Abstract
Signatures of recent positive selection often overlap across human populations, but the question of how often these overlaps represent a single ancestral event remains unresolved. If a single selective event spread across many populations, the same sweeping haplotype should appear in each population and the selective pressure could be common across populations and environments. Identifying such shared selective events could identify genomic loci and human traits important in recent history across the globe. Additionally, genomic annotations that recently became available could help attach these signatures to a potential gene and molecular phenotype selected across populations. Here, we present a catalog of selective sweeps in humans, and identify those that overlap and share a sweeping haplotype. We connect these sweep overlaps with potential biological mechanisms at several loci, including potential new sites of adaptive introgression, the glycophorin locus associated with malarial resistance, and the alcohol dehydrogenase cluster associated with alcohol dependency.
INTRODUCTION
Positive selection is the process whereby a genetic variant rapidly increases in frequency in a population due to the fitness advantage of one allele over the other. Recent positive selection has been a driving force in human evolution, and studies of loci targeted by positive selection have uncovered potential adaptive phenotypes in recent human evolutionary history (e.g., 1–3). One observation that has emerged from scans for positively selected loci is that these signatures often overlap across multiple populations, localized to discrete locations in the genome4–7. Sequencing data available from diverse human populations provide an opportunity to characterize the frequency that overlapping signatures share a common, ancestral event - and potentially a common selective pressure. Identifying shared selective events would be of fundamental interest, highlighting loci and traits important in recent history across the globe.
Functional annotations across the human genome can also potentially connect variants targeted by selection with candidate genes and an associated mechanism. For example, the influx of expression quantitative trait loci (eQTLs) across many tissue types8, and inferred regions of ancient hominin introgression9, now provide a richer foundation to investigate the potential biological targets under selection. While identifying the causal variant at a site of positive selection is notoriously difficult, if SNPs on a selected haplotype are associated with changes in expression of a nearby gene, this information could help attach the signature to a potential gene and molecular phenotype.
Here, we focus on the detection of genomic signatures compatible with selection on a newly introduced mutation that has not yet reached fixation (i.e., a hard, ongoing sweep) to explore their distribution across populations and spanning the genome. We performed a scan for positive selection using the integrated haplotype score (iHS) on 20 populations from four continental groups from Phase 3 of the 1000 Genomes Project (1KG)10. 88% of sweep events overlapped across multiple populations, correlating with population relatedness and geographic proximity. 59% of overlaps were shared (i.e., a similar sweeping haplotype was present) across populations, and 29% of overlaps were shared across continents. We connect these multi-population sweep overlaps with potential mechanisms at (i) the glycophorin cluster (GYPA, GYPB, and GYPE), where we observe sweeps across all four continental groups in a region associated with malarial resistance; (ii) sweeps across African populations at the X chromosome gene DGKK, implicated in hypospadias in males; (iii) a sweep shared in European populations tagged by a coding variant in the gene MTHFR, which is associated with homocysteine levels and a multitude of additional traits; (iv) two putative regions of adaptive introgression from Neandertals; and (v) the alcohol dehydrogenase (ADH) cluster, where a sweep in Africa is associated with alcohol dependence in African Americans.
RESULTS AND DISCUSSION
A correction to iHS adjusting for local, low recombination rates
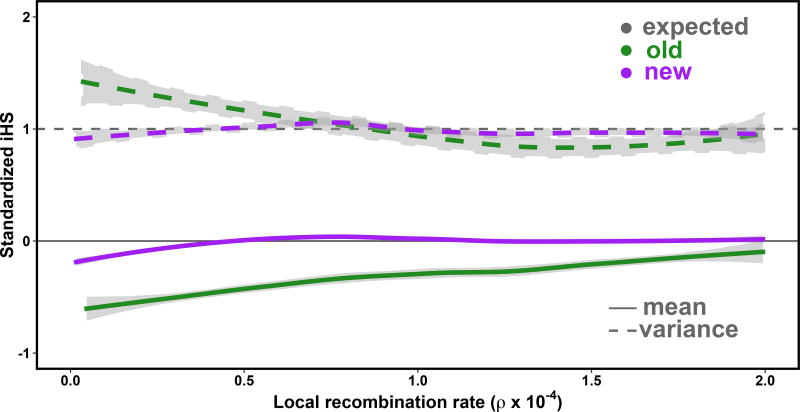
iHS was conceptualized for population data ascertained for common genetic variation 11, and may not be fully calibrated for sequencing data which includes rare variation. To examine the score in more detail, we initially applied iHS to genome sequencing data obtained from 1KG (Methods). We observed an excess of SNPs tagging strong iHS signals at lower derived allele frequencies (Supplementary Figure 1a) in frequency range where iHS is not expected to have substantial power11 (note that a negative iHS indicates extended haplotype homozygosity on the derived relative to the ancestral allele). We observed a negative correlation between the number of populations in an overlap and the local recombination rate in any population (e.g., Pearson’s correlation = −0.24, P < 8.5 × 10−10 in CHB). Simulations showed that normalizing iHS only by derived allele frequency misses additional homozygosity found at low-frequency variants in ‘cooler’ recombination rate regions, resulting in scores that are not entirely cross-comparable (Supplementary Figure 2). Normalizing iHS both by derived allele frequency and local recombination rate using a binning approach (Methods) resulted in better calibrated standardized iHS (Fig. 1), which substantially reduced the excess of high-scoring iHS values at low frequencies (Supplementary Figure 1b). This approach also eliminated correlation between low recombination rates and sweep overlaps (e.g. Pearson’s correlation = −0.04, P = 0.35 in CHB after correction). In what follows, we utilize this normalization scheme for iHS genome-wide, treating autosomes separately from the X-chromosomes (Methods, Supplementary Table 1).
Fig 1. Normalizing iHS by local recombination rate.
The mean (solid line) and variance (dashed lined) of iHS as a function of local recombination rate. iHS normalized by derived allele frequency are shown in green; normalization by both derived allele frequency and local recombination rate shown in purple. The gray lines represent a mean (= 0) and variance (= 1) for comparison.
