi: 10.3945/ajcn.112.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill JO, Wyatt HR, Peters JC. Energy balance and obesity. Circulation. 2012;126:126–32. doi: 10.1161/CIRCULATIONAHA.111.087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hjort L, Jorgensen SW, Gillberg L, Hall E, Brons C, Frystyk J, et al. 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clinical epigenetics. 2017;9:40. doi: 10.1186/s13148-017-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung A, Parks BW, Du J, Trac C, Setten R, Chen Y, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem. 2014;289:23557–67. doi: 10.1074/jbc.M114.581439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen SC, Gillberg L, Bork-Jensen J, Ribel-Madsen R, Lara E, Calvanese V, et al. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014;57:1154–8. doi: 10.1007/s00125-014-3198-8. [DOI] [PubMed] [Google Scholar]
- 31.Gillberg L, Perfilyev A, Brons C, Thomasen M, Grunnet LG, Volkov P, et al. Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia. 2016;59:799–812. doi: 10.1007/s00125-015-3852-9. [DOI] [PubMed] [Google Scholar]
- 32.de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Molecular metabolism. 2016;5:184–97. doi: 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mudaliar U, Zabetian A, Goodman M, Echouffo-Tcheugui JB, Albright AL, Gregg EW, et al. Cardiometabolic Risk Factor Changes Observed in Diabetes Prevention Programs in US Settings: A Systematic Review and Meta-analysis. PLoS medicine. 2016;13:e1002095. doi: 10.1371/journal.pmed.1002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merlotti C, Morabito A, Pontiroli AE. Prevention of type 2 diabetes; a systematic review and meta-analysis of different intervention strategies. Diabetes, obesity & metabolism. 2014;16:719–27. doi: 10.1111/dom.12270. [DOI] [PubMed] [Google Scholar]
- 35.Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–50. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 36.Perreault L, Pan Q, Mather KJ, Watson KE, Hamman RF, Kahn SE. Effect of regression from prediabetes to normal glucose regulation on long-term reduction in diabetes risk: results from the Diabetes Prevention Program Outcomes Study. Lancet. 2012;379:2243–51. doi: 10.1016/S0140-6736(12)60525-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almeida FA, Smith-Ray RL, Dzewaltowski DA, Glasgow RE, Lee RE, Thomas DS, et al. An Interactive Computer Session to Initiate Physical Activity in Sedentary Cardiac Patients: Randomized Controlled Trial. Journal of medical Internet research. 2015;17:e206. doi: 10.2196/jmir.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almeida FA, You W, Harden SM, Blackman KC, Davy BM, Glasgow RE, et al. Effectiveness of a worksite-based weight loss randomized controlled trial: the worksite study. Obesity (Silver Spring) 2015;23:737–45. doi: 10.1002/oby.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchner H, Osler ME, Krook A, Zierath JR. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 2013;23:203–9. doi: 10.1016/j.tcb.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Ling C, Ronn T. Epigenetic adaptation to regular exercise in humans. Drug discovery today. 2014;19:1015–8. doi: 10.1016/j.drudis.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS genetics. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–32. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arner P, Sahlqvist AS, Sinha I, Xu H, Yao X, Waterworth D, et al. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia. 2016;59:2393–405. doi: 10.1007/s00125-016-4074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–813. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 45.Keller M, Hopp L, Liu X, Wohland T, Rohde K, Cancello R, et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Molecular metabolism. 2017;6:86–100. doi: 10.1016/j.molmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–22. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nature communications. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson PA, de Mello VD, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics: official journal of the DNA Methylation Society. 2016;11:482–8. doi: 10.1080/15592294.2016.1178418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillberg L, Jacobsen SC, Ronn T, Brons C, Vaag A. PPARGC1A DNA methylation in subcutaneous adipose tissue in low birth weight subjects--impact of 5 days of high-fat overfeeding. Metabolism. 