Abstract
Phytase plays a prominent role in monogastric animal nutrition due to its ability to improve phytic acid digestion in the gastrointestinal tract, releasing phosphorus and other micronutrients that are important for animal development. Moreover, phytase decreases the amounts of phytic acid and phosphate excreted in feces. Bioinformatics approaches can contribute to the understanding of the catalytic structure of phytase. Analysis of the catalytic structure can reveal enzymatic stability and the polarization and hydrophobicity of amino acids. One important aspect of this type of analysis is the estimation of the number of β-sheets and α-helices in the enzymatic structure. Fermentative processes or genetic engineering methods are employed for phytase production in transgenic plants or microorganisms. To this end, phytase genes are inserted in transgenic crops to improve the bioavailability of phosphorus. This promising technology aims to improve agricultural efficiency and productivity. Thus, the aim of this review is to present the characterization of the catalytic structure of plant and microbial phytases, phytase genes used in transgenic plants and microorganisms, and their biotechnological applications in animal nutrition, which do not impact negatively on environmental degradation.
1. Introduction
Biocatalysts are important molecules that improve fermentable substrate availability and nutrient absorption, increasing the efficiency of food bioconversion. Phytases, proteases, xylanases, and amylases are typical examples of enzymatic groups that have wide applicability and contribute to increased productivity and profitability [1–3]. In monogastric animals, such as pigs, poultry, and humans, there is little or no phytase activity [4, 5]. However, these animals are fed with seeds and grains rich in phytate, which is not completely absorbed. Thus, phytate is excreted in the feces, increasing the environmental pollution by phosphorus [6, 7].
Sustainable agriculture can be achieved by adding phytase to animal feeds. Phytase improves the quality and bioavailability of some important nutrients, reducing the environmental impact of farming at the animal feed stage [8–10]. Thus, several strategies can be explored with the aim of achieving bioprocesses that increase yields and phytase stability by using microorganisms and plants. Among the microorganisms capable of producing phytase are bacteria, such as Lactobacillus, Escherichia, Bacillus, Xanthomonas, Pseudomonas, and Klebsiella spp.; fungi, such as Penicillium, Aspergillus, and Rhizopus spp.; and yeasts, such as Saccharomyces cerevisiae, Schwanniomyces castellii, Schizophyllum commune, Wickerhamomyces anomalus, and Hansenula spp. [11–13].
Plant growth and productivity can be hindered by low availability of phosphorus (P) in the soil [14]. Therefore, plants have developed mechanisms to improve soil phosphorus availability, with emphasis on the participation of purple acid phosphatases (PAPs). PAPs are acidic metallohydrolases that are involved in plant growth and pathogen defense. PAP-like isoforms have been identified in Ipomoea batatas, Solanum lycopersicum, and Hordeum vulgare. In barley, PAPs are grouped into isogenes HvPAPhy_a, HvPAPhy_b1, and HvPAPhy_b2 [15–17]. Another consideration is the technical compatibility of microbial phytase genes and plants, which enables the generation of transgenic crops. Understanding of the catalytic structure and amino acid sequences of microbial phytases and plants, via analysis of the principal genes, allows for technological advances using phytase without increasing environmental degradation.
Thus, the aim of this review is to present the characterization of the catalytic structure of plants and microbial phytases and their biotechnological applications, while proposing technical alternatives for sustainable agricultural productivity.
2. Phytase and Phosphorus Bioavailability
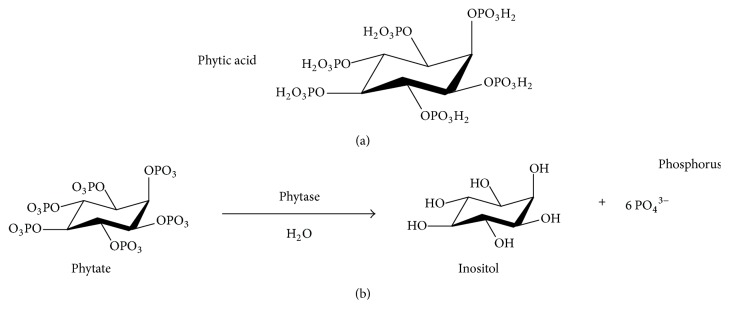
Phytate (C6H18O24P6), also known as phytic acid (myo-inositol 1,2,3,4,5,6-hexakisphosphate; PA; IP6), has a molecular weight of 660 g mol−1 (Figure 1) and is considered the main source of stored phosphorus in seeds, grains, and vegetables. Phytate is known as a food inhibitor due to its ability to chelate and thus decreases the bioavailability of some important micro- and macronutrients, such as Zn, Mg, Mn, Fe, and Ca, in monogastric animals [18, 19].
Figure 1.
Molecular structures. The phytic acid (a) and mechanism of enzyme action (b) developed by ChemBioDraw Ultra version 12 software.
Phytate is a phosphorus storage molecule and a constituent of cereals and grains. It has relatively low bioavailability due to its strong adsorption onto soil and unavailability for degradation by soil microorganisms [20, 21]. Plants have developed a variety of mechanisms to overcome this problem, such as upregulation of high-affinity phosphate transporters, improvement of internal phosphatase activity, and secretion of organic acids and phosphatases [22–25]. The latter is an important aspect as it allows mineralized organic phosphorus to be released as inorganic phosphorus into the soil [26]. Phytases are special phosphatase enzymes that catalyze the hydrolysis of phytate into lower inositol phosphates and inorganic phosphorus (Figure 1).
Human beings are limited in their ability to hydrolyze phytate; consequently, phytic acid reduces the amount of minerals required for tissue function and cellular metabolism maintenance [27]. Pigs and poultry are unable to metabolize phytate because they lack the enzyme phytase in their gastrointestinal tract; therefore, it is necessary to add inorganic phosphorus to animal feeds. However, a large amount of phytate and inorganic phosphorus are excreted into the environment because they are not fully absorbed by these animals, causing environmental impacts such as eutrophication of surface waters of lakes and rivers, harmful algal blooms, nitrous oxide production (N2O; greenhouse gas), growth of toxin-producing microorganisms, and the death of several aquatic species [18, 28]. Thus, it is desirable to use new techniques that aim to increase the quality and bioavailability of phosphorus in animal feed.
3. Characterization of Phytase
3.1. Sources of Phytase
Plants, animals, and microorganisms can produce phytase. In plants, it is present in wheat, barley, peas, soybeans, corn, rice, and spinach. The blood of vertebrates such as fish, sea turtles, and reptiles also contains phytase, which is produced by microorganisms such as yeasts, bacteria, and filamentous fungi [18, 19]. Microbial phytase is of great interest to industry due to its high level of production and extracellular activity, for example, in Aspergillus spp. [18]. Phytase (myo-inositol hexakisphosphate phosphohydrolase, EC 3.1.3.26 and EC 3.1.3.8) is characterized as a homodimeric enzyme [29, 30]. It is a phosphatase that initiates the sequential release of phytate orthophosphate groups (myo-inositol 1,2,3,4,5,6-hexakisphosphate). It belongs to the class of hydrolases and thereby hydrolyzes phytate (phytic acid) to release inositol phosphates, phosphorus, inositol, and other essential nutrients required for absorption (Figure 1). Moreover, it is an important component of a variety of metabolic processes as released phosphorus favors development, formation and mineralization of animal bone, cellular metabolism, and protein synthesis [31–35].
3.2. Catalytic Structure of Plant Phytase
Many enzymes with phytase activity can be obtained or expressed in plants, animals, and microorganisms [18, 36, 37]. They are classified into the following four groups according to their catalytic mechanisms: histidine acid phosphatases (HAPs), PAPs, Cys or β-helix phosphatases, and protein tyrosine phosphatase-like enzymes (PTP) [38].
Phytases are composed of several phosphatases; however, only some have sufficient phytase activity. The PAPs are acidic metallohydrolases, and their structures are involved in plant growth and pathogen defense. PAPs have a metal center constituting iron, zinc, and manganese [17]. The metal center coordinates the binding site residues with those of other termini. These regions are characterized by seven conserved amino acid residues in five conserved motifs (in italic): DXG, GDXXY, GNH(D/E), VXXH, and GHXH. These are involved in the dimetal nuclear center coordination [17, 39–42].