A catalog of signals of recent positive selection across human populations
We measured iHS normalized separately for the autosomes and X chromosome across 26 populations from the 1KG project (Methods, Supplementary Table 2). For each population’s iHS scan, we identified putative sweep intervals that segregated an unusual aggregation of extreme values of iHS (Methods, Supplementary Table 3). Consistent with previous reports11, the number of sweep intervals per population correlated with its effective population size (Supplementary Table 4). We defined the tag SNP for each interval as the highest scoring variant by the absolute value of iHS, as we expect the tag SNPs to be in strong linkage disequilibrium (LD) with the putative causal, selected variant of the sweep. Our sweep intervals recovered 11 of the 12 top signatures reported in the original iHS paper11, and 14 of the top 22 signatures reported elsewhere12. We observed more extreme iHS using WGS data compared to array-based genotype data. For example, previously in CEU only 6 of 256 signatures (2.3%) had an |iHS| > 5 for their most extreme score11; in our scan, 92 of 597 signatures (15%) had an |iHS| > 5. This observation is consistent with a more rapid decay of homozygosity on background haplotypes due to improved ascertainment of low-frequency and rare variants across the genome.
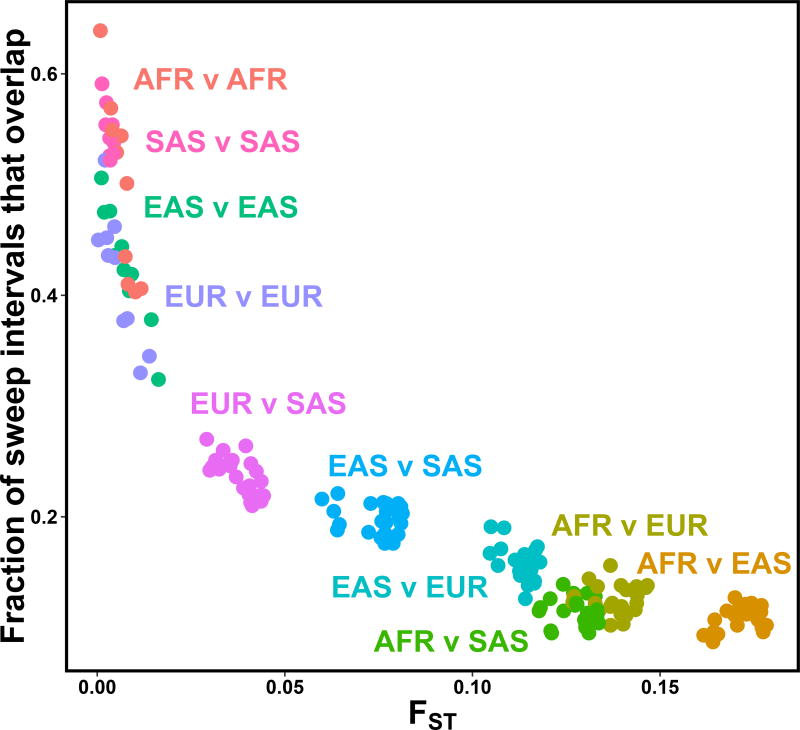
We next sought to characterize the frequency with which putative sweep intervals overlap the same genomic region across multiple populations. We excluded recently admixed populations (ASW, ACB, MXL, PEL, PUR, CLM) as events observed in those groups may reflect selection in ancestral populations predating admixture (Methods). Consistent with previous reports4–6, we noted that related populations (as measured by FST) more often overlap in their putative sweeps intervals relative to more distantly related pairs (β = −2.06, s.e. = 0.52, P = 2.5 × 10−61; Fig. 2).
Fig 2. Closely related populations have sweep overlaps more frequently.
For each population pair, the fraction of sweep intervals that overlap is plotted against pairwise estimated FST. Each population pair (dots) are colored by their continental groupings (e.g. EUR v SAS = one European population vs. one South Asian population).
Overlapping sweep intervals across human populations occur in genomic hotspots
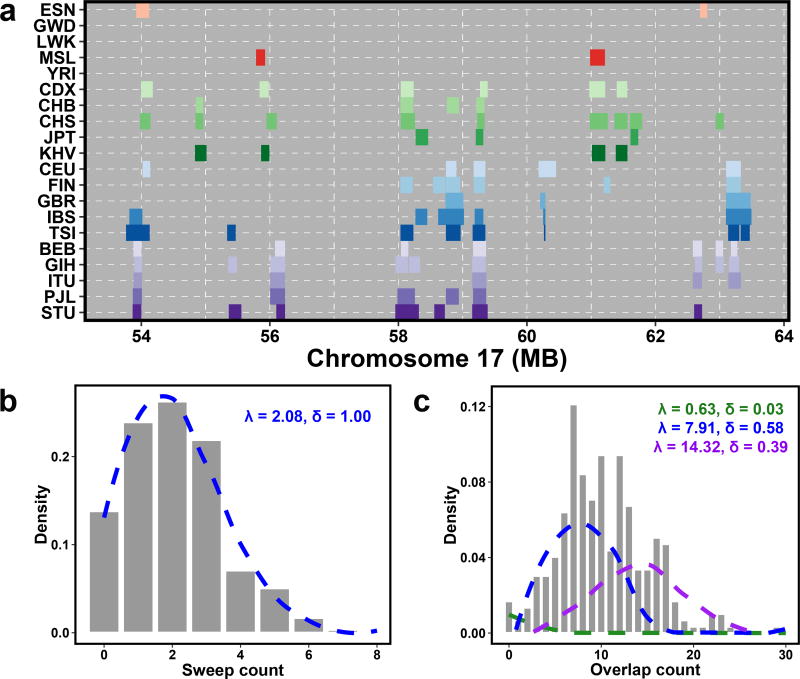
Through characterizing sweep overlaps, we observed cases where many sweeps appeared to cluster in specific genomic locations. The most striking clustering of overlapping sweep intervals occurred on chromosome 17 (Fig. 3a), with 23 overlapping events in total, of which 14 span continental groups. While previous reports have investigated how often sweeps overlap across the globe, to our knowledge the extent to which putative sweep intervals are organized across the genome has not been quantified. To model this phenomenon, we fit the observed rate of overlapping sweep events in 10Mb windows with individual or mixtures of Poisson distributions (Methods). We first modeled the count of genes in each window. We did not expect a single Poisson process to be the best fit to the gene count in each window, as genes are not uniformly distributed across the genome. Indeed, we found that a mixture model with five components best fit the frequency at which genes occur in the genome (Methods). Next, we found that the counts of sweeps in an individual population in genomic windows were best modeled by a single rate (Fig. 3b). This result was somewhat surprising, as we expected that sweeps targeting genetic variation in genes or nearby regulatory regions would follow a non-uniform rate like genes themselves. When we looked at the rate of sweep overlaps, we found that sweeps overlapping across populations were best fit by a mixture of Poisson distributions with three different rates (P = 2.7 × 10−7, χ2 test vs. two component mixture, Fig. 3c, Methods). These results suggest that some regions of the genome have a higher rate of overlaps than others, but this observation was not obviously explained by the number of genes in a window (Pearson’s correlation = 0.07, P = 0.22). We also tested the relationships of local recombination rate and estimates of background selection13 with the number of sweep overlaps in a window. We hypothesized that these features might improve a model of regions of the genome under selection rather beyond just the locations of genes, and could better explain the observed hotspots of sweep overlaps. While both features were correlated with overlap count, local recombination rate was most significant (~5% variance explained) and local background selection did not explain additional variation. These data indicate that putative sweep intervals overlapping multiple human populations may aggregate in discrete “hotspots” of activity, which are not entirely explained by the presence of genes, background selection, or local recombination rates. These hotspots could be targeting specific genic regions, or additional genomic features not assessed here.
