Abstract
The addressable pocket of a protein is often not functionally relevant in disease. This is true for the multidomain, bromodomain-containing transcriptional regulator TRIM24. TRIM24 has been posited as a dependency in numerous cancers, yet potent and selective ligands for the TRIM24 bromodomain do not exert effective anti-proliferative responses. We therefore repositioned these probes as targeting features for heterobifunctional protein degraders. Recruitment of the VHL E3 ubiquitin ligase by dTRIM24 elicits potent and selective degradation of TRIM24. Using dTRIM24 to probe TRIM24 function, we characterize the dynamic genome-wide consequences of TRIM24 loss on chromatin localization and gene control. Further, we identify TRIM24 as a novel dependency in acute leukemia. Pairwise study of TRIM24 degradation versus bromodomain inhibition reveals enhanced anti-proliferative response from degradation. We offer dTRIM24 as a chemical probe of an emerging cancer dependency, and establish a path forward for numerous selective yet ineffectual ligands for proteins of therapeutic interest.
INTRODUCTION
Dysregulation of gene control is a hallmark characteristic of cancer, and individual tumor types are commonly dependent on discrete gene control factors1. Research in clinical cancer genetics and functional cancer biology has validated a still growing list of compelling transcriptional addictions with immediate therapeutic relevance. Threatening the clinical impact of these findings is the persistent challenge in the development of direct-acting chemical inhibitors of transcription factors and transcriptional regulators2.
Transcriptional regulators challenge coordinated efforts in ligand discovery as they often function via protein-protein interactions mediated by large interfacial domains that lack the characteristic features of addressable hydrophobic pockets. Many of these proteins exhibit a multidomain structure, often further complicated by intrinsic disorder or limited biochemical characterization. It is therefore not always clear which domain to target, and commonly the ligandable domain is not responsible for the tumor-associated phenotype.
Such has been our community’s experience targeting bromodomain-containing proteins. After our first report of functional inhibition of the BET family of human co-activator proteins via bromodomain inhibition with JQ13, we and many others undertook to develop bromodomain inhibitors more broadly across the molecular phylogeny of 41 human proteins. Protein targets validated as cancer dependencies by genetic knockdown or knockout were successfully approached with discovery chemistry, only to realize that bromodomain engagement is insufficient to meaningfully influence cancer gene control. This has been the experience with BRG1/BRM14 and as considered here, TRIM24.
TRIM24 (originally transcriptional intermediary factor 1α) is a multidomain protein that has been broadly characterized as a co-regulator of transcription5. It is a member of the TRIM/RBCC protein family, defined by a conserved amino-terminal tripartite motif and variable carboxy-terminal domains6,7. The RING domain of TRIM24 has been reported to be involved with the ubiquitination and degradation of the master transcription factor, p538,9, and a conserved LxxLL motif has been implicated in context-dependent nuclear receptor co-activation or co-repression10,11. Chromatin localization of TRIM24 is thought to be mediated, at least in part, by a tandem plant homeodomain finger-bromodomain (PHD-BROMO) that can recognize the H3K4me0 and H3K23ac histone modifications as a chromatin-associated epigenetic reader protein12.
TRIM24 has recently been implicated as a cancer dependency in breast and prostate cancers. High levels of TRIM24 are associated with oncogenesis and disease progression in a wide variety of cancer lineages12–17. Ectopic expression of TRIM24 in normal human mammary epithelial cells (HMEC) caused increased cellular proliferation and oncogenic transformation18. Additionally, genetic knockdown of TRIM24 has been associated with impaired cell growth and induction of apoptosis12–17.
Potent and selective inhibitors of the TRIM24 bromodomain have been developed by multiple groups19,20. IACS-9571 (1) is a potent dimethylbenzimidazolone inhibitor of the TRIM24 bromodomain. Administration of IACS-9571 to cultivated cancer cells can displace a proportion of an exogenously expressed PHD-BROMO-TRIM24 from SAHA-induced hyperacetylated chromatin. However, overt effects on cancer proliferation as a phenotypic consequence have not been demonstrated21, suggesting that bromodomain inhibition alone may not be sufficient as an anti-cancer strategy.
We therefore have undertaken to adapt TRIM24 inhibitors to heterobifunctional TRIM24 degraders, inspired by the all-chemical strategy for target protein degradation we recently reported for BET bromodomains22. In our index study, we conjugated Cereblon (CRBN) E3 ubiquitin ligase binding phthalimides at permissive sites on BRD4-targeting ligands. Compounds such as dBET1 exhibited rapid, potent, and selective degradation of BET bromodomain proteins (BRD2-4). Interestingly, BET degraders functioned at sub-stoichiometric concentrations, exhibiting improved potency compared to BET bromodomain inhibitors, presumably via a catalytic-like target turnover mechanism. Subsequently, chemical biologists have validated these findings for BET bromodomain degradation23–25 notably extending these findings to include novel ligands for the VHL E3 ubiquitin ligase26. To date, targeted degradation has equated or even improved the phenotypic response as compared to target inhibition, but the application of this strategy to revive ineffectual ligands has not been demonstrated.
In this study we present dTRIM24 (2), a selective bifunctional degrader of TRIM24. We compare the mechanistic consequences of selective engagement of the TRIM24 bromodomain (IACS-9571) to selective degradation of the entire TRIM24 multidomain transcriptional regulator (dTRIM24). Using the dTRIM24 chemical probe, we report TRIM24 as a novel dependency in leukemia. Mechanistically, the TRIM24 degrader is more effective at displacing TRIM24 from chromatin compared to IACS-9571, and treatment with dTRIM24 consequently exerts a pronounced effect on genome-wide transcription at TRIM24 target genes. This pair of tool compounds can be used to further delineate the functions of TRIM24 and domain dependence of TRIM24 across different contexts and define a TRIM24-mediated transcriptional program in cancer.
