Abstract
INTRODUCTION
Comparative transcriptome analyses in Alzheimer’s disease (AD) and other neurodegenerative proteinopathies can uncover both shared and distinct disease pathways.
METHOD
We analyzed 940 brain transcriptomes including patients with AD, progressive supranuclear palsy (PSP)-a primary tauopathy and controls.
RESULTS
We identified transcriptional co-expression networks implicated in myelination, which were lower in PSP temporal cortex (TCX) compared to AD. Some of these associations were retained even after adjustments for brain cell population changes. These TCX myelination network structures were preserved in cerebellum (CER) but they were not differentially expressed in CER between AD and PSP. Myelination networks were down-regulated in both AD and PSP, when compared to control TCX samples.
DISCUSSION
Down-regulation of myelination networks may underlie both PSP and AD pathophysiology, but may be more pronounced in PSP. These data also highlight conservation of transcriptional networks across brain regions and the influence of cell- type changes on these networks.
Keywords: Proteinopathies, Alzheimer’s disease, progressive supranuclear palsy, myelination, co- expression networks, transcriptome, temporal cortex, cerebellum
1. Introduction
Many neurodegenerative diseases, including Alzheimer’s disease (AD), are proteinopathies with common features including abnormal deposits of endogenous proteins, which propagate through the central nervous system (CNS) and culminate in cellular dysfunction and death, leading to clinical syndromes of dementia and/or movement disorders (reviewed[1]). Despite their commonalities, key differences are thought to exist in the events that trigger one proteinopathy vs. another; as well as in the downstream pathophysiologic pathways that distinguish these neurodegenerative diseases. Gene expression profiling studies may discover genes implicated in neurodegenerative diseases and uncover the complex molecular pathways leading to these disorders[2, 3]. With few exceptions[4–8], previous studies have investigated differential gene expression (DGE) in relatively small cohorts and were limited to comparison of individual gene transcripts rather than systems-level analysis. Further, most studies assessed one disease group against controls, rather than pursuing comparison between different diseases.
We postulate that comparison of brain gene expression levels in different neurodegenerative proteinopathies can uncover molecular pathways that are common to as well as those that are distinct for these diseases. Discovery of brain transcriptional networks with differential expression between different proteinopathies may uncover molecular pathways that may differentially influence these conditions. In contrast, networks that have similar expression changes in different diseases in comparison to controls may point to common dysregulated molecular pathways.
To test this hypothesis, we focused on two distinct proteinopathies, AD[9, 10] and progressive supranuclear palsy (PSP)[11, 12]. Although brain tau protein accumulation is a neuropathologic hallmark in both, these conditions are distinguished by different predominant tau isoform aggregates[13], and the unique presence in AD[9] of senile plaques composed predominantly of amyloid β (Aβ). They also have distinct clinical presentations. AD is the most common type of dementia[10], whereas PSP is a relatively rare parkinsonian movement disorder[12].
To identify genes and networks that are differentially altered in these conditions, we performed DGE and co-expression network analysis[14] in brain transcriptome[15–17] of subjects with AD or PSP. To determine whether observed network differences are driven by changes in AD vs. PSP or different extent of change in both, we also compared each diagnostic group with elderly control samples without any neurodegenerative diagnoses. All co-expression modules (CEM) were tested for enrichment of CNS cell-types[18], to identify altered networks that may be indicative of selectively vulnerable cell populations. Furthermore, to determine the contribution of cell-population changes to our findings[19], we performed all network analyses using two models: Comprehensive Model which adjusted for levels of 5 CNS cell-specific transcripts and Simple Model which was not thus adjusted. Finally, we validated these results by protein analysis in brain tissue.
Our findings reveal conserved brain myelination networks that are altered in both AD and PSP, but to a greater extent in the latter. These results have implications for the role of myelin metabolism in the pathophysiology of these distinct neurodegenerative proteinopathies, and ultimately for identification of novel therapeutic targets and biomarkers. Further, our large-scale transcriptome data, which we made available to the research community[16], provides information regarding brain region conservation and CNS cell-enrichment of transcriptional networks, as well as the influence of cell-population changes on their expression patterns.
2. Methods
Please also refer to Supplementary Methods for details.
2.1 Subjects and Samples
In a two-stage design, Mayo Clinic brain expression genome-wide association study (eGWAS) was used as the Discovery Cohort and Mayo Clinic RNA sequencing (RNAseq) samples were used as the Replication Cohort. The Discovery Cohort[15, 16] had Whole Genome DASL array-based transcriptome measurements, whereas the Replication Cohort[16, 17] had RNAseq data obtained with 101 base-pair (bp), paired-end sequencing on Illumina HiSeq2000 instruments, as previously published. The Discovery Cohort had whole genome genotypes from the Illumina HumanHap300-Duo Genotyping BeadChips[20], and the Replication Cohort from the Illumina Infinium HumanOmni2.5-8 BeadChip, which were utilized in quality control (QC).
2.2 Analyses
2.2.1 Differential Gene Expression (DGE)
DGE analyses of brain tissue from subjects of two diagnostic categories were conducted with multivariable linear regression conducted in R. Discovery Cohort DGE analyses utilized normalized gene expression measures as dependent variable, diagnosis as independent variable of primary interest and included age at death, gender, number of APOE ε4 alleles, plate, RIN, and (RIN-RINmean)2 as biological and technical covariates. Replication Cohort DGE analyses used conditional quantile normalized (CQN)[21] gene expression measures as dependent variable, diagnosis as independent variable of primary interest and included age at death, gender, RIN, brain tissue source, and flowcell as biological and technical covariates. We also included cell specific gene levels as covariates to account for neuronal loss, gliosis and/or vascular tissue, as previously described[22]. We did this correction by including as covariates, expression levels of genes (Probe ID; ENCODE ID) that are specific for the main five cell types present in the central nervous system (CNS): ENO2 for neurons (ILMN_1765796, ENSG00000111674), GFAP for astrocytes (ILMN_1697176, ENSG00000131095), CD68 for microglia (ILMN_2267914, ENSG00000129226), OLIG2 for oligodendrocytes (ILMN_1727567, ENSG00000205927) and CD34 for endothelial cells (ILMN_1732799, ENSG00000174059). Significance accounting for multiple testing was assigned using q values which are based on false discovery rates (Benjamini-Hochberg FDR)[23].
Unique genes representing probes with a q value<0.05 for the Discovery Cohort temporal cortex DGE analyses were assessed for enrichment of pathways and gene ontology (GO) biological processes using Metacore (Thompson Reuters)[24, 25].
2.2.2 Weighted Gene Co-Expression Network Analysis (WGCNA)
WGCNA R package[14] version 1.41 was used for both cohorts, independently. In all analyses, gene expression residuals obtained after multiple linear regression with independent variables, were input to WGCNA. Network analyses were run under two different models: “Comprehensive Model”, which adjusts for all covariates described in the above section; and “Simple Model”, which adjusts for the same covariates except for the five CNS cell-specific gene expression levels.
Networks were built using two diagnostic groups to analyze their associations with diagnosis. For each pairwise diagnostic group, consensus modules were identified and tested for (GO) enrichment in WGCNA. All modules were further annotated for enrichment of genes that are primarily expressed in one of the five major cell types that exist in the CNS, i.e. neurons, oligodendrocytes, microglia, astrocytes and endothelia, as described in the next section.
Eigengene, the first signed principle component, was calculated for each module. For each gene, module membership (MM) was calculated as the correlation between the gene and the module eigengene. Genes with MM≥0.7 are considered to be “important (hub)” genes for the network. To test the association of disease phenotype with network modules, eigengenes of consensus modules were correlated with the binary disease phenotype. Unless otherwise specified, “correlation” means Pearson correlation. Preservation of different networks were assessed using WGCNA “modulePreservation” function with 100 permutations to calculate a Zsummary score, which indicates well-preservation if >10.
2.2.3 Cell-enrichment analyses
Gene expression measures from purified cell populations, isolated from human brain tissue, were obtained from Zhang et al[18]. We analyzed the 21,390 genes that remained after removal of those that had expression changes due to technical issues or duplicates. Genes with a mean gene expression level, FPKM > 5 in the target cell and a fold change > 4 when compared with each other cell type, were deemed to be enriched for that target cell. All co-expression modules (CEM) were tested for enrichment for each of the 5 human brain cell-enriched genes using a one-sided Fisher’s test.
2.3 Protein analysis
We investigated brain protein levels for the key genes from the oligodendroglial networks and other genes of interest using Western blot analysis. We assessed human TCX brain tissue from 18 controls against either 20 PSP or 20 AD cases, in addition to 2 control samples measured for every protein on every gel to control for potential blot-to-blot variability. Differential protein analysis was also conducted for key myelin genes from the oligodendroglial networks using proteome data obtained from 84 AD and 83 PSP TCX samples from the Mayo Clinic RNAseq Replication Cohort. Myelination patterns were assessed in a subset of AD, PSP and control TCX samples (4 each) using established immunohistochemistry methods[26, 27]. We evaluated immunocytochemical patterns for the myelin and oligodendrocyte proteins using rat oligodendrocyte-enriched cultures[28].
3. Results
3.1 Brain Transcriptome Profiling in the Discovery Cohort identifies transcriptional changes in neurodegenerative diseases in the temporal cortex
To determine differentially expressed (DE) genes between human brains with AD vs. PSP, we utilized whole transcriptome data from our “Mayo eGWAS Discovery Cohort”[15, 16]. Expression levels were obtained from TCX, which is typically affected by AD neuropathology but spared in PSP; and from the cerebellar cortex (CER), which is relatively spared in AD[9], while the superior cerebellar peduncle and the dentate nucleus may be affected in PSP[12]. Following QC (Supplementary Figs. 1–3)[15, 29], 359 subjects with WG-DASL microarray expression measures from TCX (181 AD, 178 without AD pathology i.e. nAD including 97 PSP) and 343 with CER (173 AD, 170 nAD including 96 PSP) were retained (Supplementary Table 1). There were 17,902 WG-DASL probes (13,928 unique genes) that were expressed in >50% of all TCX samples analyzed and 17,122 such probes (13,440 unique genes) for CER samples (Supplementary Table 2).
We performed DGE using the “Comprehensive Model”, which includes adjustment for levels of five genes that have cell-specific expression for the main cell types present in the central nervous system (CNS)[18], as previously described[22, 30]. The rationale for this cell-type adjustment was to account for brain cell population changes that can occur in CNS diseases as a result of neuronal loss or gliosis, which can then influence transcriptome profiling outcomes[19] (Supplementary Results). Indeed, we identified significantly lower ENO2, but higher GFAP, CD68 and CD34 levels in the TCX, but not the CER, of AD subjects in comparison to those without AD pathology (Supplementary Figs. 4A–B), consistent with known cellular changes that occur in affected brain regions in AD[31, 32].
DGE results for all pairwise diagnostic comparisons are presented in Supplementary Tables 3–10 and Supplementary Text. There were 3,381 transcripts (3,094 unique genes) with significant DE in the TCX AD vs. PSP analysis (Supplementary Table 4). In contrast, there were only 6 significant probes in the CER AD vs. PSP DGE analyses (Supplementary Table 6). DGE results in the Discovery Cohort suggested strong transcriptional changes in the TCX but not CER for all diagnostic comparisons.
Pathway enrichment analysis of the most significant DEGs in the AD vs. PSP TCX analysis (3,094 genes, q< 0.05) implicated 66 enriched GO and MetaCore pathways with an FDR<0.05, including established pathways such oxidative phosphorylation, where a systematic downregulation in AD TCX of genes in this pathway is observed (Supplementary_Table.11, Supplementary_Fig.5), replicating prior observations[21, 28]. In the smaller AD vs. nTau analysis (572 genes) “Protein folding and maturation_POMC processing” was a significant MetaCore pathway, also detected in the AD vs. PSP analysis. Assessment of the 745 unique genes differentially expressed in PSP vs. nTau TCX, detected 3 significant and overlapping GO processes: “axon ensheathment in central nervous system”; “central nervous system myelination” and “oligodendrocyte development”.
