Abstract
Three-dimensional (3D) tumor spheroid models have gained increased recognition as important tools in cancer research and anti-cancer drug development. However, currently available imaging approaches employed in high-throughput screening drug discovery platforms e.g. bright field, phase contrast, and fluorescence microscopies, are unable to resolve 3D structures deep inside (>50 µm) tumor spheroids. In this study, we established a label-free, non-invasive optical coherence tomography (OCT) imaging platform to characterize 3D morphological and physiological information of multicellular tumor spheroids (MCTS) growing from ~250 µm up to ~600 µm in height over 21 days. In particular, tumor spheroids of two cell lines glioblastoma (U-87 MG) and colorectal carcinoma (HCT 116) exhibited distinctive evolutions in their geometric shapes at late growth stages. Volumes of MCTS were accurately quantified using a voxel-based approach without presumptions of their geometries. In contrast, conventional diameter-based volume calculations assuming perfect spherical shape resulted in large quantification errors. Furthermore, we successfully detected necrotic regions within these tumor spheroids based on increased intrinsic optical attenuation, suggesting a promising alternative of label-free viability tests in tumor spheroids. Therefore, OCT can serve as a promising imaging modality to characterize morphological and physiological features of MCTS, showing great potential for high-throughput drug screening.
Keywords: Multicellular Tumor Spheroid (MCTS), Optical Coherence Tomography (OCT), High-throughput, Label-free imaging
Introduction
More than 90% of cancer drugs in development fail because of lack of efficacy or unexpected toxicity in clinical trials (1–3). This failure rate in the late stages of clinical trials is in large part due to the use of overly simplistic in vitro cell assays and in vivo animal models with limited predictive value during various stages of drug discovery. Recently, three-dimensional (3D) tumor spheroid models have been explored to provide novel physiological and pharmacological data that are more predictive of drug efficacy and toxicity in the clinic (2–24). These studies show that 3D tumor spheroid models are expected to have an immediate and long-lasting impact in shortening drug discovery timelines, reducing costs of investment, and bringing new medicines to more patients. To characterize 3D tumor models in high-throughput screening, bright field imaging or phase contrast imaging are routinely employed (7,9,22,24). However, these modalities can only provide 2D projections of tumor spheroids and are unable to resolve their 3D structures. Likewise, fluorescence imaging, both confocal and non-confocal, is often used to characterize 3D tumor spheroids (8,9,16,18,22); however, its low penetration of fluorescent reagents and limited light penetration in tumor spheroids over 50 µm in diameter significantly limits our understanding of the morphology and physiology inside the tumor spheroids. Light sheet fluorescence microscopy has been proposed as an alternative to create a high-resolution, 3D profile of an entire tumor spheroid (11,25). However, this method requires extensive steps to prepare the samples, involving transfer, staining and mounting and/or clearing of tumor spheroids, which dampens its strength for high-throughput screening and longitudinal imaging (11).
Optical coherence tomography (OCT) (26), a rapidly developing optical imaging modality, is capable of obtaining label-free, non-destructive 3D images at millimeter depths in biological tissues (27–30). Unlike fluorescence imaging technologies, OCT employs low-coherence interferometry to detect back-scattered signals and provides reconstructed depth-resolved images at micron-level spatial resolutions. In the field of oncology, both standard and functional OCT have been used to analyse pathological features of tumors and evaluate their margins ex vivo in tissues (31–35). OCT also facilitates in vivo studies of murine models harbouring implanted tumors and detects their margins, microenvironment, and vascular dynamics (36,37). Pilot studies have also used OCT to observe in vitro 3D tumor spheroids and evaluate their responses to photodynamic therapy based on morphological information obtained from OCT (38,39).
Quantitative assessment of tumor volume, the key factor predicting cancer progression and recurrence (40), has been challenging in both in vitro and in vivo settings. 2D bright field, phase contrast, and fluorescent imaging cannot resolve the entire depth structure of larger spheroids (>50 µm), so volumes are estimated using cross-sectional radii/diameters, a primitive method unsuitable for obtaining precise volume for irregularly-shaped spheroids (7,9,41). To achieve precise volume calculation, a spheroid must be clearly resolved in 3D space so that its structure can be empirically measured. One can employ the high resolution, high depth-imaging capabilities of OCT to satisfy this need. However, very few studies have reported the utilization of OCT for quantitative assessment of tumor spheroids. Although Sharma et al. used OCT to measure heterogeneous geometries of 3D tumor spheroids (38), an oblate spheroid shape was still assumed in order to calculate the volume of 3D tumor models.
