Abstract
We here present a new way to engineer complex proteins toward multidimensional specifications, through a simple yet scalable directed evolution strategy. By robotically picking mammalian cells that are identified, under a microscope, to express proteins that simultaneously exhibit several specific properties, we can screen through hundreds of thousands of proteins in a library in a matter of a few hours, evaluating each along multiple performance axes. We demonstrate the power of this approach by identifying a novel genetically encoded fluorescent voltage indicator, simultaneously optimizing brightness and membrane localization of the protein using our microscopy-guided cell picking strategy. We produced the high-performance opsin-based fluorescent voltage reporter Archon1, and demonstrated its utility by imaging spiking and millivolt-scale subthreshold and synaptic activity in acute mouse brain slices as well as in larval zebrafish in vivo. We also demonstrate measurement of postsynaptic responses downstream of optogenetically controlled neurons in C. elegans.
Keywords: Protein engineering, robotics, directed molecular evolution, voltage imaging, mouse, C. elegans, optogenetics, zebrafish, synaptic transmission, subthreshold activity
INTRODUCTION
Directed molecular evolution is a powerful strategy for protein engineering1,2. Most strategies for screening libraries of mutated genes for improved phenotypes either exhibit high throughput but are limited to the detection of fluorescence2, or can extract multiple protein functional parameters but exhibit modest throughput or complexity of usage that challenges end user protein engineers3–7. In order to enable the simple and high throughput directed evolution of proteins so that multiple properties can each be simultaneously optimized toward specific goals, we developed a strategy for robotically picking mammalian cells that are identified, under a microscope, to express proteins with appropriate properties. Our strategy is inexpensive and easily used by protein engineers, and enables the assessment of hundreds of thousands of protein variants in a few hours.
We applied our strategy to the creation of genetically encoded fluorescent indicators of membrane potential8. An ideal genetically encoded fluorescent voltage indicator would localize well to the plasma membrane, be bright and exhibit high signal-to-noise ratio (SNR), exhibit large and linear fluorescent changes in response to voltage fluctuations, respond to changes in voltage rapidly enough to preserve the fidelity of spiking, exhibit stable (i.e., non-photobleaching) fluorescence over timescales appropriate for conducting a biological experiment, and be compatible with optogenetic control of neural activity9. To date it has remained difficult to simultaneously optimize all of these properties – perhaps because directed evolution methods that select for one property may de-optimize other properties10, and also the optimization may need to be done in mammalian cells to ensure high performance in neurons3,11.
There are two classes of genetically encoded voltage reporters8 – one uses the fluorescence of microbial opsins (e.g., archaerhodopsin-3 (Arch)-based fluorescent voltage reporters12,13) to report neural activity, and the other couples a GFP-like fluorescent protein to a voltage sensing membrane protein (e.g., a voltage-sensitive phosphatase14 or an opsin15,16). Fluorescent opsins have been relatively dim, and suffer from poor localization, and thus exhibit low SNR (Supplementary Tables 1, 2). GFP-like fluorescent protein-containing reporters are brighter, but have to date exhibited small fractional changes in fluorescence, photobleach rapidly, and are incompatible with optogenetic control due to spectral overlap (Supplementary Tables 1, 2). We used robotic cell picking to screen mammalian cells expressing individual members of a library of fluorescent voltage sensor candidates, based upon brightness, localization, and photostability. The result, the fluorescent voltage sensor Archon1 (so named because it is based on Arch, with 13 point mutations), exhibits excellent localization, SNR, sensitivity, response speed, photostability, and full compatibility with optogenetic control. It is also severalfold brighter than previous opsin-based fluorescent voltage reporters that are red light sensitive. We demonstrate the utility of Archon1 in multiple species by imaging subthreshold (e.g., ~5 mV) synaptic responses in mouse cortical brain slices, high speed spiking and subthreshold activity in larval zebrafish brain, and neural responses downstream of optogenetically controlled neurons in C. elegans.
RESULTS
Robotic multidimensional directed evolution of proteins
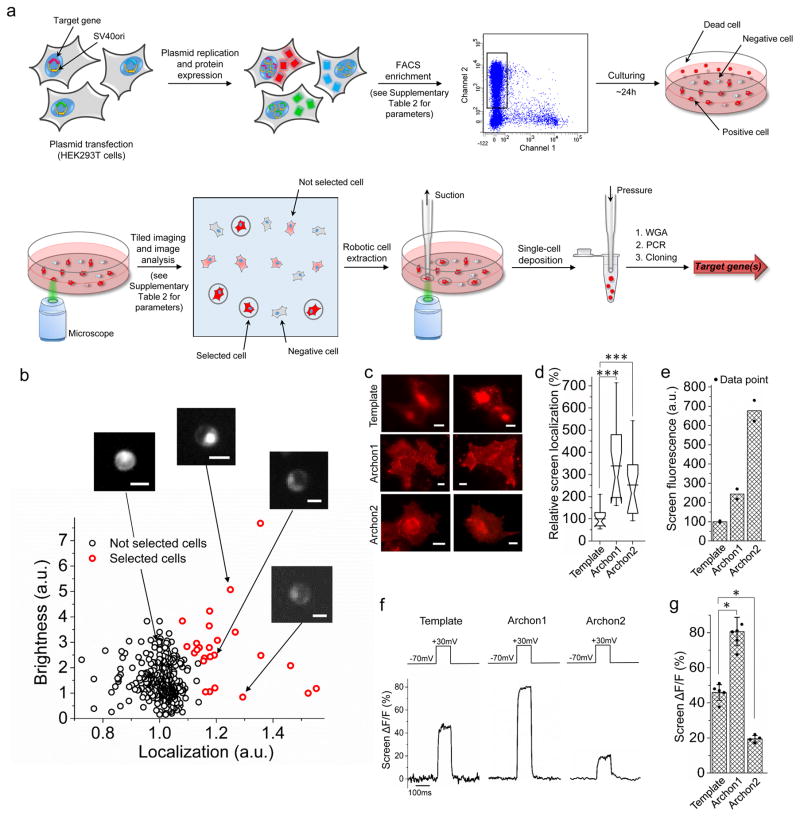
In order to be able to screen large numbers of mutant genes along multiple parameters, we combined microscopy and image analysis with robotic cell picking (Fig. 1a). We adapted a computer-vision-guided automated micropipette capable of controlled suction and positive pressure, and thus the isolation of single cells (see Methods)17,18, so that we could screen 300,000 cells expressing different constructs in ~4 hours. We transfected a gene library into HEK293T cells so that transfected cells would receive from one to four plasmids per cell (details of characterization in Supplementary Fig. 1), eliminated dead and nontransfected cells using FACS, and then performed multiple rounds of microscopy-guided cell picking to examine multiple parameters (e.g., brightness, membrane localization; Supplementary Fig. 2).
Figure 1. Multi-parameter directed evolution of proteins in mammalian cells via robotic cell picking.
(a) Pipeline for multi-parameter directed evolution of proteins in mammalian cells using robotic cell picking. Abbreviations: FACS, fluorescence-activating cell sorting (FACS); WGA, whole-genome amplification. (b) Example data and analyses reflecting the quantitative metrics used in the cell-picker step during the second round of directed evolution, for simultaneous optimization of brightness and localization. Scale bar: 10 μm. (c) Representative fluorescence images of HEK293T cells expressing the template, Archon1 and Archon2 (n = 15, 16, and 16 cells for Archon1, Archon2, and the template respectively). Dynamic ranges for the images were normalized to facilitate visual comparison. Scale bars, 5μm. Imaging conditions: 62 mW/mm2, λex = 628/31BP (bandpass, used throughout) from an LED, λem= 664LP (longpass, used throughout) used in (c,d). (d) Relative membrane localization of the indicators of c in HEK293T cells (n = 15, 16, and 16 cells for Archon1, Archon2, and the template respectively, each from 2 independent transfections; ***P < 0.0001 for Archon1 and ***P = 0.0003 for Archon2, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test with the template as control group). Box plots with notches are used throughout this paper, when n > 6, as recommended by ref40 (narrow part of notch, median; top and bottom of the notch, 95% confidence interval for the median; top and bottom horizontal lines, 25% and 75% percentiles for the data; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; horizontal line, mean). (e) FACS mean fluorescence intensity for sets of live HEK293T cells expressing these indicators (n = 2 transfected samples, each; individual data points in black dots). (f) Representative fluorescence changes for these indicators with a 100 mV voltage step, measured in HEK293T cells. Imaging conditions: λex = 637nm laser light, λem= 664LP, 800 mW/mm2 used for the template and 80–800 mW/mm2 used for Archons in (f,g), with light intensity adjusted to prevent signal saturation. (g) Population data of fluorescence changes, as in f, for these indicators (n=5, 6, and 4 cells for the template, Archon1, and Archon2, each from 2 independent transfections; individual data points in black dots; error bars, standard deviation; *P = 0.0155 for Archon1 and *P = 0.0374 for Archon2, Kruskal-Wallis analysis of variance followed by post-hoc Steel’s test with the template as control group), taken in the steady state.
To validate the overall workflow, we performed three rounds of directed molecular evolution to develop a monomeric near-infrared fluorescent protein (FP) from the RpBphP1 bacteriophytochrome19 (see Supplementary Table 3 for screening parameters; screening progress is described in Supplementary Fig. 3). The final selected variant, named miRFP, had absorbance and emission maxima at 674 nm and 703 nm, respectively, and demonstrated monomeric state both in vitro and in cultured mammalian cells (Supplementary Figs. 4–6). Furthermore, miRFP exhibited higher molecular brightness than previously developed, spectrally similar near-infrared FPs (Supplementary Table 4) and could be readily expressed in neurons in culture and in vivo and imaged using both one- and two-photon microscopy (Supplementary Figs. 7).
Multidimensional screening of genetically encoded voltage indicators
We next turned to multidimensional screening for a high-performance fluorescent voltage sensor. To obtain a molecule compatible with optogenetic control, we began with a template with red fluorescence (since optogenetic controllers are sensitive to blue light, ideally we would have a voltage reporter that would be illuminated by orange or red light). We began with the opsin core of the previously reported voltage sensor QuasAr2, with a fluorescence excitation maxima at 590 nm12. For the first round of directed molecular evolution, we generated a gene library with error-prone PCR and cloned it into the expression vector. After expression of the library in HEK cells for 48 hours, we used FACS to remove non-transfected cells and cells expressing non-fluorescent (and thus non-functional) mutants, which was >99.9% of the entire population (Supplementary Fig. 8). We then performed microscopy-guided cell picking to screen for cells containing genes whose products exhibited exemplary brightness and membrane localization, simultaneously (see Supplementary Table 3 for screen imaging parameters). We also assessed, in a subset of these cells, fluorescence photostability by taking time-lapse images under continuous illumination, but found that the variants selected already had great photostability, and as measuring photostability is time-consuming, we halted this specific part of the analysis. Selected cells were those exhibiting high-performing combinations of membrane localization and fluorescence brightness along the Pareto frontier20 (i.e., the set of cells that dominate other cells in at least one parameter), as previously used to computationally evaluate progress in the context of multiobjective evolutionary optimisation21–23 (Fig. 1b; see Methods for details of the step-by-step Pareto frontier algorithm). These genes were then rescreened for brightness, membrane localization, and voltage sensitivity (see Methods), yielding a quantitative three-parameter metric. We calculated the product of these three measured parameters (each normalized to its maximum value, to ensure the roughly equal contribution of each parameter to the product), and then the five variants with the highest such products were selected for sequencing. Sequence analysis revealed four amino acid positions, namely T56, T117, T183 and L198, that were changed in four out of the five variants, and we also identified four amino acid mutations in α-helices (T20S, L31V, K47R, and A137T) and two mutations in β-strands (S80P and D88N) which were represented at least once in the selected mutants (Supplementary Fig. 9).