RESULTS
Design of a bifunctional small molecule to degrade TRIM24
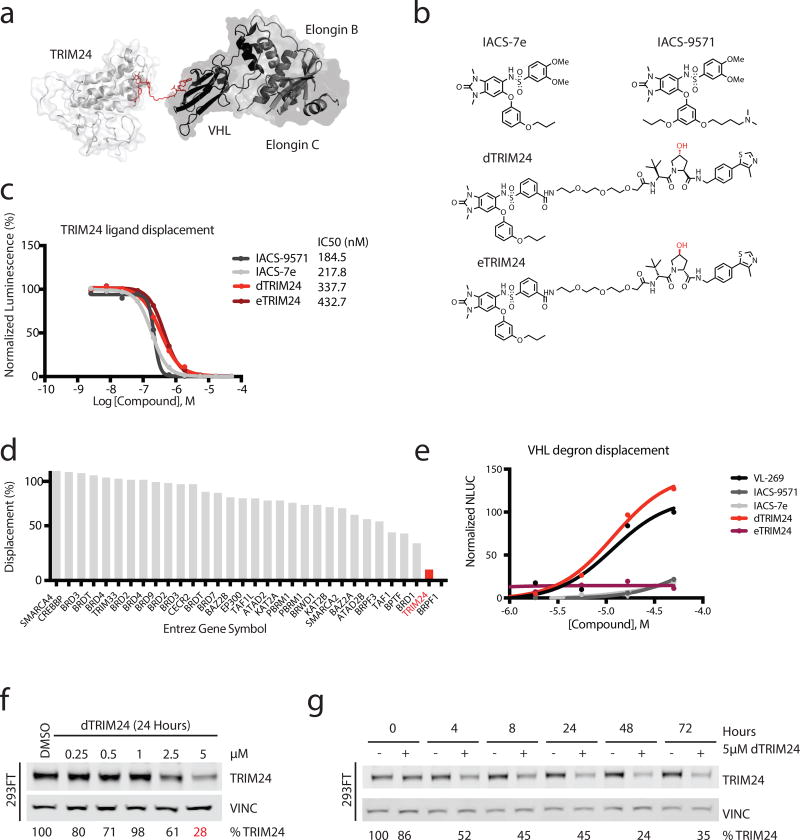
We designed a bifunctional molecule consisting of a ligand to TRIM24 chemically conjugated to a ligand to VHL. Ternary complex formation was computationally modeled to position the VHL chemical degron at a predicted permissive site on the dimethylbenzimidazolone warhead, enabled by published crystal structures of TRIM24 and VHL with their respective ligands, IACS-9571 and VL-269 (3) 23,27 (Figure 1a). Because the benzimidazolone of IACS-9571 binds to the bromodomain of TRIM24, we used the solvent exposed sulfonamide tail as an attachment point to build the degrader. We synthesized dTRIM24 by chemically conjugating IACS-7e (4)20, a close derivative of IACS-9571, to the VHL ligand, VL-269. We continued utilizing IACS-9571 in all subsequent experiments, as a potent and published reference TRIM24 bromodomain ligand. We also synthesized the enantiomer of dTRIM24, eTRIM24 (5), which contains an inactive VHL binding moiety to serve as a chemical control for degradation23,28 (Figure 1b).
Figure 1. Design and chemical characterization of dTRIM24 as a degrader of TRIM24.
(a) Crystal structure of TRIM24 PHD-BROMO bound by IACS-9571 in proximity to the crystal structure of VHL bound to VL-269 to model a strategy for building the degrader (PDB: 4YC9 and 4W9H). (b) Chemical structures of IACS-7e, IACS-9571, dTRIM24 and eTRIM24 with stereocenter of the VHL ligand component highlighted in red. (c) TRIM24 ligand displacement assay (values represent means normalized to DMSO calculated from n=2 technical replicates). n=3 independent experiments with one representative experiment shown. (d) dTRIM24 in vitro binding assay to panel of human bromodomain proteins by single point screening at 1 µM dTRIM24 in singlicate (BromoScan). (e) Cellular VHL degron displacement assay (values represent means normalized to 50 µM VL-269 calculated from n=2 technical replicates). n=2 independent experiments with one representative experiment shown. (f) Immunoblot of TRIM24 and Vinculin following 24 hours of incubation with the indicated concentrations of dTRIM24 in 293FT cells. (g) Immunoblot of TRIM24 and Vinculin following treatment of 293FT cells with 5 µM dTRIM24 for the indicated incubation times. For f–g, percentages were calculated by normalization of the band intensity to the loading control and relative to DMSO at each timepoint. n=3 independently conducted experiments with one representative experiment shown. Full immunoblots shown in Supplementary Fig. 9.
Active binding to the TRIM24 bromodomain by the bifunctional degrader was validated using a homogeneous assay for TRIM24 ligand displacement (TRIM24 AlphaScreen; Supplementary Figure 1a), in which dTRIM24 demonstrated comparable potency to bromodomain ligands (Figure 1c). Selectivity profiling across 32 human bromodomains confirmed selective and potent binding to TRIM24, comparable to the reported selectivity of the TRIM24 ligands (Figure 1d, Supplementary Figure 1b). Cellular permeability of TRIM24 was confirmed using a VHL degron displacement assay developed in our laboratory, where lead degrader molecules were assessed for the ability to compete with active FKBP-directed VHL ligands, in effect to restore bioluminescence attributable to rescued FKBP-nLuc protein stability29 (Figure 1e, Supplementary Figure 1c).
To assess the phenotypic effects of dTRIM24, we treated cells and measured TRIM24 protein abundance by immunoblot, first in 293FT cells. Both dose- and time-dependent degradation of TRIM24 were observed, with maximum degradation apparent at 5 µM (Figure 1f). Substantial degradation is evident at 4 hours, and TRIM24 depletion is maintained through 72 hours with continuous drug treatment; there is no recovery as observed previously with dBET122 (Figure 1g). Characteristic of certain chemical inducers of dimerization, high concentrations of dTRIM24 produced less TRIM24 degradation, consistent with binary complex formation out-numbering ternary complex formation (known as the hook effect30; Supplementary Figure 1d). We also synthesized a second degrader, DFCI-4107 (6), which contains the reportedly more potent ligand of TRIM24, IACS-9571 (Supplementary Figure 2a). However, DFCI-4107 does not degrade TRIM24 (Supplementary Figure 2b), attributable at least to decreased cellular permeability as measured by the VHL degron displacement assay (Supplementary Figure 2c).
TRIM24 degradation depends on VHL and the proteasome
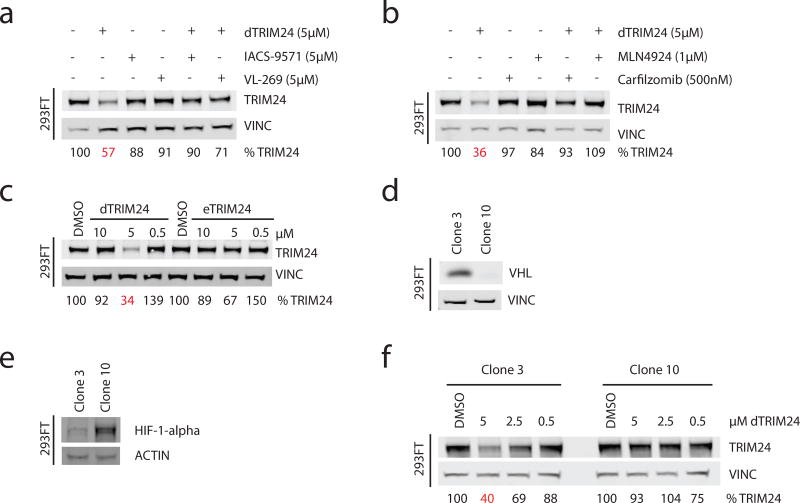
To explore if degradation of TRIM24 is dependent on binding to both TRIM24 and VHL, we co-administered dTRIM24 and equimolar amounts of IACS-9571 or VL-269 to 293FT cells. Both IACS-9571 and VL-269 firmly prevented TRIM24 degradation, demonstrating that binding to TRIM24 and VHL in cells is essential for productive degradation (Figure 2a). To establish dependency on the proteasome, we treated 293FT cells with both dTRIM24 and the catalytic proteasome inhibitor, carfilzomib. TRIM24 degradation was rescued by carfilzomib, indicative of a proteasome-dependent degradation event. Further, the neddylation inhibitor MLN4924 also rescued degradation, supporting the required function of a cullin E3 complex, such as VHL, and importantly refuting putative degradation of TRIM24 through autoubiquitination via its RING domain ligase activity (Figure 2b)31. Requisite binding to VHL was further confirmed using eTRIM24, a functional TRIM24 binder incapable of binding VHL, which failed to elicit TRIM24 degradation (Figure 2c).