Other PAP-like isoforms have been identified in I. batatas and S. lycopersicum [15–17]. PAPhy genes in H. vulgare are grouped into isogenes: HvPAPhy_a, HvPAPhy_b1, and HvPAPhy_b2 [4]. The isogene HvPAPhy possesses remarkable phytase activities in mature grains and proteins produced by Pichia pastoris. Isoform a (HvPAPhy_a) has a molecular mass of 60,29 kDa and contains 544 amino acids, with the ligand-binding sites at amino acid sequence 199, 226, 283, 365, and 402. Isoform_b1 (HvPAPhy_b1) has a molecular mass of 59,51 kDa and contains 536 amino acids, with the ligand-binding sites at amino acid sequence 194, 221, 278, 359, and 396. Isoform b2 (HvPAPhy_b2) has a molecular mass of 59,34 kDa and contains 537 amino acids, with the ligand-binding sites at amino acid sequence 194, 221, 278, 360, and 397 (Figure 2). The genes HvPAPhy_a, HvPAPhy_b1, HvPAPhy_b2, PHY_US417, PHYA, AVPIDOX, MtPHY1, MtPT1, and SK-57 are the major genes evaluated in transgenic plants and microorganisms presenting phytase activity (Table 1).
Figure 2.
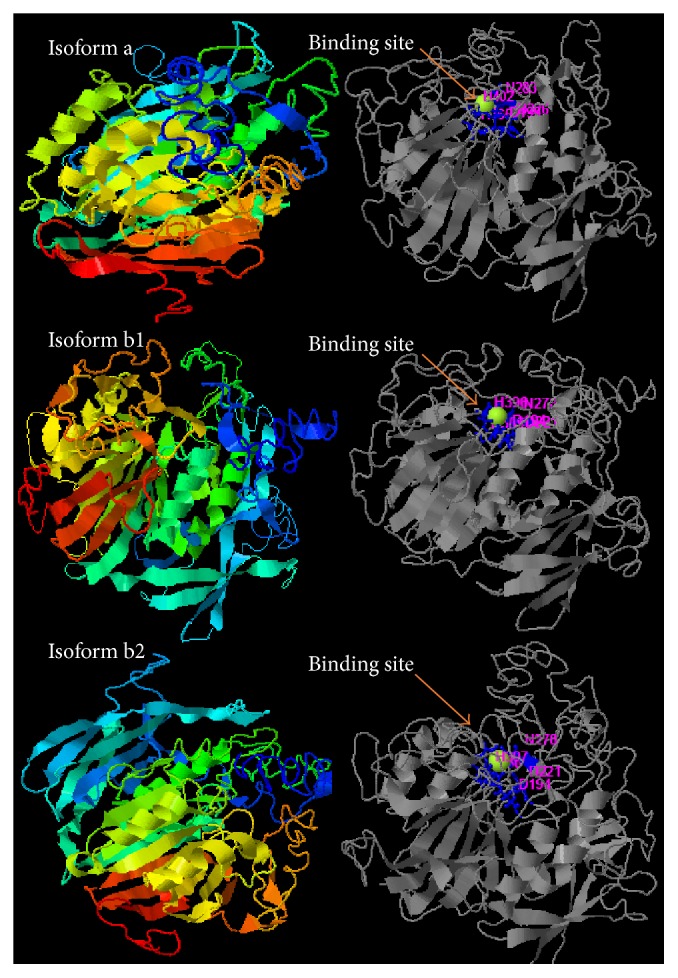
Structural model of purple acid phosphatase from barley (Hordeum vulgare) proposed by Dionisio et al. [4]. PAPhy genes are grouped in isogenes HvPAPhy_a, HvPAPhy_b1, and HvPAPhy_b2. Isogenes HvPAPhy possess significant phytase activity in the mature grains and proteins already were produced in P. pastoris. Structural model used i-TASSER server for protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [105]. FASTA sequences were obtained from https://www.ncbi.nlm.nih.gov/protein. Isoform a is constituted by 544 amino acids, 60,29 kDa, and Hphob of 49,5%. The ligand-binding site residues from isoform a are represented by amino acid sequence 199, 226, 283, 365, and 402. Isoform b1 is constituted by 536 amino acids and 59,51 kDa with the ligand-binding site residues being represented by amino acid sequence 194, 221, 278, 359, and 396. Isoform b2 is constituted by 537 amino acids and 59,34 kDa. Ligand-binding site residues are constituted by amino acid sequence 194, 221, 278, 360, and 397.
Table 1.
Major genes evaluated in transgenic plants and microorganisms with phytase activity.
| Gene | Plant target | Microbial target | Reference |
|---|---|---|---|
| HvPAPhy_a | Hordeum vulgare L. | — | Holme et al. [106] |
| PHY_US417 | Arabidopsis thaliana | — | Belgaroui et al. [14] |
| PHYA | Hordeum vulgare L. | — | Mohsin et al. [107] |
| AVP1DOX | Solanum lycopersicum | — | Yang et al. [108] |
| MtPHY1 | Medicago sativa L. | — | Ma et al. [109] |
| MtPT1 | Medicago sativa L. | — | Ma et al. [109] |
| HvPAPhya | — | Pichia pastoris | Dionisio et al. [4] |
| HvPAPhyb1 | — | Pichia pastoris | Dionisio et al. [4] |
| HvPAPhyb2 | — | Pichia pastoris | Dionisio et al. [4] |
| sk-57 | — | Pichia pastoris | Xiong et al. [73] |
3.3. Catalytic Structure of Microbial Phytase
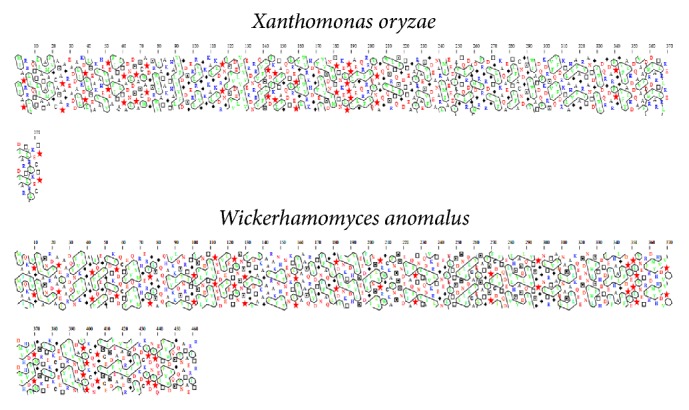
Bioinformatics approaches have contributed to increased knowledge about phytase [43]. One approach is comparative sequence analysis of phytase families, which allows for phylogenetic inferences and the prediction of functional sites [4, 34, 44]. Another is molecular modeling, which allows for inferences about enzymatic structure when no three-dimensional structure has been determined [45–47]. Based on the above-mentioned analyses, phytase B from Aspergillus niger (histidine acid phosphatases (HAP)) plays an important role in phytate hydrolysis. Its structure includes 460 amino acid residues and is composed of a large α/β-domain with a six-stranded β-sheet surrounded by several α-helices and a small α-domain. It contains five disulfide bonds at positions 52–368, 109–453, 197–422, 206–279, and 394–402, most of which are located in loops next to the surface. These bonds are due to the conformational stability of native phytases A and B and the maintenance of their catalytic activities. Moreover, researchers found that disulfide bonds have an important role in maintaining active site integrity [48]. Oakley [49] presented a structural phytase model of phytase A from A. niger, consisting of an α/β-domain, an α-domain, and an N-terminal extension. N-Acetylglucosamine residues are bound to four sites of the phytase structure (N82, N184, N316, and N353) within the active site, which is formed by an α-helix cavity. A structural model of A. niger phytase (HAP) proposed by Mishra et al. [50] is represented in Figure 3.
Figure 3.
Structural model of Aspergillus niger phytase proposed by Mishra et al. [50] available at https://www.ncbi.nlm.nih.gov/protein and i-TASSER server used for protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [105]. The domains are identified in the secondary structure elements being alpha-helices (1) and the secondary structure (2).