For the second round of directed molecular evolution, we generated a site-directed library of variants containing mutations identified in our first round, as well as mutations near the Schiff base linkage (some of which had been previously explored13,24). We repeated the screen as in the first round, including the assessment of a subset of the clones for photostability. Since the method of electrical stimulation used in the screen was not very quantitative25,26, we characterized the top 21 variants via whole-cell patch clamp and concurrent imaging. Discarding mutants with kinetics (i.e., τon or τoff) slower than 10 ms, and using the product of normalized brightness, localization and voltage sensitivity as the final ranking criterion, we obtained seven final candidates with improved brightness and membrane localization, of which two exhibited higher voltage sensitivity (Supplementary Figs. 10, 11). Prioritizing localization as the key parameter, with brightness and voltage sensitivity as secondary parameters, we chose two molecules – denoted Archon1 and Archon2 - for further investigation. Archon1 and Archon2 localized well on the plasma membrane of HEK cells (Fig. 1c,d), and exhibited 2.4- and 6.8-fold increased brightness, respectively, over the parental template (Fig. 1e). Fluorescence changes (ΔF/F) in HEK cells for Archon1 and Archon2 over 100 mV voltage swings were 81 ± 8% and 20 ± 2%, respectively, compared to 46 ± 4% for the template (mean ± standard deviation throughout; Fig. 1f,g). We investigated the contribution of specific point mutations to changes in localization, brightness, voltage sensitivity, and kinetics, and found the patterns that emerged to be complex (Supplementary Table 6), with a given mutation often improving one parameter but worsening another. Thus, simultaneous multidimensional hierarchical screening may be key for generating practical fluorescent voltage indicators that excel along multiple axes at once.
Biophysical properties of Archons
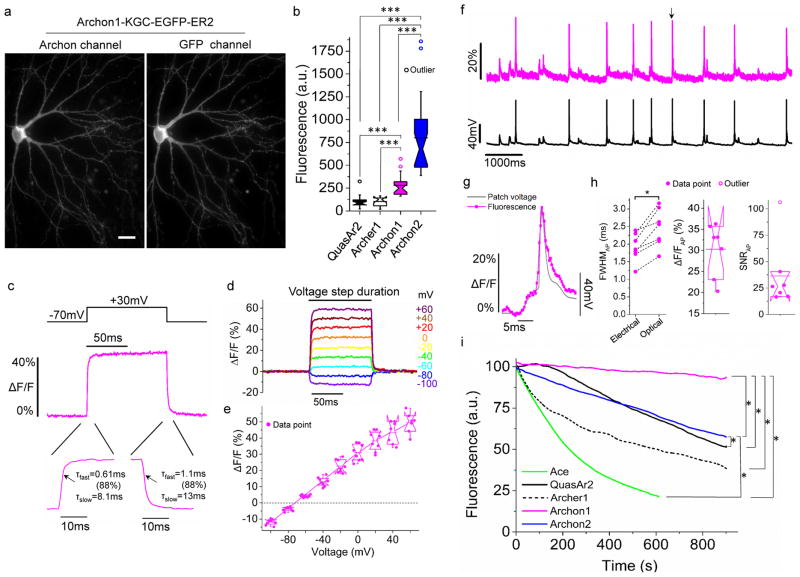
We fused Archons with EGFP for visualization, and Golgi export trafficking signal (KGC) and endoplasmic reticulum export (ER) sequences27,28 (Fig. 2a) which are widely used to help archaeal opsins express better in vivo (although Archon1 had excellent membrane localization in cultured neurons without them; Supplementary Fig. 12). Archons demonstrated excellent membrane localization in cultured mouse neurons (see Supplementary Fig. 13 for images of neurons expressing other sensors, under the same promoter and imaged after the same time period). Focusing on the Arch-derived voltage sensors (QuasAr2-mOrange and Archer1-EGFP, abbreviated as QuasAr2 and Archer1 in Fig. 2), we found greater brightness for Archons over earlier molecules (Fig. 2b) in cultured neurons. Archon expression did not alter membrane resistance, membrane capacitance, or resting potential of cultured neurons (Supplementary Fig. 14).
Figure 2. Characterization of Archons in cultured cells.
(a) Representative fluorescent images of Archon1 (left, excitation (λex) with 637 nm laser light, emission (λem) at 664LP) and GFP (right, λex = 475/34BP from an LED and λem = 527/50BP) channels in a cultured mouse hippocampal neuron (n = 32 cells from 5 independent transfections). Scale bar: 10 μm. (b) Relative fluorescence of QuasAr2, Archer1, Archon1, and Archon2 in cultured neurons (n = 18, 16, 23, and 23 cells respectively, from 4 independent transfections each, from one culture; λex = 637nm laser light at 800 mW/mm2 and λem = 664LP for Fig. 2c–g; ***P < 0.0001, Kruskal–Wallis analysis of variance followed by post-hoc Steel-Dwass test on each pair; see Supplementary Table 5 for full statistics for Fig. 2). Box plots with notches are used (see caption of Fig. 1d for description). Open circles represent outliers, data points which are less than 25th percentile or greater than 75th percentile by more than 1.5 times the interquartile range. (c) Representative fluorescence response of Archon1 in a cultured neuron, to a 100 mV change delivered in voltage-clamp. τfast and τslow indicate time constants with the fluorescence trace fit according to , with the % indicating A/(A+B). Image acquisition rate: 3.2 kHz. (d) Representative fluorescence traces of Archon1 in response to a series of voltage steps in voltage-clamp mode. Image acquisition rate: 2.3 kHz. (e) Population data corresponding to the experiment of d (n = 8 neurons from 3 cultures). Data was normalized so that −70 mV was set to 0 ΔF/F. (f) Single-trial optical recording of Archon1 fluorescence responses (magenta) during spontaneous activity, with concurrent current clamp trace (black), for a cultured hippocampal neuron. Peak marked with arrow is zoomed-in in (g). Image acquisition rate: 2.3 kHz. (g) Zoomed-in view of peak marked with arrow in (f), scaled to match peaks. (h) Quantification of electrical and optical full width at half maximum (FWHM; dashed lines connect data points from same neuron), ΔF/F, and signal-to-noise ratio (SNR), per action potential (AP) across all recordings (n = 160 APs from 7 neurons from 5 cultures). *P = 0.0156, Wilcoxon signed-rank test. (i) Photobleaching curves of Ace, QuasAr2, Archer1, Archon1 and Archon2 under continuous illumination (n= 5, 7, 5, 9, and 7 neurons from 1, 1, 1, 2, and 2 cultures, respectively; 475/34BP from an LED at 13 mW/mm2 for Ace2N-4aa-mNeon, 637nm laser light at 2.2W/mm2 for QuasAr2 and Archer1, 637nm laser light at 800mW/mm2 for Archon1 and Archon2; light intensity was adjusted to have the same initial signal-to-noise ratio (SNR) of action potentials, e.g. 25±8, 26±12, 26±10, 26±10 and 28±7 for Quasar2, Archer1, Archon1, Archon2 and Ace2N-4aa-mNeon respectively; image acquisition rate: 333Hz); *P = 0.0184 for Archon1 and Archon2, Archon1 and QuasAr2, and Archon2 and QuasAr2; *P = 0.0456 for Archon1 and Archer1, Archon1 and Ace, and Archon2 and Ace; Kruskal–Wallis analysis of variance of bleaching time followed by post-hoc Steel-Dwass test on each pair).
When we expressed Archon2 in mouse brain slices, we obtained lower SNR and membrane localization than with Archon1 (see below); therefore we focus on Archon1 in the main text (but we include the Archon2 data in supplementary figures). In cultured neurons, we found that Archon1 exhibited a ΔF/F of 43 ± 5% (Fig. 2c–e) for a 100 mV deflection, with a linear voltage dependence (Fig. 2e; compare to other indicators described in Supplementary Table 1). The speed of response of Archon1 was fast, with a biexponential response to a 100 mV voltage step with time constants of onset of 0.61 ± 0.06 ms (88% of total amplitude) and 8.1 ± 0.5 ms (remaining amplitude), and a time constant of inactivation of 1.1 ± 0.2 ms (88% of total amplitude) and 13 ± 3 ms (remaining amplitude) (Fig. 2c). Archon1 fluorescence was able to follow small, high-speed changes in voltage in cultured neurons, including few-millivolt voltage transients, as well as action potentials, with the latter broadening by a few hundred microseconds in waveform duration, and for which Archon1 exhibited a ΔF/F of 30 ± 6% and SNR of 36 (Fig. 2f–h, Supplementary Fig. 15). Archon2 exhibited faster kinetics but lower voltage sensitivity than Archon1 (Supplementary Fig. 16 and Supplementary Table 1). Both Archons demonstrated linear dependence of fluorescence intensity vs. 637 nm excitation light power, which suggests that fluorescence was a single-photon process (Supplementary Fig. 16h).
Photobleaching limits the impact of voltage imaging in neuroscience, since signal decreases result in signal eventually blending in with noise. We excited Archons with 800mW/mm2 637 nm light for 900 seconds, and found that Archon1 retained 95 ± 16% of its baseline fluorescence (Fig. 2i), far better retention of fluorescence than Archon2, QuasAr2, Archer1 and Ace (Supplementary Table 1). Thus, Archon1 may be able to support voltage imaging over timescales relevant to behavior.
As Archons are derived from Arch, a light-driven proton pump, we characterized their responses to illumination with 470/20 nm light at 15 mW/mm2 (as used to image EGFP) and 637 nm at 800 mW/mm2 (as used for voltage imaging). Under all tested wavelengths, Archons showed no steady state photocurrent (Supplementary Fig 17). Under repetitive pulses of blue illumination, the first pulse generated a transient photocurrent of −8 ± 6 pA for a few milliseconds (n = 8 cells from one culture), as did subsequent pulses (Supplementary Fig. 17). Under repetitive pulses of red illumination, Archon1 showed a brief (<5 ms) transient photocurrent of −33 ± 25 pA, while subsequent pulses of red light produced no photocurrent (Supplementary Fig. 17). Archon2 showed no photocurrents under any condition (Supplementary Fig. 17). Buoyed by these results, we measured changes in the red fluorescence of Archons under blue light intensities typically used for optogenetic control4; in particular, under blue light at 4.8 mW/mm2 and with red illumination as above, Archons showed <2% changes in fluorescence (Supplementary Figs. 15, 16) This property allowed us to use Archon1 in conjunction with the channelrhodopsin CoChR4,29 for all-optical interrogation of neurons, driving and monitoring action potentials with light (Supplementary Fig. 18). Using Archons, one could image voltage fluctuations in neuronal processes in culture, and even single dendritic spines without averaging (Supplementary Fig. 19).
Use of Archons for synaptic and spiking imaging in intact mouse cortical slices
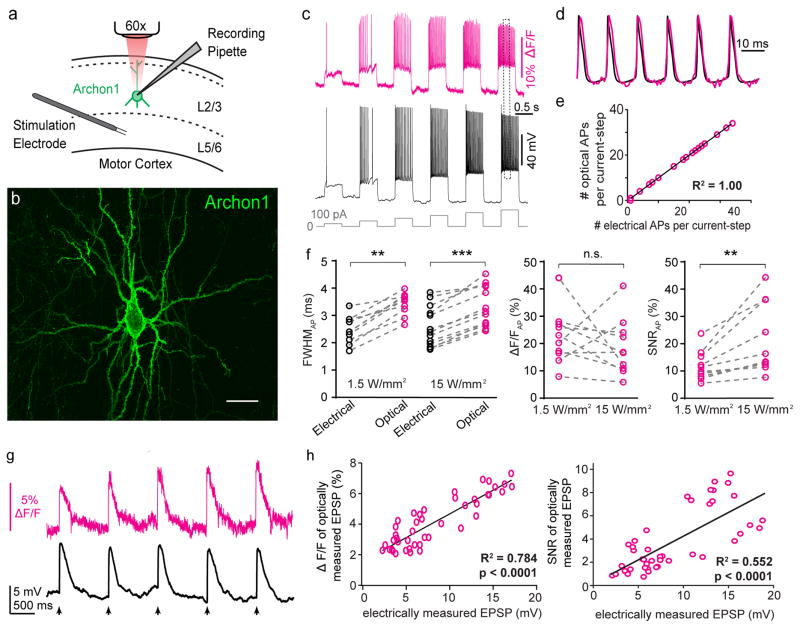
We expressed Archons in cortical pyramidal neurons via in utero electroporation (IUE; Supplementary Fig. 20). We performed voltage-clamp recordings from L2/3 pyramidal neurons in acute brain slices from 3–4 week old mice, and simultaneously monitored Archon1 fluorescence at the cell body (Fig. 3a,b). Archon1 expressed in vivo showed good membrane localization in cell bodies and processes without aggregation, and illumination with red light did not alter membrane properties (Fig. 3b and Supplementary Figs. 20–22), whereas QuasAr2 and Archer1 showed aggregation in cell bodies and processes (Supplementary Fig. 23). We used a series of voltage-steps in voltage-clamp mode to test the ability of Archons to report membrane voltage. Stepping the holding-potential (Vm) from −90 to +10 mV resulted in step-like fluorescent signals for Archon1 (Supplementary Fig. 24; Archon1: ΔF/F per 100 mV: 23.5 ± 9.3%). The on- and off-kinetics of Archons were well described by a double-exponential function, and reached steady state within a few ms (Supplementary Fig. 24). These data suggested that Archon1 should be sensitive enough to report subthreshold voltage events and fast enough to report individual action potentials in acute brain slices. To test this, we injected a series of 2 ms current pulses with increasing amplitudes, while monitoring membrane potential and fluorescent signals. Archon1 allowed reliable detection of voltage transients from both subthreshold depolarization and action potentials (APs) under 1.5 and 15 W/mm2 of excitation light (Supplementary Fig. 24). The high temporal precision and voltage sensitivity of Archon1 observed in neuron culture was borne out as excellent action potential reporting fidelity in mouse cortical neurons in brain slice (Fig. 3c–e, Supplementary Fig. 24), with excellent kinetics, dynamic range, and signal to noise when action potentials were imaged in mouse brain slices (Fig. 3f). Archon1 faithfully reported action potentials, even when elicited at the highest frequencies tested. Archon2 exhibited qualitatively similar functionality, but with reduced voltage sensitivity and SNR (Supplementary Fig. 25). Indeed, using Archon1, it was possible to image synaptic events of millivolt scale depolarization amplitude, triggered in layer 2/3 with layer 5 stimulation, with excellent signal quality (Fig. 3g). Voltage deflections of ~5 mV could be observed as ~2–3% fluorescence changes (Fig. 3h, left), and synaptic events this small could be imaged with signal to noise ratio of 2 or greater (Fig. 3h, right).