Figure 2. Characterization of the cellular mechanism of degradation of dTRIM24.
(a) Immunoblot of TRIM24 and Vinculin following chemical competition with 5 µM of ligands IACS-9571 and VL-269 with co-treatment of 5 µM dTRIM24 for 24 hours in 293FT cells. (b) Immunoblot of TRIM24 and Vinculin following incubation with carfilzomib (500nM) or MLN4924 (1 µM) with co-treatment of 5 µM dTRIM24 for 24 hours in 293FT cells. (c) Immunoblot of TRIM24 and Vinculin following treatment with the indicated concentrations of dTRIM24 or eTRIM24 for 24 hours in 293FT cells. (d) Immunoblot of VHL and Vinculin in Clone 3 and Clone 10, expanded from single 293FT cells after transient transfection with a guide targeting the VHL locus for CRISPR/Cas9-mediated VHL knockout (antibody: Cell Signaling 68547). (e) Immunoblot of HIF-1α and Actin in Clone 3 and Clone 10. (f) Immunoblot of TRIM24 and Vinculin following treatment of Clone 3 or Clone 10 for 24 hours with the indicated concentrations of dTRIM24. For a–f, percentages were calculated by normalization of the band intensity to the loading control and relative to DMSO. n=3 independently conducted experiments with one representative experiment shown. Full immunoblots shown in Supplementary Fig. 9 and Supplementary Fig. 10.
To definitively demonstrate a requirement for VHL, we created a clonal population of 293FT cells that do not express functional VHL via CRISPR/Cas9-mediated deletion. Of all clonal lines tested, Clone 10 exhibited elevated HIF-1α, consistent with a functional loss of VHL, in contrast to a sister clone, Clone 3 (Figure 2d,e). Genetic modification of the VHL locus in Clone 10 was confirmed by VHL resequencing (Supplementary Figure 3a). Using these paired 293FT reagents, as well as an established VHL null renal cell carcinoma line (786-O), we demonstrated an absolute requirement for VHL to degrade TRIM24 (Figure 2f, Supplementary Figure 3b,c). As such, we confirmed the expression of VHL at both the protein and transcript level32 in all lines used throughout the remainder of this study (Supplementary Figure 3d,e).
TRIM24 as a novel dependency in acute leukemia
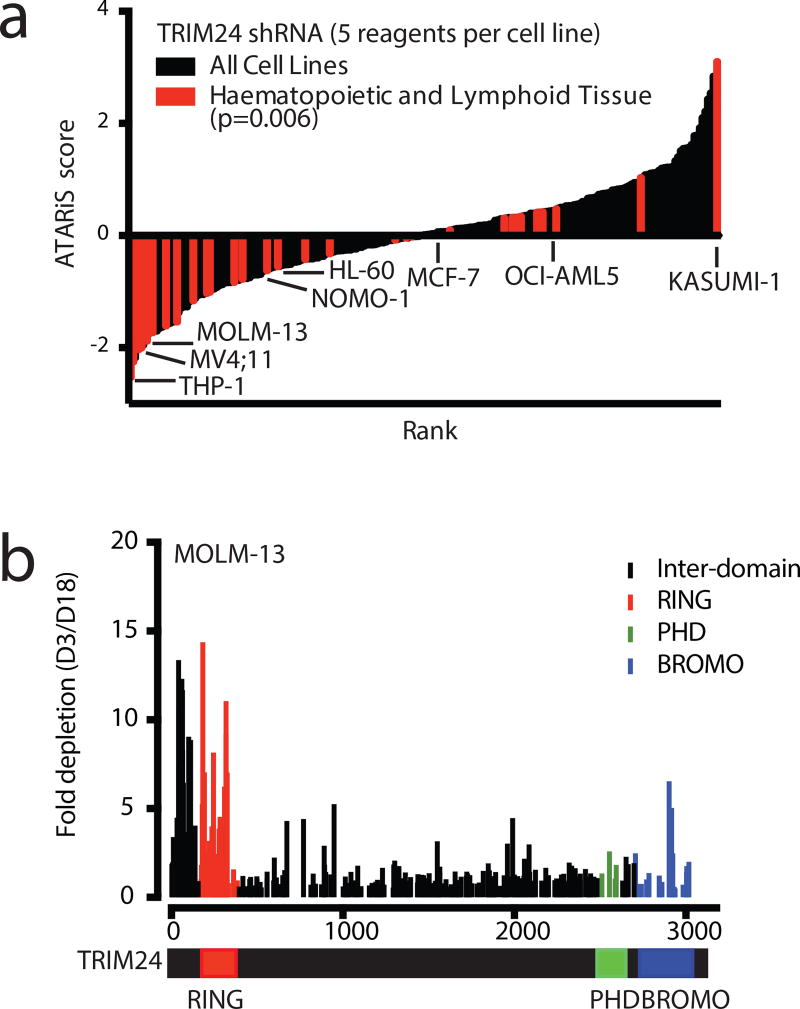
With an active TRIM24 degrader in hand, we explored the requirement for TRIM24 in models of human cancer. To identify candidate cell lines, we interrogated the Project Achilles database, an RNAi-screen aimed at identifying essential genes in a wide panel of cancer cell lines using genome-scale loss-of-function genetics, for asymmetric dependence on TRIM2433. Ranked by the ATARiS score (a reported relational metric that weighs on-target activity of shRNA reagents), we discovered that tumors of hematopoietic origin were disproportionally sensitive to knockdown of TRIM24 in comparison to other cancer cell lines (Figure 3a).
Figure 3. Genetic dependency on TRIM24 in acute leukemia.
(a) 216 cancer cell lines ranked by ATARiS score from TRIM24 genetic knockdown from the Achilles dataset34. All hematopoietic and lymphoid tissue cell lines are indicated in red and selected cell lines are labeled (p-value determined by t-test). (b) CRISPR/Cas9-mediated mutagenesis tiling TRIM24 in MOLM-13 stably expressing Cas9. After transduction with a pool of all sgRNAs, fold change from Day 3 to Day 18 was calculated for each guide by next generation sequencing at both timepoints in singlicate.
To explore a putative dependency on TRIM24 in acute leukemia, as well as to identify functional hotspots within discrete protein domains, we applied a powerful, recently reported CRISPR-scanning strategy to introduce mutations through the TRIM24 locus34. Fold change from Day 3 to Day 18 was calculated by quantifying each guide through NGS sequencing. In MOLM-13 cells, a human acute myeloid leukemia (AML) line and one of the most sensitive hematopoietic lines to TRIM24 knockdown from the Achilles dataset, TRIM24 guides targeting the 5’ region of the gene dropped out over 10-fold, indicative of a genetic dependency on TRIM24 (Figure 3b).
Because TRIM24 is a multifunctional protein, we were curious to understand which domain of TRIM24 appeared responsible for the dependency in these lines. Guides targeting the genomic region corresponding to the RING domain were efficiently depleted, while guides directed immediately downstream of the RING domain were comparatively inactive. Mutagenesis in the genomic region corresponding to the PHD finger or bromodomain did not cause depletion greater than baseline. This suggests that the RING domain could be functionally responsible for the TRIM24 dependency in this context. To validate and extend these findings, we performed the same analysis in the human MV4;11 leukemia cell line. Again, genetic abrogation of the PHD-BROMO was not depleted, whereas RING domain function was required for proliferation (Supplementary Figure 4a).