Fig 3. Overlapping sweeps tend to cluster in the genome.
(A) An example of a 10 megabase (Mb) window on chromosome 17 with multiple overlaps across many populations. (B) The distribution of sweep interval counts in 10 Mb windows across the genome for a single population (LWK). The histogram plots the observed counts, and the blue dashed line is the best-fit Poisson distribution. (C) The distribution of sweep overlaps across two or more populations in 10 Mb windows across the genome. The histogram plots the observed counts, and the dashed lines represent the results of Poisson mixture modeling. The best-fit model was the three-component model shown here.
Complex patterns of sweep sharing across populations and continents
We next sought to identify selective sweeps that are potentially shared across populations, i.e., where the putative sweeping haplotype is similar across populations. Sharing could occur in several ways, including a common ancestral event occurring before population divergence that persisted to the present day, or via gene flow of advantageous alleles between populations. To characterize haplotype similarity across populations at our genomic intervals tagged by unusual iHS, we utilized the program fastPHASE14. Using a hidden Markov model, fastPHASE models the observed distribution of haplotypes as mosaics of K ancestral haplotypes, allowing us to map a sweep tag SNP to an ancestral haplotype jointly across multiple populations at once without arbitrarily choosing a physical span to build a tree of haplotypes or otherwise measure relatedness (Methods).
Overall, out of 1,803 intervals shared across populations, 521 (29%) were shared across continents, frequently between Europe and South Asia, consistent with observed lower genetic differentiation relative to other continental comparisons (Supplementary Table 5). Indeed, consistent with our previous analysis using all intervals, FST predicted the fraction of sweep overlaps that were shared between a pair of populations (β = −2.80, s.e. = 0.31, P = 9.1 × 10−88; Supplementary Figure 3). Though more closely related populations have a higher fraction of shared sweeps, they also have more total unshared sweeps. This relationship could be due to shared selective pressures in nearby populations, false negatives in our sharing analysis, or a combination of both.
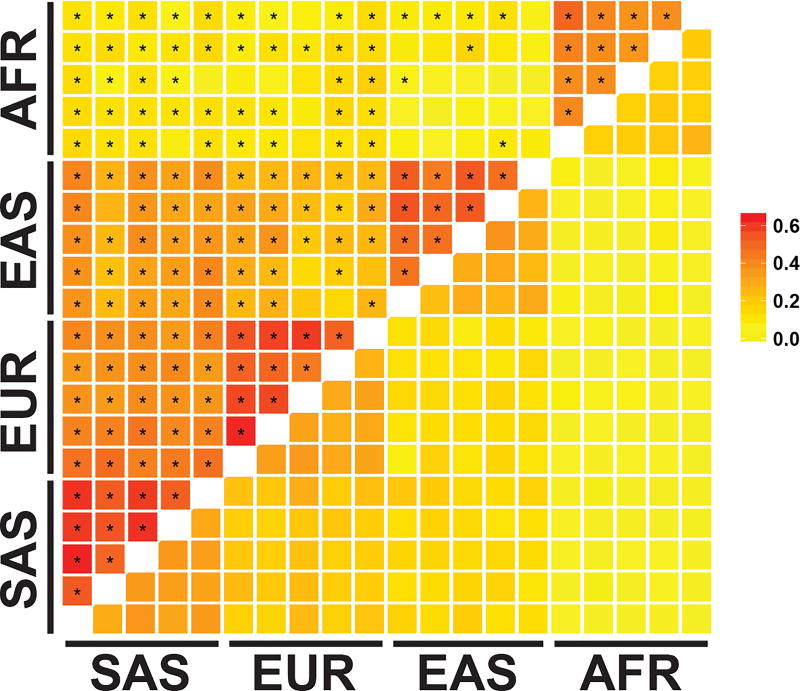
To determine if the observed extent of sweep sharing was unusual, we applied our fastPHASE haplotype labeling procedure to matched random sites across the genome. For all within-continent population pairs, and all but 4 of 75 Eurasian between-continent pairs, the degree of sweep sharing was higher than the background rate (Fig. 4, Supplementary Table 6), suggesting that the sweep sharing we observed was not driven purely by haplotype similarities across closely related populations. The number of populations in a shared sweep was inversely correlated with the length of the shared haplotype (Spearman’s rho = −0.23, P < 2.2×10−16, Supplementary Figure 4), which is compatible with more widely shared sweeping haplotypes being broken down by recombination over time. There was a borderline significant correlation between the nucleotide diversity of a sweeping haplotype and the number of populations sharing a sweep (Spearman’s rho = 0.049, P = 0.041, Supplementary Figure 5), though this weak correlation was not significant when we looked at diversity between populations within continents.
Fig 4. Enrichment of shared sweeps across population pairs.
Squares below the diagonal represent the null fraction of overlaps shared across population pairs, from randomly placed overlaps across the genome. Squares above the diagonal represent the observed fraction of sweep overlaps shared for each population pairs. Squares are marked with an asterisk if the observed fraction shared was significantly higher than the null distribution. Populations are arranged alphabetically within continental groups by population code, top to bottom, right to left.