Figure 3. Millivolt-scale imaging of neural voltage in intact brain slices.
(a) Schematic of experimental recording configuration. Archon1-expressing pyramidal neurons in layer (L) 2/3 of motor cortex were targeted by patch-clamp recording, and Archon fluorescence at the soma was imaged at 1 kHz. Excitation intensity was ~7 mW over the area of the soma (i.e., ~15 W/mm2 at 637 nm, but 10x lower intensity, 1.5 W/mm2 at 637nm, was used in Fig. 3f for comparison). A bipolar stimulation electrode was in some experiments placed in L5 to trigger excitatory synaptic events in Archon1-expressing L2/3 pyramidal neurons. (b) Representative image of Archon1 expressing neuron in L2/3 of mouse motor cortex (n = 70 slices from 3 mice). Scale bar: 25 μm. (c) A series of 500 ms current steps with increasing amplitudes (from 100 to 600 pA, in 100pA increments; gray line) were injected through the recording pipette, resulting in action potentials of varying frequency. Magenta, imaged trace; black, simultaneous whole-cell patch-clamp in current clamp mode. (d) Zoomed-in view of APs marked with dotted box in c, scaled to match peaks. (e) Graph of the number of optically detected APs vs. the number of electrically detected APs for every 500 ms-long current injection across all cells that underwent the experiment of c (n = 22 steps from 5 neurons); straight line indicates linear regression. (f) Quantification of electrical and optical full width at half maximum (FWHM, left), ΔF/F (middle), and SNR (right) for action potentials; n = 10 neurons from 6 mice; means are plotted for each cell; dashed lines connect data points from same neuron; Wilcoxon signed-rank test: **P = 0.002 and ***P = 0.0002 for FWHM, P = 0.375 (not significant, n.s.) for ΔF/F, **P = 0.002 for SNR, see Supplementary Table 5 for full statistics for Fig. 3. (g) Representative optical (magenta) and electrical (black) signals from electrically evoked excitatory postsynaptic potentials (EPSPs). Arrows indicate times of stimulation (5 stimuli at 1 Hz, followed by inter-trial intervals of >30 seconds). (h) Population data of ΔF/F (left) and SNR (right) from individual EPSPs as in (g) across all cells (n = 45 EPSPs from 4 neurons from 2 mice); straight line indicates linear regression.
Use of Archons for synaptic and spiking imaging in living animal brains
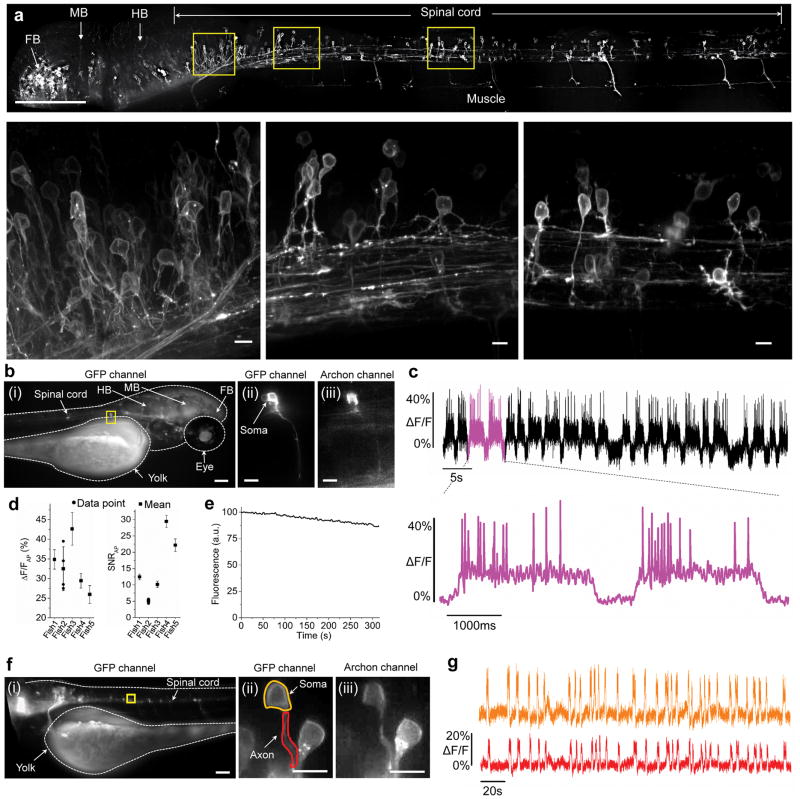
We next explored the use of Archon1 in multiple in vivo contexts. Zebrafish (Danio rerio) is a species important for the study of the development and operation of the nervous system30–34. We transiently expressed a zebrafish codon-optimized version of Archon1 (zArchon1) fused to EGFP in a subset of neurons. zArchon1, expressed in zebrafish larvae, demonstrated excellent membrane localization (Fig. 4a). zArchon1 reported action potentials with large fluorescence changes from baseline to peak (Fig. 4b,c), with excellent voltage sensitivity and SNR in larval zebrafish neurons (Fig. 4d). Notably, our voltage sensitivity and SNR are severalfold higher than those found for earlier voltage reporters in other intact neural systems (Supplementary Table 2). We assessed the photobleaching properties of zArchon1 in zebrafish larvae by applying the same illumination condition used for voltage imaging. The fluorescence of zArchon1 declined to 84 ± 8% of baseline fluorescence over 300 seconds (Fig. 4e), or 0.05%/s in zebrafish in vivo (vs. 0.01%/s in cultured mouse neurons). We could observe putative subthreshold (i.e., smaller than the amplitude of spikes) events (Supplementary Fig. 26), as well as voltage recording from neuronal processes (Fig. 4f,g). As expected from the aforementioned conclusions, imaging of responses was stable over timescales of many minutes (Supplementary Fig. 27).
Figure 4. Voltage imaging of Archon1-expressing neurons in larval zebrafish.
a Representative fluorescence image (top; GFP channel: excitation (λex) at 465 nm laser light, emission (λem) at 527/50BP) of neurons expressing zArchon1-EGFP in the spinal cord of a zebrafish larva at 3 days post fertilization (dpf) (n = 4 fish). Yellow boxes indicate neurons zoomed-in in the bottom panels. Scale bar, 125 μm. FB, forebrain; MB, midbrain; HB, hindbrain. (Bottom) From left to right: high magnification images of the neurons highlighted in the yellow boxes in the top panel. Scale bar, 5 μm. (b) Representative image (i, λex = 475/34BP from an LED, λem = 527/50BP) of neurons expressing zArchon1-GFP in the spinal cord of a 4 dpf zebrafish larva immobilized in agarose under wide-field microscopy (n = 5 fish). The yellow box indicates a neuron zoomed in in later panels. Scale bar: 100 μm. (ii) High magnification image of the neuron highlighted in the yellow box of (i) in the GFP channel. Scale bar: 10μm. (iii) As in ii, but in the Archon (λex = 637 nm laser light, λem = 664LP) channel. (c) Representative fluorescence trace (top) of zArchon1 reporting spontaneous activity of the neuron shown in b (at soma; λex = 637 nm laser light at 2.2 W/mm2, λem = 664LP; image acquisition rate: 500 Hz; n = 5 neurons from 5 fish). Bottom, the section of b highlighted in magenta, shown at an expanded time scale. (d) Population data of fluorescence changes and signal-to-noise ratios of zArchon1 during action potentials (APs; n = 21, 4, 132, 71 and 58 action potentials for fish 1–5, respectively; plotted is mean and standard deviation for each fish; raw data points are shown for fish with n <10 APs). (e) Photobleaching of zArchon1 fluorescence measured in in vivo in zebrafish larvae (n = 11 neurons from 6 fish) over 300 s of continuous illumination at 2.2 W/mm2. (f) Representative image (in the GFP channel) of neurons expressing zArchon1 in the spinal cord of a zebrafish larva at 4 dpf immobilized in agarose under wide-field microscopy (n = 3 fish). A yellow box indicates neurons zoomed in in later panels. Scale bar: 100μm. (ii) High magnification image of the neurons highlighted in the yellow box of (i) in the GFP channel. Scale bar: 10um. Highlighted regions indicate the soma (yellow) and the axon (red) of the neuron of interest. (iii) As in ii, but in the Archon channel. (g) Representative fluorescence trace of zArchon1 reporting spontaneous activity at the soma and the axon of the neuron shown in f (n = 3 neurons from 3 fish). The traces were acquired at the soma (yellow) and the axon (red) of the neuron (λex = 637 nm laser light at 2.2 W/mm2, λem = 664/LP, image acquisition rate: 250 Hz).
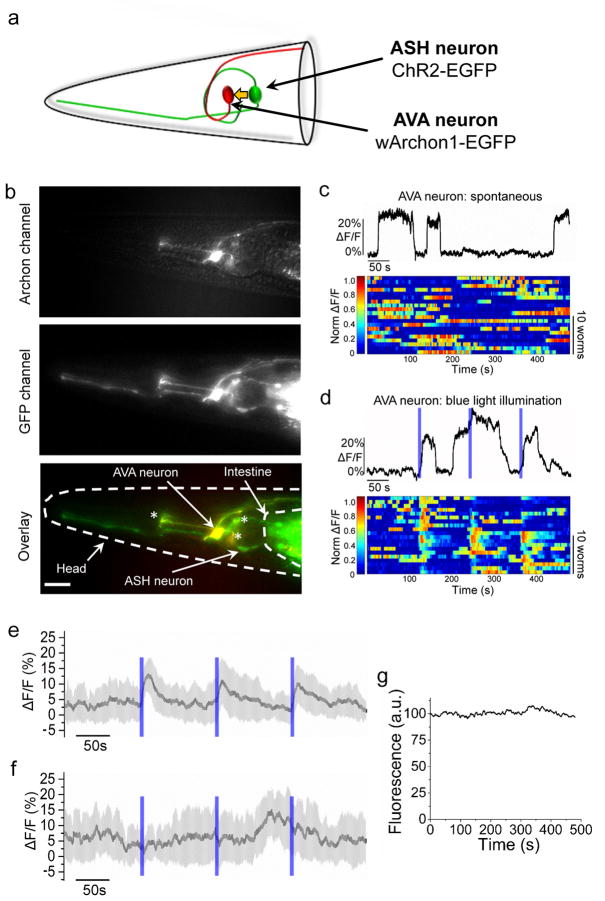
Finally, we explored the usage of Archon1 in the nematode C. elegans, a popular model organism in neuroscience. We used the rig-3 promoter to drive expression of codon-optimized Archon1 (wArchon1) fused to EGFP in AVA interneurons, involved in backward locomotion, while simultaneously expressing the blue light driven optogenetic controller channelrhodopsin-2 (ChR2) in an upstream neuron, the ASH neuron, under control of the sra-6 promoter (Fig. 5a). wArchon1 demonstrated good membrane localization both at the soma and in individual axons of AVA neurons (Fig. 5b and Supplementary Fig. 28). AVA neurons, when imaged at points at the soma or along the axon, exhibited long-lasting (tens of seconds to several minutes) high and low states similar to those previously reported in AVA calcium recordings35 (Fig. 5c), with changes in fluorescence intensity relative to baseline of magnitude of ~20–25% and SNR of ~25–35 (although the diversity of these fluctuations (Fig. 5c), in contrast to the stereotyped action potentials of vertebrate neurons, makes it difficult to arrive at a single number; n = 20 worms). The absence of blue light crosstalk with Archon function allows for combining voltage imaging using wArchon1 with optogenetic control using opsins. When blue light pulses were delivered, 51 out of 60 blue light pulses resulted in sustained elevation of wArchon1 fluorescence in AVA neurons lasting for 38±13 seconds (Fig. 5d,e; ΔF/F of ~16–21%, ~20–28 SNR; n=20 worms). In contrast, when no ChR2 was present, no effect of blue light was seen (Fig. 5f and Supplementary Fig. 29). Finally, wArchon1 exhibited essentially zero photobleaching under 8 minutes of continuous excitation with powers similar to those used in the voltage imaging experiments (Fig. 5g), thus supporting recordings of neural activity over behaviorally relevant time scales.