3.2 Co-expression network analysis in the Discovery Cohort identifies modules that are enriched for specific brain cell types
We constructed co-expression networks[14] under both the “Comprehensive” and “Simple Models”, where the latter was implemented given the observed correlation of the cell markers with one another (Supplementary Fig. 6, Supplementary Text), leading to the possibility of over-correction under the Comprehensive Model. All co-expression modules (CEM) were annotated for enrichment of cell-type expressed genes, which are primarily expressed in one of five human brain cell types[18] (Supplementary Tables 12–16), and which are sufficient to differentiate the cell populations from one another (Supplementary Figs. 7–8).
Results are provided for the TCX CEM in the Discovery Cohort under the “Comprehensive” (Supplementary Tables 17–20) and “Simple” models (Supplementary Tables 21–24) for all pairwise diagnostic groups. CEM naming conventions are shown in Supplementary Table 25. In the AD+PSP cohort, 44 TCX consensus CEM were identified under the “Comprehensive Model” of which 12 had significant enrichment for one of the 5 brain-cell enriched gene sets (Supplementary Table 18). Using the “Simple Model”, 31 such TCX CEM were identified, of which 10 had enrichment for brain cell-enriched genes (Supplementary Table 22). The TCX CEM generated under the Comprehensive and Simple Models were well preserved (Supplementary Figs. 9–10). Table 1 shows those TCX CEM in the AD+PSP Discovery Cohort that had significant brain cell-enrichment under both analytic models (Supplementary Figs. 9–11).
Table 1. Temporal Cortex Co-expression networks in the Discovery Cohort with significant cell type enrichment.
WGCNA consensus modules are generated from the Discovery Cohort’s WG-DASL transcript data obtained from the temporal cortex. Results are shown both for the simple module (no correction for cell type variation) and the Comprehensive Model (corrects for cell type variation). Modules that have significant cell type enrichment under both models are shown. The modules that significantly associate with diagnosis in the AD vs. PSP comparisons are bolded. Disease Association Beta=Coefficient of association with AD diagnosis, i.e. positive beta=increased in AD. Cell Type Enrichment P value is Bonferroni-corrected. Top GO Biological Process (BP)= Gene Ontology BP with the most significant enrichment in the module are shown. Top GO Enrichment P value is Bonferroni-corrected.
| Model | Module Name | Module Size | Cell Type Enrichment | Disease Association | Top GO Biological Process | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Type | OR | P value | Beta | P value | ID | Name | Enrichment P value | |||
| Simple | AD+PSP.TCX17.CSsimple | 213 | Astrocyte | 54.4 | 5.83E-80 | 0.06 | 3.34E-01 | GO:0007399 | nervous system development | 3.25E-06 |
| AD+PSP.TCX10.Cssimple | 404 | Microglia | 55.6 | 7.34E-158 | 0.11 | 7.66E-02 | GO:0006955 | immune response | 1.98E-53 | |
| AD+PSP.TCX21.Cssimple | 153 | Microglia | 4.3 | 8.74E-03 | 0.12 | 5.09E-02 | NA | NA | NA | |
| AD+PSP.TCX1.CSsimple | 2046 | Neuron | 9.8 | 1.00E-100 | −0.21 | 5.50E-04 | GO:0007268 | synaptic transmission | 2.15E-60 | |
| AD+PSP.TCX14.CSsimple | 314 | Neuron | 9.5 | 9.06E-27 | −0.16 | 8.50E-03 | GO:0007268 | synaptic transmission | 2.93E-12 | |
| AD+PSP.TCX5.CSsimple | 654 | Neuron | 4.9 | 1.06E-19 | −0.29 | 1.04E-06 | NA | NA | NA | |
| AD+PSP.TCX26.CSsimple | 102 | Neuron | 7.4 | 2.56E-06 | −0.14 | 1.96E-02 | GO:0098655 | cation transmembrane transport | 3.55E-02 | |
| AD+PSP.TCX11.CSsimple | 340 | Oligodendrocyte | 96.4 | 2.78E-81 | 0.27 | 5.58E-06 | GO:0042552 | myelination | 1.03E-07 | |
| AD+PSP.TCX29.CSsimple | 58 | Oligodendrocyte | 47.1 | 2.01E-13 | 0.32 | 4.43E-08 | NA | NA | NA | |
|
| ||||||||||
| Comprehensive | AD+PSP.TCX14.CS | 264 | Astrocyte | 31.5 | 1.86E-59 | −0.11 | 6.41E-02 | GO:0007399 | nervous system development | 6.32E-07 |
| AD+PSP.TCX26.CS | 120 | Microglia | 153.6 | 9.19E-106 | −0.17 | 4.56E-03 | GO:0006955 | immune response | 2.72E-34 | |
| AD+PSP.TCX42.CS | 41 | Neuron | 8.9 | 2.58E-03 | 0.12 | 4.38E-02 | NA | NA | NA | |
| AD+PSP.TCX27.CS | 111 | Neuron | 10.3 | 8.18E-12 | −0.01 | 8.31E-01 | GO:0007268 | synaptic transmission | 1.33E-02 | |
| AD+PSP.TCX16.CS | 219 | Neuron | 6.7 | 7.15E-13 | 0.01 | 9.12E-01 | NA | NA | NA | |
| AD+PSP.TCX12.CS | 305 | Neuron | 6.6 | 7.31E-16 | 0.02 | 7.32E-01 | GO:0007268 | synaptic transmission | 1.57E-13 | |
| AD+PSP.TCX8.CS | 377 | Neuron | 7.1 | 5.76E-22 | 0.03 | 6.26E-01 | GO:0007268 | synaptic transmission | 1.51E-17 | |
| AD+PSP.TCX2.CS | 752 | Neuron | 14.1 | 4.96E-93 | 0.08 | 1.97E-01 | GO:0007268 | synaptic transmission | 4.44E-20 | |
| AD+PSP.TCX41.CS | 41 | Oligodendrocyte | 40.6 | 3.98E-08 | 0.06 | 3.02E-01 | NA | NA | NA | |
| AD+PSP.TCX40.CS | 44 | Oligodendrocyte | 20.1 | 2.29E-03 | 0.20 | 8.00E-04 | NA | NA | NA | |
| AD+PSP.TCX10.CS | 308 | Oligodendrocyte | 81.6 | 1.70E-72 | 0.19 | 1.43E-03 | GO:0042552 | myelination | 2.37E-09 | |
We tested whether CEM are preserved between two brain regions i.e. TCX and CER (Supplementary Text, Supplementary Tables 26–33). Similar to the TCX modules, CER CEM built under the Simple vs. Comprehensive Models were well-preserved (Supplementary Figs. 12–14). Further, CER vs. TCX CEM from the Comprehensive Model were well-preserved (Supplementary Figs. 15–17).
3.3 Myelination co-expression modules have replicable neurodegenerative disease association in the temporal cortex
We tested the association of network modules with neuropathologic diagnoses. Under the Simple Model, there were 17 CEM that had significant DE in the TCX of AD vs. PSP subjects in the Discovery Cohort (Supplementary Table 22), of which 7 also had brain cell-enrichment (Fig. 1A, Table 1). In contrast, under the Comprehensive Model, there were only 9 TCX CEM with disease association (Supplementary Table 18), of which 4 had brain cell-enrichment (Fig. 1B, Table 1). None of the CER CEM with brain cell-enrichment had significant association with disease (Table 2), despite being well-preserved with the TCX modules.
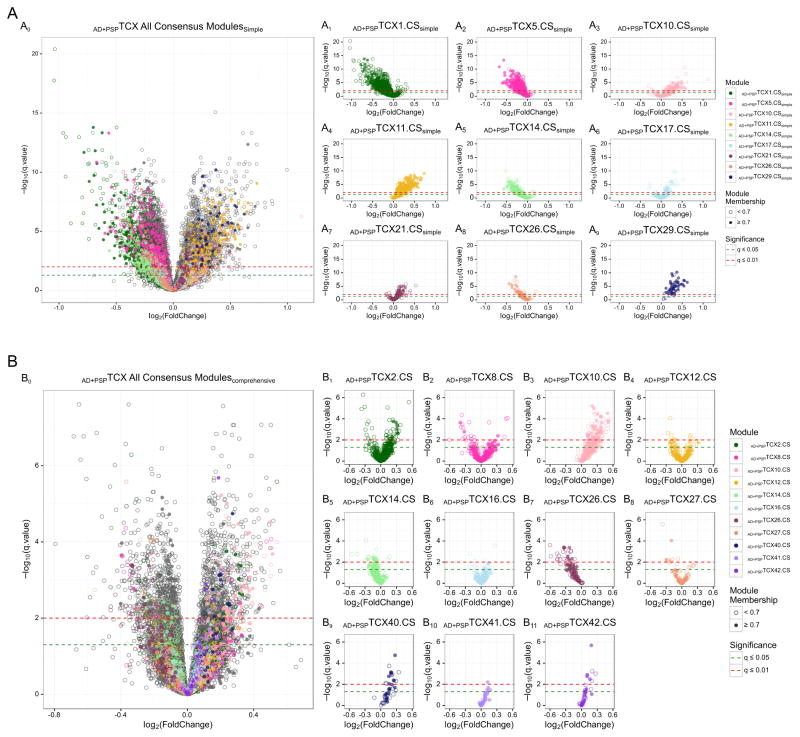
Fig. 1. Volcano plots of fold change vs. significance for differential gene expression (DGE) in the temporal cortex (TCX).
Results are shown for the primary analysis of AD vs. PSP TCX DGE in the Discovery Cohort, under the Simple (A0–A9) and Comprehensive (B0–B11) models. Each circle represents a transcript, which are colored differently according to the CEM they pertain to. Transcripts with strong module membership (MM) values≥0.7 are shown as filled circles; or empty circles if MM<0.7. Results are shown for all transcripts analyzed (A0, B0) and also separately for those CEM with consistent brain cell-enrichment across both models. DGE results that are significant at q≤0.05 or q≤0.01 are shown above the green and red dotted lines, respectively.
Table 2. Cerebellum Co-expression networks in the Discovery Cohort with significant cell type enrichment.