Though the majority of between-continent shared sweeps were found across non-African populations, we did observe examples of shared sweeps between African and non-African populations. In total, 9.4% of observed sweep overlaps between African and non-African population pairs were called as shared (491 total), compared with 4.0% of control overlaps (99% CI: 3.7–4.4%). For example, on chromosome 1 at ~47Mb, a sweeping haplotype shared across African and European populations fell in a cluster of cytochrome P450 genes (Supplementary Figure 6, Supplementary Note).
Shared and overlapping sweeps in a region implicated in malarial resistance
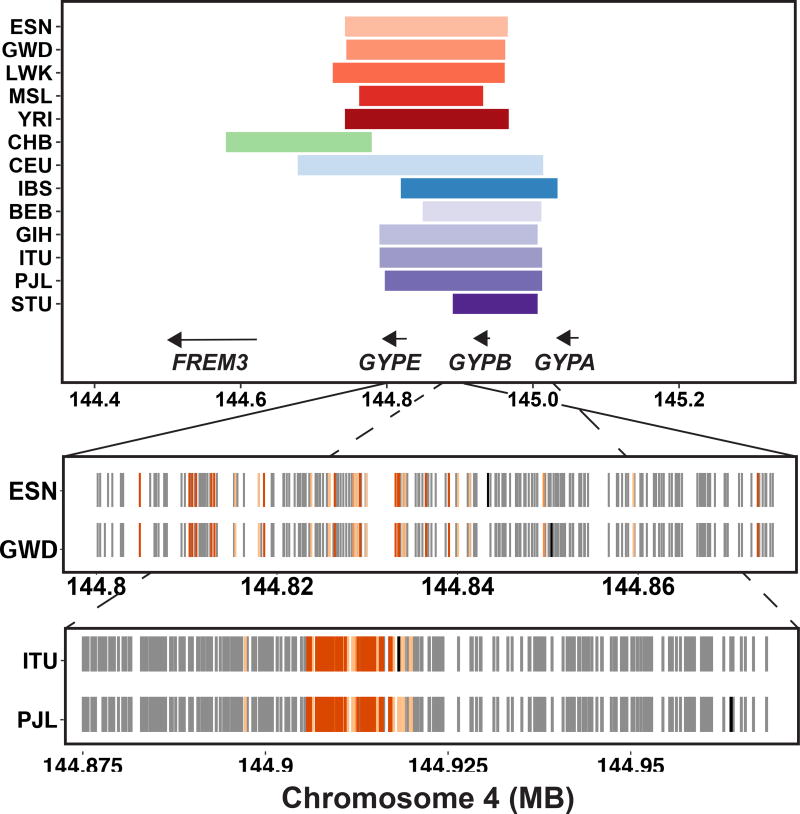
With a catalog of shared and overlapping selective sweeps in hand, we next aimed to identify specific regions of sweep sharing that connected the interval to a gene, pathway, or phenotype when considered alongside additional genomic annotations. With a sweep overlap across thirteen populations from all four continental groups, the glycophorin gene cluster (GYPA/GYPB/GYPE) came to our attention for its repeated targeting by positive selection and its prior implication in malaria resistance (Fig. 5). This genomic region has been noted as a target of positive selection in humans15–18, and as a target of ancient balancing selection shared between humans and chimpanzees19. In IBS and South Asians, the sweep appeared to be on a shared haplotype, while the African populations, CHB, and CEU each had unique sweeping haplotypes (Fig. 5). This complex locus contains a segmental duplication, making mapping and phasing of short-read data difficult. However, we observed residual unusual iHS in the surrounding region, and in an iHS scan of only those variants passing the 1KG “strict” mask. We identified multiple signatures of positive selection on distinct haplotypes in all four continental groups (Supplementary Table 5), with some linked to one or more potentially causal variant(s), either coding or structural (Supplementary Note). The frequency and diversity of apparent adaptive pressures at this locus underscores the role of selection on host-pathogen interactions over recent and longer evolutionary time-scales in modern humans, and the potential importance of this locus in particular in that process.
Fig 5. Signatures of positive selection at the GYP locus on chromosome 4.
We observed signatures of positive selection in 13 populations at the GYP locus, including at least one population from each studied continental group. The top panel shows the sweep intervals of populations with sweeps at this locus, and the positions of the GYP genes. The bottom panels show the sweeping haplotypes for two African (ESN, GWD) and two South Asian (ITU, PJL) populations’ within-continent shared sweeps. The gray tick marks in each populations’ row indicate the presence of a derived allele on the sweeping haplotype most common in that population, with a black tick indicating the position of each population’s SNP with the most extreme iHS value. Also shown in orange are the significant eQTLs for GYPE (light orange) or both GYPB and GYPE (dark orange) in LD with these population’s shared haplotype (D’ = 1). The eQTLs for GYPB and GYPE are from whole blood.
Intersection of signatures of positive selection with the GWAS catalog
Previous work has indicated an enrichment of extreme iHS at GWAS signals for autoimmune diseases20, and we hypothesized that this or other traits might be enriched for GWAS signatures linked to our signatures of positive selection. We tested for enrichment of linked GWAS SNPs overall and for specific traits (autoimmune disorders, height, and schizophrenia) by generating random SNP sets from the HapMap3 variant set (Methods), and identifying the number of sweeps in each population linked to a GWAS variant compared to the random SNP sets. We saw no clear, compelling evidence of enrichment for GWAS SNPs overall or any of the traits tested. In total, 186 sweep tag SNPs from all 20 populations (out of 11,655; 1.6%) were in strong LD with at least one genome-wide significant GWAS SNP (r2 ≥ 0.9, Supplementary Table 7), compared to a mean of 193 in our random SNP sets (95% C.I.: 168–220). However, this intersection did identify candidates for a potential phenotype under selection at some loci. In one example, a sweep overlap across all five African populations falls at the gene DGKK on the X chromosome (Supplementary Note, Supplementary Figure 7). Variants in this gene have been associated in Europeans with hypospadias21,22, a prevalent birth defect of ectopic positioning of the opening of the urethra in males. A second example occurred at the MTHFR gene on chromosome 1, where a nonsynonymous variant (A222V, rs1801133) has been extensively studied for its association with homocysteine levels23,24. A sweep overlap at this locus with three European populations (CEU, GBR, IBS) and JPT was called as shared across all four populations (Supplementary Note, Supplementary Figure 8).