Figure 5. All-optical electrophysiology in C. elegans.
(a) Schematic of AVA neuron expressing wArchon1-EGFP (red) and ASH neuron expressing ChR2-EGFP (green), in the head of C. elegans. A yellow arrow indicates synaptic connection from ASH onto AVA. (b) Fluorescence images of the C. elegans head expressing wArchon1-EGFP in an AVA neuron (under rig-3 promoter) and ChR2-GFP (under sra-6 promoter) in the ASH neuron (top, Archon channel; middle, GFP channel; bottom, overlay), as well as pharyngeal neurons that express wArchon1-EGFP under control of the rig-3 promoter (asterisks; n = 20 worms). Scale bar: 20 μm. (c; top) A representative trace of wArchon1 fluorescence reporting spontaneous activity in the soma of an AVA neuron (n = 20 cells from 20 worms). (Bottom) Individual traces of wArchon1 fluorescence reporting spontaneous activity in an AVA neuron (n=20 neurons in 20 worms). (d; top) A representative trace of wArchon1 fluorescence in soma of an AVA neuron under three pulses of blue light stimulation (0.2 mW/mm2, λex=475/34BP light from an LED, 6 s; blue bars). (Bottom) Individual traces of wArchon1 fluorescence in an AVA neuron under blue light illumination (n=20 neurons in 20 worms). (e) Averaged wArchon1 fluorescence changes for traces presented in panel d. Shaded area is standard deviation. (f) Averaged wArchon1 fluorescence changes for traces recorded under same conditions as in panel d using the worms expressing only wArchon1-EGFP in AVA neurons. Shaded area is standard deviation. (g) Photobleaching curve of wArchon1 expressed in AVA neurons under continuous 637 nm excitation illumination (n = 10 cells from 10 worms, λex = 637 nm laser light at 800 mW/mm2, λem = 664LP).
DISCUSSION
Here we present a methodology for multidimensional directed evolution of proteins via high-throughput screening of large gene libraries in mammalian cells – namely, microscopy-guided robotic cell picking. We show that robotic cell picking with single cell precision can be used for directed evolution of proteins, assembling such a system out of commercially available parts that can be installed on any microscope equipped with a motorized stage17, and optimizing the software to enable single cell picking according to modularly incorporated image processing algorithms. Since robotic cell screening and selection can be performed on adherent cultured mammalian cells, it enables straightforward interrogation of multiple properties of expressed proteins with easily adjustable spatial and temporal resolutions. These features of our cell picker will enable it to be deployed easily into a wide variety of protein engineering contexts, without requiring the custom fabrication involved with other strategies such as microfluidics5–7 or laser-released micropallets36. For example, microfluidic devices require custom microfabrication, and can require redesign and reimplementation when screening assays or criteria are changed, which in turn can be challenging for many protein engineers7,37. Here, by choosing a strategy that essentially automates the manual steps done by a molecular engineer, and by implementing it with off-the-shelf parts that are easy to set up, and easily customized software, we aim to not only enable powerful multidimensional screens but to democratize the process of performing them.
We used our multidimensional screening approach to generate the opsin-based fluorescent voltage reporter Archon1, which exhibits good membrane localization in neurons of multiple species (mouse, C. elegans, zebrafish), severalfold improved brightness over previous opsin-based reporters, severalfold improvements in voltage sensitivity to single APs and in photobleaching over GFP-based reporters, and compatibility with optogenetic control. Our custom cell-picker code allowed us to select cells based on microscopy-obtained quantitative metrics for multiple parameters (i.e., brightness, localization) on multiple cells within a single screening session. The obtained quantitative metrics were then used to identify cells (and variants) possessing an optimal combination of multiple parameters along the Pareto frontier (for our two-dimensional evaluation of brightness and localization) or simply by ranking molecules by the product of normalized parameters (for our three-dimensional evaluation of brightness, localization, and voltage sensitivity) – both attempts to give roughly equal priority to each parameter, so that the cells picked reflected molecules simultaneously optimized along multiple axes at once. As a result of this simultaneity, the final variant, Archon1, exhibited improved characteristics for each property selected for, over its precursor template. We demonstrated the utility of Archon1 by imaging single spikes and millivolt-scale subthreshold or synaptic activity in acute mouse brain slices and larval zebrafish in vivo, as well as postsynaptic responses downstream of optogenetic control in C. elegans. The ability to survey neural activity in such well-defined systems, e.g., brain circuits from the mouse, or entire transparent organisms, may greatly synergize with new strategies that allow for mapping of physiological data onto fine wiring and connectivity.
Imaging of Archon1 requires excitation light intensity higher than required for GFP-like-protein-based fluorescent reporters, but the light is red in wavelength and thus less absorbed by tissues than bluer wavelengths. Archon1 supported imaging with about an order of magnitude lower light intensity in comparison to the best performing earlier Arch-based voltage sensors. To achieve light intensity above 0.1 W/mm2, we here used commercially available red laser diodes, with pricing comparable to LED setups, and found that they provided sufficient light power to image Archon1 in a variety of neural systems as explored in this paper. Thus, in price, such setups might be comparable to blue LED setups used for imaging GCaMP under 1-photon microscopy.
Imaging of neuronal activity using voltage sensors opens up the exciting possibility for simultaneous recordings of large populations of neurons with single cell single spike resolution in vivo. Several ongoing trends, as they mature, will help make voltage imaging even more accessible. Currently available scientific-grade cameras can perform fast imaging (at 500–1000 Hz) over pixel counts smaller by an order of magnitude than those commonly used for calcium imaging (at 10–20 Hz); new cameras capable of fast imaging at cellular resolution over broader fields of view will continue to enhance the power of voltage imaging. Adding in optics capable of large volume imaging with precise optical sectioning (e.g., through light-sheet scanning33,38, or through computational optical sectioning39) may also be of use. Densely labeled neurons may challenge population imaging because assigning photons to individual neurons will be more complex; this may be alleviated by restricting sensor expression to the somata29, so that the light emitting sources are made sparser.
ONLINE METHODS
Molecular cloning and mutagenesis
The Ace2N-4aa-mNeon, Archer1-KGC-EGFP-ER2 and Quasar2-mOrange-KGC-ER2 genes were synthesized de novo by GenScript, based on the sequences reported in the original publications12,13,16. Plasmids encoding mIFP, iRFP670, iRFP682, iRFP702, iRFP and iRFP720 were acquired from Addgene. The RpBphP1/PAS-GAF domains and QuasAr2-ER2 genes were synthesized de novo (Genscript) with mammalian codon optimization and subcloned into the pN1 vector (Clontech) using AgeI/NotI sites. Synthetic DNA oligonucleotides used for cloning were purchased from Integrated DNA Technologies. PrimeStar Max mastermix (Clontech) was used for high-fidelity PCR amplifications. Restriction endonucleases were purchased from New England BioLabs and used according to the manufacturer’s protocols. Ligations were performed using T4 DNA ligase (Fermentas) or InFusion HD kits (Clontech). Small-scale isolation of plasmid DNA was performed with Mini-Prep kits (Qiagen); large-scale DNA plasmid purification was done with GenElute™ HP Endotoxin-Free Plasmid Maxiprep Kits (Sigma-Aldrich). Random mutagenesis was performed with GeneMorph II Random Mutagenesis Kits (Stratagene), using conditions that resulted in a mutation frequency of up to 15 mutations per 1,000 base pairs. The QuasAr2 random library with a mutation frequency of 10–15 mutations per 1,000 base pairs was prepared by GenScript. Site-directed libraries of Arch mutants were synthesized de novo as gBlocks (EpochLifescience), amplified with corresponding primers using PCR and subcloned into the pN1 vector. Obtained gene libraries in expression vectors were electroporated into NEB10-beta E.coli host cells (New England BioLabs). Serial dilutions (10−4 and 10−5) of the electroporated cells were plated on LB/agar medium supplemented with 100 mg·mL−1 kanamycin to estimate electroporation efficiency. The remainder of the cells was grown overnight in LB medium supplemented with 100 mg mL−1 of kanamycin for subsequent plasmid DNA isolation.
To express voltage sensors in primary hippocampal neurons, the corresponding genes were PCR amplified and swapped with the ArchT-GFP gene in pAAV-CaMKIIα-ArchT-GFP plasmid (Addgene plasmid #37807) using BamHI and EcoRI sites. For in vivo expression in mouse brain the Archon1/2-KGC-EGFP-ER2, QuasAr2-mOrange-KGC-ER2, and Archer1-KGC-EGFP-ER2 constructs were cloned into the pCAG-WPRE vector using KpnI and BsrGI sites. For transient expression in zebrafish larvae, we designed the expression vector by cloning the 4 non-repetitive upstream activating sequences (4nrUAS) as previously described41 together with a beta actin core minimal promoter followed by the 1.2 kb long 3′UTR sequence of Danio rerio synaptotagmin IIa (syt2a). The expression cassette was flanked by Tol2 transposon ends. The Archon-KGC-EGFP-ER2 and miRFP genes were codon-optimized for expression in zebrafish using the online resource at http://www.bioinformatics.org/, synthesized de novo and cloned into the designed pTol2-4nrUAS vector using SpeI and AscI sites. For expression in C. elegans, a codon-optimized fusion of Archon1 to EGFP (codon optimization was done using the online resource at http://www.bioinformatics.org/) was subcloned into a pSM vector backbone using KpnI and SacI, and the rig-3 promoter was inserted upstream using FseI and AscI sites.