WGCNA consensus modules are generated from the Discovery Cohort’s WG-DASL transcript data obtained from the cerebellum. The remaining definitions are as per Table 1.
| Model | Module Name | Module Size | Cell Type Enrichment | Disease Association | Top GO Biological Process | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Type | OR | P value | Beta | P value | ID | Name | Enrichment P value | |||
| Simple | AD+PSP.CER31.CSsimple | 83 | Astrocyte | 11.0 | 5.08E-06 | 0.02 | 6.86E-01 | NA | NA | NA |
| AD+PSP.CER16.CSsimple | 288 | Astrocyte | 20.4 | 2.06E-41 | 0.07 | 2.47E-01 | GO:0007399 | nervous system development | 2.60E-04 | |
| AD+PSP.CER17.CSsimple | 239 | Microglia | 91.6 | 1.26E-139 | 0.01 | 8.24E-01 | GO:0002376 | immune system process | 1.49E-42 | |
| AD+PSP.CER12.CSsimple | 407 | Neuron | 2.4 | 4.39E-02 | −0.02 | 7.37E-01 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 2.98E-02 | |
| AD+PSP.CER4.CSsimple | 854 | Neuron | 2.9 | 5.77E-08 | −0.02 | 7.81E-01 | GO:0016070 | RNA metabolic process | 1.25E-05 | |
| AD+PSP.CER35.CSsimple | 65 | Neuron | 11.2 | 1.07E-07 | −0.03 | 6.62E-01 | GO:0048265 | response to pain | 3.56E-02 | |
| AD+PSP.CER21.CSsimple | 180 | Oligodendrocyte | 129.6 | 5.39E-72 | 0.01 | 8.51E-01 | GO:0042552 | myelination | 8.00E-08 | |
|
| ||||||||||
| Comprehensive | AD+PSP.CER13.CS | 284 | Astrocyte | 18.9 | 4.01E-37 | 0.06 | 3.10E-01 | GO:0042063 | gliogenesis | 9.54E-06 |
| AD+PSP.CER25.CS | 70 | Astrocyte | 42.6 | 1.43E-26 | 0.06 | 3.63E-01 | GO:0010634 | positive regulation of epithelial cell migration | 1.71E-03 | |
| AD+PSP.CER19.CS | 138 | Microglia | 119.9 | 2.57E-107 | 0.02 | 7.81E-01 | GO:0006955 | immune response | 2.21E-33 | |
| AD+PSP.CER3.CS | 1089 | Neuron | 2.6 | 3.87E-08 | −0.02 | 6.96E-01 | GO:0016071 | mRNA metabolic process | 3.86E-07 | |
| AD+PSP.CER6.CS | 703 | Neuron | 2.4 | 1.24E-03 | 0.01 | 8.77E-01 | GO:0007399 | nervous system development | 2.42E-02 | |
| AD+PSP.CER23.CS | 92 | Neuron | 13.0 | 3.69E-13 | −0.03 | 5.96E-01 | GO:0007268 | synaptic transmission | 4.39E-05 | |
| AD+PSP.CER20.CS | 131 | Oligodendrocyte | 149.4 | 3.37E-66 | 0.003 | 9.66E-01 | GO:0042552 | myelination | 1.31E-09 | |
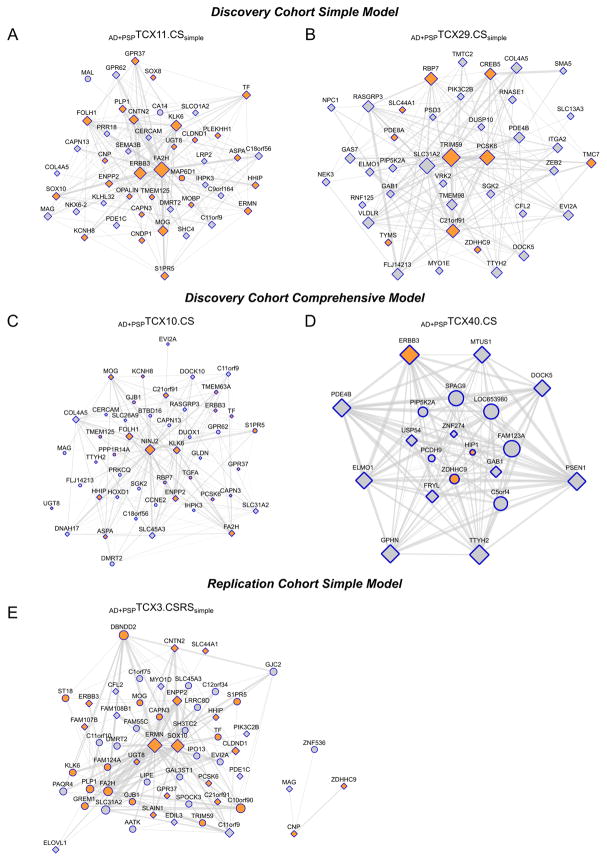
Inspection of the TCX CEM with disease association and cell-enrichment under both analytic models revealed that only the modules enriched in oligodendrocyte transcripts, implicated in myelination, had strong preservation under the Simple (Fig. 2A–B, AD+PSPTCX11.CSsimple, AD+PSPTCX29.CSsimple) and Comprehensive Models (Fig. 2C–D, AD+PSPTCX10.CS, AD+PSPTCX40.CS). Hereforth, we refer to these CEM and others with oligodendrocyte-transcript enrichment as “myelination modules”. All four myelination modules had higher levels of expression in TCX of AD subjects compared to PSP (Table 1, Fig. 1A4, 1A9, 1B3, 1B9). This remarkably consistent association of myelination networks with disease irrespective of adjustment for five brain cell-type specific expression markers suggested that these networks may be differentially regulated in AD vs. PSP for reasons other than brain cell population changes. No other TCX brain cell-enriched modules had consistent direction of association with disease under both analytic models.
Fig. 2. Oligodendrocyte networks in the Discovery and Replication Cohorts with disease association.
Temporal cortex (TCX) oligodendrocyte-specific gene enriched networks in the Discovery Cohort under the Simple (A, B) and Comprehensive Models (C, D); and in the Replication Cohort under the Simple Model are shown for the primary AD vs. PSP analysis. These CEMs have significantly different levels between AD and PSP. None of the corresponding modules in the Replication Cohort under the Comprehensive Model were significantly associated with disease. The circles or squares represent the nodes for the genes within the CEM. For each module, the top 150 connections according to TOM weight are shown for genes with a MM > 0.7. The size of a node correlates with the number of connections for that node with others within the network. Gene transcripts that are enriched within oligodendrocytes are shown in orange. Transcripts with significant differential expression at q<0.05 are shown as a square. Thickness of the connection lines is determined by the weight of the connection.
For these reasons, we focused on the myelination modules in the independent “Mayo Clinic RNAseq” Replication Cohort[16]. Following QC of this cohort (Supplementary Figs. 18–21), expression measures were retained for 80 AD and 82 PSP subjects, as well as 76 elderly controls. The 13,273 TCX RNAseq transcripts (13,211 unique genes) which overlapped with those from the Discovery Cohort were utilized in all downstream analyses. In the co-expression network AD vs. PSP analyses of the Replication Cohort, one myelination module was identified under the Simple Model (Table 3, Fig. 2E, AD+PSPTCX3.CSRSsimple); and three such modules under the Comprehensive Model (AD+PSPTCX2.CSRS, AD+PSPTCX8.CSRS, AD+PSPTCX26.CSRS).
Table 3. Temporal Cortex Co-expression networks in replication cohort with significant oligodendrocyte-specific gene enrichment.
WGCNA consensus modules are generated from the Replication Cohort’s RNAseq transcript data obtained from the temporal cortex. This dataset is independent from the Discovery Cohort. Results are shown both for the simple module (no correction for cell types) and the Comprehensive Model (corrects for cell types). Modules that are significantly enriched for oligodendrocyte-specific genes are shown. There are a total of 99 oligodendrocyte-enriched genes. The number of these that pertain to each module are shown. Disease Association Beta=Coefficient of association with AD diagnosis where AD is in the diagnostic comparison, or else for PSP.
| Model | Diagnostic Comparison | Module Name | Module Size | Number of Oligodendrocyte Genes in Module | Oligodendrocyte Enrichment OR | Oligodendrocyte Enrichment P value | Disease Association Beta | Disease Association P value |
|---|---|---|---|---|---|---|---|---|
| Simple | AD+Con | AD+Con.TCX10.CSRS.simple | 398 | 15 | 5.95 | 2.45E-07 | −0.094 | 9.19E-01 |
| AD+Con.TCX4.CSRS.simple | 924 | 73 | 40.48 | 2.44E-63 | −0.008 | 2.44E-01 | ||
|
| ||||||||
| AD+PSP | AD+PSP.TCX3.CSRS.simple | 1542 | 93 | 125.11 | 6.12E-80 | 0.279 | 3.31E-04 | |
|
| ||||||||
| PSP+Con | PSP+Con.TCX5.CSRS.simple | 737 | 73 | 52.71 | 9.60E-69 | −0.221 | 5.19E-03 | |
| PSP+Con.TCX12.CSRS.simple | 253 | 15 | 9.68 | 5.35E-08 | −0.176 | 2.74E-02 | ||
|
| ||||||||
| Comprehensive | AD+Con | AD+Con.TCX7.CSRS | 526 | 17 | 5.15 | 4.49E-05 | −0.228 | 4.12E-03 |
| AD+Con.TCX24.CSRS | 65 | 15 | 46.61 | 9.42E-17 | −0.143 | 7.40E-02 | ||
| AD+Con.TCX26.CSRS | 52 | 5 | 14.81 | 6.03E-03 | −0.025 | 7.54E-01 | ||
| AD+Con.TCX2.CSRS | 886 | 56 | 19.35 | 5.81E-38 | −0.042 | 6.05E-01 | ||
|
| ||||||||
| AD+PSP | AD+PSP.TCX2.CSRS | 946 | 49 | 13.44 | 4.62E-28 | 0.009 | 9.07E-01 | |
| AD+PSP.TCX8.CSRS | 628 | 25 | 7.02 | 5.42E-10 | 0.003 | 9.66E-01 | ||
| AD+PSP.TCX26.CSRS | 69 | 15 | 43.23 | 2.84E-16 | −0.050 | 5.29E-01 | ||
|
| ||||||||
| PSP+Con | PSP+Con.TCX2.CSRS | 1291 | 74 | 29.01 | 5.46E-52 | −0.100 | 2.12E-01 | |
| PSP+Con.TCX22.CSRS | 112 | 14 | 21.9 | 1.35E-11 | −0.064 | 4.26E-01 | ||
The Simple Model myelination module (AD+PSPTCX3.CSRSsimple) was highly preserved with the corresponding modules from the Discovery Cohort (Supplementary Fig. 22–23) and had significantly higher transcript levels in AD subjects compared to PSP (Table 3). The direction and effect size of disease association for myelination modules between these independent cohorts was remarkably similar (Tables 1 and 3). In contrast, none of the 3 myelination CEM in the Replication Cohort under the Comprehensive Model showed disease association in the AD vs. PSP analysis (Table 3), unlike the corresponding analysis in the Discovery Cohort (Table 1).
To distinguish whether the higher levels of myelination networks in AD vs. PSP TCX are due to an upregulation in AD or more downregulation in PSP brains, we compared the elderly control brain tissue in the Replication Cohort against either AD or PSP TCX transcriptome. Under the Simple Model, we identified two myelination CEM each in the AD+control and PSP+control analyses. Both PSP+Control (Table 3, PSP+ConTCX5.CSRSsimple, PSP+ConTCX12.CSRSsimple) TCX CEM were significantly lower in PSP. Under the Comprehensive Model, there were 4 AD+control and 2 PSP+control myelination CEM. Interestingly all these modules showed lower myelination network levels in both neurodegenerative disease groups in comparison to controls, one of which achieved statistical significance (Table 3, AD+ConTCX7.CSRS).
Together with the results from the Discovery Cohort, these findings suggest that myelination networks are downregulated in both AD and PSP compared to controls, but are more downregulated in PSP. For the Replication Cohort, the network associations with PSP are more pronounced in the Simple Model, whereas those for AD are more pronounced in the Comprehensive Model. Nevertheless, levels of the oligodendrocyte marker OLIG2 are not significantly different between diagnostic groups (Supplementary Fig. 4C). Therefore downregulation of myelination networks in these diseases cannot be entirely explained by oligodendrocyte cell population changes (Supplementary Text).
3.4 Myelination co-expression modules harbor neurodegenerative disease risk genes with replicable differential expression
To investigate the genes from the myelination modules further, we focused on a subset that have the highest module membership (MM); in addition to genes that are implicated in the pathophysiology of AD[33, 34], PSP[35], or in myelin biology[36] (Supplementary Results, Table 4). We evaluated the AD vs. PSP differential expression of these individual transcripts in the Discovery and Replication Cohorts, under the Simple and Comprehensive Models. We also performed DGE analyses for all other pairwise diagnostic comparisons (Supplementary Table 34).
Table 4. Differential temporal cortex gene expression of oligodendrocyte module genes in Discovery and Replication Cohorts.