However, PhyA from Xanthomonas oryzae pv. oryzae (beta propeller phytase (BPHY)) plays a role in the degradation of phytic acid. It encodes a 373-amino acid protein including a 28-amino acid-predicted signal peptide. Its active site is located on the top of the β-propeller having a high conservation of amino acid residues involved in the metal ions binding to a phytase identified from Bacillus amyloliquefaciens (BaPhy), which has high- and low-affinity calcium sites responsible for enzymatic activity. PhyA from X. oryzae is similar to a phytase identified from Bacillus amyloliquefaciens (BaPhy), which has high- and low-affinity calcium sites responsible for enzymatic activity. However, differences in enzymatic activity between phytases can be attributed to differences in phosphate binding affinity [12].
Other types of phytases (HAP) PhyK and AppA are found in plants and microorganisms. PhyK belongs to a group of phytases synthesized by plant-associated bacteria, such as Xanthomonas campestris, Pseudomonas syringae, and Erwinia carotovora. This structural model was proposed by Böhm et al. [51] to explain the binding interaction between an enzyme and a substrate. The structural model observed and proposed by Shivange et al. [52] described the phytase of a mutant Yersinia mollaretii. Shivange et al. [52] described five amino acid substitutions, D52N, T77K, K139E, G187S, and V298M, next to the active site loop (S42-T47) and catalytically important residues involved in substrate binding (R37, R41, E241, and D327). Structural analysis of phytase from different environmental sources is important for understanding its specific role in the microenvironment [51]. Figures 4 and 5 represent structural models of X. oryzae phytase proposed by Wilkins et al. [53] and W. anomalus phytase proposed by Kaur et al. [54].
Figure 4.
Structural model of Xanthomonas oryzae phytase proposed by Wilkins et al. [53] available at https://www.ncbi.nlm.nih.gov/protein and i-TASSER server used for protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [105]. The domains are identified in the secondary structure elements being alpha-helices (1) and the secondary structure (2).
Figure 5.
Structural model of Wickerhamomyces anomalus phytase proposed by Kaur et al. [54] available at https://www.ncbi.nlm.nih.gov/protein and i-TASSER server used for protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [105]. The domains are identified in the secondary structure elements being alpha-helices (1) and the secondary structure (2).
Wickerhamomyces anomalus has already been described as Pichia anomala and Hansenula anomala. This species exhibits antimicrobial activities and flavoring features that are responsible for its frequent association with food, beverages, and feed products [13]. Joshi and Satyanarayana [55] described phytase from P. anomala as a HAP that is thermostable and used as a feed additive. The PPHY gene of P. anomala synthesizes a cell-bound phytase with an ORF of 1389 bp encoding a 462-amino acid protein [53]. Kaur et al. [54] described that the amino acid sequence analysis from P. anomala has varying similarity to those of phosphatases from Pichia stipitis (62%), Candida dubliniensis (51%), Candida albicans (51%), and Arxula adeninivorans (35%) and phytases from Debaryomyces castellii (50%) and Pichia fabianii (39%). However, amino acid sequence analysis from W. anomalus [54] showed only 5% similarity to phytase from A. niger [49].
Structural models of A. niger, W. anomalus, and X. oryzae presented here revealed varying percentages of hydrophobicity between microbial groups. The hydrophobicity of phytase from A. niger (44.8%) and that of phytase from W. anomalus (48.1%) are similar; however, that of X. oryzae is high and different (51.7%) (Figures 3–5). Similarly, the hydrophobicity of HvPAPhy_a is 49.5% (Figure 2). Phytases from A. niger, W. anomalus, and H. vulgare have low hydrophobicity compared with those from X. oryzae. The degree of hydrophobicity is an important aspect and reflects the proportions of β-sheets and α-helices in the protein structure. Additionally, the relative amounts of hydrophobic and hydrophilic amino acids partially explain the enzymatic stability. HAP phytase from Sporotrichum thermophile showed that surface charge distribution and a high density of hydrophilic amino acids on the surface contribute to the thermal stability of phytase [56]. Moreover, using bioinformatics analysis, Bertrand et al. [57] reported that hydrophobic amino acids are not distributed randomly but form clusters. Thus, it has been suggested that the position of a cluster corresponds to real positions in the secondary structures [57]. Vertical clusters are often associated with β-sheets, whereas horizontal ones often correspond to α-helices [57]. The hydrophobicity of phytases from plants and microorganisms analyzed was represented in this study by Hydrophobicity Clusters Analysis according to Bertrand et al. [57], with the aim of contributing to the analysis of phytase thermostability (Figures 6 and 7).
Figure 6.
Secondary structure assignment of microbial phytase by Hydrophobicity Clusters Analysis (HCA) proposed by Bertrand et al. [57]. FASTA data proposed by Kaur et al. [54] are available at https://www.ncbi.nlm.nih.gov/protein. Hydrophobic amino acids are not distributed randomly but form clusters. The clusters are correspondent for the real positions in the regular secondary structures. Vertical clusters are often associated with beta strands and horizontal clusters correspond to alpha-helices. Xanthomonas oryzae, Hphob = 51,7%, and Wickerhamomyces anomalus, Hphob = 48,1%.
Figure 7.
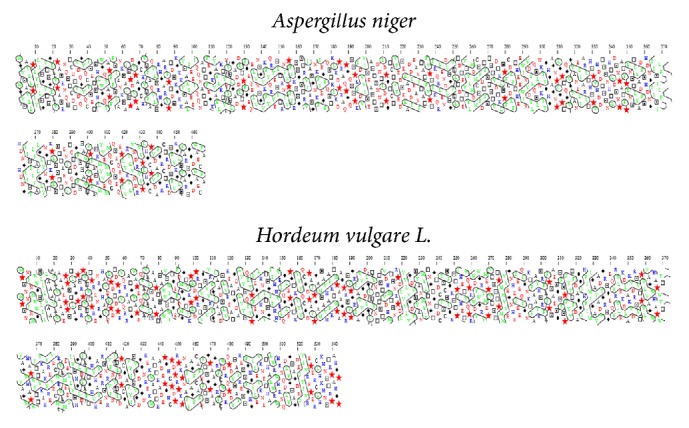
Secondary structure assignment of phytase from Aspergillus niger and Hordeum vulgare L. by Hydrophobicity Clusters Analysis (HCA) proposed by Bertrand et al. [57]. FASTA data proposed by Mishra et al. [50] and Dionisio et al. [4] are available at https://www.ncbi.nlm.nih.gov/protein. Hydrophobic amino acids are not distributed randomly but form clusters. The clusters are correspondent for the real positions in the regular secondary structures. Vertical clusters are often associated with beta strands and horizontal clusters correspond to alpha-helices. Aspergillus niger, Hphob = 44,8%, and Hordeum vulgare L. isoform a, Hphob = 49,5%.
4. Bioprocess and Strategy of Production
4.1. Transgenic Plants and Phytase
Plant growth and productivity can be hindered by low availability of phosphorus in the soil [14]. Therefore, plants have developed mechanisms that include upregulation of high-affinity phosphate transporters and improvement of internal phosphatase activity [22–24]. Transgenic plants have been used with the aim of increasing the availability of phosphorus in the soil. Genes such as the β-propeller phytases from Bacillus subtilis have been inserted in transgenic plants of Arabidopsis and tobacco [14, 58].
A recent study revealed overexpression of an extracellular form of the phytase PHY_US417 of B. subtilis secreted by transgenic Arabidopsis lines (ePHY) and showed increased phosphate acquisition [14]. Furthermore, phytase genes PhyA and AppA from A. niger and Escherichia coli have been inserted into Brassica napus and increased phosphorus bioavailability in soil and seed phytate [59]. Other reports described a reduction of 50% and 48% in phosphorus secretion by broilers and piglets, respectively, after feeding with transgenic soybean (Glycine max) and canola (B. napus) seeds with reduced phytase activities [60, 61]. This strategy has also achieved success in decreasing seed phytate levels by endogenous phytase gene expression during the early stages of seed development in transgenic soybean [62]. However, it remains necessary to advance knowledge about the molecular mechanisms regulating phytase formation during grain development and germination [4].