2014;63:263–71. doi: 10.1016/j.metabol.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–8. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 51.Pfeiffer S, Kruger J, Maierhofer A, Bottcher Y, Kloting N, El Hajj N, et al. Hypoxia-inducible factor 3A gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Scientific reports. 2016;6:27969. doi: 10.1038/srep27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Main AM, Gillberg L, Jacobsen AL, Nilsson E, Gjesing AP, Hansen T, et al. DNA methylation and gene expression of HIF3A: cross-tissue validation and associations with BMI and insulin resistance. Clinical epigenetics. 2016;8:89. doi: 10.1186/s13148-016-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim AY, Park YJ, Pan X, Shin KC, Kwak SH, Bassas AF, et al. Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nature communications. 2015;6:7585. doi: 10.1038/ncomms8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houde AA, Legare C, Biron S, Lescelleur O, Biertho L, Marceau S, et al. Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL-cholesterol levels in severely obese men and women. BMC medical genetics. 2015;16:29. doi: 10.1186/s12881-015-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nilsson E, Matte A, Perfilyev A, de Mello VD, Kakela P, Pihlajamaki J, et al. Epigenetic Alterations in Human Liver From Subjects With Type 2 Diabetes in Parallel With Reduced Folate Levels. J Clin Endocrinol Metab. 2015;100:E1491–501. doi: 10.1210/jc.2015-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirola CJ, Scian R, Gianotti TF, Dopazo H, Rohr C, Martino JS, et al. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine (Baltimore) 2015;94:e1480. doi: 10.1097/MD.0000000000001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez Gianotti T, Castano GO, et al. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 58.Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–63. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 59.Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell reports. 2013;3:1020–7. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 60.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 61.Laker RC, Lillard TS, Okutsu M, Zhang M, Hoehn KL, Connelly JJ, et al. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1alpha gene and age-dependent metabolic dysfunction in the offspring. Diabetes. 2014;63:1605–11. doi: 10.2337/db13-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brons C, Jacobsen S, Nilsson E, Ronn T, Jensen CB, Storgaard H, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab. 2010;95:3048–56. doi: 10.1210/jc.2009-2413. [DOI] [PubMed] [Google Scholar]
- 63.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2015;44:1277–87. doi: 10.1093/ije/dyu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–79. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mudry JM, Lassiter DG, Nylen C, Garcia-Calzon S, Naslund E, Krook A, et al. Insulin and Glucose Alter Death-Associated Protein Kinase 3 (DAPK3) DNA Methylation in Human Skeletal Muscle. Diabetes. 2017;66:651–62. doi: 10.2337/db16-0882. [DOI] [PubMed] [Google Scholar]
- 66.Rutkowski JM, Stern JH, Scherer PE. The cell biology of fat expansion. J Cell Biol. 2015;208:501–12. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saltiel AR. Insulin resistance in the defense against obesity. Cell Metab. 2012;15:798–804. doi: 10.1016/j.cmet.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Altshuler-Keylin S, Kajimura S. Mitochondrial homeostasis in adipose tissue remodeling. Science signaling. 2017:10. doi: 10.1126/scisignal.aai9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emont MP, Yu H, Wu J. Transcriptional control and hormonal response of thermogenic fat. J Endocrinol. 2015;225:R35–47. doi: 10.1530/JOE-15-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu J, Jun H, McDermott JR. Formation and activation of thermogenic fat. Trends Genet. 2015;31:232–8. doi: 10.1016/j.tig.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat Rev Endocrinol. 2014;10:24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 72.Ghosh S, Yates AJ, Fruhwald MC, Miecznikowski JC, Plass C, Smiraglia D. Tissue specific DNA methylation of CpG islands in normal human adult somatic tissues distinguishes neural from non-neural tissues. Epigenetics: official journal of the DNA Methylation Society. 2010;5:527–38. doi: 10.4161/epi.5.6.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serra D, Mera P, Malandrino MI, Mir JF, Herrero L. Mitochondrial fatty acid oxidation in obesity. Antioxidants & redox signaling. 2013;19:269–84. doi: 10.1089/ars.2012.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng Z, Schmelz EM, Liu D, Hulver MW. Targeting mitochondrial alterations to prevent type 2 diabetes-Evidence from studies of dietary redox-active compounds. Molecular nutrition & food research. 2014;58:1739–49. doi: 10.1002/mnfr.201300747. [DOI] [PubMed] [Google Scholar]