Finally, we attempted to extend these findings to breast cancer, where TRIM24 has been suggested as a tumor dependency, specifically in the MCF-7 human breast adenocarcinoma cell line9,12. Although dTRIM24 achieved selective and near complete degradation of TRIM24 in MCF-7 cells (Supplementary Figure 5a–c, Supplementary Dataset 1), both dTRIM24 and IACS-9571 did not affect cell growth (Supplementary Figure 5d). Further, we failed to observe an effect on p53 protein levels, as the RING domain of TRIM24 has been implicated as a ubiquitin ligase for p53 in MCF-7 cells (Supplementary Figure 5e). As we also did not observe sensitivity to CRISPR/Cas9-mediated disruption of TRIM24 in 2D-cultured cells (Supplementary Figure 5f), we restricted further mechanistic studies to the previously unrecognized findings in leukemia.
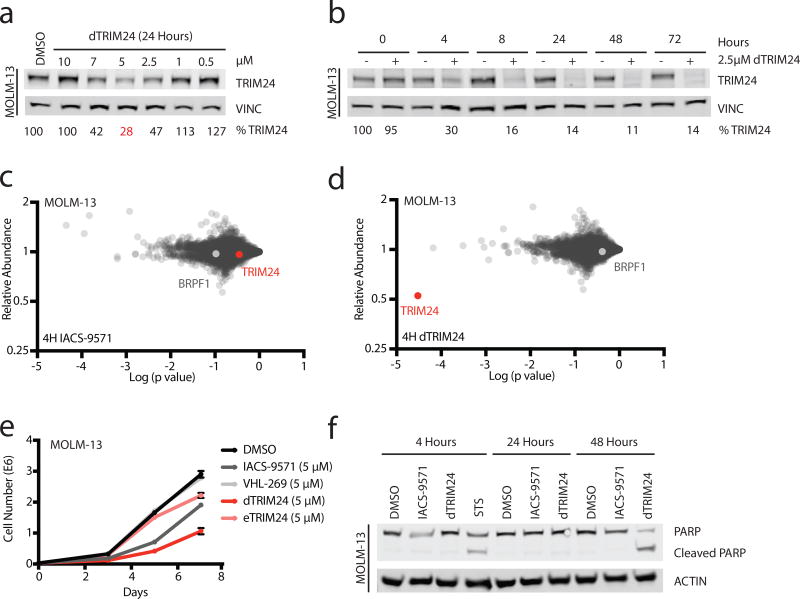
The above genetic studies suggest that the PHD-BROMO of TRIM24 may be dispensable, but the RING domain is essential for proliferation. As there are no reported RING domain inhibitors, we explored whether MOLM-13 leukemia cells would be susceptible to TRIM24 bromodomain-mediated degradation. We observed dose- and time-dependent degradation of TRIM24 with dTRIM24 treatment (Figure 4a,b). Cellular selectivity of dTRIM24 was confirmed in this context using quantitative proteomics. Immediate effects of dTRIM24 were compared to IACS-9571 after 4 hours of treatment. No proteins were significantly depleted by IACS-9571, whereas a striking selectivity for TRIM24 degradation was observed for dTRIM24 (Figure 4c,d, Supplementary Dataset 2).
Figure 4. Antiproliferative effect of selective TRIM24 degradation in acute leukemia.
(a) Immunoblot for TRIM24 and Vinculin following 24 hours of incubation of MOLM-13 cells with the indicated concentrations of dTRIM24. (b) Immunoblot for TRIM24 and Vinculin following treatment of MOLM-13 cells with 2.5 µM dTRIM24 for the indicated incubation times. For a–b, percentages were calculated by normalization of the band intensity to the loading control and relative to DMSO at each timepoint. n=3 independently conducted experiments with one representative experiment shown. (c) Proteomic analysis in MOLM-13 cells treated for 4 hours with DMSO or 2.5 µM of IACS-9571. Fold-change in the abundance of 5897 proteins from IACS-9571 to DMSO treatment versus p-value (t-test; triplicate analysis). (d) As described in (c), with cells treated with 2.5 µM of dTRIM24. (e) Growth over time of MOLM-13 cells treated with 5 µM of indicated compounds over 7 days (values represent means +/− standard deviation, n=3 independently conducted experiments). (f) Induction of apoptosis in MOLM-13 cells indicated by PARP cleavage at 48 hours with dTRIM24, comparable to PARP cleavage induced by treatment with the positive control staurosporine (STS) for 4 hours. n=2 independently conducted experiments with one representative experiment shown. Full immunoblots shown in Supplementary Fig. 10.
Notably, no additional bromodomain-containing proteins were depleted, reporting favorably on the selectivity of the IACS-like warhead employed in this context. We find this remarkable, as IACS-9571 exhibits off-target activity for the BRPF1 bromodomain, which was not degraded. In prior work to degrade BRD9, we observed that selective bromodomain ligands can evoke off-target degradation effects (in that context the degradation of BRD435). The dTRIM24 experience highlights that non-selective probes can elicit comparatively more selective degradation effects, perhaps extending the use of this chemical strategy toward improved target-specific chemical probes.
We next assessed the anti-proliferative consequences of chemical degradation versus inhibition of TRIM24. Growth over time was determined for MOLM-13 cells treated with dTRIM24, IACS-9571, VL-269, and eTRIM24. dTRIM24 suppressed growth to a greater extent than did IACS-9571, accompanied by apoptosis measured as enhanced PARP cleavage (Figure 4e,f). In agreement with a sustained proliferative defect observed following dTRIM24 treatment, we found near complete degradation of TRIM24 in dTRIM24-treated cells throughout the duration of the experiment (Supplementary Figure 4b). Lastly, we asked whether TRIM24 dependency was common across leukemia cell lines. We detected degradation of TRIM24 in an expanded panel of leukemia lines (Supplementary Figure 4c) and evaluated these cell lines for their proliferative capacity following loss of TRIM24. Our results nicely recapitulated those obtained from Project Achilles (Figure 3a) with those cell lines displaying sensitivity to genetic loss of TRIM24 also exhibiting sensitivity to loss of TRIM24 protein (Supplementary Figure 4d).