Genes that are members of the oligodendrocyte modules in the Discovery AD+PSP Cohort, Comprehensive Model and that have a) the highest module membership (MM); b) implication in AD; c) implication in PSP; or d) implication in myelin biology are shown. The results for these genes are shown for this analysis, as well as Discovery AD+PSP Cohort, Simple Model; and Replication AD+PSP cohort, Comprehensive and Simple Models. The modules the genes reside in, MM values, and differential gene expression coefficients (DxBeta), uncorrected p values (DxpValue) and FDR-based q values (DxqValue) are shown. Top panel results are from the Discovery cohort. Bottom panel results are from the Replication Cohort. Beta=Coefficient of association with AD diagnosis. Significant DGE results are bolded. Nominally significant results are italicized. WG-DASL Illumina probe IDs are shown for the discovery and ENSEMBL gene IDs utilized in the RNAseq analyses are shown for the replication cohort results. Transcripts that are enriched in oligodendrocytes have 1 under the Oligodendrocyte column.
| Discovery Cohort (WG-DASL) | AD vs. PSP (n=278) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Simple Model | Comprehensive Model | |||||||||||
|
| ||||||||||||
| Gene | Probe | Oligodendrocyte | Module | MM | DxBeta | DxpValue | DxqValue | Module Name | MM | DxBeta | DxpValue | DxqValue |
| NINJ2a | ILMN_1731745 | 1 | AD+PSP.TCX11.CS.simple | 0.82 | 0.37 | 8.14E-08 | 1.90E-06 | AD+PSP.TCX10.CS | 0.90 | 0.24 | 4.65E-04 | 6.46E-03 |
|
|
|
|||||||||||
| COL4A5a | ILMN_1742534 | 0 | AD+PSP.TCX11.CS.simple | 0.85 | 0.41 | 4.63E-11 | 5.00E-09 | AD+PSP.TCX10.CS | 0.89 | 0.27 | 1.08E-05 | 4.95E-04 |
|
|
|
|||||||||||
| SLC45A3a | ILMN_1726114 | 0 | AD+PSP.TCX11.CS.simple | 0.86 | 0.49 | 2.72E-11 | 3.31E-09 | AD+PSP.TCX10.CS | 0.88 | 0.36 | 4.96E-07 | 6.21E-05 |
|
|
|
|||||||||||
| SLC31A2a | ILMN_1758938 | 0 | AD+PSP.TCX29.CS.simple | 0.84 | 0.43 | 1.47E-07 | 3.04E-06 | AD+PSP.TCX10.CS | 0.86 | 0.37 | 1.35E-05 | 5.73E-04 |
|
|
|
|||||||||||
| DMRT2a | ILMN_1751785 | 0 | AD+PSP.TCX11.CS.simple | 0.88 | 0.41 | 4.72E-05 | 3.11E-04 | AD+PSP.TCX10.CS | 0.85 | 0.23 | 1.71E-02 | 7.45E-02 |
|
|
|
|||||||||||
| PDE4Ba | ILMN_2296439 | 0 | AD+PSP.TCX29.CS.simple | 0.83 | 0.38 | 1.63E-08 | 5.20E-07 | AD+PSP.TCX40.CS | 0.87 | 0.25 | 2.37E-04 | 4.17E-03 |
|
|
|
|||||||||||
| KLK6a | ILMN_1780255 | 1 | AD+PSP.TCX11.CS.simple | 0.89 | 0.29 | 3.42E-05 | 2.41E-04 | AD+PSP.TCX10.CS | 0.85 | 0.22 | 1.43E-03 | 1.39E-02 |
|
|
|
|||||||||||
| S1PR5a | ILMN_2073184 | 1 | AD+PSP.TCX11.CS.simple | 0.89 | 0.28 | 2.92E-05 | 2.13E-04 | AD+PSP.TCX10.CS | 0.85 | 0.16 | 1.46E-02 | 6.71E-02 |
|
|
|
|||||||||||
| FA2Ha | ILMN_1791531 | 1 | AD+PSP.TCX11.CS.simple | 0.94 | 0.41 | 1.23E-06 | 1.61E-05 | AD+PSP.TCX10.CS | 0.85 | 0.24 | 5.42E-04 | 7.11E-03 |
|
|
|
|||||||||||
| ENPP2a | ILMN_1780799 | 1 | AD+PSP.TCX11.CS.simple | 0.83 | 0.34 | 2.39E-08 | 7.18E-07 | AD+PSP.TCX10.CS | 0.85 | 0.22 | 3.38E-04 | 5.23E-03 |
|
|
|
|||||||||||
| BACE1b | ILMN_2320349 | 0 | AD+PSP.TCX0.CS.simple | 0.03 | 0.14 | 4.49E-05 | 2.99E-04 | AD+PSP.TCX10.CS | 0.71 | 0.12 | 9.24E-04 | 1.02E-02 |
|
|
|
|||||||||||
| MOGd | ILMN_2310001 | 1 | AD+PSP.TCX11.CS.simple | 0.90 | 0.39 | 6.20E-06 | 5.94E-05 | AD+PSP.TCX10.CS | 0.83 | 0.24 | 2.92E-03 | 2.30E-02 |
|
|
|
|||||||||||
| PLP1d | ILMN_1790106 | 1 | AD+PSP.TCX11.CS.simple | 0.85 | 0.18 | 8.33E-05 | 4.99E-04 | AD+PSP.TCX10.CS | 0.78 | 0.12 | 5.70E-03 | 3.56E-02 |
|
|
|
|||||||||||
| PSEN1b | ILMN_1809193 | 0 | AD+PSP.TCX2.CS.simple | 0.60 | 0.29 | 5.04E-07 | 8.03E-06 | AD+PSP.TCX40.CS | 0.83 | 0.17 | 3.40E-03 | 2.55E-02 |
|
|
|
|||||||||||
| SLCO1A2c | ILMN_1806979 | 0 | AD+PSP.TCX11.CS.simple | 0.80 | 0.42 | 5.51E-06 | 5.40E-05 | AD+PSP.TCX10.CS | 0.75 | 0.31 | 6.77E-04 | 8.28E-03 |
|
|
|
|||||||||||
| PLLPd | ILMN_2082865 | 1 | AD+PSP.TCX11.CS.simple | 0.73 | 0.09 | 1.88E-02 | 4.25E-02 | AD+PSP.TCX10.CS | 0.67 | 0.02 | 5.86E-01 | 7.45E-01 |
|
|
|
|||||||||||
| CNPd | ILMN_1811758 | 1 | AD+PSP.TCX11.CS.simple | 0.84 | 0.17 | 4.34E-05 | 2.91E-04 | AD+PSP.TCX10.CS | 0.69 | 0.07 | 6.49E-02 | 1.83E-01 |
|
|
|
|||||||||||
| BIN1b | ILMN_2309245 | 0 | AD+PSP.TCX11.CS.simple | 0.73 | 0.10 | 2.05E-03 | 6.80E-03 | AD+PSP.TCX10.CS | 0.63 | 0.09 | 1.29E-03 | 1.29E-02 |
|
|
|
|||||||||||
| CR1b | ILMN_1742601 | 0 | AD+PSP.TCX10.CS.simple | 0.42 | 0.45 | 6.01E-06 | 5.78E-05 | AD+PSP.TCX10.CS | 0.61 | 0.19 | 5.73E-02 | 1.69E-01 |
|
|
|
|||||||||||
| MOBPc | ILMN_2298464 | 1 | AD+PSP.TCX11.CS.simple | 0.71 | 0.31 | 1.07E-02 | 2.66E-02 | AD+PSP.TCX10.CS | 0.59 | 0.13 | 3.08E-01 | 5.02E-01 |
| Replication Cohort (RNAseq) | AD vs. PSP (n=162) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Simple Model | Comprehensive Model | |||||||||||
|
| ||||||||||||
| Gene | GeneID | Oligodendrocyte | Module Name | MM | DxBeta | DxpValue | DxqValue | Module Name | MM | DxBeta | DxpValue | DxqValue |
| NINJ2a | ENSG00000171840 | 1 | AD+PSP.TCX3.CSRS.simple | 0.82 | 0.35 | 2.29E-02 | 3.83E-02 | AD+PSP.TCX8.CSRS | 0.87 | 0.03 | 8.40E-01 | 9.41E-01 |
|
|
||||||||||||
| COL4A5a | ENSG00000188153 | 0 | AD+PSP.TCX3.CSRS.simple | 0.89 | 0.44 | 5.06E-03 | 1.00E-02 | AD+PSP.TCX8.CSRS | 0.86 | 0.04 | 7.27E-01 | 8.94E-01 |
|
|
||||||||||||
| SLC45A3a | ENSG00000158715 | 0 | AD+PSP.TCX3.CSRS.simple | 0.88 | 0.37 | 3.95E-02 | 6.22E-02 | AD+PSP.TCX2.CSRS | 0.87 | 0.12 | 4.90E-01 | 7.57E-01 |
|
|
||||||||||||
| SLC31A2a | ENSG00000136867 | 0 | AD+PSP.TCX3.CSRS.simple | 0.89 | 0.25 | 6.00E-02 | 9.01E-02 | AD+PSP.TCX2.CSRS | 0.83 | −0.02 | 8.84E-01 | 9.59E-01 |
|
|
||||||||||||
| DMRT2a | ENSG00000173253 | 0 | AD+PSP.TCX3.CSRS.simple | 0.84 | 0.25 | 1.33E-01 | 1.81E-01 | AD+PSP.TCX2.CSRS | 0.81 | 0.07 | 6.33E-01 | 8.44E-01 |
|
|
||||||||||||
| PDE4Ba | ENSG00000184588 | 0 | AD+PSP.TCX3.CSRS.simple | 0.66 | 0.40 | 5.52E-09 | 1.09E-07 | AD+PSP.TCX16.CSRS | 0.63 | 0.14 | 1.24E-02 | 1.13E-01 |
|
|
||||||||||||
| KLK6a | ENSG00000167755 | 1 | AD+PSP.TCX3.CSRS.simple | 0.87 | 0.31 | 6.21E-02 | 9.29E-02 | AD+PSP.TCX2.CSRS | 0.85 | 0.10 | 5.01E-01 | 7.64E-01 |
|
|
||||||||||||
| S1PR5a | ENSG00000180739 | 1 | AD+PSP.TCX3.CSRS.simple | 0.85 | 0.24 | 1.63E-01 | 2.16E-01 | AD+PSP.TCX2.CSRS | 0.83 | 0.03 | 8.33E-01 | 9.39E-01 |
|
|
||||||||||||
| FA2Ha | ENSG00000103089 | 1 | AD+PSP.TCX3.CSRS.simple | 0.87 | 0.26 | 1.10E-01 | 1.53E-01 | AD+PSP.TCX2.CSRS | 0.83 | 0.05 | 7.17E-01 | 8.87E-01 |
|
|
||||||||||||
| ENPP2a | ENSG00000136960 | 1 | AD+PSP.TCX3.CSRS.simple | 0.89 | 0.40 | 6.38E-03 | 1.23E-02 | AD+PSP.TCX2.CSRS | 0.82 | 0.12 | 3.24E-01 | 6.36E-01 |
|
|
||||||||||||
| BACE1b | ENSG00000186318 | 0 | AD+PSP.TCX3.CSRS.simple | 0.70 | 0.08 | 2.29E-01 | 2.90E-01 | AD+PSP.TCX2.CSRS | 0.72 | 0.05 | 3.90E-01 | 6.92E-01 |
|
|
||||||||||||
| MOGd | ENSG00000204655 | 1 | AD+PSP.TCX3.CSRS.simple | 0.87 | 0.33 | 5.52E-02 | 8.38E-02 | AD+PSP.TCX8.CSRS | 0.84 | 0.00 | 9.93E-01 | 9.97E-01 |
|
|
||||||||||||
| PLP1d | ENSG00000123560 | 1 | AD+PSP.TCX3.CSRS.simple | 0.79 | 0.34 | 4.63E-02 | 7.16E-02 | AD+PSP.TCX2.CSRS | 0.73 | 0.11 | 4.56E-01 | 7.36E-01 |
|
|
||||||||||||
| PSEN1b | ENSG00000080815 | 0 | AD+PSP.TCX3.CSRS.simple | 0.91 | 0.30 | 6.40E-04 | 1.62E-03 | AD+PSP.TCX8.CSRS | 0.82 | 0.02 | 7.97E-01 | 9.25E-01 |
|
|
||||||||||||
| SLCO1A2c | ENSG00000084453 | 0 | AD+PSP.TCX3.CSRS.simple | 0.81 | 0.46 | 1.02E-02 | 1.87E-02 | AD+PSP.TCX8.CSRS | 0.70 | 0.06 | 6.71E-01 | 8.64E-01 |
|
|
||||||||||||
| PLLPd | ENSG00000102934 | 1 | AD+PSP.TCX3.CSRS.simple | 0.79 | 0.30 | 4.61E-02 | 7.14E-02 | AD+PSP.TCX8.CSRS | 0.91 | 0.00 | 9.72E-01 | 9.89E-01 |
|
|
||||||||||||
| CNPd | ENSG00000173786 | 1 | AD+PSP.TCX3.CSRS.simple | 0.90 | 0.36 | 1.05E-02 | 1.91E-02 | AD+PSP.TCX8.CSRS | 0.92 | 0.03 | 7.50E-01 | 9.02E-01 |
|
|
||||||||||||
| BIN1b | ENSG00000136717 | 0 | AD+PSP.TCX3.CSRS.simple | 0.80 | 0.07 | 3.13E-01 | 3.80E-01 | AD+PSP.TCX2.CSRS | 0.81 | −0.02 | 7.80E-01 | 9.18E-01 |
|
|
|
|||||||||||
| CR1b | ENSG00000203710 | 0 | AD+PSP.TCX3.CSRS.simple | 0.61 | 1.11 | 3.87E-06 | 2.02E-05 | AD+PSP.TCX27.CSRS | 0.72 | 0.40 | 9.29E-02 | 3.38E-01 |
|
| ||||||||||||
| MOBPc | ENSG00000168314 | 1 | AD+PSP.TCX3.CSRS.simple | 0.78 | 0.22 | 2.06E-01 | 2.64E-01 | AD+PSP.TCX26.CSRS | 0.83 | −0.01 | 9.64E-01 | 9.86E-01 |
MM values close to 1 reflect high connectivity of the gene to the module[14]. The MM values of these genes were generally high (>0.70) and similar in all analyses (Table 4). Under the Simple Model, all 20 genes had higher levels in AD compared to PSP TCX in both cohorts, with remarkably consistent effect estimates. All 20 transcript associations were significant in the Discovery and 8 in the Replication Cohort, including disease implicated genes PSEN1, SLCO1A2 and CR1. Under the Comprehensive Model, in the Discovery Cohort, all 20 genes had higher TCX level estimates in AD subjects vs. PSP, 14 of which were statistically significant. In the Replication Cohort, none of the associations were retained under the Comprehensive Model, suggesting that cell-type adjustment may be accounting for a larger portion of the diagnostic differences for these genes in this cohort.