- 75.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–7. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 76.Baccarelli AA, Byun HM. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clinical epigenetics. 2015;7:44. doi: 10.1186/s13148-015-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sunny NE, Bril F, Cusi K. Mitochondrial Adaptation in Nonalcoholic Fatty Liver Disease: Novel Mechanisms and Treatment Strategies. Trends in endocrinology and metabolism: TEM. 2016 doi: 10.1016/j.tem.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Boudina S, Graham TE. Mitochondrial function/dysfunction in white adipose tissue. Exp Physiol. 2014;99:1168–78. doi: 10.1113/expphysiol.2014.081414. [DOI] [PubMed] [Google Scholar]
- 79.Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends in endocrinology and metabolism: TEM. 2012;23:435–43. doi: 10.1016/j.tem.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–71. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15:595–605. doi: 10.1016/j.cmet.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jitrapakdee S, Wutthisathapornchai A, Wallace JC, MacDonald MJ. Regulation of insulin secretion: role of mitochondrial signalling. Diabetologia. 2010;53:1019–32. doi: 10.1007/s00125-010-1685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol. 2012;8:92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 84.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. The lancet Diabetes & endocrinology. 2015;3:526–34. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuo M. ATP-binding cassette proteins involved in glucose and lipid homeostasis. Biosci Biotechnol Biochem. 2010;74:899–907. doi: 10.1271/bbb.90921. [DOI] [PubMed] [Google Scholar]
- 86.Kruit JK, Wijesekara N, Westwell-Roper C, Vanmierlo T, de Haan W, Bhattacharjee A, et al. Loss of both ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation, and impaired beta-cell function. Diabetes. 2012;61:659–64. doi: 10.2337/db11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends in endocrinology and metabolism: TEM. 2010;21:268–76. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–14. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 90.Liu L, Zheng LD, Zou P, Brooke J, Smith C, Long YC, et al. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle. 2016;15:2033–41. doi: 10.1080/15384101.2016.1192732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou P, Liu L, Zheng L, Stoneman RE, Cho A, Emery A, et al. Targeting FoxO1 with AS1842856 suppresses adipogenesis. Cell Cycle. 2014;13:3759–67. doi: 10.4161/15384101.2014.965977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Havel PJ. Update on adipocyte hormones: regulation of energy balance and carbohydrate/lipid metabolism. Diabetes. 2004;53(Suppl 1):S143–51. doi: 10.2337/diabetes.53.2007.s143. [DOI] [PubMed] [Google Scholar]
- 93.Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nature reviews Immunology. 2006;6:772–83. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]
- 94.Houde AA, Legare C, Hould FS, Lebel S, Marceau P, Tchernof A, et al. Cross-tissue comparisons of leptin and adiponectin: DNA methylation profiles. Adipocyte. 2014;3:132–40. doi: 10.4161/adip.28308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee YS, Kim JW, Osborne O, Oh DY, Sasik R, Schenk S, et al. Increased adipocyte O2 consumption triggers HIF-1alpha, causing inflammation and insulin resistance in obesity. Cell. 2014;157:1339–52. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–60. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–45. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pasanen A, Heikkila M, Rautavuoma K, Hirsila M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3alpha is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42:1189–200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 99.Girgis CM, Cheng K, Scott CH, Gunton JE. Novel links between HIFs, type 2 diabetes, and metabolic syndrome. Trends Endocrin Met. 2012;23:372–80. doi: 10.1016/j.tem.