Transgenic wheat and microalgae have been employed for the expression of phytase activity. Transgenic microalgae, such as Chlamydomonas reinhardtii, have been revealed as an interesting model system for the production of a fungal phytase (A. niger PhyA E228K, mE228K) due to their suitability for genetic transformation and scalability. Lines with high expression levels of in vitro phytase activity were obtained [63]. Expression of the PhyA gene from Aspergillus japonicus in wheat transgenic plants exhibited an increase in phytase activity and iron and zinc contents and a decrease in phytic acid content in seeds. Such reports strengthen an effective method of increasing the bioavailability of iron and zinc and propose a novel way to combat nutritional deficiency [64].
Thus, transgenic plants may be an alternative that warrants further exploration due to the technical compatibility of the genes of plants and microorganisms. It is a promising technology because it promotes efficiency and agricultural productivity, aiming to improve animal nutrition without causing environmental degradation.
4.2. Microbial Phytase
Enzyme production has been observed in various species, especially A. niger [20, 65, 66]. It is possible to observe variations in the yield of enzymatic production due to the physiological peculiarity of each microorganism and varying bioprocess conditions [67]. A. niger wild-type strain NRRL3135 produces high amounts of phytase in mineral salts medium, containing glucose and corn starch as the carbon source. However, with the addition of phosphate in the culture medium formulation, there is no evidence of phytase production. A. niger can produce two kinds of phytase, PhyA and PhyB, depending only on the pH variation from 2.5 to 5.5, respectively. Moreover, it is possible to use genetic engineering tools to introduce the PhyA gene driven by a specific promoter into another strain of A. niger, producing a high yield [68].
Bacillus amyloliquefaciens produces extracellular phytase at 37°C in a medium supplemented with wheat bran and casein hydrolysate by submerged fermentation. Other species also produce high amounts of phytase under specific fermentation conditions. B. subtilis can be cultivated at 60°C and pH between 7 and 7.5 to produce high amounts of phytase. Moreover, B. subtilis has been used in genetic modifications aimed at the expression of specific phytase genes. Other examples are phytases from Aspergillus awamori and Aspergillus fumigatus employed by genetic methods modified and evaluated by SDS-PAGE and Western blotting. Thus, these works reveal that the recombinant strain exhibits overexpression, high activity, and thermostability at pH between 4 and 7 at 60°C [69, 70].
Pichia pastoris is a methylotrophic, nonpathogenic yeast and is widely used to produce various heterologous proteins because it has the capacity to perform important and necessary posttranslational modifications of the protein of interest. Transcription of the genes inserted into the vector is driven by the AOX1 promoter mediated by the action of methanol in the medium or in the presence of methanol as a sole carbon source [71, 72]. The phytase gene of 1347 bp from A. niger SK-57 can be obtained by PCR and expressed in P. pastoris using the AOX1 promoter. Under these conditions, P. pastoris produced 6.1 g·l−1 of purified phytase with an activity of 865 U·ml−1 and pH optima between 2.5 and 5.5 at 60°C. Thus, P. pastoris can be an alternative microorganism for use as a production system for commercial phytase [73]. Another strain of methylotrophic yeast, Hansenula polymorpha, can be genetically modified to express phytase effectively under submerged fermentation conditions with the use of the formate dehydrogenase promoter [11].
Moreover, phytase can be obtained from the mushroom Schizophyllum sp. Cultivation is usually carried out in solid-state fermentation at 30°C in a culture medium consisting of wheat bran suitable for a few days at pH between 6 and 7 [74]. Gram-negative bacteria of the Chromobacterium sp. genus have been isolated from a variety of habitats in Brazilian ecosystems and have already presented phytase activity. Chromobacterium sp. usually can be cultivated at 28°C in a specific medium containing phytate. Bacteria of the genera Escherichia, Rahnella, and Pseudomonas also produce phytase [75]. Different bioprocesses for microbial phytase production are described in Table 2.
Table 2.
Microbial phytase obtained by submerged and solid-state fermentation.
| Microorganisms | Process | Yield | Reference |
|---|---|---|---|
| Aspergillus niger mutant | SSF | 154.0 U·l−1 | Bhavsar et al. [79] |
| Chromobacterium sp. | SF | 7.4–12.4 U·ml−1 | Costa et al. [75] |
| Schizophyllum sp. | SSF | 55.5 U·ml−1 | Salmon et al. [74] |
| Bacillus subtilis | SF | 8.5–9.0 U·mg−1 | Fu et al. [69] |
| Bacillus laevolacticus | SF | 2.957 U·ml−1 | Fu et al. [69] |
| Aspergillusawamori and fumigatus | SF | 12.86 and 20.75 U·ml−1 | Martin et al. [70] |
|
| |||
| Hansenula polymorpha | SF | 13.5 g l−1 | Mayer et al. [11] |
SSF: solid-state fermentation; SF: submerged fermentation; U ml−1: units per milliliter; U mg−1: units per milligram; g l−1: gram per liter.
5. Animal Supplementation and Sustainable Agriculture
In monogastric animals, there is little or no evidence of phytase activity in the digestive tract [4, 5]. In these animals, food is not completely digested, and cellular bioavailability is low. Most of the phytate present in the feed is excreted, increasing the environmental phosphate concentration in areas where livestock grazing is intense [6, 7]. Low-phosphate bioavailability and phosphorus deficiency in animal feed based on dry grain can be supplemented by adding inorganic phosphate. Due to the high market demand for meat, this strategy of phosphorus supplementation is used increasingly in animal feed. However, the practice of adding inorganic phosphorus is unsustainable, because phosphorus is not a renewable resource in nature, and its continued harvesting would result in depletion of this environmental resource in a few decades [6, 9, 67, 76, 77]. Thus, the addition of phytase to animal feed is an interesting and sustainable alternative, due to its ability to increase phosphate bioavailability in diets based on dry grain and to decrease the environmental impacts caused by phosphorus accumulation in the environment [4, 31, 78]. Adding phytase to animal feed also reduces the operational production cost as the inclusion of inorganic phosphate is no longer required. Moreover, phytase also increases the availability of other minerals such as zinc, iron, and manganese, which are important for animal metabolism [7, 67, 79–82]. Animal feed supplementation provides beneficial responses in terms of animal performance such as increased weight of ruminants, broilers, and fishes [82–84]. Other beneficial effects can be achieved by incorporating mixed enzymatic formulations that result in an increase in the coefficients of ileal nutrient digestibility and standardized ileal digestibility by amino acid absorption and energy production [85–88].
Multienzymatic complexes can also provide efficient nutrient absorption. Previous reports described secondary effects associated with the action of multienzymatic complexes added to stimulate hormone synthesis [89, 90]. Bedford and Cowieson [91] analyzed the multifactorial effects of exogenous enzymes on the intestinal microbiota and the improvement of animal performance. Phytase has positive contributions for use in animal feed supplementation; however, it is still necessary to evaluate the specific effects of carbohydrases and proteases on multienzymatic complexes [92–96]. Thus, animal characteristics, such as species or age, are important sources of variation in these studies. However, the main cause is directly associated with characteristics of animal feed in both qualitative and quantitative aspects. The nonstarch portion is diversified and large, which favors increased digestive viscosity and hinders enzymatic action. Studies have reported that this complicates the understanding of the effects of these multienzymatic complexes on animal feeds. Thus, their use and performance can be underestimated mainly due to their variation in the concentrations of enzyme and substrate [97].
Organic acids are other additives that can be supplemented in phytase formulations. Increased animal performance is caused by one or more actions: (1) a decrease in gastric pH favoring pepsin activity in the fed state; (2) immune system improvement due to microbiological selectivity, which reduces Gram-negative bacteria in favor of Lactobacillus proliferation in the gastrointestinal tract; and (3) improvement of both mineral and protein digestibility from food [98]. The benefits of organic acids show high interspecific diversity and are largely dependent on the profile of the gastrointestinal tract. In pigs, the gastrointestinal pH is above 6.0, and acidification is beneficial to phytase activity, due to its acidic pH optimum [99]. Thus, enzymatic formulations containing phytase generally include the organic acids, acetic, citric, fumaric, lactic, formic, and propionic acids [98–100], and the following enzymes: xylanases [87, 92–95], cellulase [101], β-glucanase [102], β-mannanase [96, 103], and proteases [104].