Evidence for adaptive introgression from Neandertals in non-African populations
Examples of positive selection on introgressed genetic variation have shown that positive selection acted on genetic variation from ancient hominins at some loci. While some of these examples are confined to a single population (e.g. EPAS1 in Tibetans25), most are common across multiple populations26,27, and thus we hypothesized that a subset of our shared sweeps could be examples of adaptive introgression. We identified 141 candidate sweeps in partial to strong LD with inferred introgressed haplotypes9 (r2 ≥ 0.6, Methods, Supplementary Table 8), including previously described adaptive targets such as the HYAL2 locus in East Asians28 and OAS1 in Europeans29. We did not observe an overall enrichment of these introgressed haplotypes in our iHS intervals (P = 0.59, χ2 test, Methods), suggesting that introgression alone was not predictive of an unusual iHS signature. Of these 141 loci, we illustrate two candidate sweeps shared across multiple populations (Supplementary Note): (i) a shared sweep between Europeans and South Asians on chromosome 3 near CT64, a non-coding RNA primarily expressed in the testes (Supplementary Figure 9); and (ii) a sweep at ~41Mb on chromosome 1, where all five South Asian populations have evidence of an introgressed haplotype at low to moderate frequency (18–30%) (Supplementary Figure 10).
Overlapping and shared sweeps enriched in the ethanol oxidation pathway
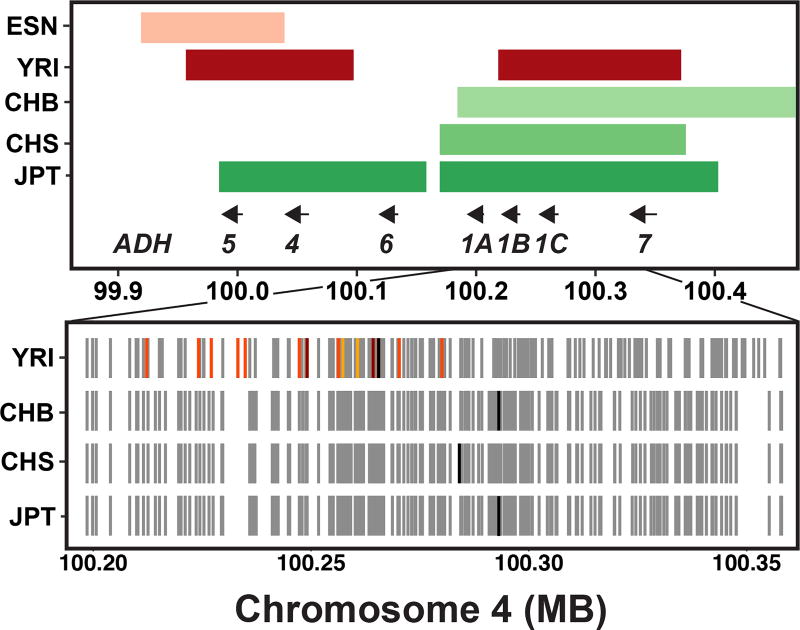
We next sought to explore possible biological pathways targeted by shared selective events. As a large fraction of causal variants under positive selection are potentially non-coding30–32, we hypothesized that regulatory variation in the form of eQTLs could indicate a potential causal, functional variant and/or gene target. We identified genes with cis-eQTLs from all tissue types in the GTEx V6p dataset that were linked with shared sweeps (r2 ≥ 0.9) and tested for overrepresentation of biological pathways in this set of genes using ConsensusPathDB33. Excluding the human leukocyte antigen (HLA) genes (Methods), the most significant pathway was ethanol oxidation (P = 2 × 10−5, q-value = 0.047), with 7 of 10 genes included in our shared sweeps gene set. This pathway includes the ADH gene cluster, which contains a previously described East Asian selective event targeting rs1229984-T34, a derived nonsynonymous variant in ADH1B associated with increased ADH1B enzyme activity35 and decreased risk of AD in East Asians36. A recent report also found evidence for an independent selective event for rs1229984 in Europeans37. Within this region, we observed the East Asian sweep, and independent sweeping haplotypes in YRI and ESN (Fig. 6).
Fig 6. Signatures of positive selection at the ADH locus on chromosome 4.
The top panel shows the sweep intervals of populations with sweeps at this locus, and the positions of the seven ADH cluster genes. The bottom panel shows the sweeping haplotypes of the four populations with sweeps in this region, with grey tick marks indicating the derived alleles present on the most common sweeping haplotype in that population. The black tick marks indicate the position of the SNP with the most extreme iHS in each population. For YRI, the positions of significant ADH4 and ADH1C eQTLs in subcutaneous adipose tissue (light orange), GWAS SNPs from Gelernter et al., 2014 (dark orange), and SNPs that are eQTLs for both genes and are GWAS SNPs (red) in LD with YRI’s tag SNP (r2 > 0.9) are shown.
As genome-wide association studies have identified genetic variation in the ADH locus associated with alcohol dependence (AD)38,36, we tested whether these associations were linked to the sweeping haplotype. In the YRI sweep interval spanning ADH1B, the derived allele of the leading iHS SNP (rs12639833-T, iHS = −5.133) was significantly associated with decreased risk for AD in African Americans38. This AD GWAS in African Americans identified independent associations at a nonsynonymous variant in ADH1B (rs2066702) and a synonymous variant in ADH1C (rs2241894)38. The sweep tag in YRI is in perfect LD with ADH1C lead variant rs2241894 (r2 = 1), lies in an intron of ADH1C, and is a significant eQTL for ADH1C (esophagus mucosa) and ADH4 (esophagus muscularis, skeletal muscle). Several other SNPs in strong LD with rs12639833 have extreme negative iHS, are eQTLs for increased ADH1C and ADH4 expression (including in the liver)8,39, and were significantly associated with decreased risk for AD in African Americans (Supplementary Table 9). Alcohol dehydrogenases oxidize ethanol to acetaldehyde, a process that is thought to occur primarily in the liver40. These data suggest a similar mechanism is at play in individuals of West African ancestry as in East Asians, where the selected allele increases ADH enzyme activity35, resulting in an adverse physical response from alcohol consumption41, and reduced risk for alcohol dependence36. Taken collectively, these patterns suggest that (i) alcohol oxidation pathways broadly have been subject to recent positive selection in humans, (ii) that genes in this pathway have been repeatedly targeted, with multiple events segregating at these sites, (iii) the selective pressure appears to operate across the major continental groups included in this study, and (iv) sweeping haplotypes at the ADH locus tag functional variation associated with protection against alcohol dependence.