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 100.Heikkila M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell Mol Life Sci. 2011;68:3885–901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang C, Qu A, Matsubara T, Chanturiya T, Jou W, Gavrilova O, et al. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes. 2011;60:2484–95. doi: 10.2337/db11-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue--link to whole-body phenotypes. Nat Rev Endocrinol. 2015;11:90–100. doi: 10.1038/nrendo.2014.185. [DOI] [PubMed] [Google Scholar]
- 103.Tchkonia T, Thomou T, Zhu Y, Karagiannides I, Pothoulakis C, Jensen MD, et al. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17:644–56. doi: 10.1016/j.cmet.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–9. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kantidze OL, Razin SV. 5-hydroxymethylcytosine in DNA repair: A new player or a red herring? Cell Cycle. 2017;16:1499–501. doi: 10.1080/15384101.2017.1346761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dubois-Chevalier J, Oger F, Dehondt H, Firmin FF, Gheeraert C, Staels B, et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014;42:10943–59. doi: 10.1093/nar/gku780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–65. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 109.Pattamaprapanont P, Garde C, Fabre O, Barres R. Muscle Contraction Induces Acute Hydroxymethylation of the Exercise-Responsive Gene Nr4a3. Frontiers in endocrinology. 2016;7:165. doi: 10.3389/fendo.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ponnaluri VK, Ehrlich KC, Zhang G, Lacey M, Johnston D, Pradhan S, et al. Association of 5-hydroxymethylation and 5-methylation of DNA cytosine with tissue-specific gene expression. Epigenetics: official journal of the DNA Methylation Society. 2017;12:123–38. doi: 10.1080/15592294.2016.1265713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rohde K, Keller M, Stumvoll M, Dietrich A, Bluher M, Bottcher Y. DNA 5-hydroxymethylation in human adipose tissue differs between subcutaneous and visceral adipose tissue depots. Epigenomics. 2015;7:911–20. doi: 10.2217/epi.15.50. [DOI] [PubMed] [Google Scholar]
- 112.Nicoletti CF, Nonino CB, de Oliveira BA, Pinhel MA, Mansego ML, Milagro FI, et al. DNA Methylation and Hydroxymethylation Levels in Relation to Two Weight Loss Strategies: Energy-Restricted Diet or Bariatric Surgery. Obes Surg. 2016;26:603–11. doi: 10.1007/s11695-015-1802-8. [DOI] [PubMed] [Google Scholar]
- 113.Richmond RC, Sharp GC, Ward ME, Fraser A, Lyttleton O, McArdle WL, et al. DNA Methylation and BMI: Investigating Identified Methylation Sites at HIF3A in a Causal Framework. Diabetes. 2016;65:1231–44. doi: 10.2337/db15-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–6. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zheng LD, Linarelli LE, Brooke J, Smith C, Wall SS, Greenawald MH, et al. Mitochondrial Epigenetic Changes Link to Increased Diabetes Risk and Early-Stage Prediabetes Indicator. Oxidative medicine and cellular longevity. 2016;2016:5290638. doi: 10.1155/2016/5290638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cote S, Gagne-Ouellet V, Guay SP, Allard C, Houde AA, Perron P, et al. PPARGC1alpha gene DNA methylation variations in human placenta mediate the link between maternal hyperglycemia and leptin levels in newborns. Clinical epigenetics. 2016;8:72. doi: 10.1186/s13148-016-0239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, et al. Blood lipids influence DNA methylation in circulating cells. Genome biology. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silva-Martinez GA, Rodriguez-Rios D, Alvarado-Caudillo Y, Vaquero A, Esteller M, Carmona FJ, et al. Arachidonic and oleic acid exert distinct effects on the DNA methylome. Epigenetics: official journal of the DNA Methylation Society. 2016;11:321–34. doi: 10.1080/15592294.2016.1161873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.de la Rocha C, Perez-Mojica JE, Zenteno-De Leon S, Cervantes-Paz B, Tristan-Flores FE, Rodriguez-Rios D, et al. Associations between whole peripheral blood fatty acids and DNA methylation in humans. Scientific reports. 2016:6. doi: 10.1038/srep25867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hall E, Volkov P, Dayeh T, Bacos K, Ronn T, Nitert MD, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC medicine. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105:991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- 122.de Mello VD, Matte A, Perfilyev A, Mannisto V, Ronn T, Nilsson E, et al. Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action. Epigenetics: official journal of the DNA Methylation Society. 2017 doi: 10.1080/15592294.2017.1294305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hansen NS, Strasko KS, Hjort L, Kelstrup L, Houshmand-Oregaard A, Schrolkamp M, et al. Fetal Hyperglycemia Changes Human Preadipocyte Function in Adult Life. J Clin Endocrinol Metab. 2017;102:1141–50. doi: 10.1210/jc.2016-3907. [DOI] [PubMed] [Google Scholar]
- 124.Hamidi T, Singh AK, Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7:247–65. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 125.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harbor perspectives in biology. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clarke-Harris R, Wilkin TJ, Hosking J, Pinkney J, Jeffery AN, Metcalf BS, et al. PGC1alpha promoter methylation in blood at 5–7 years predicts adiposity from 9 to 14 years (EarlyBird 50) Diabetes. 2014;63:2528–37. doi: 10.2337/db13-0671. [DOI] [PubMed] [Google Scholar]
- 127.Ronn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against Type 2 diabetes. Epigenomics. 2015;7:451–60. doi: 10.2217/epi.15.7. [DOI] [PubMed] [Google Scholar]
- 128.Huang YT, Maccani JZ, Hawley NL, Wing RR, Kelsey KT, McCaffery JM. Epigenetic patterns in successful weight loss maintainers: a pilot study. Int J Obes (Lond) 2015;39:865–8. doi: 10.1038/ijo.2014.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bysani M, Perfilyev A, de Mello VD, Ronn T, Nilsson E, Pihlajamaki J, et al. Epigenetic alterations in blood mirror age-associated DNA methylation and gene expression changes in human liver. Epigenomics. 2017;9:105–22. doi: 10.2217/epi-2016-0087. [DOI] [PubMed] [Google Scholar]
- 130.Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mouse-Human Experimental Epigenetic Analysis Unmasks Dietary Targets and Genetic Liability for Diabetic Phenotypes. Cell Metab. 2015;21:138–49. doi: 10.1016/j.cmet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Crujeiras AB, Diaz-Lagares A, Sandoval J, Milagro FI, Navas-Carretero S, Carreira MC, et al. DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: a genome-wide analysis from non-obese and obese patients. Scientific reports. 2017;7:41903. doi: 10.1038/srep41903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxidants & redox signaling. 2013;19:240–2. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- 133.Zheng L, Brooke J, Smith C, Almeida FA, Cheng Z. Mitochondrial epigenetic changes and progression from metabolically healthy obesity to metabolically unhealthy obesity: a cross-sectional study. Lancet Diabetes Endocrinol. 2016:4. [Google Scholar]
- 134.Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, Javierre BM, et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67:463–70. doi: 10.1007/s13105-011-0084-4. [DOI] [PubMed] [Google Scholar]
- 135.Moleres A, Campion J, Milagro FI, Marcos A, Campoy C, Garagorri JM, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J. 2013;27:2504–12. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 136.Martin-Nunez GM, Cabrera-Mulero R, Rubio-Martin E, Rojo-Martinez G, Olveira G, Valdes S, et al. Methylation levels of the SCD1 gene promoter and LINE-1 repeat region are associated with weight change: an intervention study. Molecular nutrition & food research. 2014;58:1528–36. doi: 10.1002/mnfr.201400079. [DOI] [PubMed] [Google Scholar]
- 137.Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekstrom TJ, Tegner J, et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics: official journal of the DNA Methylation Society. 2014;9:1557–69. doi: 10.4161/15592294.2014.982445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–11. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 139.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome biology. 2015;16:8. doi: 10.1186/s13059-014-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dahlman I, Sinha I, Gao H, Brodin D, Thorell A, Ryden M, et al. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int J Obes (Lond) 2015;39:910–9. doi: 10.1038/ijo.2015.31. [DOI] [PubMed] [Google Scholar]