To determine whether the transcriptional changes observed in the TCX was also reflected in protein levels, we sought to validate these findings by performing western blot analyses in TCX samples (Supplementary Table 35, Supplementary Fig. 24). Given the limited dynamic range for quantitation of blots labeled with HRP-tagged antibodies, as in our study, these western blots should be considered as semi-quantitative. Despite significant variability, all myelin proteins had lower levels in PSP TCX (Supplementary Fig. 24. A–F) consistent with the transcript results, although these trends did not reach statistical significance, likely due to the relatively small sample size of this protein analysis cohort. All myelin proteins except MOG and PLLP had lower level estimates in AD TCX (Supplementary Fig. 24. G–L), but not statistically significant, highlighting the need to evaluate much larger cohorts for protein validations.
Using proteome data from a larger cohort of 84 AD and 83 PSP TCX samples, obtained with Liquid Chromatography Coupled to Tandem Mass Spectrometry (LC-MS/MS) analysis, we identified significantly lower protein levels for myelin proteins MBP and CNP in PSP compared with AD TCX; and lower estimates that did not reach statistical significance for MOG, PLP1 and BIN1 (Supplementary Table.36). As expected, GFAP, APP and MAPT protein levels were lower in PSP compared to AD TCX.
We assessed myelination patterns, as well as microgliosis and astrogliosis in a subset of AD, PSP and control TCX samples (Supplementary Table 37, Supplementary Fig. 25). As expected, there is variability in the level of pathology. Given this and the small number of samples assessed, statistical differences in quantitative neuropathology cannot be detected. Nevertheless, the pattern of reduced myelination can be appreciated in select AD and PSP vs. control TCX samples. We evaluated immunocytochemical patterns for the myelin and oligodendrocyte proteins in rat primary oligodendrocyte-enriched cultures and demonstrated high cell type selectivity and regional specificity of selected antibodies for their cellular targets (Supplementary Table 38, Supplementary Fig.26–27, Supplementary_Text).
4. Discussion
In this study, we identified highly conserved myelination networks that are altered in both PSP and AD brains but to a greater extent in the former. This study is distinct from prior transcriptome studies in neurodegenerative diseases[3, 5, 6, 37] in several ways. We provide comparison of multiple neurodegenerative conditions, in addition to controls; use two independent cohorts; study two brain regions; use two different approaches for measuring gene expression; assess cell population variability; and perform protein validations (Supplementary Discussion). The underlying premise of our approach is that comparative analyses of different neurodegenerative diseases can uncover transcripts and molecular networks that are disease-specific as well as those that underlie shared aspects of disease pathology. To our knowledge, this is the first study, which has performed a systematic comparison of brain transcriptomes from AD vs. a primary tauopathy, PSP. Our conclusions are based on a collective dataset of 940 brain transcriptomes.
Our study yields insights into the role of myelination in the pathophysiology of two neurodegenerative diseases. To our knowledge oligodendrocyte/myelination pathways have not been studied comparatively in AD vs. PSP at a systems-biology level. There is evidence from neuropathology that oligodendrocyte/myelination dysfunction could contribute to both AD and PSP. Oligodendroglial tau deposits are a key aspect of PSP neuropathology[12]. Myelin loss was demonstrated in AD white matter (WM)[38], and focal intracortical demyelination associated with Aβ plaques was observed in AD gray matter (GM)[39]. Further, human brain myelination has distinct aspects that may predispose it to vulnerabilities resulting in neuropsychiatric illness[36, 40]. Myelination in humans an evolutionarily late event, which is distinguished from that of other species by its extent in both gray and white matter (GM, WM) and by developmental myelination extending well into middle ages[36].
We find that brain gene expression networks enriched in oligodendrocyte transcripts involved in myelination are downregulated in PSP compared to AD. This downregulation is observed in two independent cohorts and is retained in the Discovery Cohort, even after adjusting for cell-specific markers (Comprehensive Model) to account for any cell-population changes. The similarity in the findings in the Discovery Cohort under both the Simple and Comprehensive Models suggest that the transcriptional changes are unlikely to be solely due to cell-population changes. This is further corroborated by the fact that TCX, where these transcriptional changes are observed, is a region typically unaffected by PSP pathology[12]. Myelination networks are also downregulated in PSP TCX in comparison to controls, providing further support that these transcriptional changes are unlikely to be due to gross changes in pathology.
The depression of these findings under the Comprehensive Model in the Replication, but not the Discovery Cohort, may be multifactorial. First, the latter has >50% greater sample size. Second, RNAseq measures in the Replication Cohort may provide a more precise measurement of gene levels that may have led to better adjustment for cell-type changes or over-correction due to their stronger inter-correlation.
Importantly, there is downregulation of myelin proteins in PSP TCX in protein data from 167 brain samples assessed by LC-MS/MS, as well as a smaller cohort evaluated by semi-quantitative western blots analysis. Myelination patterns and cellular specificity of the antibodies used to assess myelination proteins are demonstrated by immunohistochemistry and immunocytochemistry in human brains and rat primary oligodendrocyte-enriched cultures. Thus, our transcriptome findings are also corroborated by protein data.
Our study paradigm allowed us to distinguish that myelination networks may also be downregulated in AD, but to a lesser extent than in PSP, rather than simply being upregulated in AD vs. PSP. This lesser alteration in AD TCX is intriguing, especially given that AD, unlike PSP, has significant pathology in TCX[9]. This finding further implies that the myelination network changes are unlikely to be a mere consequence of pathology. The enhanced vulnerability of myelination networks in PSP, in comparison to AD, leads to a number of compelling hypotheses. Both AD and PSP are characterized by aggregates of tau, which is a microtubule associated protein (MAP) and a constituent of both neurons and oligodendrocytes[13, 40]. Microtubules (MT) and tau are integral to oligodendroglial function and myelination, which are disrupted when tau is either overexpressed or downregulated. Hence, alteration of myelination networks in both AD and PSP is consistent with these data.
A key question is why this alteration is more enhanced in PSP in a brain region far less affected than in AD. One explanation may be the difference in the type of tau aggregate, with PSP harboring 4R-tau aggregates, composed of a tau isoform with 4 MT-binding domains, whereas AD has both 3R- and 4R-tau aggregates. In cultured oligodendrocytes, 4R-tau becomes increased and 3R-tau decreased with development[40]. We can therefore postulate that myelination pathways may be more vulnerable to 4R-tauopathies, such as PSP. Another reason may be the presence of genetic risk factors in PSP with a role in myelination. Indeed, variants in/near MOBP are implicated in risk of PSP[35], and CBD[41], another 4R-tauopathy. MOBP encodes the CNS-expressed myelin-associated oligodendrocytic basic protein, which is a member of the myelination networks identified herein.
Another finding from our study is the remarkable conservation of brain transcriptional networks that are independently constructed in two brain regions, TCX and CER. This finding is consistent with the prior observations in healthy control brains[42], and suggest that the broad architecture of the brain transcriptional networks is unlikely to be driven by cell population differences in disease-affected vs. –unaffected tissue. Additionally, although we focused on myelination networks in this study, we identified modules enriched in astrocytic, microglial and neuronal transcripts, which show consistencies with prior transcriptome studies[6, 37, 43]. The detailed findings from our analyses that we present here, as well as the accessibility of our large-scale data[16] should establish this study as a highly useful resource.
In summary, our study identifies downregulation of myelination networks as a potential pathophysiologic component of both PSP and AD. Our findings are based on postmortem brain tissue which reflects a “snapshot” of gene expression networks for end-stage disease. Nevertheless, this work can be instrumental in launching future biomarker or therapeutic discovery efforts. Neuroimaging studies in living patients support white matter[44, 45] and specifically myelin alterations[46] in preclinical AD. Key molecules within myelination networks identified in our study can serve to develop novel molecular imaging tools for tracking myelin neuropathology in longitudinal cohorts followed for incident AD and other neurodegenerative diseases. Such cohorts should also enable detection of longitudinal changes in gene and protein expression levels for these molecules, which can help establish their temporal relationship with cognitive and other clinical outcomes. These future studies can provide fundamental new insight into the role of myelin dysregulation in the cascade of pathophysiological processes in AD[47] and other neurodegenerative conditions. Additionally, the specific expression network alterations uncovered in our study can be tested in model systems for their potential as therapeutic targets. There are known interactions between oligodendrocytes and the other CNS cell-types; including inflammatory and astrocytic activation in myelin breakdown and remyelination[36]. We find that established and candidate AD genes[34], such as PSEN1, BIN1 and CR1, reside in myelination modules. Given these and the effects of both tau[12, 40] and Aβ[38, 39] on myelination, we posit that myelin may indeed be the “glue” that holds together key biological functions in the adult brain, the disruption of which results in neuropsychiatric conditions such as AD and PSP.