In summary, we identified overlapping and shared signatures of positive selection across human populations, using a modified version of the iHS statistic. We observed more extreme iHS in sequencing data compared to SNP array genotype data, which could be a consequence of more rapid decay of homozygosity on unselected haplotypes due to the presence of rare variants. We found that closely related populations are more likely to share sweeping haplotype signatures, though we identified examples of sharing across genetically distant populations. These loci immediately raise questions of how these examples arose, whether by gene flow after divergence or a common ancestral event. Though only a small amount of gene flow between African and non-African populations is thought to have occurred since their divergence, the introduction of an adaptively advantageous allele at very low frequency could lead to the signature we observe here. But in considering the collection of putative shared sweeps we highlighted here, it seems apparent that each locus is unique, segregating patterns of genetic variation, suggestive of a range of compatible (and potential quite complex) population models that could help explain these data. Future work to model the potential scenarios leading to shared sweeps will likely require modeling of individual regions to elucidate the evolutionary history of specific events. We also found that the rate of sweep overlaps is not uniform across the genome, but in some locations overlaps cluster together, contributing to the complexity of the sweeps in those regions. These features made identifying the tag SNP for a sweep and calling sharing between sweep overlaps difficult in these regions. That said, we hope that our catalog of unusually long haplotypes shared across human populations will help to elucidate genes - and ultimately phenotypes - that are still evolving across the wide range of environments human have experienced in recent history.
MATERIALS AND METHODS
iHS scan
We downloaded phased genotype files for phase 3 of the 1000 Genomes Project from the 1KG FTP (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/). These data were converted to Beagle-formatted files, and filtered to include only biallelic SNVs (excluded indels) with a minor allele frequency (MAF) greater than 1%. A fine-scale recombination map was downloaded from the 1KG FTP (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130507_omni_recombination_rates/), and scaled to units of ρ (=4Ner) for each population. Effective population size was estimated for each population by calculating nucleotide diversity (π) in a sliding window (100kb) across the genome, and estimating Ne from the median values π (Ne = π/(4*µ)). Ancestral alleles were identified using the human-chimp-macaque alignment from Ensembl (accessed from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/). SNPs were filtered for only those where the ancestral allele was supported by both the chimp and macaque alignments.
Unstandardized iHS were calculated using WHAMM (v0.14c), using a modified version of iHS calculation code (v1.3) that increased speed of the calculation, and initially standardized by derived allele frequency as described in the original iHS paper11, with 50 allele frequency bins. In the final standardization, we binned autosomal SNPs into 500 bins (50 allele frequency bins × 10 local recombination rate bins), or 150 bins for chromosome X (50 allele frequency bins × 3 local recombination rate bins). These standardization files are available in Supplementary Table 1.
Regions of the genome putatively undergoing recent hard sweeps - what we refer to in the main text as iHS intervals - were identified by counting the number of SNPs with |iHS| > 2 in 100kb windows (windows incrementing by one SNP, i.e., overlapping windows). We took the union of the top 1% of windows, by the total number or by fraction of SNPs with |iHS| > 2 in the window, as our intervals. We performed this interval calling separately for each of the 20 populations included in this study. The SNP we use to label (i.e., tag) each sweep interval was identified as the SNP with the most extreme iHS, and the sweep frequency as the tag SNP derived allele frequency if the iHS was less than zero, and ancestral allele frequency if iHS was greater than zero. We limited our analyses of individual sweep loci to those with a tag SNP of MAF > 15%, to focus on signatures unlikely to have extreme iHS due to very low frequency.
Neutral simulations
We performed neutral simulations of a population with a CHB-like demographic history using the forward simulation software SLiM42 (v. 2.4.2). We simulated chromosomes with the following demographic model43: an ancestral population size of 13,000 diploids with a mutation rate of 1 × 10−7 was burned in for 130,000 generations, then a bottleneck reduced the population size to 2,500 diploids at 32,490 generations, followed by exponential growth at a rate of 1.002082, and finally 216 diploids were sampled at generation 34,500. We simulated 85 3MB regions at a recombination rate of r = 1×10−8 or 1×10−12, and calculated unstandardized iHS for all variants. Only variants in the central 200 kb were used for the comparison in Supplementary Figure 2, to maximize the number of low-frequency (<5% MAF) variants that reached the end of their haplotype within the simulated chromosomes (for a total of 159,487 variants at r = 1 × 10−8, and 151,710 variants at r = 1 × 10−12). The mean of unstandardized iHS for variants in an allele frequency bin were compared between the difference recombination rates with the Mann-Whitney test (wilcox.test), and the variances were compared with the F test (var.test) in R44.
Sweep overlaps
To identify sweep overlaps, we compared the iHS intervals for each population and identified regions of the genome where two or more populations had a sweep interval. We calculated the fraction of sweep overlaps for each population pair as the mean of the fraction of sweep intervals in one population that overlap with a sweep interval in the second population (i.e. (fraction in pop A + fraction in pop B) / 2). We estimated FST for each pair of populations across all variants (n=2,627,240) in the 1000 Genomes VCF files on chromosome 2 using the Weir and Cockerham estimator implemented in VCFtools (v. 0.1.12b)45. We performed linear regression on (fraction of overlaps ~ FST) for each population pair, and estimated the standard error of the slope using a block jackknife for unequal group size46, deleting one population pair group (e.g. EUR v EUR, or AFR v EUR) for each resample.