Protein characterization in vitro
Protein expression and purification were performed as described previously42, with a few modifications. The pBAD/HisB vectors (Life Technologies/Invitrogen) encoding iRFP670, iRFP682, iRFP702, iRFP, iRFP720 and miRFP were co-transformed with pWA23h plasmid, encoding heme oxygenase1 from Bradyrhizobium ORS278 (hmuO) under the rhamnose promoter, into the BW25113 E.coli strain (CGSC#7636 in The Coli Genetic Stock Center). Bacterial cells were grown in RM medium supplemented with ampicillin, kanamycin, 0.002% arabinose, and 0.02% rhamnose for 15–18 h at 37°C and then for 24 h at 18°C. Proteins were purified using TALON Metal Affinity Resin (Clontech) according to the manufacturer’s protocol with one minor modification: in the wash buffer, 100mM EDTA was used instead of 400 mM imidazole. The fluorescence spectra were measured using a Fluorolog 3 spectrofluorometer (Jobin Yvon) and a SpectraMax-M5 plate reader (Molecular Devices). For absorbance measurements, a Lambda 35 UV/Vis spectrometer (Perkin Elmer) was used. Background light scattering was removed by subtracting a fitted λ−4 curve from the measured spectrum. For determination of quantum yield, the fluorescence signal of purified proteins was compared with that of the equally absorbing iRFP. To determine the extinction coefficient, we compared the absorbance value for the protein at the main peak centered in the red part of the spectrum with the absorbance value of the short wavelength peak centered at 370–390 nm assuming the latter to have the extinction coefficient of the free biliverdin IXα, which is 39,900 M−1cm−1 (ref43). pH titrations were done using a series of commercially available pH buffers (HYDRION). Size exclusion chromatography was performed by GenScript on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences) using a gel filtration standard (#1511901; BIO-RAD). Two-photon absorption (2PA) spectra and cross sections of the proteins were measured in PBS buffer, pH = 7.4 at concentrations ~1–5·10−5 M in 1 mm glass spectroscopy cuvettes (Starna cells) using femtosecond fluorescence. In particular, two-photon excitation (TPE) spectra were collected using an MOM Sutter Instrument two-photon fluorescent microscope coupled with an Insight DeepSee (Newport) femtosecond laser tunable from 680 to 1300 nm. A Plan NeoFluar 2.5x/0.075 Zeiss objective was used to excite and collect fluorescence which was passed through a HQ705/100 filter (Chroma) before reaching the PMT. To correct the TPE spectra for the wavelength-to-wavelength variations of laser properties (pulse duration and beam shape), Styryl 9M (Aldrich) in chloroform was used as a reference standard44. The TPE fluorescence had quadratic dependence on excitation power in the whole spectral range as presented in Supplementary Fig. 7. Absolute 2PA cross section was obtained using Styryl 9M (Aldrich) in chloroform as a standard44. Fluorescence intensity, F, excited at 900 nm, was measured as a function of excitation power, P, for both the sample and the reference in the same conditions through a ET675/20 filter (Chroma), with the transmission center at 667 nm in the MOM setup (18° incidence angle). From the fit of these dependencies to a quadratic function F=aP2, a values were obtained and then normalized44 to the concentrations (obtained spectrophotometrically, using a BioMate™ S3 spectrophotometer) and to the differential quantum efficiencies at 667 nm (obtained with a spectrofluorimeter, PC1 ISS). In Supplementary Fig. 7, we present the two-photon action spectrum (σ2 φ) for miRFP and compare it to that of EGFP (measured before45).
Gene library transfection
Conventional calcium phosphate transfection was modified to deliver a small number of plasmids per single cell to enable efficient single cell phenotyping and genotyping (Supplementary Fig. 1). HEK293FT cells were selected as the expression host due to several reasons: i) they are suitable for calcium phosphate transfection; ii) they are widely regarded as high expressors for a variety of protein payloads (and excellent for optogenetic tool characterization); iii) they are known to have perhaps the lowest mutation rate among commonly used mammalian cell lines towards exogenous DNA46; iv) they are robust and easy to work with. Cells were authenticated by the manufacturer and tested for mycoplasma contamination to their standard levels of stringency and were here used because they are common cell lines for testing new tools. As an expression vector, we used the commercially available pN1 plasmid (Clontech) which can be replicated in HEK293FT cells due to the SV40 ori of replication47. The replication of plasmids enables a higher level of protein expression upon single copy plasmid delivery, facilitating optical detection of recombinant protein. We used the CMV promoter to drive expression of target genes because it is known to be perhaps the strongest promoter among those commonly used in HEK293 cells48.
HEK293FT cells (Invitrogen) were maintained between 10% and 70% confluence at 37°C with 5% CO2 in DMEM medium (Cellgro) supplemented with 10% heat inactivated FBS (Corning), 1% penicillin/streptomycin (Cellgro), and 1% sodium pyruvate (BioWhittaker). Transfection of HEK293FT cells with gene libraries was performed using a commercially available calcium phosphate (CaPhos) transfection kit (LifeTechnologies) according to the manufacturer’s protocol with minor modifications as follows. HEK293FT cells from the exponential growth phase were seeded at a density to be approximately 70% confluent on the day of transfection. Culture medium was replaced with fresh medium ~30–60 minutes before adding DNA-CaPhos co-precipitate so that the medium was of pH ~7.4. 2× CaCl2/DNA solution was added quickly to an equal volume of 2× HBS solution at room temperature, mixed gently for 20–30 sec by pipetting up and down, and added dropwise to the cell culture. Culture medium was carefully replaced with fresh medium 24 hours after transfection. The “empty” pUC19 plasmid was used as “dummy” DNA to keep the total amount of DNA constant for all transfection conditions, and to avoid variation in DNA-CaPhos co-precipitate formation49,50.
FACS screening
To sort the gene library-transfected HEK293FT cells using flow cytometry, cells were harvested from a culture dish ~48 h after gene library transfection by applying trypsin for 5–10 mins (Cellgro) and then washed twice by centrifuging the cell suspension for 5 minutes at 500 rpm and re-suspending cells in PBS (Cellgro). The washed cells were then re-suspended in PBS supplemented with 4% FBS (Corning) and 10mM EDTA at a density of 1–2·106 cells/ml and filtered through a 30μm filter (Falcon) to prevent clogging on the FACS machine. The filtered cells were sorted by FACSAria (BD Biosciences) running BDFACS Diva8.0 software and equipped with standard 405, 488, 561 and 640 nm solid-state lasers. Debris, dead cells and cell aggregates were gated out using forward and side scatter before desired fluorescence signals were detected (see Supplementary Figure 3a for gating strategy used). For screening RpBphP1/PAS-GAF libraries, excitation at 640 nm and emission at 670/30 nm and 710/50 nm were used; for QuasAr2 libraries excitation was at 640 nm and emission was at 710/50 nm (see Supplementary Table 3 for details). Approximately 1.5 times more cells than the size of each library were screened per FACS sorting session and 10–100k cells exhibiting higher fluorescent intensity than that of the positive control (HEK293FT cells transfected with a plasmid encoding the template protein) were collected in a 5 ml tube. Collected cells were plated on a 3 cm cell culture dish coated with Matrigel (BD Biosciences) for further screening and sorting using our custom cell picker (see below). Sorter cells were analyzed by fluorescence wide-field microscopy to evaluate the abundance of positive cells (on average 80% of cells were positive).
Multi-parameter screening and single cell isolation using cell picker
After 24 hour incubation of collected cells in a culture dish, the cell medium was gently replaced with fresh media to remove non-attached cells. Attached cells in the dish were then subjected to microscope-guided cell screening using our single cell manipulation system (CellSorter, CellSorter INC)18, controlled by software updated for this study to version CellSorter4.017,18. This version was modified from previous versions to be compatible with any microscope, motorized stage, camera and/or other optional hardware (e.g. excitation source) via the open-source micro-manager software (micro-manager.org). In addition, CellSorter4.0 software enables cell analysis, detection, and picking according to external image segmentation and analysis algorithms (in particular, our membrane localization algorithm, described in the section “Protein characterization in mammalian cells”), and acquisition of timelapse movies of selected fields of view (helpful for evaluating fluorophore photostability, which we did on a subset of the cells simultaneously screened on brightness and localization). The “single-mode” mode of operation (see below) that enables picking of one cell per tube, essential for the directed evolution here performed, was also created for the current study. The cell sorter consists of a pulled glass micropipette with an opening of 50 μm in diameter, a motorized micromanipulator (Marzhauser SM 3.25), and a pressure controller that manipulates the pressure inside the micropipette. Both the micromanipulator and pressure controller are operated by the CellSorter4.0 software. The cell sorter was installed on an inverted microscope (Nikon Eclipse Ti equipped with 10x NA 0.3, 20x and 40x objective lenses, a SPECTRA-X light engine (Lumencor) with 390/22 nm, 438/24 nm, 475/28 nm, 510/25 nm, 585/29 nm, and 631/28 nm exciters (Semrock), a 5.5 Zyla camera (Andor), and automated stage (Ludl), controlled by NIS-Elements AR software to obtain fluorescent images of the entire population of cells in a culture dish. To isolate cells with desired properties (e.g. high fluorescence intensity, exclusive plasma membrane localization) from a petri dish, we followed the workflow of cell picking described in Supplementary Figure 2. Briefly, (1) fluorescent images of cells in a culture dish are acquired using the microscope; (2) 10–50 cells exhibiting desired properties (e.g., high fluorescence intensity, exclusive plasma membrane localization) are selected per dish (10–20k cells per 3cm dish); (3) the coordinates of selected cells are compiled and fed to the CellSorter software; (4) the CellSorter software orders the micromanipulator to position the tip of the micropipette 5–10 μm above a first target cell; (5) negative pressure is applied through the micropipette to detach and pick up the target cell from the dish; (6) the micropipette moves the cell to a rack where PCR tubes are placed and releases it into a designated PCR tube pre-filled with PBS by applying positive pressure. “Single-mode” (isolation of a single cell per tube) or “multi-mode” (collection of all cells into a single tube) cell picking can be performed, by selecting the appropriate option in the software.
To simultaneously optimize brightness and localization in a single cell picking step, acquired images were analyzed by custom MATLAB code to quantify brightness (mean fluorescence signal) as well as membrane localization (the ratio of mean fluorescence on the membrane to that of the cytoplasm; cells with ratio ≤1 were excluded from further analysis for Pareto front identification, since that would mean zero fluorescence enrichment on the membrane vs. cytoplasm), for every detected single cell. Thus, our image analysis reported the coordinates on the dish, as well as two values corresponding to brightness and localization degree, for every detected and analyzed cell. These quantitative metrics were then used to identify cells with optimal combinations of both brightness and membrane localization via the concept of Pareto optimality. Briefly, we first identified the cell with the highest brightness, and the cell with the best localization. Then, we identified a set of cells that have at least one parameter better than its corresponding parameter for the aforementioned pair of cells, in a fashion so that there are no cells outside this set that would have simultaneously both parameters better than any cells within the identified set. Next, we counted the number of cells in the identified set plus the first pair of identified cells. If this number was lower than number of cells we wanted to pick, we repeated this selection calculation on the set of the detected cells, excluding the set of cells already selected in the first iteration, until the total number of cells identified equals the number of cells we wanted to pick. In turn, the number of cells we wanted to pick was defined by the number of clones we thought we could feasibly screen after gene recovery.
Target gene recovery
Cells individually collected in PCR tubes by the cell picker were subjected to whole genome amplification using a commercially available whole genome amplification kit (WGA, New England BioLabs) followed PCR amplification. Amplicons with a size corresponding to that of the target gene were purified by agarose gel electrophoresis and cloned into an expression vector, and the purified plasmids were individually transfected and expressed in HEK cells for assessing desired characteristics of each gene. To account for the mutagenic activity of HEK293T cells towards exogenous DNA and potential incorporation of multiple plasmids into a given cell (see Supplementary Fig. 1 for details), at least 5 genes were screened from each HEK cell isolated.
Protein characterization in mammalian cells
HEK293FT (Invitrogen) and HeLa (ATCC CCL-2) cells were maintained between 10% and 70% confluence at 37°C with 5% CO2 in DMEM medium (Cellgro) supplemented with 10% heat inactivated FBS (Corning), 1% penicillin/streptomycin (Cellgro), and 1% sodium pyruvate (BioWhittaker). Cells were authenticated by the manufacturer and tested for mycoplasma contamination to their standard levels of stringency and were here used because they are common cell lines for testing new tools. We used HeLa cells simply as a testbed for protein expression, and not for any reasons of investigating the properties of HeLa cells in their own right. HEK293FT and HeLa cells were transiently transfected using TransIT-X2 (Mirus Bio LLC) according to the manufacturer’s protocol and analyzed 48 h after transfection. Cells were imaged using a Nikon Eclipse Ti inverted microscope equipped with a SPECTRA X light engine (Lumencor) with 475/28 nm and 631/28 nm exciters (Semrock), and a 5.5 Zyla camera (Andor), controlled by NIS-Elements AR software, and using 10x NA 0.3 (Supplementary Fig. 3c, 4b,c) and 40x NA 1.15 (Fig. 1c,d and Supplementary Fig. 3d, 4d, 6, 10b,c) objective lenses. To compare brightness of mIFP, miRFP and RpBphP1 intermediate mutants we calculated the mean near-infrared fluorescence intensity of ~100% confluent HEK293FT cell cultures expressing corresponding proteins (Supplementary Fig. 4b,c). For plasma membrane localization analysis, voltage sensor variants were co-transfected with membrane-anchored YFP (Fig. 1d, Supplementary Fig. 10b). To quantify protein localization, we wrote MATLAB code that automatically detected cells in each image and calculated the degree of similarity (i.e., co-localization) between the normalized images acquired in the green and red channels by averaging the difference between the green channel intensity profile and the red channel intensity profile, and then taking the reciprocal of this ratio (so that a higher value means better co-localization). For Supplementary Fig. 4d, raw photobleaching curves were normalized to the spectrum of the red LED of SPECTRA X light engine, the transmission profile of the excitation filter and dichroic mirror, and the absorbance spectrum of respective FP. For flow cytometry analysis HEK293T cells were stained with SYTOX Green (Life Technologies) and analyzed using 488 and 640 nm laser lines and the respective 515/20BP (BP, bandpass) and 710/50BP emission filters on a BD LSR II analyzer (Fig. 1e and Supplementary Fig. 10a).