Mechanistic effects of TRIM24 degradation on gene control
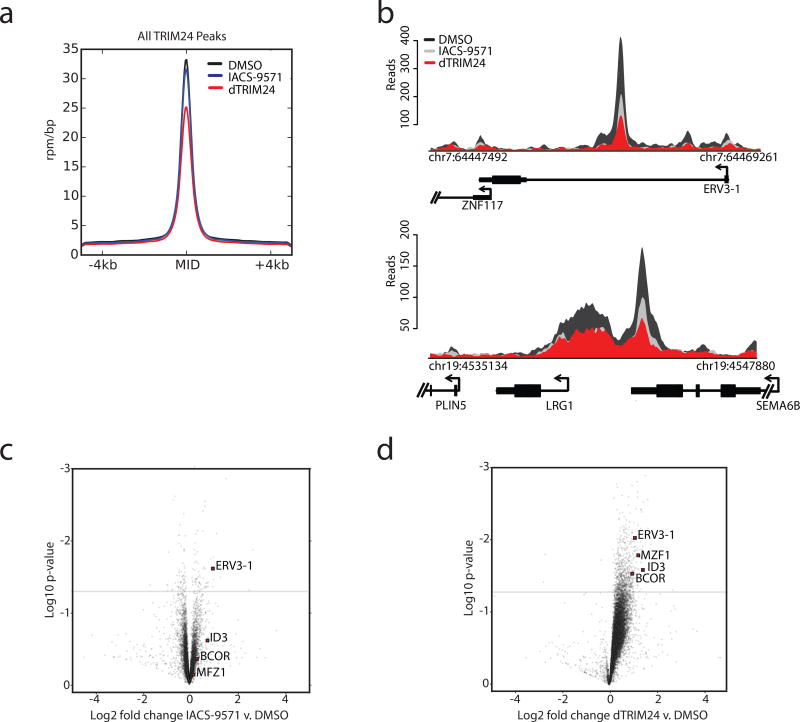
We next investigated the effect of TRIM24 degradation on chromatin association in MOLM-13 cells, a human leukemia cell line sensitive to TRIM24 loss. MOLM-13 cells were treated with dTRIM24, and chromatin was extracted and analyzed for TRIM24 occupancy by chromatin immunoprecipitation using anti-TRIM24 immunoglobulins followed by DNA sequencing (ChIP-seq). To ascertain quantitative effects on TRIM24 localization with chemical perturbation, we performed these studies using our established spike-in control method of reference genome normalization (ChIP-Rx)36. The effect of dTRIM24 was compared to IACS-9571 in parallel experiments. As expected, dTRIM24 treatment caused a more pronounced decrease in TRIM24 chromatin association than IACS-9571 genome-wide, as evident by measures of average genome occupancy of TRIM24 at all observed peaks and exemplary loci (Figure 5a,b).
Figure 5. Global displacement of TRIM24 from chromatin and transcriptional response to TRIM24 degradation.
(a) Average genome-wide reads for TRIM24 centered on a ± 4 kb window around the TRIM24 peaks with the indicated treatment conditions (2.5 µM) for 24 hours. The y-axis shows ChIP-Rx signal (rpm/bp) and the x-axis indicates position. (b) ChIP-seq binding density of TRIM24 at exemplary asymmetrically loaded TRIM24 loci following the indicated treatments for 24 hours. The y-axis indicates ChIP-seq signal (rpm/bp) and the x-axis indicates genomic position. For a–b n=1 independently conducted experiment. (c) Gene expression change represented by Log2 fold change of transcripts versus Log10 p-value after 24 hours of 2.5 µM IACS-9571 treatment. Expression values were normalized to cell number using ERCC spike-in controls. (d) As described in (c), with dTRIM24 treatment. For c–d, data represents biological triplicates.
Gene control is often influenced by the relative abundance of a transcriptional regulator or chromatin mark at a given locus37. To understand the role of TRIM24 in genome-wide transcription control, and to characterize dTRIM24 function, we looked at transcriptional response with dTRIM24 treatment, genome-wide (RNA-seq). Gene expression analysis revealed that degradation of TRIM24 via dTRIM24 was observed to markedly deregulate a defined set of gene products, whereas TRIM24 bromodomain inhibition with IACS-9571 was comparatively ineffectual (Figure 5c,d). Further, the consequence of dTRIM24 was predominantly transcriptional upregulation of target genes, consistent with the proposed role of TRIM24 in transcriptional repression38–40 (Figure 5d). Putative tumor suppressors are among the most transcriptionally upregulated, including BCOR, ERV3, MZF1, and ID341–48. These genes are frequently inactivated in blood cancers, and their derepression in this context may contribute to the effects of TRIM24 mediated dependency in MOLM-13 cells.
We explored the comparative transcriptional consequences in AML lines, and noticed that sensitivity to dTRIM24 is correlated with the effect of TRIM24 on gene control (Supplementary Figure 6a,b). Of the gene sets perturbed in the sensitive lines, we saw the greatest enrichment in processes related to chromatin, transcription and proliferation, as expected from epigenetic perturbation (Supplementary Figure 6c). When looking more closely into the genes driving the asymmetric deregulation, we noticed that MYC target genes are disproportionally downregulated in the AML lines sensitive to dTRIM24 treatment (Supplementary Figure 7a,b) and master transcription factors in AML, such as MYC, MYB, and GATA2 are themselves downregulated (Supplementary Figure 7c). While these are likely secondary effects to the immediate transcriptional response of TRIM24 degradation, they nonetheless may contribute to the altered transcriptional state correlated with sensitivity.
The enrichment in the modulation of transcriptional and proliferative gene sets in the sensitive lines prompted us to investigate the effect of dTRIM24 on the cell cycle. In only the sensitive lines, we see a G1/S cell cycle arrest (Supplementary Figure 8a,b). There is evidence of apoptosis with a sub G1 population emerging, which is consistent with PARP cleavage seen in the MOLM-13 cells (Figure 4f). These results suggest a difference in downstream response to TRIM24 loss that may, in part, account for differences in sensitivity versus insensitivity to dTRIM24. These functional studies qualify dTRIM24 as a chemical probe useful for more thorough mechanistic interrogation of TRIM24 in human biology in subsequent research.
DISCUSSION
Small molecule inducers of protein degradation have recently emerged as powerful tools for probing the activity of target proteins. Degraders combine the temporal advantages of small-molecule probes with the ability to control protein levels typically limited to genetic approaches. Here, we have synthetized and characterized a first bifunctional degrader of TRIM24. We outlined a chemical strategy focused on the mechanism of degradation of endogenous TRIM24 by taking advantage of the ubiquitin ligase activity of VHL, an E3 ligase that we harnessed for rationally designed degradation. We have shown that dTRIM24 induces rapid and sustained proteasomal degradation of TRIM24. dTRIM24 selectively degrades TRIM24 across the proteome, which is dependent on binding to both the bromodomain of TRIM24 as well as VHL.
The dependency of human acute leukemia cells on TRIM24 is a provocative finding, as leukemias are often characterized by transcriptional addiction to oncogenic transcription factors such as MYC, mechanism-based dependencies such as BRD4, or pathognomonic fusion oncoproteins (e.g. MLL fusions). Here, we report the unrecognized dependence of acute leukemia on TRIM24. The epigenomic, transcriptional state of leukemias likely predisposes to sensitivity to the aberrant transcriptional activity induced by rapid TRIM24 depletion. In this context, we believe TRIM24 depletion could be a therapeutic route in this disease. Building on these findings, future research with pharmacologically optimized investigational drugs based on dTRIM24 will enable the validation of this target in translational murine model studies of leukemia. In parallel, access to dTRIM24 within the research community will help to resolve the unexpected observation that studied breast cancer cell lines are apparently neither chemically nor genetically dependent on TRIM24.
This consistency between genetic dependence and chemical dependence in leukemia and breast cell lines points to chemical degradation as a strategy that can recapitulate genetic depletion effects. Because TRIM24 functional genetics studies across a wide variety of cancer cell lineages point to TRIM24 as a dependency, we anticipate that dTRIM24 can be used to rapidly and specifically deplete TRIM24 protein to probe its essentiality in these various contexts.