Rates across the genome
To assess the rate of sweep intervals across the genome, we subdivided the genome into ten megabase non-overlapping windows (n=297 in total) and counted the number of sweep intervals for each individual population, and the number of overlaps across 2 or more populations, in each window. To ensure the sweep intervals called for each population were independent, we merged adjacent sweep intervals into one interval if their tag SNPs were in modest LD or greater (r2 > 0.4). If a sweep interval or overlap spanned two windows, we counted it once in the window with more than half of its physical distance. We used all Ensemble HG19 gene annotations (from http://genome.ucsc.edu/cgi-bin/hgTables), merged into non-overlapping intervals with BEDTools v2.19.147. We used the local recombination rates described above, and background selection estimates (B statistic) were downloaded from http://www.phrap.org/software_dir/mcvicker_dir/bkgd.tar.gz13. We performed forward regression with the medians of B and local recombination rate in our windows as potential predictors of overlap count in each window, along with the gene count in the window. The final model included only median local rho (R2 = 0.048, P = 5.2×10−3), as median B explained no additional variation. For Poisson mixture modeling of the overlap rate, we fit mixtures of independent Poisson distributions to the data by minimizing the negative log likelihood with the non-linear minimizer function (nlm) in R44. We compared mixture models by calculating the Bayesian information criterion and performing a likelihood ratio test.
Identifying sweeping haplotypes with fastPHASE
For each sweep overlap, we identified the physical region spanning all tag SNPs, and an additional 5kb to either side. We ran fastPHASE on this region, using the -u option to identify each 1KG population as a subpopulation, -B to indicate known haplotypes, and -Pzp to output cluster probabilities for each individual at each SNP. We tested a range of values of K (number of haplotype clusters) and T (number of random EM algorithm starts) on a subset of sweep overlaps, and found broadly similar results across the range (Supplementary Figure 11). We used K = 10 clusters and T = 10 for all overlaps in the final analysis. From the output cluster probabilities, we identified the sequence of haplotype clusters for each SNP position in each individual as the most likely haplotype cluster at each SNP. We then identified the haplotype cluster sequences of all chromosomes carrying the selected tag allele, and the most common of those to be the reference sweeping haplotype sequence.
To identify if a pair of populations as “shared”, we required an identical reference haplotype sequence to span the selected tag allele in both populations. To form shared clusters, we grouped together all populations that were called as shared with at least one other population. To calculate the null rate of haplotype sharing across population pairs, we selected random regions of the genome of the same size (within 10kb), distance to nearest gene (within 50kb), and local recombination rate (within an order of magnitude of rho) as our observed sweep overlap regions. For each sweep overlap, we identified 10 matched windows, for a total of 30,450 regions across the genome (ranging from 153–2588 random overlaps per population pair). We identified tag SNPs for each population in the random regions matching the distance from the other populations’ tag SNPs and derived allele frequency (within 5%) of the observed overlap. We then ran fastPHASE on the randomly selected regions and performed the shared haplotype-calling procedure as for observed overlap windows described above. To compare the observed fraction of overlaps called as shared to the null haplotype sharing for each pair of populations, we performed 1000 bootstraps by sampling with replacement the number of observed overlaps from the null. Population pairs where the shared sweep fraction of observed overlaps was higher than the shared fraction of random overlaps for all 1000 samples are marked with an asterisk in Fig. 4. We performed linear regression on (fraction of shared overlaps ~ FST) for each population pair, and estimated the standard error of the slope using a block jackknife for unequal group size46, deleting one population pair group (e.g. EUR v EUR, or AFR v EUR) for each resample.
We measured the length of the shared sweeping haplotype in each population as the maximum length of the reference shared haplotype, identified as above, in that population. We calculated nucleotide diversity in each population as the average number of pairwise differences between chromosomes carrying the tag allele in the region of the shared haplotype, divided by the length of the shared haplotype in that population. We tested for correlations between the number of populations sharing a sweep and the mean shared haplotype length and mean nucleotide diversity using Spearman’s rho in R (cor.test). We also tested, in each population individually, the correlation between nucleotide diversity of the shared haplotype in that population and the number of populations in the shared sweep.
eQTLs linked to sweep haplotypes
To connect shared sweeps to potential causal genes, we utilized the GTEx v6p eQTL dataset8 downloaded from the GTEx portal (http://www.gtexportal.org/). For each population’s tag SNPs, we identified LD proxies (r2 ≥ 0.9, calculated in the same population) within 1 Mb of the sweep interval, and intersected these SNPs with all significant GTEx eQTLs from all tissue types. eQTLs in the GTEx V6p data set were identified using a cohort of mostly white individuals (84.3%), with a smaller fraction of African Americans (13.7%). For sweep overlaps that were called as shared, we identified a shared SNP set as the intersection of LD proxy sets for all populations in a shared group. We created a gene list of all genes with eQTLs from any tissue that intersected with shared SNP sets, excluding HLA genes. We chose to exclude HLA genes, owing to its genomic complexity and its enrichment for signatures of recent positive selection. To test for enrichment of this gene set with biological pathways, we used over-representation analysis of all pathway databases in ConsensusPathDB (http://cpdb.molgen.mpg.de/)33 with the background set of all genes.
Intersection of sweeps with Neandertal haplotypes, Neandertal PheWAS, and GWAS SNPs
We downloaded the Neandertal haplotype calls reported in 48 from http://akeylab.gs.washington.edu/vernot_et_al_2016_release_data/. This dataset contains inferred introgressed Neandertal haplotypes on the autosomes of the non-African individuals from 1KG Phase 3. To calculate LD between introgressed haplotypes and sweep tag SNPs, we pooled overlapping haplotypes across individuals and created a genotype of 0 or 1 based on presence/absence of the overlapping introgressed haplotype in each individual. We then calculated LD between this presence/absence genotype and the tag SNPs within 1 Mb of the introgressed haplotype separately for each population. We considered haplotypes with R2 > 0.6 with sweep tag SNPs as candidates for adaptive introgression. To examine potential enrichment of introgressed haplotypes in LD with sweep tag SNPs, we compared the fraction of introgressed haplotypes in LD with sweep tag SNPs to the fraction of distance and frequency matched SNPs in LD with sweep tag SNPs (r2 > 0.6) using a χ2 test. We downloaded the Neandertal PheWAS data at https://phewascatalog.org/neanderthal 49, and intersected all reported associations with variants in strong LD (r2 ≥ 0.9) with each sweep tag SNP in each population.