- 141.Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab. 2016;23:369–78. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 142.Wang S, Song JY, Yang YD, Zhang YN, Wang HJ, Ma J. HIF3A DNA Methylation Is Associated with Childhood Obesity and ALT. PloS one. 2015:10. doi: 10.1371/journal.pone.0145944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arner P, Sinha I, Thorell A, Ryden M, Dahlman-Wright K, Dahlman I. The epigenetic signature of subcutaneous fat cells is linked to altered expression of genes implicated in lipid metabolism in obese women. Clinical epigenetics. 2015;7:93. doi: 10.1186/s13148-015-0126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baumeier C, Saussenthaler S, Kammel A, Jahnert M, Schluter L, Hesse D, et al. Hepatic DPP4 DNA Methylation Associates With Fatty Liver. Diabetes. 2017;66:25–35. doi: 10.2337/db15-1716. [DOI] [PubMed] [Google Scholar]
- 145.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–43. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kirchner H, Sinha I, Gao H, Ruby MA, Schonke M, Lindvall JM, et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Molecular metabolism. 2016;5:171–83. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Volkov P, Bacos K, Ofori JK, Esguerra JLS, Eliasson L, Ronn T, et al. Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes. 2017;66:1074–85. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 148.Garcia-Cardona MC, Huang F, Garcia-Vivas JM, Lopez-Camarillo C, Del Rio Navarro BE, Navarro Olivos E, et al. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int J Obes (Lond) 2014;38:1457–65. doi: 10.1038/ijo.2014.30. [DOI] [PubMed] [Google Scholar]
- 149.Huang RC, Garratt ES, Pan H, Wu Y, Davis EA, Barton SJ, et al. Genome-wide methylation analysis identifies differentially methylated CpG loci associated with severe obesity in childhood. Epigenetics: official journal of the DNA Methylation Society. 2015;10:995–1005. doi: 10.1080/15592294.2015.1080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gu T, Gu HF, Hilding A, Sjoholm LK, Ostenson CG, Ekstrom TJ, et al. Increased DNA methylation levels of the insulin-like growth factor binding protein 1 gene are associated with type 2 diabetes in Swedish men. Clinical epigenetics. 2013;5:21. doi: 10.1186/1868-7083-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilson LE, Harlid S, Xu Z, Sandler DP, Taylor JA. An epigenome-wide study of body mass index and DNA methylation in blood using participants from the Sister Study cohort. Int J Obes (Lond) 2017;41:194–9. doi: 10.1038/ijo.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman AK, Aslibekyan S, et al. Association of Body Mass Index with DNA Methylation and Gene Expression in Blood Cells and Relations to Cardiometabolic Disease: A Mendelian Randomization Approach. PLoS medicine. 2017;14:e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW, Jr, Goring HH, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24:5330–44. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Soriano-Tarraga C, Jimenez-Conde J, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet. 2016;25:609–19. doi: 10.1093/hmg/ddv493. [DOI] [PubMed] [Google Scholar]
- 155.Mamtani M, Kulkarni H, Dyer TD, Goring HH, Neary JL, Cole SA, et al. Genome- and epigenome-wide association study of hypertriglyceridemic waist in Mexican American families. Clinical epigenetics. 2016;8:6. doi: 10.1186/s13148-016-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai PC, Kumar P, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clinical epigenetics. 2016;8:13. doi: 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang B, Gao W, Li J, Yu C, Cao W, Lv J, et al. Methylation loci associated with body mass index, waist circumference, and waist-to-hip ratio in Chinese adults: an epigenome-wide analysis. Lancet. 2016;388(Suppl 1):S21. [Google Scholar]
- 158.Ali O, Cerjak D, Kent JW, James R, Blangero J, Carless MA, et al. Methylation of SOCS3 is inversely associated with metabolic syndrome in an epigenome-wide association study of obesity. Epigenetics: official journal of the DNA Methylation Society. 2016;11:699–707. doi: 10.1080/15592294.2016.1216284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 2015;23:1493–501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, et al. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation. 2014;130:565–72. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]