Induced transmembrane voltage (ITV) in HEK cells
To screen for voltage sensitivity, HEK293FT cells expressing mutants selected with our cell picker system were subjected to a reproducible electric field between two platinum electrodes as described previously12,51 with two minor modifications. First, to measure the actual value of the electric field applied across the cell culture environment, we used an oscilloscope (Agilent Technologies) connected in parallel to the platinum electrodes deployed. Second, to increase the throughput of ITV screening, the positions of the platinum electrodes were controlled by the CellSorter micromanipulator and associated software. In brief, HEK293FT cells were plated on 24 well plates and transfected with 500 ng of target plasmid DNA per well using TransIT-X2 (Mirus Bio LLC) following the manufacturer’s protocol. Cell imaging was performed on an inverted Eclipse Ti-E (Nikon) equipped with a CMOS camera (Zyla5.5, Andor), LEDs (Spectra, Lumencor), a 637 nm Laser (637 LX, OBIS) focused on the back focal plane of a 40x NA 1.15 objective (Nikon), and a filter set with 664LP (LP, longpass; emission) and 650nm (dichroic) filters (Semrock). The pair of platinum electrodes, with a gap of 4 mm, and mounted on our automated micromanipulator, was sequentially placed in the wells, and trains of electrical pulses (20–100 V/cm, 50 ms, 2 Hz) generated by a DG2041A Arbitrary Waveform Function Generator (RIGOL) and amplified with a high voltage amplifier (Model 2205, Trek) were applied across the cell culture to induce changes in the membrane voltage. Fluorescent images were recorded at a 200 Hz frame rate in 2x2 binning mode for 20 s.
Whole-cell electrophysiology and fluorescence recording in HEK cells
Voltage sensitive variants selected from the ITV screening were subjected to whole-cell electrophysiology in HEK293FT cells. To evaluate voltage sensor candidates, HEK293FT cells were transfected with 100 ng of target plasmid DNA using the calcium phosphate protocol described above. 24 hours post transfection, HEK293FT cells were re-plated on round coverslips (0.15 mm thick, 25 mm in diameter, coated with 2% growth factor reduced Matrigel in DMEM for 1 h at 37°C) at a density of 20,000 cells per well in a 24-well plate and incubated for a day at 37°C. Whole-cell patch clamp recording was performed between 48 and 72 h post transfection in Tyrode’s solution consisting of (in mM) 125 NaCl, 2 KCl, 3 CaCl2, 1 MgCl2, 10 HEPES, 30 glucose, pH7.3 (NaOH adjusted) at 320 mOsm; the intracellular solution consisted of (in mM) 135 K-gluconate, 8 NaCl, 10 HEPES, 4 Mg-ATP, 0.4 Na-GTP, 0.6 MgCl2, 0.1 CaCl2, pH 7.25 (KOH adjusted) at 295mOsm. A gap-junction blocker, 2-aminoethoxydiphenyl borate (50 μM, Sigma), was added to eliminate electrical coupling between cells. All-trans-retinal was not supplemented for any HEK cell recordings. Borosilicate glass pipettes (WPI) with an outer diameter of 1 mm and a wall thickness of 0.2 mm were pulled using a Flaming/Brown micropipette puller (P-97, Sutter Instruments) to obtain a tip resistance of 3–10 MΩ. Pipettes were positioned by a Sutter MP285 manipulator during whole-cell patching. To ensure accurate measurements, data was acquired from HEK293FT cells with access resistance <15 MΩ, having resting potentials between −10 and −40 mV, membrane resistance >0.3 GΩ, and holding current (for a holding potential of −70 mV) within ±100 pA. For Fig. 1f,g and Supplementary Figure 10, patch-clamp recordings were acquired via an Axopatch 700B amplifier (Molecular Devices) and Digidata 1440 digitizer (Molecular Devices) in Tyrode’s solution maintained at 32°C during experiments using a warmed holding platform (64-1663D, Warner Instruments) controlled by a temperature controller (TC-324B, Warner Instruments). Fluorescence imaging was performed on an inverted fluorescence microscope (Nikon Ti), equipped with a red laser (637 nm, 100 mW, Coherent, OBIS 637LX, pigtailed) expanded by a beam expander (Thorlabs) and focused onto the back focal plane of a 40x NA 1.15 objective lens (Nikon). Images were taken by an EMCCD camera (iXon, Andor) at a 500 Hz frame rate in 2x2 binning mode for 2 s. The voltage sensitivity was imaged in voltage-clamp mode with a holding potential of −70 mV for 1s and then applying voltage steps from −70 mV to +30 mV for 100 ms. For Supplementary Figure 17, photocurrents were recorded at room temperature in voltage-clamp mode with a holding potential of −70 mV in response to 500 ms light pulses using a Multiclamp 700B and Digidata 1550A digitizer (Molecular Devices), and a PC running pClamp10 (Molecular Devices).
Primary neuron culture and transfection
All procedures involving animals at MIT were conducted in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Massachusetts Institute of Technology Committee on Animal Care. Hippocampal neurons were prepared from postnatal day 0 or 1 Swiss Webster (Taconic) mice (both male and female mice were used) as previously described4,52 with the following modifications: dissected hippocampal tissue was digested with 50 units of papain (Worthington Biochem) for 6–8 min, and the digestion was stopped with ovomucoid trypsin inhibitor (Worthington Biochem). Cells were plated at a density of 20,000–30,000 per glass coverslip coated with Matrigel (BD Biosciences). Neurons were seeded in 100μl plating medium containing MEM (Life Technologies), glucose (33mM, Sigma), transferrin (0.01%, Sigma), Hepes (10mM, Sigma), Glutagro (2mM, Corning), Insulin (0.13%, Millipore), B27 supplement (2%, Gibco), and heat inactivated FBS (7.5%, Corning). After cell adhesion, additional plating medium was added. AraC (0.002 mM, Sigma) was added when glia density was 50–70% of confluence. Neurons were grown at 37°C and 5% CO2 in a humidified atmosphere.
Cultured neurons were transfected at 4–5 days in vitro (DIV) with a commercial calcium phosphate transfection kit (Life Technologies) as previously described4. Briefly, 500 ng of plasmid DNA per well was used for transfection followed by additional washing with acidic MEM buffer (pH 6.7–6.8) after 30–60 min of calcium phosphate precipitate incubation to remove residual precipitates53. All measurements on neurons were taken between DIV 14 and 18 DIV (~9–14 d post transfection) to allow for sodium channel maturation. No all-trans-retinal was supplemented for any cultured neuron recordings.
Fluorescence microscopy of primary neurons
Fluorescent imaging of voltage sensors expressed in cultured hippocampal neurons for Fig. 2, Supplementary Fig. 12–16 and Supplementary Fig. 18, 19 was performed using a Nikon Eclipse Ti inverted microscope equipped with a 40x NA 1.15 water immersion objective (Nikon), a 637 nm Laser (637 LX, OBIS) focused on the back focal plane of the objective, a SPECTRA X light engine (Lumencor) with 475/28 nm, 585/29 nm, and 631/28 nm exciters (Semrock), a 470 nm LED (ThorLabs) and a 5.5 Zyla camera (Andor), controlled by NIS-Elements AR software.
Electrophysiology in primary hippocampal neurons
Whole-cell patch clamp recordings of cultured neurons for Fig. 2 and Supplementary Fig. 14–16 were acquired via an Axopatch 700B amplifier (Molecular Devices) and Digidata 1440 digitizer (Molecular Devices). Neurons were patched between ~DIV14 and DIV18. Neurons were bathed in Tyrode’s solution (125 NaCl, 2 KCl, 3 CaCl2, 1 MgCl2, 10 HEPES, 30 glucose, pH7.3 (NaOH adjusted)) at 32°C during measurements. Borosilicate glass pipettes with an outer diameter of 1 mm and a wall thickness of 0.2mm with resistance of 3–10 MΩ were filled with internal solution containing 135 K-gluconate, 8 NaCl, 10 HEPES, 4 Mg-ATP, 0.4 Na-GTP, 0.6 MgCl2, 0.1 CaCl2, pH 7.25 (KOH adjusted) at 295 mOsm. Measurements from primary neuron cultures were performed on the electrophysiology setup described in the “Whole-cell electrophysiology and fluorescence recording in HEK cells” section. Patch-clamp data was acquired only if the resting potential was below −45mV and access resistance was <25MΩ. Access resistance was compensated at 30–70%. Fluorescence imaging was performed on an inverted fluorescence microscope (Nikon Ti), equipped with a red laser (637nm, 100mV, Coherent, OBIS 637LX, Pigtailed) expanded by a beam expander (Thorlabs) and focused onto the back focal plane of a 40x NA 1.15 objective lens (Nikon). Synaptic blockers (NBQX, 10 μM; d(−)-2-amino-5-phosphonovaleric acid, 25 μM; gabazine, 20 μM; Tocris) were added to the imaging medium for measurements of single-cell electrophysiology. For voltage sensor kinetics studies presented in Fig. 2c, Supplementary Fig. 15,16 and Supplementary Table 1 images were acquired with an EMCCD camera (iXon, Andor) at a 3.2 kHz frame rate using an optical mask (Optomask, Andor). For other concurrent imaging and electrophysiology recordings the acquisition rate was reduced to 2.3kHz to achieve longer recording times without camera overheating and to reduce data storage.
In utero electroporation
Embryonic day (E) 15.5 timed-pregnant female C57BL/6 and CD1 (Charles River; for Fig. 3, Supplementary Figs. 20–22, 24, and 25) or Swiss Webster (Taconic; Supplementary Fig. 7, 23) mice were deeply anesthetized with 2% isoflurane. Uterine horns were exposed and periodically rinsed with warm sterile PBS. A plasmid encoding Archon1, Archon2 or miRFP (pCAG-Archon1/2-KGC-EGFP-ER2-WPRE, pAAV-Syn-miRFP; 1μg/μl) diluted with PBS was injected into the lateral ventricle of one cerebral hemisphere of an embryo. Five voltage pulses (50 V, 50 ms duration, 1 Hz) were delivered using round plate electrodes (CUY21 electroporator, NEPA GENE, Japan; ECM™ 830 electroporator, Harvard Apparatus). Injected embryos were placed back into the dam, and allowed to mature to delivery. All experimental manipulations were performed in accordance with protocols approved by the Harvard Standing Committee on Animal Care or Massachusetts Institute of Technology Committee on Animal Care (according to location of the respective experiments), following guidelines described in the US National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Acute brain slice preparation
Acute brain slices were obtained from C57BL/6 and CD1 (Charles River) mice at P20 – P30, using standard techniques. Mice were used without regard for sex. No statistical methods were used to estimate sample size for animal studies throughout. No randomization or blinding were used for animal studies throughout. Mice were anaesthetized by isoflurane inhalation and perfused transcardially with ice-cold artificial cerebrospinal fluid (ACSF) containing (in mM) 125 NaCl, 2.5 KCl, 25 NaHCO3, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4 and 11 glucose (295 mOsm/kg). Cerebral hemispheres were removed, placed in cold choline-based cutting solution (consisting of (in mM): 110 choline chloride, 25 NaHCO3, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, 1.25 NaH2PO4, 25 glucose, 11.6 ascorbic acid, and 3.1 pyruvic acid), blocked and transferred into a slicing chamber containing ice-cold choline-based cutting solution. Coronal slices (300 μm thick) were cut with a Leica VT1000s vibratome, transferred to a holding chamber containing ACSF, and recovered for 30 min at 34°C, followed by another 30 min at room temperature. Slices were subsequently maintained at room temperature until use. Both cutting solution and ACSF were constantly bubbled with 95% O2/5% CO2.