In summary, we have designed and characterized a cell-permeable chemical degrader of TRIM24. These studies propose TRIM24 as a novel target for acute leukemia drug development. We anticipate that dTRIM24 will be a useful tool to further probe the function of TRIM24 by rapid chemical depletion in hematopoietic cancers and other biological contexts. Moreover, this research highlights the value of chemical equity capable of binding target proteins of interest, irrespective of functional consequence. Even inactive ligands may be repositioned as highly functioning targeted degraders. In that manner, this chemistry offers a new path to undruggable targets.
ONLINE METHODS
Cell lines and lentivirus production
MV4;11 (RPMI supplemented with 10% FBS (Sigma-Aldrich), MOLM-13 (RPMI supplemented with 10% FBS), and OCI-AML5 (MEMalpha Glutamax supplemented with 20% FBS and 10 µg/mL hGM-CSF) cell lines were kindly provided by the laboratory of Prof. Scott A. Armstrong. HL-60 (IMDM supplemented with 20% FBS), NOMO-1 (RPMI supplemented with 10% FBS), and THP-1 (RPMI supplemented with 10% FBS and BME) cells were provided by the Biological Samples Platform of the Broad Institute. 293FT and MCF-7 (DMEM supplemented with 10% FBS) cells were purchased from ATCC. KASUMI-1 (RPMI supplemented with 20% FBS) cells were kindly provided by the laboratory of Prof. Kimberly Stegmaier. Cells were cultured at 37 °C with 5% CO2 and supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin. Cells matched their expected cell-type morphology. Cells tested negative for mycoplasma using the MycoAlert kit (Lonza). Lentivirus production was performed in 293FT cells by co-transfection of pMD2.G, psPAX2 (Addgene #12259 and 12260, respectively), and a given lentiviral expression plasmid using Lipofectamine 2000 (Invitrogen). Viral supernatants were collected 48 and 72 after transfection, filtered through a 0.22 µm membrane and concentrated 20-fold with Lenti-X Concentrator. All cells lines were transduced by spinoculation at 2000 rpm for 1 hour at room temperature with 8 µg/mL polybrene.
TRIM24 degrader modeling
The coordinates of VHL's 4W9H structure were manually dragged in order to orient its ligand binding pocket head-to-head with the ligand binding pocket of TRIM24 (4YC9), with the amide of the VHL ligand approximately 11 Å away from the methyl ether of the TRIM24 ligand. All extraneous objects were removed, chain IDs were renamed, and remaining molecules merged. Theoretical electron density of the merged structure was calculated using pdb2mrc program in the EMAN package49 and dTRIM24 was fit in Coot using real-space-refinement50.
TRIM24 ligand displacement assay
The TRIM24 expression construct pRSFDuet-1.TRIM24.PHD-Bromo(824-1011)6xHis was kindly provided by D. Patel and colleagues, MSKCC12. The plasmid was transformed into BL21(DE3)pLysS competent E. coli cells; 500 mL cultures were inoculated and grown until A600 reached 0.7, then chilled at 4°C with addition of 0.05 mM ZnCl2. Once cooled, 0.5 mM IPTG was added and cultures were shaken overnight at 28°C. Cultures were pelleted, resuspended in lysis buffer (20 mM Tris (pH 8.0), 500 mM NaCl, 20 mM imidazole with protease inhibitors), and disrupted by sonication. After centrifugation, supernatant was purified by FPLC with a HisTrap nickel column (GE) with a 3-step imidazole gradient (0.33 M, 0.66 M, 1 M). Fractions containing protein were collected, pooled and concentrated by centrifugation in a 10K MWCO filter tube (Millipore). Samples were then further purified by gel filtration on FPLC with a HiPrep Sephacryl 16/60 column (GE). Samples were concentrated again by centrifugation with a 10K MWCO filter, diluted 1:1 with freezing buffer (50 mM Tris (pH 8.0), 300 mM NaCl, 5% glycerol, 0.05 mM ZnCl), flash frozen in liquid nitrogen and stored at −80C. Histone H3K23ac peptide synthesis was performed on solid phase with Rink amide resin (Novabiochem) on a CEM Liberty peptide synthesizer with standard coupling conditions using FMOC-protected amino acids. The amino-terminal tail of histone H3 was synthesized (1–33) with side-chain acetylation on lysine residue 23. A final MTT-protected lysine was added at position 34 for facile biotinylation via selective deprotection and subsequent coupling of N-biotinyl-NH-PEG2-COOH (Novabiochem). The final peptide was cleaved from resin with 95% TFA solution, and purified by reversed-phase HPLC and lyophilization. The AlphaScreen assay was performed according to manufacturer’s protocol with minor modifications (PerkinElmer, USA). This assay was carried out in room temperature buffer containing 50 mM HEPES, 300mM NaCl, 0.1% w/v BSA, 0.05% w/v Tween-20 and 2 mM DTT at pH 7.9. All steps were carried out under low light conditions. 10 µL of a solution of a master mix 100 nM His-TRIM24 (PHD-Bromo), 40 µg/mL nickel chelate acceptor beads and 100 nM biotinylated-H3K23ac peptide was added to an AlphaPlate-384 (PerkinElmer, USA). Plates were briefly spun at 150g. Next, 100 nL compound dissolved in DMSO from stock plates was added by pin transfer using a Janus Workstation (PerkinElmer, USA) for final concentrations from 2.5 nM to 50 µM. After 30 minutes of incubation, 10 µL of streptavidin-coated donor beads (40 µg/mL) was added to the plate. The plate was spun down once again, and incubated for 1 hour at room temperature. Fluorescence signal was read on the Envision 2104 (PerkinElmer, USA) using the manufacturer’s protocol.
VHL degron displacement assay
A lentiviral construct comprising firefly luciferase (Fluc) and FKBP12(F36V) fused to nanoluciferase (Nluc) was used to produce virus and transduce 293FT cells. For intracellular VHL competition assays, cells were dispensed into white 384-well culture plates (Thermo) in 20 µL at 4000 cells/well. The cells were incubated overnight and pinned with 100 nL of competitor compound in DMSO followed by 100 nL of 20 µM dTag-12 (10) using a JANUS workstation (PerkinElmer). Cells were incubated with compounds for 5.5 hours at 37°C, 5% CO2 and then allowed to cool to room temperature for 30 min. Following incubation, 25 µL/well Fluc buffer (200 mM Tris, 15 mM MgSO4, 100 µM EDTA, 1 mM DTT, 1 mM ATP, 200 µM Coenzyme A, 400 µM D-Luciferin, 0.1% Triton X-100) was added to each plate, incubated for 15 min at RT, and then read on an Envision 2104 (PerkinElmer) for luminescence. 25 µL of Nluc buffer (25 mM Na4PPi, 10 mM NaOAc, 15 mM EDTA, 500 mM NaSO4, 500 mM NaCl, 16 µM coelenterazine, 50 µM 4-(6-methyl-1,3-benzothiazol-2-yl)aniline [sc-276812]) was then added to each well and the plate incubated for 15 min at RT and then read again. Nluc values were normalized by setting values from dTag-12-only wells to 0% and those treated with dTag-12 and 50 µM VL-269 (VHL ligand) to 100%. Data were further analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(agonist) vs. normalized response –variable slope’ analysis module.