We downloaded the GWAS catalog from https://www.ebi.ac.uk/gwas on 10/12/16. We identified all genome-wide significant associations (P < 5 × 10−8) in strong LD (r2 ≥ 0.9) with each sweep tag SNP in each population. To test for enrichment of GWAS variants generally and of specific phenotype classes, we performed permutation tests with 10,000 random SNP sets from the HapMap3 variant set (from ftp://ftp.ncbi.nlm.nih.gov/hapmap/phase_3/) matched for allele frequency and distance to gene with the GWAS variants of interest. We then compared the empirical distribution of intersection of these matched SNP sets with the sweep tag SNPs and proxies to the number of observed GWAS intersections. To control for potentially linked GWAS variants, we simply counted the number of sweeps in each population that intersected a GWAS or control set variant.
Indels, structural variants, and annotations
Indels were not included in our iHS scan, but could be the causal variant on a sweeping haplotype. To identify candidates for causal indels, we calculated LD with sweep tag SNPs for all indels in the 1000 Genomes phase 3 VCF files within 1 Mb of the sweep interval in each population. To identify potential functional coding variants among indels and SNPs on sweeping haplotypes, we used ANNOVAR to annotate coding variation50. In the glycophorin region, we tested for LD between sweep tag SNPs and structural variant calls from phase 3 of the 1KG (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/integrated_sv_map/; February 24, 2015 release).
Supplementary Material
Acknowledgments
We would like to thank C. Brown, E. Leffler, and three anonymous reviewers for helpful comments that improved the quality of the manuscript. This work was supported by grants from the National Institutes of Health (NIDDK R01DK101478 to BFV, T32GM008216 to KEJ) and a fellowship from the Alfred P. Sloan Foundation (BR2012-087 to BFV).
Footnotes
Data Availability
The standardized iHS values for all 26 populations in 1KG Phase 3 are available at http://coruscant.itmat.upenn.edu/data/JohnsonEA_iHSscores.tar.gz.
Code Availability
WHAMM v0.14c and iHS calculator v1.3 are available at http://coruscant.itmat.upenn.edu/whamm/dlnotes.html. Custom code used to process and analyze output from the iHS scan and fastPHASE is available at https://github.com/kelsj/sharedSweeps.
AUTHOR CONTRIBUTIONS
K.E.J. and B.F.V. planned the study. K.E.J. assembled input data and performed the experiments. K.E.J. and B.F.V. interpreted the data and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Tishkoff Sa, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamberov YG, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fumagalli M, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–7. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 4.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metspalu M, et al. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am. J. Hum. Genet. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, et al. Detecting and characterizing genomic signatures of positive selection in global populations. Am. J. Hum. Genet. 2013;92:866–881. doi: 10.1016/j.ajhg.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguet F, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernot B, Akey JM. Resurrecting Surviving Neandertal Linages from Modern Human Genomes. Science (80-. ) 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 10.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:0446–0458. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baum J, Ward RH, Conway DJ. Natural selection on the erythrocyte surface. Mol. Biol. Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- 16.Wang HY, Tang H, Shen CKJ, Wu CI. Rapidly Evolving Genes in Human. I. The Glycophorins and Their Possible Role in Evading Malaria Parasites. Mol. Biol. Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 17.Ko WY, et al. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am. J. Hum. Genet. 2011;88:741–754. doi: 10.1016/j.ajhg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leffler EM, et al. Resistance to malaria through structural variation of red blood cell invasion receptors. 2017;356:1–29. doi: 10.1126/science.aam6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leffler EM, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj T, et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am. J. Hum. Genet. 2013;92:517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Zanden LFM, et al. Common variants in DGKK are strongly associated with risk of hypospadias. Nat. Genet. 2011;43:48–50. doi: 10.1038/ng.721. [DOI] [PubMed] [Google Scholar]
- 22.Geller F, et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet. 2014;46:957–963. doi: 10.1038/ng.3063. [DOI] [PubMed] [Google Scholar]
- 23.Paré G, et al. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma Homocysteine in a healthy population a genome-wide evaluation of 13 974 participants in the women’s genome health study. Circ. Cardiovasc. Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Meurs JBJ, et al. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am. J. Clin. Nutr. 2013;98:668–76. doi: 10.3945/ajcn.112.044545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huerta-Sánchez E, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racimo F, Marnetto D, Huerta-Sánchez E. Signatures of Archaic Adaptive Introgression in Present-Day Human Populations. Mol. Biol. Evol. msw216. 2016 doi: 10.1093/molbev/msw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Q, Hu Y, Xu S, Wang J, Jin L. Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in east asians. Mol. Biol. Evol. 2014;31:683–695. doi: 10.1093/molbev/mst260. [DOI] [PubMed] [Google Scholar]
- 29.Sams AJ, et al. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman SR, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enard D, Messer PW, Petrov Da. Genome-wide signals of positive selection in human evolution. Genome Res. 2014;24:885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu F, et al. Population genomic analysis of 962 whole genome sequences of humans reveals natural selection in non-coding regions. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–8. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, et al. Evidence of positive selection on a class I ADH locus. Am. J. Hum. Genet. 2007;80:441–456. doi: 10.1086/512485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol. Psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galinsky KJ, et al. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelernter J, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry. 2014;19:41–9. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pashos EE, et al. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 2017;20:558–570. e10. doi: 10.1016/j.stem.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SL, Chau GY, Yao CT, Wu CW, Yin SJ. Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: Significance of first-pass metabolism. Alcohol. Clin. Exp. Res. 2006;30:1132–1142. doi: 10.1111/j.1530-0277.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 41.Matsuo K, et al. Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: Analysis of 2,299 Japanese subjects. Cancer Epidemiol. Biomarkers Prev. 2006;15:1009–1013. doi: 10.1158/1055-9965.EPI-05-0911. [DOI] [PubMed] [Google Scholar]
- 42.Haller BC, Messer PW. SLiM 2: Flexible, interactive forward genetic simulations. Mol. Biol. Evol. 2017;34:230–240. doi: 10.1093/molbev/msw211. [DOI] [PubMed] [Google Scholar]
- 43.Terhorst J, Kamm JA, Song YS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2016;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Development Core Team. R Found. Stat. Comput. Vienna Austria. Vol. 0. 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 45.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busing FMTA, Meijer E, Leeden R. van der. Delete-m jackknife for unequal m. Stat. Comput. 1999;9:3–8. [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vernot B, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–9. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonti CN, et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science (80-. ) 2016;351:737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.