Therefore, the addition of supplementary phytase to the feed of monogastric animals shows many advantages, these being associated or not with multienzymatic complexes or with organic acids, providing an adequate supply of phosphorus and other minerals without damaging the environment.
6. Perspectives
Phytase plays an important role by releasing phosphorus and other nutrients from phytate added to monogastric animal feed and decreases the amount of this element in the environment. Its use allows the consumption of phosphorus available in grains used in animal nutrition. The catalytic structure, production, and biotechnological applications of phytase are increasingly evaluated in research to increase the understanding of three-dimensional structures and to optimize bioprocesses with the aim of increasing production and activity. Plants and different microorganisms can produce phytase, the yields being related to the bioprocess employed. Future perspectives indicate the use of recombinant plants or microorganisms with phytase activity to improve the use of phosphorus and other minerals and to propose a new strategy to improve animal feed. Thus, understanding the catalytic structure of phytase from plants or different microorganisms is an important aspect that allows for sustainable agriculture and attention to the environment.
Acknowledgments
This review was funded by Federal University of Tocantins with financial support of the CNPq.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Alkorta I., Garbisu C., Llama M. J., Serra J. L. Industrial applications of pectic enzymes: a review. Process Biochemistry. 1998;33(1):21–28. doi: 10.1016/S0032-9592(97)00046-0. [DOI] [Google Scholar]
- 2.Naidu G. S. N., Panda T. Production of pectolytic enzymes - A review. Bioprocess Engineering. 1998;19(5):355–361. doi: 10.1007/s004490050532. doi: 10.1007/s004490050532. [DOI] [Google Scholar]
- 3.Lynd L. R., Weimer P. J., Van Zyl W. H., Pretorius I. S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiology and Molecular Biology Reviews. 2002;66(3):506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dionisio G., Madsen C. K., Holm P. B., et al. Cloning and characterization of purple acid phosphatase phytases from wheat, barley, maize, and rice. Plant Physiology. 2011;156(3):1087–1100. doi: 10.1104/pp.110.164756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhametzianova A. D., Akhmetova A. I., Sharipova M. R. Microorganisms as phytase producers. Mikrobiologiia. 2012;81(3):291–300. [PubMed] [Google Scholar]
- 6.Martins B. A. B., Borgatti L. M. D. O., Souza L. W. D. O., Robassini S. L. D. A., Albuquerque R. D. Bioavailability and poultry fecal excretion of phosphorus from soybeanbased diets supplemented with phytase. Revista Brasileira de Zootecnia. 2013;42(3):174–182. doi: 10.1590/S1516-35982013000300005. [DOI] [Google Scholar]
- 7.Humer E., Schwarz C., Schedle K. Phytate in pig and poultry nutrition. Journal of Animal Physiology and Animal Nutrition. 2015;99(4):605–625. doi: 10.1111/jpn.12258. [DOI] [PubMed] [Google Scholar]
- 8.Guimarães L. H. S., Peixoto-Nogueira S. C., Michelin M., et al. Screening of filamentous fungi for production of enzymes of biotechnological interest. Brazilian Journal of Microbiology. 2006;37(4):474–480. doi: 10.1590/S1517-83822006000400014. [DOI] [Google Scholar]
- 9.Lelis G. R., Albino L. F. T., Calderano A. A., et al. Diet supplementation with phytase on performance of broiler chickens. Revista Brasileira de Zootecnia. 2012;41(4):929–933. doi: 10.1590/S1516-35982012000400014. [DOI] [Google Scholar]
- 10.Adrio J. L., Demain A. L. Microbial enzymes: tools for biotechnological processes. Biomolecules. 2014;4(1):117–139. doi: 10.3390/biom4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer A. F., Hellmuth K., Schlieker H., et al. An expression system matures: A highly efficient and cost-effective process for phytase production by recombinant strains of Hansenula polymorpha. Biotechnology and Bioengineering. 1999;63(3):373–381. doi: 10.1002/(SICI)1097-0290(19990505)63:3<373::AID-BIT14>3.0.CO;2-T. doi: 10.1002/(SICI)1097-0290(19990505)63:3<373::AID-BIT14>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee S., Sankaranarayanan R., Sonti R. V. PhyA, a Secreted Protein of Xanthomonas oryzae pv. oryzae, Is Required for Optimum Virulence and Growth on Phytic Acid as a Sole Phosphate Source. Molecular Plant-Microbe Interactions. 2003;16(11):973–982. doi: 10.1094/MPMI.2003.16.11.973. [DOI] [PubMed] [Google Scholar]
- 13.Schneider J., Rupp O., Trost E., et al. Genome sequence of Wickerhamomyces anomalus DSM 6766 reveals genetic basis of biotechnologically important antimicrobial activities. FEMS Yeast Research. 2012;12(3):382–386. doi: 10.1111/j.1567-1364.2012.00791.x. [DOI] [PubMed] [Google Scholar]
- 14.Belgaroui N., Berthomieu P., Rouached H., Hanin M. The secretion of the bacterial phytase PHY-US417 by Arabidopsis roots reveals its potential for increasing phosphate acquisition and biomass production during co-growth. Plant Biotechnology Journal. 2016;14(9):1914–1924. doi: 10.1111/pbi.12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durmus A., Eicken C., Sift B. H., et al. The active site of purple acid phosphatase from sweet potatoes (Ipomoea batatas). Metal content and spectroscopic characterization. European Journal of Biochemistry. 1999;260(3):709–716. doi: 10.1046/j.1432-1327.1999.00230.x. [DOI] [PubMed] [Google Scholar]
- 16.Bozzo G. G., Raghothama K. G., Plaxton W. C. Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. European Journal of Biochemistry. 2002;269(24):6278–6286. doi: 10.1046/j.1432-1033.2002.03347.x. [DOI] [PubMed] [Google Scholar]
- 17.Antonyuk S. V., Olczak M., Olczak T., Ciuraszkiewicz J., Strange R. W. The structure of a purple acid phosphatase involved in plant growth and pathogen defence exhibits a novel immunoglobulin-like fold. Biology and Medicine. 2014;1:101–109. doi: 10.1107/S205225251400400X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta R. K., Gangoliya S. S., Singh N. K. Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. Journal of Food Science and Technology. 2015;52(2):676–684. doi: 10.1007/s13197-013-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohn L., Meyer A. S., Rasmussen S. K. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. Journal of Zhejiang University SCIENCE B. 2008;9(3):165–191. doi: 10.1631/jzus.b0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullaney E. J., Ullah A. H. J. The term phytase comprises several different classes of enzymes. Biochemical and Biophysical Research Communications. 2003;312(1):179–184. doi: 10.1016/j.bbrc.2003.09.176. [DOI] [PubMed] [Google Scholar]
- 21.Gerke J. Phytate (Inositol Hexakisphosphate) in Soil and Phosphate Acquisition from Inositol Phosphates by Higher Plants. A Review. Plants. 2015;4(4):253–266. doi: 10.3390/plants4020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker A., Ceasar S. A., Palmer A. J., et al. Replace, reuse, recycle: Improving the sustainable use of phosphorus by plants. Journal of Experimental Botany. 2015;66(12):3523–3540. doi: 10.1093/jxb/erv210. [DOI] [PubMed] [Google Scholar]
- 23.Schachtman D. P., Reid R. J., Ayling S. M. Phosphorus uptake by plants: from soil to cell. Plant Physiology. 1998;116(2):447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheible W.-R., Rojas-Triana M. Sensing, signalling, and control of phosphate starvation in plants: molecular players and applications. Phosphorus Metabolism in Plants. 2015;48:25–64. doi: 10.1002/9781118958841.ch2. [DOI] [Google Scholar]