Electrophysiology and voltage imaging in acute brain slice
Individual slices were transferred to a recording chamber mounted on an upright microscope (Olympus BX51WI, see below) and continuously superfused (2–3 ml/min) with ACSF warmed to ~32°C by passing it through a feedback-controlled in-line heater (SH-27B; Warner Instruments). Cells were visualized through a 60x water-immersion objective with either infrared differential interference contrast (DIC) optics, or epifluorescence to identify GFP-positive cells. Whole-cell voltage- and current-clamp recordings were obtained from GFP-positive pyramidal neurons in layer 2/3 of motor cortex, using patch pipettes (tip resistance 2.2–3.5 MΩ) pulled from borosilicate glass (G150F-3, Warner Instruments). For current-clamp recordings the pipette solution consisted of in (mM): 130 K-gluconate, 10 KCl, 4 NaCl, 10 HEPES, 4 Mg2-ATP, 0.3 Tris-GTP, 14 Tris-phosphocreatine (290 mOsm/l; pH 7.28 adjusted with KOH), and for voltage-clamp recordings a cesium-based pipette solution was used (135 CsMeSO3, 1 EGTA(CsOH), 10 HEPES, 3.3 QX-314(Cl-), 4 Mg2-ATP, 0.3 Na-GTP, Na2-phosphocreatine; 295 mOsm/l; pH 7.35 adjusted with CsOH). For 2-photon imaging of recorded cells 20 μM AlexaFluor594 was added to the respective internal solution. Voltage-clamp recordings were performed in the presence of tetrodotoxin (TTX, 0.5 μM) and cadmium (50 μM). EPSPs were evoked by positioning a tungsten bipolar electrode (FHC) in layer 5, and delivering a train of 5 pulses (0.1 ms, 1 Hz). Individual trials were separated by > 30s. Stimulation strength was adjusted to evoke sub-threshold EPSPs, and only cells in which clean, short-latency EPSPs could be evoked were used for voltage imaging.
Archon fluorescence was excited via a red laser (637 nm, 140 mW, Coherent Obis 637-140 LX), which was focused onto the back focal plane of the objective (Olympus LUMFL N 60x/1.10 W). Neutral density filters were used such that the power recorded after the objective was ~7 mW. The laser spot was ~25 μm in diameter at the sample plane; the resulting intensity was ~15 W/mm2. Fluorescence was collected through the same objective, passed through a 705/100 nm emission filter, and imaged onto an EMCCD camera (Andor iXON Ultra 888) at 1000 frames/s. In order to acquire images at this frame rate, we restricted the EMCCD region of interest to a 99 x 300 pixel window (binned 3 x 3), and individual sweeps were no longer than 30 s. Membrane currents and potentials were amplified and low-pass filtered at 3 kHz using a Multiclamp 700B amplifier (Molecular Devices), digitized at 10 kHz and acquired using National Instruments acquisition boards and a custom version of ScanImage written in MATLAB (Mathworks) (https://github.com/bernardosabatinilab/SabalabSoftware_Nov2009.git). For two-photon images presented in Supplementary Fig. 22, individual neurons expressing Archon1 were filled through a recording pipette with an internal solution containing Alexa Fluor 594 (20 μM), and both Alexa Fluor 594 and GFP fluorescence were visualized using a Ti-Sapphire laser (Coherent) tuned to 850 nm.
Visualization of Archon expression in fixed brain tissue
Deeply anesthetized mice were perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.3) and brains were postfixed for 4 h at 4°C. 50 μm sections were cut on a vibratome, stained with fluorescent Nissl stain (NeuroTrace 640/660; Molecular Probes), and mounted in ProLong Antifade Diamond reagent (Invitrogen). Images were taken first with a slide-scanning wide-field microscope (VS120, Olympus), and high-resolution images of regions of interest were subsequently acquired with a Leica LS8 confocal microscope (Harvard NeuroDiscovery Center). Confocal images represent maximum intensity projections of 15 to 40 μm image stacks.
Transient expression in zebrafish embryos
All experiments were conducted in accordance with MIT Committee on Animal Care and Janelia Farm IACUC protocols (as appropriate for the respective locations of specific experiments). Zebrafish, Danio rerio species, were raised and bred at 28°C according to standard methods. DNA plasmids encoding zArchon1-KGC-GFP-ER2 or miRFP were co-injected with Tol2 transposase mRNA into embryos of the pan-neuronal expressing Gal4 line, tg(elavl3:GAL4-VP16)nns6(ref54). The embryos used in the study were homozygous nacre. Briefly, DNA and Tol2 transposase mRNA, synthesized using pCR2FA as a template (ref55) (mMESSAGE mMACHINE® SP6 Transcription Kit, Thermofisher), were diluted to a final concentration of 25ng/μl in 0.4mM KCl solution containing 0.05% phenol red solution (Sigma Aldrich) to monitor the injection quality. The mixture was kept on ice to minimize degradation of mRNA during the injection. The mixture was injected into embryos at 1–4 cell stages as described previously56. Larvae, tg(elavl3:GAL4-VP16)nns6(ref54), were screened for green fluorescence in the brain and spinal cord at 2–3 days post fertilization (dpf; animals were used without regard to sex) and subsequently imaged at 3–4 dpf.
Voltage imaging in zebrafish embryos
Zebrafish larvae at 3–4 dpf were used to image neurons expressing zArchon1. To prevent motion artifacts during imaging, larvae were paralyzed by applying a paralytic agent, pancronium bromide, at a final concentration of 0.20 mg/ml (Sigma Aldrich), to stop muscle motion. Larvae were placed in a dish with the paralytic agent until they stopped moving, taking about one minute on average. Paralyzed larvae were immobilized in 1.5% ultra-low-melting agarose (Sigma Aldrich) prepared in E3 medium following standard protocols57. The embedded larvae were mounted on an inverted epifluorescent microscope (Nikon Eclipse Ti) for voltage imaging. The fluorescence of zArchons was excited by a red laser (OBIS 637 LX) focused onto the back focal plane of a 40x 1.15NA water-immersion objective (Nikon). Illumination intensities of 1.1–2.2 W/mm2 were used for voltage imaging recorded using a sCMOS camera (Zyla 5.5, Andor) with image acquisition rates of 250 – 500 Hz. No chemical or physical stimuli were applied before or during recording of spontaneous activity.
Lightsheet zebrafish imaging
Lightsheet imaging for Supplementary Fig. 7 was performed on a Zeiss Z.1 lightsheet microscope. The fixed sample was embedded in 1% agarose in a capillary and mounted on the freely rotating stage of the Z.1 lightsheet microscope. For image acquisition, the sample with the surrounding agarose gel was extruded from the glass capillary. Lightsheets were generated by two illumination objectives (10x, NA 0.2), and the fluorescence signal detected by a 20x water immersion objective (NA 1.0). The laser line used for excitation was 638 nm. Optical filters used to separate and clean the fluorescence response included a Chroma T647lpxr as a dichroic, and a Chroma ET665lp for miRFP. Tiled datasets were taken with the Zeiss ZEN Software, and subsequently merged and processed with FIJI, and Arivis Vision4D.
Voltage imaging in C. elegans
The Archon1-KGC-EGFP-ER2 gene was codon-optimized for expression in C. elegans using the online resource at http://www.bioinformatics.org/. Worms were maintained and grown following standard protocols58. SWF4 (flvEx3[rig-3::wArchon1-KGC-EGFP-ER2, sra-6::ChR2-GFP, elt-2::nGFP]) and SWF5 (flvEx4[rig-3::wArchon1-KGC-EGFP-ER2, sra-6::ChR2-GFP), elt-2::nGFP]) were two independent lines generated by injecting the indicated plasmids into N2 background worms and picking those with strongest expression of the wArchon1-EGFP fusion. Results from these two lines were indistinguishable. SWF7 (flvEx5[rig-3::wArchon1-KGC-EGFP-ER2, elt-2::nGFP]), generated by injecting the indicated plasmid into N2 background worms, was used for control experiments examining the action of wArchon1 in the absence of channelrhodopsin-2.
The SWF4, SWF5 and SWF7 transgenic worms (used without regard to sex) at L4 stage of development were put onto NGM plates with OP50 lawns supplemented with 100 μM all-trans-retinal (Sigma-Aldrich, USA) no less than 16 hours prior to experiments. Worms were mounted on 5% agarose pads on microscope slides, immobilized with 5mM tetramisole and imaged using a Nikon Eclipse Ti inverted microscope equipped with a 40x NA 1.15 water immersion objective (Nikon), a 637nm Laser (637 LX, OBIS) focused on the back focal plane of the objective, a SPECTRA X light engine (Lumencor) with 475/28 nm excitation filter (Semrock), and a 5.5 Zyla camera (Andor), controlled by NIS-Elements AR software. Fluorescence of wArchon1 was imaged with 635nm excitation at 800 mW/mm2 and 664LP emission filter (Semrock); GFP fluorescence was imaged with a 475/34BP excitation filter and a 527/50BP emission filter (Semrock). Optogenetic stimulation was performed with 475/34 nm illumination at 0.2 mW/mm2.
Data analysis and statistics
Data were analyzed offline using NIS-Elements Advance Research software, Origin (OriginLab), c, Excel (Microsoft), ImageJ, Igor Pro (Wavemetrics), BoxPlotR and MATLAB. Analysis of all presented fluorescence traces was performed as following: cells and a neighboring cell-free region were selected manually and fluorescence measurements were performed for each region of interest (ROI), and then fluorescence from an Archon-free region was subtracted from cell fluorescence to correct for background; except Fig. 3 and Supplementary Fig. 24, 25, in which Archon fluorescence was extracted by a maximum-likelihood pixel-weighting algorithm described previously59. In addition, for Fig. 3 and Supplementary Fig. 24, 25 Archon fluorescence traces were corrected for photobleaching by subtracting baseline fluorescence traces that were low-pass filtered and fit to a double exponential or an exponential function. All fluorescent traces were presented without noise filtering except for the zoomed-in trace in Fig. 4c (bottom), which was filtered for noise using a moving average window. Fluorescence changes to voltage steps were calculated as ΔF/F=(Fss−Fbl)/Fbl, where Fss (steady-state fluorescence) is the mean fluorescence intensity averaged over 50–70 ms during a voltage step after the fluorescence signal reaches its plateau, and Fbl (baseline fluorescence) is the mean fluorescence intensity averaged over 100 ms before the voltage step. Fluorescence changes during the action potential (AP) were calculated as ΔF/F=(Fpeak−Fbl)/Fbl, where Fpeak (peak fluorescence) is the max fluorescence intensity during an AP, Fbl (baseline fluorescence) is the mean fluorescence intensity averaged over the 100 to 200 ms before an AP. The SNR for an AP was calculated by dividing the peak fluorescence of an AP by the standard deviation of the baseline fluorescence over a 100 to 200 ms window preceding the AP. These SNRs were averaged to determine the AP SNR for a given cell. AP width was calculated at 50% of peak AP fluorescence by linearly interpolating the average AP fluorescence for a cell. This width was compared to the electrical AP waveform width after the electrical signal was down-sampled to the frame rate of the camera.
For kinetics analysis (Fig. 2c, Supplementary Fig. 10, 16, 24, 25, and Supplementary Table 1, 6), fluorescent traces were averaged across cells and the fluorescence rise segment and fluorescence decay segment were extracted from the averaged trace in MATLAB by inspection. Only the first 50ms in the fluorescence rise and fluorescence decay segments were used in the downstream bi-exponential fitting. Next, the fluorescence rise (inverted for convenience of using the single equation below for both rise and decay) or decay segment, F(t), was fitted to the following bi-exponential function in MATLAB: F(t) = A×(C×exp(−t/t1)+(1−C)×exp(−t/t2)), where t1 was the time constant of the fast component and t2 was the time constant of the slow component. The percentage of the total magnitude that was associated with the fast component (%t1) was defined as “C” above. For Supplementary Fig. 24, 25, to measure the time constant of actual voltage changes in voltage-clamp experiments, a series of hyperpolarizing voltage steps (6 repetitions of −5 mV) were applied immediately before the voltage imaging protocol (without capacitance and series resistance compensation), and the decay constants of the first transient of each step was analyzed in the same way as described above.