Immunoblotting
Cells were lysed for 15 minutes on ice using RIPA buffer with added protease inhibitor cocktail (Roche) and 0.1% benzonase (Novagen). The lysate was spun at 16,000g for 15 minutes at 4°C. The protein concentration of the lysate was quantified using the BCA assay (Pierce). The primary antibodies used were TRIM24 (Proteintech 14208-1-AP), Actin (Santa Cruz SC-8432), Vinculin (Santa Cruz SC-25336), VHL (SantaCruz SC-17780), VHL (Cell Signaling 68547), HIF-1α (Cell Signaling 3716S), PARP (Cell Signaling 9542S) and p53 (Cell Signaling 2527S). Fluorescence-labeled secondary antibodies (LI-COR) and the OdysseyCLxImager (LI-COR) were used to image and visualize the immunoblot.
Creation of VHL deficient 293FT line
293FT cells were transiently transfected with the pSpCas9(BB)-2A-GFP (PX458) expression vector containing an sgRNA targeting the 5’ region of the VHL gene (TGACTAGGCTCCGGACAACC). After 48 hours, GFP positive cells were sorted into single cell clones and expanded. Clones were analyzed for VHL deficiency by immunoblotting for VHL, sequencing for genome modification at the VHL locus, as well as functionally validating VHL deficiency by immunoblotting for HIF-1α stabilization.
Quantitative mass spectrometry
Three replicates of 5E6 cells were treated with compound for 4 hours and mass spectrometry was performed and analyzed as described previously22. 5897 proteins were quantified from MOLM-13 cells and 6622 proteins were quantified from MCF-7 cells. Means for protein abundance from three technical replicates were calculated and were plotted versus p-value determined by two-tailed T-test.
Cell proliferation
All lines were seeded at 30,000 cells/well, except for the KASUMI-1 cells, which were seeded at double the density due to their slower doubling time. At endpoints, cells were suspended and mixed with Viacount reagent (Millipore) at 1:3. The mixture was incubated for 5 minutes, and viable cells were counted on the Guava easycyte flow cytometer (Millipore). Means from three technical replicates of cell counts were calculated and three biological replicates were plotted with standard deviation.
CRISPR/Cas9-mediated saturating mutagenesis
sgRNAs targeting every PAM sequencing in exonic regions of the TRIM24 gene were designed. Guides that mapped to only TRIM24 were purchased in a pooled format (CustomArray, Inc.) and cloned using Gibson assembly into the LRG (lenti-sgRNA-EFS-GFP) plasmid. This pool of plasmids was transformed into electrocompetent cells (VWR, #71003-038). Cells stably expressing constitutive Cas9 were infected with pooled lentivirus made from the cloned library, at a transduction efficiency of 40%. 15E6 cells were transduced to maintain over 1000X coverage of the library at Day 0. 15E6 cells were collected at Day 3 and Day 18 and their genomic DNA was extracted using QIAamp DNA Blood Maxi Kit (Qiagen, #51194). Guides from these populations of cells were sequenced as described previously51, by taking advantage of a two-step PCR: the first step amplified the extracted DNA, and the second step added adapters and barcodes for sequencing by Illumina NextSeq 500 using single end 75bp reads. Read counts for each guide were used to calculate fold change of each guide by dividing Day 3/Day 18.
RNA-sequencing with ERCC spike in mix
Biological triplicates of 5E5 cells were treated with DMSO, 2.5 µM IACS-9571 or 2.5 µM dTRIM24 for 24 hours (the MOLM-13 cells were treated for 24 hours (Early Time Point), and 48 hours (Late Time Point)). RNA was isolated using the RNeasy kit (Qiagen) and cell count normalized. ERCC spike-in control (Life Technologies) was added to the samples. Library preparation was performed using Illumina Stranded mRNA prep and the sequencing was run using Illumina NextSeq 500 with single end 75bp reads. Fastq files were aligned to Hg19 using HiSat2 with default parameters. Aligned reads were assembled against a reference transcriptome (refSeq release 58). The cuffquant module of Cufflinks was used to determine transcript abundance. Transcripts were normalized and fpkm was calculated by using the cuffnorm module.
Chromatin immunoprecipitation followed by highly parallel sequencing with reference exogenous genome (ChIP-RX)
ChIP-seq was performed as described previously52 with minor modifications. 1×108 cells were treated with DMSO, 2.5 µM IACS-9571 or 2.5 µM dTRIM24. Mouse 3T3 cells were used as spike-in controls and were added to each treatment condition at 10% of the human cells. Cells were cross-linked with 1.1% formaldehyde (10X crosslink solution: 11% formaldehyde, 50 mM HEPES pH 7.3, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0) followed by 3 washes with PBS. Nuclei were isolated as described previously51. In order to enrich for chromatin fragments between 200 and 700 bp, samples were sonicated at 4°C for 15 minutes. Sonicated lysates were cleared by centrifugation at 20,000 g for 10 min and incubated spinning end over end overnight at 4°C with magnetic beads prebound with antibody (TRIM24, Bethyl A300-851A). Beads were washed three times with sonication buffer, one time with sonication buffer with 500 mM NaCl added, one time with LiCl wash buffer (20 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and once with TE. DNA was eluted in elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, and 1% SDS). Cross-links were reversed overnight at 65°C. RNA and protein were digested with 0.2 mg/mL RNase A for 2 hr. followed by 0.2 mg/mL Proteinase K for 1 hr. DNA was purified with column based ChIP DNA purification kit (Active Motif) according to manufacture instructions. Libraries for Illumina sequencing were prepared using ThruPLEX DNA-seq Kit (Rubicon) using 50 ng of immunoprecipitated DNA as the starting point. Libraries were sequenced on an Illumina NextSeq 500 (single end 150 bp reads) and aligned to either mm9 or hg19 respectively.
ChIP-seq data processing
Calculating read density
We calculated the normalized read density of a ChIP-Seq dataset in any genomic region using the Bamliquidator (version 1.0) read density calculator (https://github.com/BradnerLab/pipeline/wiki/bamliquidator). Briefly, ChIP-Seq reads aligning to the region were extended by 200bp and the density of reads per base pair (bp) was calculated. For ChIP-Rx, the density of reads in each region was normalized to the total number of reads originating from the reference exogenous genome (reference adjusted reads per million, rrpm) producing units of rrpm per base pair (rrpm/bp).
Identifying enriched regions
We used the MACS version 1.4.1 (Model based analysis of ChIP-seq) peak finding algorithm to identify regions of ChIP-seq enrichment over background52. A p value threshold of enrichment of 1E-9 was used for all datasets.
Scaling cell count normalized ChIP-Rx datasets
Scale factors (million mapped reads of reference exogenous genome) for each ChIP-Rx dataset were calculated as previously described35 for mouse genomic reads. Briefly, ChIP-Rx datasets, which were produced from equal cell number inputs, were scaled by scale factors to create Y-axis arbitrary units (reference-adjusted reads per million, rrpm) to normalize difference in occupancy per cell.
Meta representations of TRIM24 occupancy in DMSO and dTRIM24
Visualization regions were created as 10kb wide regions centered on binding sites identified and ranked by the ROSE software package. These regions were further binned into 50bp bins and read density was calculated as above for both background input and IP samples and background signal was subtracted before plotting.