- 25.Vance C. P., Uhde-Stone C., Allan D. L. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist. 2003;157(3):423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 26.Raghothama K. G. Phosphate acquisition. Annual Review of Plant Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 27.Annor G. A., Tano Debrah K., Essen A. Mineral and phytate contents of some prepared popular Ghanaian foods. SpringerPlus. 2016;5(1, article no. 581):1–8. doi: 10.1186/s40064-016-2202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKie V. A., McCleary B. V. A novel and rapid colorimetric method for measuring total phosphorus and phytic acid in foods and animal feeds. Journal of AOAC International. 2016;99(3):738–743. doi: 10.5740/jaoacint.16-0029. [DOI] [PubMed] [Google Scholar]
- 29.Hegeman C. E., Grabau E. A. A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiology. 2001;126(4):1598–1608. doi: 10.1104/pp.126.4.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guimarães L. H. S., Terenzi H. F., Jorge J. A., Leone F. A., Polizeli M. D. L. T. M. Characterization and properties of acid phosphatases with phytase activity produced by Aspergillus caespitosus. Biotechnology and Applied Biochemistry. 2004;40(2):201–207. doi: 10.1042/BA20030208. [DOI] [PubMed] [Google Scholar]
- 31.Brinch-Pedersen H., Sørensen L. D., Holm P. B. Engineering crop plants: Getting a handle on phosphate. Trends in Plant Science. 2002;7(3):118–125. doi: 10.1016/S1360-1385(01)02222-1. [DOI] [PubMed] [Google Scholar]
- 32.Lim M.-H., Lee O.-H., Chin J.-E., et al. Simultaneous degradation of phytic acid and starch by an industrial strain of Saccharomyces cerevisiae producing phytase and α-amylase. Biotechnology Letters. 2008;30(12):2125–2130. doi: 10.1007/s10529-008-9799-x. [DOI] [PubMed] [Google Scholar]
- 33.Lelis G. R., Albino L. F. T., Tavernari F. C., Rostagno S. Suplementação dietética de fitase em dietas para frangos de corte. Revista Eletrônica Nutritime. 2009;6(2):875–889. [Google Scholar]
- 34.Kumar V., Singh G., Verma A. K., Agrawal S. In silico characterization of histidine acid phytase sequences. Enzyme Research. 2012;2012 doi: 10.1155/2012/845465.845465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Lin Y., Zheng X., et al. Combined strategies for improving expression of Citrobacter amalonaticus phytase in Pichia pastoris. BMC Biotechnology. 2015;15(1, article no. 88) doi: 10.1186/s12896-015-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dvořáková J. Phytase: Sources, Preparation and Exploitation. Folia Microbiologica. 1998;43(4):323–338. doi: 10.1007/BF02818571. [DOI] [PubMed] [Google Scholar]
- 37.Oh T.-K., Oh S., Kim S., et al. Expression of Aspergillus nidulans phy gene in Nicotiana benthamiana produces active phytase with broad specificities. International Journal of Molecular Sciences. 2014;15(9):15571–15591. doi: 10.3390/ijms150915571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graminho E. R., Takaya N., Nakamura A., Hoshino T. Purification, biochemical characterization, and genetic cloning of the phytase produced by Burkholderia sp. Strain a13. The Journal of General and Applied Microbiology. 2015;61(1):15–23. doi: 10.2323/jgam.61.15. [DOI] [PubMed] [Google Scholar]
- 39.Klabunde T., Sträter N., Krebs B., Witzel H. Structural relationship between the mammalian Fe(III)Fe(II) and the Fe(III)Zn(II) plant purple acid phosphatases. FEBS Letters. 1995;367(1):56–60. doi: 10.1016/0014-5793(95)00536-I. [DOI] [PubMed] [Google Scholar]
- 40.Boudalis A. K., Aston R. E., Smith S. J., et al. Synthesis and characterization of the tetranuclear iron(iii) complex of a new asymmetric multidentate ligand. A structural model for purple acid phosphatases. Dalton Transactions. 2007;(44):5132–5139. doi: 10.1039/b709293h. [DOI] [PubMed] [Google Scholar]
- 41.Schenk G., Elliott T. W., Leung E., et al. Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle. BMC Structural Biology. 2008;8, article no. 6 doi: 10.1186/1472-6807-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang R., Chan K.-H., Yeung E., Lim B. L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiology. 2009;151(1):199–209. doi: 10.1104/pp.109.143180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar V., Singh G., Sangwan P., Verma A. K., Agrawal S. Cloning, Sequencing, and in silico analysis of β-Propeller Phytase. Biotechnology Research International. 2014;2014:11. doi: 10.1155/2014/841353.841353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar V., Agrawal S. Short Communication: An insight into protein sequences of PTP-like cysteine phytases. Nusantara Bioscience. 2014;6(1):102–106. [Google Scholar]
- 45.Fu D., Huang H., Meng K., et al. Improvement of Yersinia frederiksenii phytase performance by a single amino acid substitution. Biotechnology and Bioengineering. 2009;103(5):857–864. doi: 10.1002/bit.22315. [DOI] [PubMed] [Google Scholar]
- 46.Viader-Salvadó J. M., Gallegos-López J. A., Carreón-Treviño J. G., Castillo-Galván M., Rojo-Domínguez A., Guerrero-Olazarán M. Design of thermostable beta-propeller phytases with activity over a broad range of pHs and their overproduction by pichia pastoris. Applied and Environmental Microbiology. 2010;76(19):6423–6430. doi: 10.1128/AEM.00253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu D., Li Z., Huang H., et al. Catalytic efficiency of HAP phytases is determined by a key residue in close proximity to the active site. Applied Microbiology and Biotechnology. 2011;90(4):1295–1302. doi: 10.1007/s00253-011-3171-0. [DOI] [PubMed] [Google Scholar]
- 48.Kumar K., Dixit M., Khire J. M., Pal S. Atomistic details of effect of disulfide bond reduction on active site of Phytase B from Aspergillus niger: A MD Study. Bioinformation. 2013;9(19):963–967. doi: 10.6026/97320630009963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oakley A. J. The structure of Aspergillus niger phytase PhyA in complex with a phytate mimetic. Biochemical and Biophysical Research Communications. 2010;397(4):745–749. doi: 10.1016/j.bbrc.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Mishra I. G., Bhurat K. B., Tripathi N., Tantwai K., Tiwari S. GenBank AFJ79736.1. 2012. Molecular cloning and characterization of phytase gene from Aspergillus niger; pp. 1–490. http://www.ncbi.nlm.nih.gov/protein/AFJ79736. [Google Scholar]
- 51.Böhm K., Herter T., Müller J. J., Borriss R., Heinemann U. Crystal structure of Klebsiella sp. ASR1 phytase suggests substrate binding to a preformed active site that meets the requirements of a plant rhizosphere enzyme. FEBS Journal. 2010;277(5):1284–1296. doi: 10.1111/j.1742-4658.2010.07559.x. [DOI] [PubMed] [Google Scholar]
- 52.Shivange A. V., Serwe A., Dennig A., Roccatano D., Haefner S., Schwaneberg U. Directed evolution of a highly active Yersinia mollaretii phytase. Applied Microbiology and Biotechnology. 2012;95(2):405–418. doi: 10.1007/s00253-011-3756-7. [DOI] [PubMed] [Google Scholar]