Supplementary Material
Supplementary Fig. 1. Mayo Clinic eGWAS quality control (QC) flowchart. Flowchart outlining QC procedures and parameters for WG-DASL temporal cortex (TCX) and cerebellum (CER) datasets. No sample exclusions were necessary after QC of WG-DASL expression data. All subjects in the Mayo Clinic eGWAS were also part of the Mayo Clinic LOAD GWAS, which was performed on a larger cohort (N=2465). WG-DASL = Whole Genome cDNA mediated Annealing, Selection, extension, and Ligation assay, PCA = Principal components analysis.
Supplementary Fig. 2. Mayo Clinic eGWAS principal components analysis. Scatter plot of principal components 1 and 2 for temporal cortex (A) and cerebellum (B) expression measures. Samples are colored according to WG-DASL plate.
Supplementary Fig.3: QQ plots for expression residuals of select genes: Data points are colored according to plate. All probes shown have detectable signal in >50% of the samples. These plots demonstrate that including plate as a covariate adequately controls for the batch effect of plate.
Supplementary Fig. 4. Brain cell-specific gene expression levels by diagnosis in the Discovery and Replication cohorts: Gene expression level residuals adjusted for covariates are plotted by diagnosis. Brain cell-specific expression levels for the genes utilized in the Comprehensive analytic model and the brain cells for which they are specific are as follows: ENO2 for neurons, GFAP for astrocytes, CD68 for microglia, OLIG2 for oligodendrocytes and CD34 for endothelial cells. Box plots depict the median (line through box); and bottom and top lines of the box are the 1st and 3rd quartiles, respectively. The bottom and top of the whiskers are 1.5 x inter-quartile range beyond the bottom and top of the boxes. The dots are outliers beyond this range. Box plots of gene expression levels for the different diagnoses are shown. Pairwise diagnostic comparisons that are significantly different are shown with a horizontal line above the significant comparison and an asterisk. Results are shown for the Discovery Cohort, temporal cortex (A); cerebellum (B); Replication Cohort, temporal cortex (C); and cerebellum (D).
Supplementary Fig. 5: Oxidative phosphorylation pathway from MetaCore (Thompson Reuters) where 29 genes in the pathway are expressed at significantly lower levels in the temporal cortex of AD subjects when compared with PSP. Downregulated genes are indicated with blue thermometers, where the height of the blue bar reflects the regression beta (see also Supplementary_Tables.4 and 11).
Supplementary Fig. 6. Pairwise correlations of brain gene expression levels for five brain cell-specific genes across diagnostic groups: Data are shown for the following five CNS cell-specific genes: ENO2 for neurons, GFAP for astrocytes, CD68 for microglia, OLIG2 for oligodendrocytes and CD34 for endothelial cells. These genes are utilized for performing adjustments in DGE analyses and in the WGCNA “Comprehensive Model” analyses. Residual gene levels after adjustment of covariates were plotted. All pairwise correlations for the levels of these genes measured in the temporal cortex of the Discovery Cohort (A–J); and the Replication Cohort (L–U) are shown. The correlations were conducted within the main diagnostic groups, separately. Pearson correlation coefficients and p-values are shown (K, V).
Supplementary Fig. 7. Human brain cell type-enriched genes cluster dendrogram. Log2FPKM expression measures of 1270 cell-type enriched genes, from isolated populations of cells were assessed. The dist function was used to calculate Euclidian distances between samples and the hclust function (method =“average”) to perform cluster analysis. Samples are labeled to indicate cell type (N=Neuron, A=Astrocyte, M=Microglia, E=Endothelia, O=Oligodendroglia), Age (Years) and Sex (F = Female, M= Male), and color coded to further differentiate cell types.
Supplementary Fig. 8. Human brain cell type-enriched genes correlation heatmap. Log2FPKM expression measures of 1270 cell-type enriched genes, from isolated populations of cells were assessed. The cor function was used to create a correlation matrix and the melt function (reshape2) was used to manipulate the data matrix for plotting using ggplot2. Samples are labeled to indicate cell type (N=Neuron, A=Astrocyte, M=Microglia, E=Endothelial, O=Oligodendroglia), Age (Years) and Sex (F = Female, M= Male).
Supplementary Fig. 9. Preservation of temporal cortex (TCX) co-expression modules (CEM) from subjects with AD and PSP (AD+PSP) under the Simple vs. Comprehensive Models, using Simple Model as the reference (Discovery Cohort): WGCNA “modulePreservation” function with 100 permutations was applied to calculate preservation statistic Zsummary for a specific module’s genes in comparison groups. Zsummary score (y-axis) vs. CEM size (x-Axis) are plotted. Typically, a Zsummary score >10 indicates that the module genes were well-preserved. All of the TCX CEM under the Simple Model are well preserved with those under the Comprehensive Model.
Supplementary Fig. 10. Preservation of TCX AD+PSP CEM under the Simple vs. Comprehensive Models, using Comprehensive Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9, except that CEM under the Comprehensive Model are used as the reference.
Supplementary Fig. 11. Overlap of transcripts in the TCX AD+PSP CEM under the Simple and Comprehensive Models (Discovery Cohort): Matrix for the number of overlapping transcripts in TCX CEM under the Simple (y-axis) and Comprehensive (x- axis) models. Modules with enrichment of brain cell-enriched transcripts are highlighted in yellow. Coloring palette to the right is according to the number of transcripts in overlapping modules.
Supplementary Fig. 12. Preservation of cerebellum (CER) AD+PSP CEM under the Simple vs. Comprehensive Models, using Simple Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 13. Preservation of CER AD+PSP CEM under the Simple vs. Comprehensive Models, using Comprehensive Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 14. Overlap of transcripts in the CER AD+PSP CEM under the Simple and Comprehensive Models (Discovery Cohort): Matrix for the number of overlapping transcripts in CER CEM under the Simple (y-axis) and Comprehensive (x-axis) models. Modules with enrichment of brain cell-enriched transcripts are highlighted in yellow. Coloring palette to the right is according to the number of transcripts in overlapping modules.
Supplementary Fig. 15. Preservation of CER vs. TCX AD+PSP CEM built under the Comprehensive Model, using CER as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 16. Preservation of CER vs. TCX AD+PSP CEM built under the Comprehensive Model, using TCX as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 17. Overlap of transcripts in the CER and TCX AD+PSP CEM under Comprehensive Model (Discovery Cohort): Matrix for the number of overlapping transcripts in the TCX CEM (y-axis) and CER CEM (x-axis) built in AD+PSP subjects under the Comprehensive Model. Red-colored cell indicates the most number of overlapping genes of the corresponding row; orange-colored cell indicates the second most; yellow-colored cell indicates the third most; the rest of the cells are white.
Supplementary Fig. 18. Mayo Clinic RNAseq QC flowchart.
Flowchart outlining quality control (QC) procedures and parameters for RNaseq TCX dataset. QC parameters and sample numbers excluded are indicated. Samples that passed both expression and genetic data QC were used for analysis. SD = standard deviation, PCA = Principal components analysis.
Supplementary Fig. 19. Mayo Clinic RNAseq Sex Check.
Mean expression (counts per million) for genes mapped to the Y chromosome were plotted for all TCX samples (N=278). Female samples cluster to the left side of the plot and males to the right. Two samples labeled as male were identified in the female cluster (red triangles) and excluded from further analysis.
Supplementary Fig. 20. Mayo Clinic RNAseq PCA plot figure legend.
Scatter plot of principal components 1 and 2 for temporal cortex RNAseq expression measures. CQN normalized gene counts, which are used in all downstream analyses, are plotted in this figure.
Supplementary Fig. 21. Mayo Clinic RNAseq GWAS heterozygosity.
Scatter plot for sample heterozygosity vs. genotype missingness rate across all subjects with genome-wide genotype data. Horizontal red lines indicate 3 standard deviations from the mean. Blue vertical lines indicate 0% and 1% missingness rates. Since more subjects than just those with TCX RNAseq measures underwent GWAS, this plot includes subjects both with and without TCX RNAseq measures. Subjects 127, 1930, 1169 and 11416 are those with TCX RNAseq that were excluded based on this QC metric.
Supplementary Fig. 22. Preservation of Replication (RNAseq) vs. Discovery (DASL) AD+PSP CEM built under the Simple Model, using RNAseq as the reference. The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 23. Overlap of transcripts in the Discovery (DASL) and Replication (RNAseq) AD+PSP CEM under the Simple Model. Matrix for the number of overlapping transcripts in the TCX Discovery Cohort CEM (y-axis) and Replication Cohort CEM (x-axis) built in AD+PSP subjects under the Simple Model. Red-colored cell indicates the most number of overlapping genes of the corresponding row; orange-colored cell indicates the second most; yellow-colored cell indicates the third most; the rest of the cells are white.
Supplementary Fig. 24. Western blot analyses of oligodendroglial network proteins: Temporal cortex levels of proteins from the oligodendroglial networks (PLP, CNP, MOG, PLLP, BACE1, PSEN1, MOBP) are shown. Other proteins that are integral to or implicated in myelin function (MBP), AD (APP, PSEN2, Tau), PSP (Appoptosin), astrocytes (GFAP), microglia (IBA-1), neurons (β3-tubulin) and GAPDH control are also measured. Each PSP and Control subject was unique. In addition 2 samples (Replicates) were measured on every gel to control for blot-to-blot variability. A–E depict the 5 sets of western blots for the complete protein validation cohort. F. Table depicting summary measures and results of multivariable linear regression analyses using all data.
Supplementary Fig. 25: Representative photomicrographs of immunohistochemically stained temporal cortex from a control (A–C), PSP (D–F), and AD brains (G–I). The MBP antibody was used to demonstrate differences in myelination across subjects (A, D, G). The Iba-1 antibody was used to label microglia with a noted difference in abundance and morphology. The Iba-1 stain was also used to evaluate the density of oligodendrocytes as their nuclei are round with compact chromatid labeled by the hematoxylin counterstain (B,E,H). The GFAP antibody was used to labels astrocytes cell bodies and processes (C, F, I).
Supplementary Fig. 26: Cellular characterization of primary rat oligodendrocyte-enriched cultures: Enriched wild-type rat oligodendrocyte cultures were grown for 4 days in B27-supplemented Neurobasal A and stained live on ice for 15 minutes with oligodendrocyte specific markers A2B5, O4 and O1. Cells were subsequently fixed, permeabilized, labeled for internal markers MBP, GFAP, IBA-1, β3tubulin and processed for immunocytochemistry including DAPI-staining for cell counting and to ensure cell integrity. Late stage oligodendrocyte marker MBP indicates presence of predominantly mature oligodendrocytes with flat “pancake-like” membrane sheaths (myelin sheaths), which are also detectable with markers O1 and O4. Cell counts of enriched OL cultures at day 4 by immunocytochemistry demonstrated ~80% O4-positive oligodendrocytes, 10% GFAP-positive astrocytes, 4% IBA-1-positive microglia and 6% b3tubulin-positive neurons compared to the total number of DAPI-positive cells.