Cell cycle assay
500,000 cells/well were treated with dTRIM24 or DMSO for 24 hours. Cells were then washed with cold PBS (pH 7.4) and fixed with cold 70% ethanol for 30 minutes. Cells were resuspended in cell cycle staining solution (100 µg/mL of RNase A (Roche, 10109169001), 40 µg/mL propidium iodide (Life Technologies, P1304MP)) and incubated for 15 minutes at room temperature in the dark. Cell cycle data was acquired by flow cytometry on a Guava easycyte flow cytometer (Millipore) using the InCyte software.
Statistical methods
For all experiments, number of replicates, error bars, and p-values are described in the respective figure legends. All biological experiments were performed at least twice. Student’s T-Tests performed were two-tailed. Assumptions are not contradicted by the data.
Data availability statement
ChIP- and RNA-sequencing data reported in this paper has been deposited to the NCBI GEO and are available under the accession number: GSE100573. Normalized and scaled quantitative mass spectrometry-based proteomics data are provided in Supplementary Datasets 1–2.
Chemical synthesis
The synthesis of dTRIM24 and other compounds used in this study is described in the Supplementary Note.
Supplementary Material
Acknowledgments
We thank the members of the Bradner and Gray labs for engaging scientific conversations. We also thank Jinhua Wang for her help with chemical characterization. Lastly, we would like to thank Scott Armstrong and Kimberly Stegmaier (Dana-Farber Cancer Institute) for kindly providing us with materials. This research was supported by a Starr Cancer Consortium Grant (J.E.B.) and NIH/NCI P01CA066996 (J.E.B.). L.N.G is supported by an NSF GRFP fellowship (2016222867). D.L.B. was supported by the Claudia Adams Barr Program in Innovative Basic Cancer Research and is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2196-14). Quantitative proteomics studies were performed by Ryan Kunz of the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School.
Footnotes
AUTHOR CONTRIBUTIONS
L.N.G designed and performed experiments, analyzed data, and wrote the manuscript. D.L.B. designed experiments, designed and synthesized molecules, and edited the manuscript. M.A.L. performed ChIP-Rx and analyzed genomic data. J.R. analyzed genomic data. J.P., C.J.O., and T.G.S. designed and performed biochemical assays. G.E.W. and M.A.E. designed and performed CRISPR/Cas9 screens. M.X. performed computational and statistical analyses. S.D.P. and H.S.S. performed structural analysis. N.P.K. guided experimental analyses. J.A.P. provided technical advice, data interpretation, and edited the manuscript. J.Q. aided in the formulation of the chemical degradation strategy. J.E.B. and N.S.G. designed the experimental strategy, wrote the manuscript, and held overall responsibility for the study.
COMPETING FINANCIAL INTERESTS
D.L.B. and J.P. are now employees of Novartis. N.S.G. is a Scientific Founder and member of the Scientific Advisory Board of C4 Therapeutics. J.E.B. is a Scientific Founder of C4 Therapeutics. J.E.B. is now an executive and shareholder in Novartis AG.
References
- 1.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darnell JE. Transcription factors as targets for cancer therapy. Nature Reviews Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vangamudi B, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer research. 2015;75:3865–3878. doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Douarin B, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. The EMBO Journal. 1995;14:2020. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 7.Reymond A, et al. The tripartite motif family identifies cell compartments. The EMBO journal. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allton K, et al. Trim24 targets endogenous p53 for degradation. Proceedings of the National Academy of Sciences. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol Cell Biol. 2014;34:2695–2709. doi: 10.1128/MCB.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khetchoumian K, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 11.Le Douarin B, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. The EMBO Journal. 1996;15:6701. [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai WW, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Z, et al. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS One. 2013;8:e63887. doi: 10.1371/journal.pone.0063887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groner AC, et al. TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer Cell. 2016;29:846–858. doi: 10.1016/j.ccell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, et al. Overexpression of TRIM24 correlates with tumor progression in non-small cell lung cancer. PloS one. 2012;7:e37657. doi: 10.1371/journal.pone.0037657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, et al. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PloS one. 2014;9:e85462. doi: 10.1371/journal.pone.0085462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. Knockdown of tripartite motif containing 24 by lentivirus suppresses cell growth and induces apoptosis in human colorectal cancer cells. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2014;22:39–45. doi: 10.3727/096504014X14078436005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathiraja TN, et al. TRIM24 links glucose metabolism with transformation of human mammary epithelial cells. Oncogene. 2015;34:2836–2845. doi: 10.1038/onc.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett J, et al. Discovery of a Chemical Tool Inhibitor Targeting the Bromodomains of TRIM24 and BRPF. J Med Chem. 2016;59:1642–1647. doi: 10.1021/acs.jmedchem.5b00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer WS, et al. Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor. J Med Chem. 2016;59:1440–1454. doi: 10.1021/acs.jmedchem.5b00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan Y, et al. Development of novel cellular histone-binding and chromatin-displacement assays for bromodomain drug discovery. Epigenetics Chromatin. 2015;8:37. doi: 10.1186/s13072-015-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter GE, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature chemical biology. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chemistry & biology. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zengerle M, Chan K-H, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS chemical biology. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gadd MS, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nature Chemical Biology. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crews CM, et al. WO 2013106646. Compounds and methods for the inhibition of vcb e3 ubiquitin ligase. 2013
- 28.Buckley DL, et al. HaloPROTACS: Use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS chemical biology. 2015;10:1831–1837. doi: 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H-T, et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chemical Biology. 2017 doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douglass EF, Jr, Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. Journal of the American Chemical Society. 2013;135:6092. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 32.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remillard D, et al. Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angewandte Chemie International Edition. 2017;56:5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando DA, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell reports. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herquel B, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:8212–8217. doi: 10.1073/pnas.1101544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herquel B, et al. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nature structural & molecular biology. 2013;20:339–346. doi: 10.1038/nsmb.2496. [DOI] [PubMed] [Google Scholar]
- 40.Tisserand J, et al. Tripartite motif 24 (Trim24/Tif1alpha) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor alpha (Raralpha) inhibition. J Biol Chem. 2011;286:33369–33379. doi: 10.1074/jbc.M111.225680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaboli M, et al. Mzf1 controls cell proliferation and tumorigenesis. Genes & development. 2001;15:1625–1630. doi: 10.1101/gad.902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grossmann V, et al. Comprehensive Cytogenetic and Molecular Genetic Characterization of T-PLL Identifies for the First Time BCOR Mutations in a Lymphatic Disease. Am Soc Hematology. 2012 [Google Scholar]
- 43.Kewitz S, Staege MS. Expression and regulation of the endogenous retrovirus 3 in Hodgkin’s lymphoma cells. Frontiers in oncology. 2013;3 doi: 10.3389/fonc.2013.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nature Reviews. Cancer. 2014;14:77. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 45.Love CL, et al. ID3 Is a Novel Tumor Suppressor Gene in Burkitt Lymphoma. Am Soc Hematology. 2012 [Google Scholar]
- 46.Perrotti D, et al. Overexpression of the zinc finger protein MZF1 inhibits hematopoietic development from embryonic stem cells: correlation with negative regulation of CD34 and c-myb promoter activity. Molecular and cellular biology. 1995;15:6075–6087. doi: 10.1128/mcb.15.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter J, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nature genetics. 2012;44:1316. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP- and RNA-sequencing data reported in this paper has been deposited to the NCBI GEO and are available under the accession number: GSE100573. Normalized and scaled quantitative mass spectrometry-based proteomics data are provided in Supplementary Datasets 1–2.