- 53.Wilkins K. E., Booher N. J., Wang L., Bogdanove A. J. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Frontiers in Plant Science. 2015;6(JULY, article no. 536):1–15. doi: 10.3389/fpls.2015.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaur P., Singh B., Böer E., et al. Pphy-A cell-bound phytase from the yeast Pichia anomala: Molecular cloning of the gene PPHY and characterization of the recombinant enzyme. Journal of Biotechnology. 2010;149(1-2):8–15. doi: 10.1016/j.jbiotec.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 55.Joshi S., Satyanarayana T. Characteristics and applicability of phytase of the yeast Pichia anomala in synthesizing haloperoxidase. Applied Biochemistry and Biotechnology. 2015;176(5):1351–1369. doi: 10.1007/s12010-015-1650-y. [DOI] [PubMed] [Google Scholar]
- 56.Maurya A. K., Parashar D., Satyanarayana T. Bioprocess for the production of recombinant HAP phytase of the thermophilic mold Sporotrichum thermophile and its structural and biochemical characteristics. International Journal of Biological Macromolecules. 2017;94:36–44. doi: 10.1016/j.ijbiomac.2016.09.102. [DOI] [PubMed] [Google Scholar]
- 57.Bertrand N., Ménager H., Maufrais C., et al. Bioinformatics advance acess. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lung S.-C., Chan W.-L., Yip W., Wang L., Yeung E. C., Lim B. L. Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilization. Journal of Plant Sciences. 2005;169(2):341–349. doi: 10.1016/j.plantsci.2005.03.006. [DOI] [Google Scholar]
- 59.Wang Y., Ye X., Ding G., Xu F. Overexpression of phyA and appA Genes Improves Soil Organic Phosphorus Utilisation and Seed Phytase Activity in Brassica napus. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0060801.e60801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denbow D. M., Grabau E. A., Lacy G. H., Kornegay E. T., Russell D. R., Umbeck P. F. Soybeans Transformed with a Fungal Phytase Gene Improve Phosphorus Availability for Broilers. Poultry Science. 1998;77(6):878–881. doi: 10.1093/ps/77.6.878. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z. B., Kornegay E. T., Radcliffe J. S., Wilson J. H., Veit H. P. Comparison of phytase from genetically engineered Aspergillus and canola in weanling pig diets. Journal of Animal Science. 2000;78(11):2868–2878. doi: 10.2527/2000.78112868x. [DOI] [PubMed] [Google Scholar]
- 62.Singh P., Punjabi M., Jolly M., Rai R. D., Sachdev A. Characterization and expression of codon optimized soybean phytase gene in E. coli. Indian Journal of Biochemistry and Biophysics. 2013;50(6):537–547. [PubMed] [Google Scholar]
- 63.Erpel F., Restovic F., Arce-Johnson P. Development of phytase-expressing chlamydomonas reinhardtii for monogastric animal nutrition. BMC Biotechnology. 2016;16(1, article no. 29) doi: 10.1186/s12896-016-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abid N., Khatoon A., Maqbool A., et al. Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transgenic Research. 2017;26(1):109–122. doi: 10.1007/s11248-016-9983-z. [DOI] [PubMed] [Google Scholar]
- 65.Ibrahim C. O. Development of applications of industrial enzymes from Malaysian indigenous microbial sources. Bioresource Technology. 2008;99(11):4572–4582. doi: 10.1016/j.biortech.2007.07.040. [DOI] [PubMed] [Google Scholar]
- 66.Shivanna G. B., Venkateswaran G. Phytase production by Aspergillus niger CFR 335 and Aspergillus ficuum SGA 01 through submerged and solid-state fermentation. The Scientific World Journal. 2014;2014 doi: 10.1155/2014/392615.392615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haefner S., Knietsch A., Scholten E., Braun J., Lohscheidt M., Zelder O. Biotechnological production and applications of phytases. Applied Microbiology and Biotechnology. 2005;68(5):588–597. doi: 10.1007/s00253-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 68.Wodzinski R. J., Ullah A. H. J. Phytase. Advances in Applied Microbiology. 1996;(42):263–302. doi: 10.1016/s0065-2164(08)70375-7. [DOI] [PubMed] [Google Scholar]
- 69.Fu S., Sun J., Qian L., Li Z. Bacillus phytases: Present scenario and future perspectives. Applied Biochemistry and Biotechnology. 2008;151(1):1–8. doi: 10.1007/s12010-008-8158-7. [DOI] [PubMed] [Google Scholar]
- 70.Martin J. A., Murphy R. A., Power R. F. G. Purification and physico-chemical characterisation of genetically modified phytases expressed in Aspergillus awamori. Bioresource Technology. 2006;97(14):1703–1708. doi: 10.1016/j.biortech.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 71.Cereghino J. L., Cregg J. M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiology Reviews. 2000;24(1):45–66. doi: 10.1016/s0168-6445(99)00029-7. [DOI] [PubMed] [Google Scholar]
- 72.Coimbra E. C., Gomes F. B., Campos J. F., et al. Production of L1 protein from different types of HPV in Pichia pastoris using an integrative vector. Brazilian Journal of Medical and Biological Research. 2011;44(12):1209–1214. doi: 10.1590/s0100-879x2011007500141. [DOI] [PubMed] [Google Scholar]
- 73.Xiong A.-S., Yao Q.-H., Peng R.-H., Han P.-L., Cheng Z.-M., Li Y. High level expression of a recombinant acid phytase gene in Pichia pastoris. Journal of Applied Microbiology. 2005;98(2):418–428. doi: 10.1111/j.1365-2672.2004.02476.x. [DOI] [PubMed] [Google Scholar]
- 74.Salmon D. N., Piva L. C., Binati R. L., et al. Formulated products containing a new phytase from Schyzophyllum sp. phytase for application in feed and food processing. Brazilian Archives of Biology and Technology. 2011;54(6):1069–1074. doi: 10.1590/S1516-89132011000600001. [DOI] [Google Scholar]
- 75.Costa P. S., Nascimento A. M. A., Lima-Bittencourt C. I., Chartone-Souza E., Santos F. R., Vilas-Boas A. Chromobacterium sp. from the tropics: Detection and diversity of phytase activity. Brazilian Journal of Microbiology. 2011;42(1):84–88. doi: 10.1590/S1517-83822011000100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steen I. Phosphorus availability in the 21st century: management of a non-renewable resource. Phosphorus and Potassium. 1998;217:25–31. [Google Scholar]
- 77.Jegannathan K. R., Nielsen P. H. Environmental assessment of enzyme use in industrial production-a literature review. Journal of Cleaner Production. 2013;42:228–240. doi: 10.1016/j.jclepro.2012.11.005. [DOI] [Google Scholar]
- 78.Tan H., Mooij M. J., Barret M., et al. Identification of novel phytase genes from an agricultural soil-derived metagenome. Journal of Microbiology and Biotechnology. 2014;24(1):113–118. doi: 10.4014/jmb.1307.07007. [DOI] [PubMed] [Google Scholar]
- 79.Bhavsar K., Ravi Kumar V., Khire J. M. High level phytase production by Aspergillus niger NCIM 563 in solid state culture: Response surface optimization, up-scaling, and its partial characterization. Journal of Industrial Microbiology and Biotechnology. 2011;38(9):1407–1417. doi: 10.1007/s10295-010-0926-z. [DOI] [PubMed] [Google Scholar]
- 80.Konietzny U., Greiner R. Bacterial phytase: Potential application, in vivo function and regulation of its synthesis. Brazilian Journal of Microbiology. 2004;35(1-2):11–18. doi: 10.1590/S1517-83822004000100002. [DOI] [Google Scholar]
- 81.Casartelli E., Junqueira O., Laurentiz A., Filardi R., Lucas Júnior J., Araujo L. Effect of phytase in laying hen diets with different phosphorus sources. Revista Brasileira de Ciência Avícola. 2005;7(2):93–98. doi: 10.1590/S1516-635X2005000200005. [DOI] [Google Scholar]
- 82.Troesch B., Jing H., Laillou A., Fowler A. Absorption studies show that phytase from Aspergillus niger significantly increases iron and zinc bioavailability from phytate-rich foods. Food and Nutrition Bulletin. 2013;34(2):S90–101. doi: 10.1177/15648265130342S111. [DOI] [PubMed] [Google Scholar]