Supplementary Fig. 27: Immunocytochemical patterns of oligodendrocyte and myelin proteins in rat primary oligodendrocyte-enriched cultures: Enriched wild type rat oligodendrocyte cultures were grown for 5 days in B27-supplemented Neurobasal A and stained live on ice for 20 minutes with oligodendrocyte-specific markers O4 (O4/MBP). Cells were subsequently fixed, permeabilized, labeled for internal markers MBP, GFAP, IBA-1, β3tubulin, CD68-like protein, Olig-2, CNP, PLLP, and MOG and processed for immunocytochemistry including DAPI-staining for cell counting and to ensure cell integrity (A–R). Columns A–P and B–Q represent single-labeled OL cultures whereas columns C–R represent the corresponding overlays including DAPI staining (e.g. C = overlay of A+B). Row A–C shows double-labeled cultures using oligodendrocyte-specific markers O4 (A) and MBP (B) respectively, and the overlay (C) of O4/MBP (green/red) plus nuclear marker DAPI (blue). Consecutive rows represent double labeled OL cultures for markers GFAP (astrocytes, green) and CNP (oligodendrocytes, red) plus DAPI (blue) (row D–F); Olig-2 (oligodendrocytes, green) and neuronal marker β3tubulin (red) plus DAPI (blue) (row G–I); microglial markers IBA-1 (green) and CD68-like marker (red) plus DAPI (blue) (row J–L). Rows M–O and P–R represent single-labeled cultures using oligodendrocyte-specific marker PLP (M, green) plus DAPI (blue) (row M–O) respectively, and PLLP (P, green) (oligodendrocytes) plus DAPI (blue) (row P–R).
Highlights.
Brain myelination transcriptional networks are downregulated in PSP and AD.
Myelination networks are higher in AD vs. PSP but lower compared to controls.
Network structures, but not expression changes, are preserved between TCX and CER.
Brain cell type changes can influence and need adjustment in transcriptome studies.
RESEARCH IN CONTEXT.
Systemic review
Reviewing the literature for gene expression profiling publications of neuro-proteinopathies, showed that most studies are limited to small cohorts and individual gene transcript rather than systems-level analysis. Further, most studies assess one disease group against controls, rather than comparative transcriptome analyses of different diseases.
Interpretation
Comparative transcriptome analyses in Alzheimer’s disease (AD) and other neurodegenerative proteinopathies can uncover both shared and distinct disease pathways. Our analysis of 940 brain transcriptomes including patients with AD, progressive supranuclear palsy (PSP) and controls identified down-regulation of myelination networks in both AD and PSP, but more pronounced in the latter.
Future directions
Future studies should investigate in ante-mortem cohorts, longitudinal changes in myelination network molecules to determine their role in the pathophysiological processes in AD and other neurodegenerative diseases with a goal to establish them as novel biomarkers. The myelination network molecules should be tested in model systems for their potential as therapeutic targets.
Acknowledgments
We thank the patients and their families for their participation, without whom these studies would not have been possible.
This work was supported by National Institute on Aging [R01 AG032990, RF AG051504 to N.E.T.; U01 AG046139 to N.E.T. and S.G.Y.; P50 AG0016574 to D.W.D, N.E.T, N.R.G.-R., R.C.P. and S.G.Y.]; U01 AG006786 to R.C.P; National Institute of Neurological Disorders and Stroke [R01 NS080820 to N.E.T]; Mayo Clinic Center for Individualized Medicine [Gerstner Family Career Development Award to M.E.M].
We thank the Petrucelli lab for the gift of the anti-tau antibody.
For samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona: The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
Dr. Petersen has acted as a consultant for Roche, Inc., Merck, Inc., Biogen, Inc. and Genentech, Inc. Dr. Graff-Radford has multicenter treatment study grants from Lilly, TauRx and consulted for Cytox.
Abbreviations
- Aβ
Amyloid β
- AD
Alzheimer’s disease
- AD+PSPTCX
AD vs. PSP temporal cortex analyses
- AD+PSPCER
AD vs. PSP cerebellum analyses
- AD+ConTCX
AD vs. Control temporal cortex analyses
- AD+ConCER
AD vs. Control cerebellum analyses
- ALS
Amyotrophic lateral sclerosis
- CBD
Corticobasal degeneration
- CEM
Co-expression modules
- CER
Cerebellum
- CNS
Central nervous system
- DE
Differential expression/differentially expressed
- DGE
Differential gene expression
- eGWAS
Expression genome-wide association study(ies)
- eQTL
Expression quantitative trait locus/loci.
- FDR
False discovery rate
- FPKM
fragments per kilobase per million
- FTLD
Frontotemporal lobar degeneration
- GO
Gene Ontology
- GWAS
Genome-wide association study(ies)
- IGAP
International Genetics of AD Project
- LBD
Lewy body disease
- MM
Module membership
- MS
Multiple sclerosis
- MSA
Multiple system atrophy
- nAD
CNS diseases (non-AD, includes PSP and nTau)
- nTau
CNS diseases without primary tau or AD pathology (non-Tau)
- PD
Parkinson’s disease
- PSP
Progressive supranuclear palsy
- PSP+ConTCX
PSP vs. Control temporal cortex analyses
- PSP+ConCER
PSP vs. Control cerebellum analyses
- QC
Quality control
- RNAseq
RNA sequencing
- TCX
Temporal cortex
- VaD
Vascular dementia
- WGCNA
Weighted gene co-expression network analysis
- WG-DASL
Whole genome cDNA-mediated Annealing, Selection, Extension, and Ligation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 2.Ertekin-Taner N, De Jager PL, Yu L, Bennett DA. Alternative Approaches in Gene Discovery and Characterization in Alzheimer’s Disease. Curr Genet Med Rep. 2013;1:39–51. doi: 10.1007/s40142-013-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper-Knock J, Kirby J, Ferraiuolo L, Heath PR, Rattray M, Shaw PJ. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol. 2012;8:518–30. doi: 10.1038/nrneurol.2012.156. [DOI] [PubMed] [Google Scholar]
- 4.Tollervey JR, Wang Z, Hortobagyi T, Witten JT, Zarnack K, Kayikci M, et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011;21:1572–82. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–20. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries CE, Kohli MA, Nathanson L, Whitehead P, Beecham G, Martin E, et al. Integrated whole transcriptome and DNA methylation analysis identifies gene networks specific to late-onset Alzheimer’s disease. J Alzheimers Dis. 2015;44:977–87. doi: 10.3233/JAD-141989. [DOI] [PubMed] [Google Scholar]
- 8.Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18:1175–82. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 10.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 11.Hauw JJ, Daniel SE, Dickson D, Horoupian DS, Jellinger K, Lantos PL, et al. Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy) Neurology. 1994;44:2015–9. doi: 10.1212/wnl.44.11.2015. [DOI] [PubMed] [Google Scholar]
- 12.Dickson DW, Ahmed Z, Algom AA, Tsuboi Y, Josephs KA. Neuropathology of variants of progressive supranuclear palsy. Curr Opin Neurol. 2010;23:394–400. doi: 10.1097/WCO.0b013e32833be924. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Mandelkow E. Tau in physiology and pathology. Nat Rev Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data. 2016;3:160089. doi: 10.1038/sdata.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen M, Burgess JD, Ballard T, Serie D, Wang X, Younkin CS, et al. Gene expression, methylation and neuropathology correlations at progressive supranuclear palsy risk loci. Acta Neuropathol. 2016;132:197–211. doi: 10.1007/s00401-016-1576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–8. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–16. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–63. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genome-wide experiments. Proceeding of the National Academy of Sciences. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, et al. A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst. 2017;4:60–72. e4. doi: 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–50. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 26.Murray ME, Przybelski SA, Lesnick TG, Liesinger AM, Spychalla A, Zhang B, et al. Early Alzheimer’s disease neuropathology detected by proton MR spectroscopy. J Neurosci. 2014;34:16247–55. doi: 10.1523/JNEUROSCI.2027-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray ME, Vemuri P, Preboske GM, Murphy MC, Schweitzer KJ, Parisi JE, et al. A quantitative postmortem MRI design sensitive to white matter hyperintensity differences and their relationship with underlying pathology. J Neuropathol Exp Neurol. 2012;71:1113–22. doi: 10.1097/NEN.0b013e318277387e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watzlawik JO, Kahoud RJ, Wootla B, Painter MM, Warrington AE, Carey WA, et al. Antibody Binding Specificity for Kappa (Vkappa) Light Chain-containing Human (IgM) Antibodies: Polysialic Acid (PSA) Attached to NCAM as a Case Study. J Vis Exp. 2016 doi: 10.3791/54139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nature genetics. 2009;41:192–8. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gusareva ES, Carrasquillo MM, Bellenguez C, Cuyvers E, Colon S, Graff-Radford NR, et al. Genome-wide association interaction analysis for Alzheimer’s disease. Neurobiol Aging. 2014;35:2436–43. doi: 10.1016/j.neurobiolaging.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16:358–72. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- 32.Mhatre SD, Tsai CA, Rubin AJ, James ML, Andreasson KI. Microglial malfunction: the third rail in the development of Alzheimer’s disease. Trends Neurosci. 2015;38:621–36. doi: 10.1016/j.tins.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–68. doi: 10.1016/S1474-4422(16)00127-7. [DOI] [PubMed] [Google Scholar]
- 35.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haroutunian V, Katsel P, Roussos P, Davis KL, Altshuler LL, Bartzokis G. Myelination, oligodendrocytes, and serious mental illness. Glia. 2014;62:1856–77. doi: 10.1002/glia.22716. [DOI] [PubMed] [Google Scholar]
- 37.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihara M, Polvikoski TM, Hall R, Slade JY, Perry RH, Oakley AE, et al. Quantification of myelin loss in frontal lobe white matter in vascular dementia, Alzheimer’s disease, and dementia with Lewy bodies. Acta Neuropathol. 2010;119:579–89. doi: 10.1007/s00401-009-0635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitew S, Kirkcaldie MT, Halliday GM, Shepherd CE, Vickers JC, Dickson TC. Focal demyelination in Alzheimer’s disease and transgenic mouse models. Acta Neuropathol. 2010;119:567–77. doi: 10.1007/s00401-010-0657-2. [DOI] [PubMed] [Google Scholar]
- 40.Richter-Landsberg C. Protein aggregate formation in oligodendrocytes: tau and the cytoskeleton at the intersection of neuroprotection and neurodegeneration. Biol Chem. 2016;397:185–94. doi: 10.1515/hsz-2015-0157. [DOI] [PubMed] [Google Scholar]
- 41.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–82. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gold BT, Johnson NF, Powell DK, Smith CD. White matter integrity and vulnerability to Alzheimer’s disease: preliminary findings and future directions. Biochim Biophys Acta. 2012;1822:416–22. doi: 10.1016/j.bbadis.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, West JD, Flashman LA, Wishart HA, Santulli RB, Rabin LA, et al. Selective changes in white matter integrity in MCI and older adults with cognitive complaints. Biochim Biophys Acta. 2012;1822:423–30. doi: 10.1016/j.bbadis.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dean DC, 3rd, Hurley SA, Kecskemeti SR, O’Grady JP, Canda C, Davenport-Sis NJ, et al. Association of Amyloid Pathology With Myelin Alteration in Preclinical Alzheimer Disease. JAMA Neurol. 2017;74:41–9. doi: 10.1001/jamaneurol.2016.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jack CR, Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Mayo Clinic eGWAS quality control (QC) flowchart. Flowchart outlining QC procedures and parameters for WG-DASL temporal cortex (TCX) and cerebellum (CER) datasets. No sample exclusions were necessary after QC of WG-DASL expression data. All subjects in the Mayo Clinic eGWAS were also part of the Mayo Clinic LOAD GWAS, which was performed on a larger cohort (N=2465). WG-DASL = Whole Genome cDNA mediated Annealing, Selection, extension, and Ligation assay, PCA = Principal components analysis.
Supplementary Fig. 2. Mayo Clinic eGWAS principal components analysis. Scatter plot of principal components 1 and 2 for temporal cortex (A) and cerebellum (B) expression measures. Samples are colored according to WG-DASL plate.
Supplementary Fig.3: QQ plots for expression residuals of select genes: Data points are colored according to plate. All probes shown have detectable signal in >50% of the samples. These plots demonstrate that including plate as a covariate adequately controls for the batch effect of plate.