For Supplementary Fig. 19e–f, ROIs for the Archon2 signal were identified using a novel algorithmic approach utilizing non-negative matrix factorization (NMF)60 on the power spectral density of each pixel’s time history. Intuitively, pixels that do not represent Archon2 activity will have a time history that is a mix of noise and camera artifacts, whereas pixels that do capture Archon2 activity will have a distinct signature in the frequency domain that captures the Archon2 dynamics. To automatically separate both the spatial and time history of these two types of pixels, a rank-2 NMF is calculated on the 3D dataset (X, Y and frequency), reshaped as a 2D matrix (space and frequency). What is required from a human is to specify one pixel that is known to be demonstrating Archon2 signal. For all other pixels, the algorithm compares the weight of the NMF component known to correspond to noise versus the NMF component known to correspond to Archon2 activity. For robustness, this algorithm is applied to overlapping partitions of the data, and then each pixel receives a set of votes as either noise or Archon2 signal. The result of this voting system is a mask that can applied to the entire dataset, removing 98% of the pixels. The remaining data is then spatially clustered via connected components and available for existing time-domain interrogations. The code is available as a MATLAB script at https://github.com/dgoodwin208/nmfroi.
All statistics were performed in JMP (SAS), except that Wilcoxon rank sum tests were performed in MATLAB (MathWorks). The Wilcoxon rank sum test was used for comparing two groups. The Kruskal–Wallis test was used for comparing multiple groups. For the Kruskal–Wallis test, Steel’s post hoc test was used to compare each group to a control group; Steel-Dwass’ post hoc test was used to compare each pair of groups. We did not perform a power analysis, since our goal was to create a new technology; as noted in ref. 61, and recommended by the NIH, “In experiments based on the success or failure of a desired goal, the number of animals required is difficult to estimate…” As noted in the aforementioned paper, “The number of animals required is usually estimated by experience instead of by any formal statistical calculation, although the procedures will be terminated [when the goal is achieved].” These numbers reflect our past experience in developing neurotechnologies.
All attempts at replication of the experiments were successful.
Data availability
The data that support the findings of this study are available from the corresponding author on request. Sequences of the reported proteins are available at Genbank at the following accession codes: miRFP MG250278; zmiRFP, MG250279; Archon1, MG250280; Archon2, MG250281; zArchon1, MG250282; zArchon2, MG250283; wArchon1, MG250284; wArchon2, MG250285. All genes of interest will be posted to Addgene for distribution upon publication.
Code availability
Computer code used to generate results for this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We thank G. Paradis and M. Saturno-Condon for help with flow cytometry, F. Chen and L. Kang for help with confocal imaging, N. Ji for assistance with C. elegans imaging, and B. Trout and C. Sudrik for help with spectroscopic analysis of iRFPs. We are grateful to X. Han and K. Hansen (Boston University) for the pCAG-WPRE expression vector, and F. Subach (Moscow Institute of Physics and Technology) for the pWA23h plasmid. We are grateful to E. Costa, D. Estandian, and A. Wassie and L. Cai for useful discussions. C.S. acknowledges the Lefler Center for the Study of Neurodegenerative Disorders for support. E.S.B was supported by the HHMI-Simons Faculty Scholars Program, the IET Harvey Prize, the MIT Media Lab, the New York Stem Cell Foundation-Robertson Award, the Open Philanthropy Project, Human Frontier Science Program RGP0015/2016, and NIH grants 1R43MH109332, 1R24MH106075, 2R01DA029639, 1R01EY023173, 1R01NS087950, 1R01MH103910, 1R01GM104948, and NIH Director’s Pioneer Award 1DP1NS087724. O.S. was supported by a Simons Fellowship. H.-J.S. was supported by a Samsung Fellowship. D.G. was supported by an NSF Fellowship. Y.-G.Y. was supported by a Samsung Fellowship. L.F. was supported by a Simons Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS
K.D.P., E.E.J., and E.S.B. initiated the project, made high-level designs and plans, and interpreted data. K.D.P., E.E.J., B.S. and O.S. developed the hierarchical multiparameter screening approach. K.D.P. and E.E.J. developed miRFP and together with C.L., M.D., T.H., H.S., and S.A. performed its characterization. K.D.P. and E.E.J. developed Archons and together with D.P. characterized them in cultured cells. C.S., D.H., J.S., and B.L.S. performed electrophysiology experiments in acute brain slices. K.D.P, E.E.J, D.G., E.P. and C.L. analyzed neuronal culture data. N. P. and Y.Y assisted on imaging setups. E.E.J. and K.D.P with help from L.F. performed experiments on zebrafish injected by C.Y., T.K., and M.B.A. K.D.P., S.W.F. and J.L.R performed experiments on C. elegans. C.R. and F.E. designed vectors for zebrafish expression. C.L. and E.E.J. performed statistical analysis. K.D.P., E.E.J., C.S., C.L., and E.S.B. wrote the paper with contributions from all authors. E.S.B. oversaw all aspects of the project.
COMPETING FINANCIAL INTERESTS
B.S. is a founder of the CellSorter startup company. K.D.P., E.E.J., and E.S.B. are inventors on patent applications regarding the molecules here reported. B.S., K.D.P., E.E.J., and E.S.B. are inventors on a patent application regarding the screening method here developed.
References
- 1.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 2.Subach FV, Piatkevich KD, Verkhusha VV. Directed molecular evolution to design advanced red fluorescent proteins. Nat Methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, et al. Microfluidic cell sorter-aided directed evolution of a protein-based calcium ion indicator with an inverted fluorescent response. Integr Biol (Camb) 2014;6:714–25. doi: 10.1039/c4ib00039k. [DOI] [PubMed] [Google Scholar]
- 6.Dean KM, et al. Microfluidics-based selection of red-fluorescent proteins with decreased rates of photobleaching. Integr Biol. 2015;7:263–73. doi: 10.1039/c4ib00251b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiedler BL, et al. Droplet Microfluidic Flow Cytometer for Sorting on Transient Cellular Responses of Genetically-Encoded Sensors. Anal Chem. 2017;89:711–719. doi: 10.1021/acs.analchem.6b03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin MZ, Schnitzer MJ. Genetically encoded indicators of neuronal activity. Nat Neurosci. 2016;19:1142–1153. doi: 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyden ES. Optogenetics and the future of neuroscience. Nat Neurosci. 2015;18:1200–1. doi: 10.1038/nn.4094. [DOI] [PubMed] [Google Scholar]
- 10.Ai HW, Baird MA, Shen Y, Davidson MW, Campbell RE. Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications. Nat Protoc. 2014;9:910–28. doi: 10.1038/nprot.2014.054. [DOI] [PubMed] [Google Scholar]
- 11.Chow BY, Chuong AS, Klapoetke NC, Boyden ES. Synthetic physiology: Strategies for adapting tools from nature for genetically targeted control of fast biological processes. Methods Enzymol. 2011;497:425–443. doi: 10.1016/B978-0-12-385075-1.00018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–33. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flytzanis NC, et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat Commun. 2014;5:4894. doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–9. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner MJ, Li JZ, Gong Y, Schnitzer MJ. FRET-opsin protein voltage sensors. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Sciencexpress. 2015;350:1–11. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Környei Z, et al. Cell sorting in a Petri dish controlled by computer vision. Sci Rep. 2013;3:1088. doi: 10.1038/srep01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salánki R, et al. Automated single cell sorting and deposition in submicroliter drops. Appl Phys Lett. 2014;105:83703. [Google Scholar]
- 19.Giraud E, et al. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 20.He L, Friedman AM, Bailey-Kellogg C. A divide-and-conquer approach to determine the Pareto frontier for optimization of protein engineering experiments. Proteins Struct Funct Bioinforma. 2012;80:790–806. doi: 10.1002/prot.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles JD, Corne DW. Approximating the nondominated front using the Pareto Archived Evolution Strategy. Evol Comput. 2000;8:149–172. doi: 10.1162/106365600568167. [DOI] [PubMed] [Google Scholar]
- 22.Currin A, Swainston N, Day PJ, Kell DB. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem Soc Rev. 2015;44:1172–239. doi: 10.1039/c4cs00351a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIsaac RS, et al. Directed evolution of a far-red fluorescent rhodopsin. Proc Natl Acad Sci. 2014;105:6374–9. doi: 10.1073/pnas.1413987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotnik T, Pucihar G, Miklavčič D. Induced transmembrane voltage and its correlation with electroporation- mediated molecular transport. J Membr Biol. 2010;236:3–13. doi: 10.1007/s00232-010-9279-9. [DOI] [PubMed] [Google Scholar]
- 26.Del Re AM, Woodward JJ. Inhibition of gap junction currents by the abused solvent toluene. Drug Alcohol Depend. 2005;78:221–224. doi: 10.1016/j.drugalcdep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Gradinaru V, et al. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–9. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shemesh O, et al. Temporally precise single-cell resolution optogenetics. Nat Neurosci. 2017;20:1796–1806. doi: 10.1038/s41593-017-0018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedrich RW, Jacobson GA, Zhu P. Circuit Neuroscience in Zebrafish. Curr Biol. 2010;20:371–381. doi: 10.1016/j.cub.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 31.Stewart AM, Braubach O, Spitsbergen J, Gerlai R, Kalueff AV. Zebrafish models for translational neuroscience research: From tank to bedside. Trends Neurosci. 2014;37:264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–20. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 34.Wyart C, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordus A, et al. Feedback from Network States Generates Variability in a Probabilistic Olfactory Circuit. Cell. 2015;161:215–227. doi: 10.1016/j.cell.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobes NC, et al. Laser-based directed release of array elements for efficient collection into targeted microwells. Analyst. 2013;138:831–838. doi: 10.1039/c2an36342a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean KM, et al. Microfluidics-based selection of red-fluorescent proteins with decreased rates of photobleaching. Integr Biol. 2015;7:263–73. doi: 10.1039/c4ib00251b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouchard MB, et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat Photonics. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prevedel R, et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat Methods. 2014;11:727–30. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krzywinski M, Altman N. Points of Significance: Visualizing samples with box plots. Nat Methods. 2014;11:119–120. doi: 10.1038/nmeth.2813. [DOI] [PubMed] [Google Scholar]
- 41.Subedi A, et al. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods. 2014;66:433–440. doi: 10.1016/j.ymeth.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat Commun. 2013;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarov NS, Drobizhev M, Rebane A. Two-photon absorption standards in the 550–1600 nm excitation wavelength range. Opt Express. 2008;16:4029–4047. doi: 10.1364/oe.16.004029. [DOI] [PubMed] [Google Scholar]
- 45.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Two-photon absorption properties of fluorescent proteins. Nat Methods. 2011;8:393–399. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebkowski JS, DuBridge RB, Antell Ea, Greisen KS, Calos MP. Transfected DNA is mutated in monkey, mouse, and human cells. Mol Cell Biol. 1984;4:1951–1960. doi: 10.1128/mcb.4.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahon MJ. Vectors bicistronically linking a gene of interest to the SV40 large T antigen in combination with the SV40 origin of replication enhance transient protein expression and luciferase reporter activity. Biotechniques. 2011;51:119–126. doi: 10.2144/000113720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin JY, et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:3–6. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okazaki M, Yoshida Y, Yamaguchi S, Kaneno M, Elliott JC. Affinity binding phenomena of DNA onto apatite crystals. Biomaterials. 2001;22:2459–2464. doi: 10.1016/s0142-9612(00)00433-6. [DOI] [PubMed] [Google Scholar]
- 51.Pucihar G, Kotnik T, Miklavcic D. Measuring the induced membrane voltage with Di-8-ANEPPS. J Vis Exp. 2009;88:4–6. doi: 10.3791/1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 54.Kimura Y, Satou C, Higashijima SI. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development. 2008;135:3001–3005. doi: 10.1242/dev.024802. [DOI] [PubMed] [Google Scholar]
- 55.Kwan KM, et al. The Tol2kit: A multisite gateway-based construction Kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 56.Fisher S, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 57.Renaud O, Herbomel P, Kissa K. Studying cell behavior in whole zebrafish embryos by confocal live imaging: application to hematopoietic stem cells. Nat Protoc. 2011;6:1897–904. doi: 10.1038/nprot.2011.408. [DOI] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 61.Dell RB, Holleran S, Ramakrishnan R. Sample size determination. ILAR J. 2002;43:207–13. doi: 10.1093/ilar.43.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request. Sequences of the reported proteins are available at Genbank at the following accession codes: miRFP MG250278; zmiRFP, MG250279; Archon1, MG250280; Archon2, MG250281; zArchon1, MG250282; zArchon2, MG250283; wArchon1, MG250284; wArchon2, MG250285. All genes of interest will be posted to Addgene for distribution upon publication.