- 83.Selle P. H., Ravindran V. Microbial phytase in poultry nutrition. Animal Feed Science and Technology. 2007;135(1-2):1–41. doi: 10.1016/j.anifeedsci.2006.06.010. [DOI] [Google Scholar]
- 84.Cao L., Wang W., Yang C., et al. Application of microbial phytase in fish feed. Enzyme and Microbial Technology. 2007;40(4):497–507. doi: 10.1016/j.enzmictec.2007.01.007. [DOI] [Google Scholar]
- 85.Madrid J., Martínez S., López C., Hernández F. Effect of phytase on nutrient digestibility, mineral utilization and performance in growing pigs. Livestock Science. 2013;154(1-3):144–151. doi: 10.1016/j.livsci.2013.03.003. [DOI] [Google Scholar]
- 86.Bedford M. R., Scott T. A., Silversides F. G., Classen H. L., Swift M. L., Pack M. The effect of wheat cultivar, growing environment, and enzyme supplementation on digestibility of amino acids by broilers. Canadian Journal of Animal Science. 1998;78(3):335–342. doi: 10.4141/A98-012. [DOI] [Google Scholar]
- 87.Selle P. H., Ravindran V., Partridge G. G. Beneficial effects of xylanase and/or phytase inclusions on ileal amino acid digestibility, energy utilisation, mineral retention and growth performance in wheat-based broiler diets. Animal Feed Science and Technology. 2009;153(3-4):303–313. doi: 10.1016/j.anifeedsci.2009.06.011. [DOI] [Google Scholar]
- 88.Kiarie E., Owusu-Asiedu A., Simmins P. H., Nyachoti C. M. Influence of phytase and carbohydrase enzymes on apparent ileal nutrient and standardized ileal amino acid digestibility in growing pigs fed wheat and barley-based diets. Livestock Science. 2010;134(1-3):85–87. doi: 10.1016/j.livsci.2010.06.105. [DOI] [Google Scholar]
- 89.Singh J., Kaur P. Optimization of process parameters for cellulase production from Bacillus sp. JS14 in solid substrate fermentation using response surface methodology. Brazilian Archives of Biology and Technology. 2012;55(4):505–512. doi: 10.1590/S1516-89132012000400004. [DOI] [Google Scholar]
- 90.Jos E. G., Talita C. E., Alana E. S., et al. Production, characterization and evaluation of in vitro digestion of phytases, xylanases and cellulases for feed industry. African Journal of Microbiology Research. 2014;8(6):551–558. doi: 10.5897/AJMR2013.6299. [DOI] [Google Scholar]
- 91.Bedford M. R., Cowieson A. J. Exogenous enzymes and their effects on intestinal microbiology. Animal Feed Science and Technology. 2012;173(1-2):76–85. doi: 10.1016/j.anifeedsci.2011.12.018. [DOI] [Google Scholar]
- 92.Lindberg J. E., Lyberg K., Sands J. Influence of phytase and xylanase supplementation of a wheat-based diet on ileal and total tract digestibility in growing pigs. Livestock Science. 2007;109(1-3):268–270. doi: 10.1016/j.livsci.2007.01.114. [DOI] [Google Scholar]
- 93.Kim J. C., Sands J. S., Mullan B. P., Pluske J. R. Performance and total-tract digestibility responses to exogenous xylanase and phytase in diets for growing pigs. Animal Feed Science and Technology. 2008;142(1-2):163–172. doi: 10.1016/j.anifeedsci.2007.07.004. [DOI] [Google Scholar]
- 94.Woyengo T. A., Sands J. S., Guenter W., Nyachoti C. M. Nutrient digestibility and performance responses of growing pigs fed phytase- and xylanase-supplemented wheat-based diets. Journal of Animal Science. 2008;86(4):848–857. doi: 10.2527/jas.2007-0018. [DOI] [PubMed] [Google Scholar]
- 95.Atakora J. K. A., Moehn S., Sands J. S., Ball R. O. Effects of dietary crude protein and phytase-xylanase supplementation of wheat grain based diets on energy metabolism and enteric methane in growing finishing pigs. Animal Feed Science and Technology. 2011;166-167:422–429. doi: 10.1016/j.anifeedsci.2011.04.030. [DOI] [Google Scholar]
- 96.Mok C. H., Lee J. H., Kim B. G. Effects of exogenous phytase and β-mannanase on ileal and total tract digestibility of energy and nutrient in palm kernel expeller-containing diets fed to growing pigs. Animal Feed Science and Technology. 2013;186(3-4):209–213. doi: 10.1016/j.anifeedsci.2013.10.008. [DOI] [Google Scholar]
- 97.Ravindran V. Feed enzymes: The science, practice, and metabolic realities. Journal of Applied Poultry Research. 2013;22(3):628–636. doi: 10.3382/japr.2013-00739. [DOI] [Google Scholar]
- 98.Khodambashi Emami N., Zafari Naeini S., Ruiz-Feria C. A. Growth performance, digestibility, immune response and intestinal morphology of male broilers fed phosphorus deficient diets supplemented with microbial phytase and organic acids. Livestock Science. 2013;157(2-3):506–513. doi: 10.1016/j.livsci.2013.08.014. [DOI] [Google Scholar]
- 99.Jongbloed A. W., Mroz Z., Van Der Weij-Jongbloed R., Kemme P. A. The effects of microbial phytase, organic acids and their interaction in diets for growing pigs. Livestock Production Science. 2000;67(1-2):113–122. doi: 10.1016/S0301-6226(00)00179-2. [DOI] [Google Scholar]
- 100.Valencia Z., Chavez E. R. Phytase and acetic acid supplementation in the diet of early weaned piglets: Effect on performance and apparent nutrient digestibility. Nutrition Research. 2002;22(5):623–632. doi: 10.1016/S0271-5317(02)00371-8. [DOI] [Google Scholar]
- 101.Knowlton K. F., Taylor M. S., Hill S. R., Cobb C., Wilsont K. F. Manure nutrient excretion by lactating cows fed exogenous phytase and cellulase. Journal of Dairy Science. 2007;90(9):4356–4360. doi: 10.3168/jds.2006-879. [DOI] [PubMed] [Google Scholar]
- 102.Kiarie E., Owusu-Asiedu A., Péron A., Simmins P. H., Nyachoti C. M. Efficacy of xylanase and Β-glucanase blend in mixed grains and grain co-products-based diets for fattening pigs. Livestock Science. 2012;148(1-2):129–133. doi: 10.1016/j.livsci.2012.05.020. [DOI] [Google Scholar]
- 103.Cho J. H., Kim I. H. Effects of beta-mannanase supplementation in combination with low and high energy dense diets for growing and finishing broilers. Livestock Science. 2013;154(1-3):137–143. doi: 10.1016/j.livsci.2013.03.004. [DOI] [Google Scholar]
- 104.Barekatain M. R., Antipatis C., Choct M., Iji P. A. Interaction between protease and xylanase in broiler chicken diets containing sorghum distillers' dried grains with solubles. Animal Feed Science and Technology. 2013;182(1-4):71–81. doi: 10.1016/j.anifeedsci.2013.04.002. [DOI] [Google Scholar]
- 105.I-Tasser online protein structure and function predictions. http://zhanglab.ccmb.med.umich.edu/I-TASSER/
- 106.Holme I. B., Dionisio G., Madsen C. K., Brinch-Pedersen H. Barley HvPAPhy_a as transgene provides high and stable phytase activities in mature barley straw and in grains. Plant Biotechnology Journal. 2017;15(4):415–422. doi: 10.1111/pbi.12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mohsin S., Maqbool A., Ashraf M., Malik K. A. Extracellular Secretion of Phytase from Transgenic Wheat Roots Allows Utilization of Phytate for Enhanced Phosphorus Uptake. Molecular Biotechnology. 2017;59(8):334–342. doi: 10.1007/s12033-017-0020-0. [DOI] [PubMed] [Google Scholar]
- 108.Yang H., Zhang X., Gaxiola R. A., Xu G., Peer W. A., Murphy A. S. Over-expression of the Arabidopsis proton-pyrophosphatase AVP1 enhances transplant survival, root mass, and fruit development under limiting phosphorus conditions. Journal of Experimental Botany. 2014;65(12):3045–3053. doi: 10.1093/jxb/eru149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma X.-F., Tudor S., Butler T., et al. Transgenic expression of phytase and acid phosphatase genes in alfalfa (Medicago sativa) leads to improved phosphate uptake in natural soils. Molecular Breeding. 2012;30(1):377–391. doi: 10.1007/s11032-011-9628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]