Supplementary Fig. 4. Brain cell-specific gene expression levels by diagnosis in the Discovery and Replication cohorts: Gene expression level residuals adjusted for covariates are plotted by diagnosis. Brain cell-specific expression levels for the genes utilized in the Comprehensive analytic model and the brain cells for which they are specific are as follows: ENO2 for neurons, GFAP for astrocytes, CD68 for microglia, OLIG2 for oligodendrocytes and CD34 for endothelial cells. Box plots depict the median (line through box); and bottom and top lines of the box are the 1st and 3rd quartiles, respectively. The bottom and top of the whiskers are 1.5 x inter-quartile range beyond the bottom and top of the boxes. The dots are outliers beyond this range. Box plots of gene expression levels for the different diagnoses are shown. Pairwise diagnostic comparisons that are significantly different are shown with a horizontal line above the significant comparison and an asterisk. Results are shown for the Discovery Cohort, temporal cortex (A); cerebellum (B); Replication Cohort, temporal cortex (C); and cerebellum (D).
Supplementary Fig. 5: Oxidative phosphorylation pathway from MetaCore (Thompson Reuters) where 29 genes in the pathway are expressed at significantly lower levels in the temporal cortex of AD subjects when compared with PSP. Downregulated genes are indicated with blue thermometers, where the height of the blue bar reflects the regression beta (see also Supplementary_Tables.4 and 11).
Supplementary Fig. 6. Pairwise correlations of brain gene expression levels for five brain cell-specific genes across diagnostic groups: Data are shown for the following five CNS cell-specific genes: ENO2 for neurons, GFAP for astrocytes, CD68 for microglia, OLIG2 for oligodendrocytes and CD34 for endothelial cells. These genes are utilized for performing adjustments in DGE analyses and in the WGCNA “Comprehensive Model” analyses. Residual gene levels after adjustment of covariates were plotted. All pairwise correlations for the levels of these genes measured in the temporal cortex of the Discovery Cohort (A–J); and the Replication Cohort (L–U) are shown. The correlations were conducted within the main diagnostic groups, separately. Pearson correlation coefficients and p-values are shown (K, V).
Supplementary Fig. 7. Human brain cell type-enriched genes cluster dendrogram. Log2FPKM expression measures of 1270 cell-type enriched genes, from isolated populations of cells were assessed. The dist function was used to calculate Euclidian distances between samples and the hclust function (method =“average”) to perform cluster analysis. Samples are labeled to indicate cell type (N=Neuron, A=Astrocyte, M=Microglia, E=Endothelia, O=Oligodendroglia), Age (Years) and Sex (F = Female, M= Male), and color coded to further differentiate cell types.
Supplementary Fig. 8. Human brain cell type-enriched genes correlation heatmap. Log2FPKM expression measures of 1270 cell-type enriched genes, from isolated populations of cells were assessed. The cor function was used to create a correlation matrix and the melt function (reshape2) was used to manipulate the data matrix for plotting using ggplot2. Samples are labeled to indicate cell type (N=Neuron, A=Astrocyte, M=Microglia, E=Endothelial, O=Oligodendroglia), Age (Years) and Sex (F = Female, M= Male).
Supplementary Fig. 9. Preservation of temporal cortex (TCX) co-expression modules (CEM) from subjects with AD and PSP (AD+PSP) under the Simple vs. Comprehensive Models, using Simple Model as the reference (Discovery Cohort): WGCNA “modulePreservation” function with 100 permutations was applied to calculate preservation statistic Zsummary for a specific module’s genes in comparison groups. Zsummary score (y-axis) vs. CEM size (x-Axis) are plotted. Typically, a Zsummary score >10 indicates that the module genes were well-preserved. All of the TCX CEM under the Simple Model are well preserved with those under the Comprehensive Model.
Supplementary Fig. 10. Preservation of TCX AD+PSP CEM under the Simple vs. Comprehensive Models, using Comprehensive Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9, except that CEM under the Comprehensive Model are used as the reference.
Supplementary Fig. 11. Overlap of transcripts in the TCX AD+PSP CEM under the Simple and Comprehensive Models (Discovery Cohort): Matrix for the number of overlapping transcripts in TCX CEM under the Simple (y-axis) and Comprehensive (x- axis) models. Modules with enrichment of brain cell-enriched transcripts are highlighted in yellow. Coloring palette to the right is according to the number of transcripts in overlapping modules.
Supplementary Fig. 12. Preservation of cerebellum (CER) AD+PSP CEM under the Simple vs. Comprehensive Models, using Simple Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 13. Preservation of CER AD+PSP CEM under the Simple vs. Comprehensive Models, using Comprehensive Model as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 14. Overlap of transcripts in the CER AD+PSP CEM under the Simple and Comprehensive Models (Discovery Cohort): Matrix for the number of overlapping transcripts in CER CEM under the Simple (y-axis) and Comprehensive (x-axis) models. Modules with enrichment of brain cell-enriched transcripts are highlighted in yellow. Coloring palette to the right is according to the number of transcripts in overlapping modules.
Supplementary Fig. 15. Preservation of CER vs. TCX AD+PSP CEM built under the Comprehensive Model, using CER as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 16. Preservation of CER vs. TCX AD+PSP CEM built under the Comprehensive Model, using TCX as the reference (Discovery Cohort): The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 17. Overlap of transcripts in the CER and TCX AD+PSP CEM under Comprehensive Model (Discovery Cohort): Matrix for the number of overlapping transcripts in the TCX CEM (y-axis) and CER CEM (x-axis) built in AD+PSP subjects under the Comprehensive Model. Red-colored cell indicates the most number of overlapping genes of the corresponding row; orange-colored cell indicates the second most; yellow-colored cell indicates the third most; the rest of the cells are white.
Supplementary Fig. 18. Mayo Clinic RNAseq QC flowchart.
Flowchart outlining quality control (QC) procedures and parameters for RNaseq TCX dataset. QC parameters and sample numbers excluded are indicated. Samples that passed both expression and genetic data QC were used for analysis. SD = standard deviation, PCA = Principal components analysis.
Supplementary Fig. 19. Mayo Clinic RNAseq Sex Check.
Mean expression (counts per million) for genes mapped to the Y chromosome were plotted for all TCX samples (N=278). Female samples cluster to the left side of the plot and males to the right. Two samples labeled as male were identified in the female cluster (red triangles) and excluded from further analysis.
Supplementary Fig. 20. Mayo Clinic RNAseq PCA plot figure legend.
Scatter plot of principal components 1 and 2 for temporal cortex RNAseq expression measures. CQN normalized gene counts, which are used in all downstream analyses, are plotted in this figure.
Supplementary Fig. 21. Mayo Clinic RNAseq GWAS heterozygosity.
Scatter plot for sample heterozygosity vs. genotype missingness rate across all subjects with genome-wide genotype data. Horizontal red lines indicate 3 standard deviations from the mean. Blue vertical lines indicate 0% and 1% missingness rates. Since more subjects than just those with TCX RNAseq measures underwent GWAS, this plot includes subjects both with and without TCX RNAseq measures. Subjects 127, 1930, 1169 and 11416 are those with TCX RNAseq that were excluded based on this QC metric.
Supplementary Fig. 22. Preservation of Replication (RNAseq) vs. Discovery (DASL) AD+PSP CEM built under the Simple Model, using RNAseq as the reference. The descriptions are as per Supplementary Fig. 9.
Supplementary Fig. 23. Overlap of transcripts in the Discovery (DASL) and Replication (RNAseq) AD+PSP CEM under the Simple Model. Matrix for the number of overlapping transcripts in the TCX Discovery Cohort CEM (y-axis) and Replication Cohort CEM (x-axis) built in AD+PSP subjects under the Simple Model. Red-colored cell indicates the most number of overlapping genes of the corresponding row; orange-colored cell indicates the second most; yellow-colored cell indicates the third most; the rest of the cells are white.
Supplementary Fig. 24. Western blot analyses of oligodendroglial network proteins: Temporal cortex levels of proteins from the oligodendroglial networks (PLP, CNP, MOG, PLLP, BACE1, PSEN1, MOBP) are shown. Other proteins that are integral to or implicated in myelin function (MBP), AD (APP, PSEN2, Tau), PSP (Appoptosin), astrocytes (GFAP), microglia (IBA-1), neurons (β3-tubulin) and GAPDH control are also measured. Each PSP and Control subject was unique. In addition 2 samples (Replicates) were measured on every gel to control for blot-to-blot variability. A–E depict the 5 sets of western blots for the complete protein validation cohort. F. Table depicting summary measures and results of multivariable linear regression analyses using all data.
Supplementary Fig. 25: Representative photomicrographs of immunohistochemically stained temporal cortex from a control (A–C), PSP (D–F), and AD brains (G–I). The MBP antibody was used to demonstrate differences in myelination across subjects (A, D, G). The Iba-1 antibody was used to label microglia with a noted difference in abundance and morphology. The Iba-1 stain was also used to evaluate the density of oligodendrocytes as their nuclei are round with compact chromatid labeled by the hematoxylin counterstain (B,E,H). The GFAP antibody was used to labels astrocytes cell bodies and processes (C, F, I).
Supplementary Fig. 26: Cellular characterization of primary rat oligodendrocyte-enriched cultures: Enriched wild-type rat oligodendrocyte cultures were grown for 4 days in B27-supplemented Neurobasal A and stained live on ice for 15 minutes with oligodendrocyte specific markers A2B5, O4 and O1. Cells were subsequently fixed, permeabilized, labeled for internal markers MBP, GFAP, IBA-1, β3tubulin and processed for immunocytochemistry including DAPI-staining for cell counting and to ensure cell integrity. Late stage oligodendrocyte marker MBP indicates presence of predominantly mature oligodendrocytes with flat “pancake-like” membrane sheaths (myelin sheaths), which are also detectable with markers O1 and O4. Cell counts of enriched OL cultures at day 4 by immunocytochemistry demonstrated ~80% O4-positive oligodendrocytes, 10% GFAP-positive astrocytes, 4% IBA-1-positive microglia and 6% b3tubulin-positive neurons compared to the total number of DAPI-positive cells.
Supplementary Fig. 27: Immunocytochemical patterns of oligodendrocyte and myelin proteins in rat primary oligodendrocyte-enriched cultures: Enriched wild type rat oligodendrocyte cultures were grown for 5 days in B27-supplemented Neurobasal A and stained live on ice for 20 minutes with oligodendrocyte-specific markers O4 (O4/MBP). Cells were subsequently fixed, permeabilized, labeled for internal markers MBP, GFAP, IBA-1, β3tubulin, CD68-like protein, Olig-2, CNP, PLLP, and MOG and processed for immunocytochemistry including DAPI-staining for cell counting and to ensure cell integrity (A–R). Columns A–P and B–Q represent single-labeled OL cultures whereas columns C–R represent the corresponding overlays including DAPI staining (e.g. C = overlay of A+B). Row A–C shows double-labeled cultures using oligodendrocyte-specific markers O4 (A) and MBP (B) respectively, and the overlay (C) of O4/MBP (green/red) plus nuclear marker DAPI (blue). Consecutive rows represent double labeled OL cultures for markers GFAP (astrocytes, green) and CNP (oligodendrocytes, red) plus DAPI (blue) (row D–F); Olig-2 (oligodendrocytes, green) and neuronal marker β3tubulin (red) plus DAPI (blue) (row G–I); microglial markers IBA-1 (green) and CD68-like marker (red) plus DAPI (blue) (row J–L). Rows M–O and P–R represent single-labeled cultures using oligodendrocyte-specific marker PLP (M, green) plus DAPI (blue) (row M–O) respectively, and PLLP (P, green) (oligodendrocytes) plus DAPI (blue) (row P–R).