Necrotic core development in 3D tumor spheroids represents a unique feature mimicking tumor necrosis in vivo that is unlikely reproduced in 2D cell cultures. When a tumor spheroid grows larger than 500 µm in diameter, a three-layer concentric structure, including an outer layer of proliferating cells, a middle layer of quiescent cells, and a necrotic core, is typically observed in the spheroid (5,6,10,15,19). The complexity of this multi-layer structure may be caused by lack of oxygen and nutrients, a feature not seen in 2D tumor cell cultures (6,19,25,42). Live/dead fluorescence assay is commonly used to assess the viability of cells and distribution of the necrotic core in 3D tumor spheroids. However, its staining with fluorescent dyes prohibits the specimens from continuing development and from being used for longitudinal studies of spheroid growth kinetics.
The purpose of this study was to employ OCT to accurately quantify the 3D structures of tumor spheroids and detect necrotic tissue using label-free intrinsic light attenuation contrast. We have recently developed high resolution OCT imaging platforms for various in vitro bioapplications, including examining angiogenic sprouting in microengineered devices (43) and evaluating progressive neuronal changes in in vitro rat hippocampal slice cultures over time (44). In this study, we employed a label-free OCT-based imaging platform to visualize 3D structures of multicellular tumor spheroids (MCTSs) in 96-well plates and obtained high resolution structural and physiological information from the spheroids over a period of 21 days. In particular, we quantified sizes, volumes and growth curves for each individual spheroid, and showed that OCT can provide accurate and robust quantification of these parameters, especially for irregular spheroids. Moreover, we demonstrated the ability of OCT to identify necrotic cores within tumor spheroids based solely on optical intrinsic attenuation contrast, offering the opportunity to track the change of necrotic regions over time. Our results demonstrated the value of this OCT-based imaging platform in MCTSs and laid the foundation for future high-throughput screening of tumor spheroids for drug testing.
Materials and Methods
Preparation of tumor spheroids
Two tumor cell lines, glioblastoma (U-87 MG) and colorectal carcinoma (HCT116), were purchased from the American Type Culture Collection (ATCC, Rockville, MD) in 2016 and 2014, respectively. Both cell lines were cultured following recommended protocols by suppliers. U-87 MG cells were maintained in EMEM (Lonza, Walkersville, MD) supplemented with 10% FBS (ThermoFisher Scientific, Atlanta, GA), 1% penicillin-streptomycin (Pen-Strep, Lonza), and additionally, 1× non-essential amino acid (NEAA, ThermoFisher Scientific) and 1% sodium pyruvate (ThermoFisher Scientific). HCT116 cells were maintained in McCoy’s 5A medium (ThermoFisher Scientific) supplemented with 10% FBS, 1% Pen-Strep, 1× NEAA and 1% sodium pyruvate. Both cell lines were cultured under standard conditions (37 °C, 5% CO2, 95% humidity). Media were changed twice weekly and tumor cells were subcultured when they reached 90% confluence. A Mycoplasma test was performed on live HCT 116 cell culture in May, 2017 using a Mycoplasma detection kit (MycoFluor™, ThermoFisher Scientific). No Mycoplasma contamination was observed. No Mycoplasma test was done on U87 MG cell culture after the purchase.
MCTSs were prepared following protocols adapted from Vinci et al (9). For growth media, 10% FBS was replaced by 1× B27, 40 ng/mL EGF recombinant human protein and 40 ng/mL FGF-Basic recombinant human protein (ThermoFisher Scientific). U-87 MG (P3 – P7 after collection or thawing) and HCT 116 (P12 – P20 after thawing) cells were seeded in ultra-low-attachment 96-well U-bottom plates (Corning, Corning, NY) with an initial concentration of 0.3 × 104 cells/ml for both cell lines and 200 µL/well cell suspensions. After seeding, the whole plate was centrifuged at 1200 rpm for 5–7 min to facilitate cell gathering at the bottom of each well. Tumor cells were incubated under standard condition and 50% of media were replaced twice weekly.
OCT Imaging System
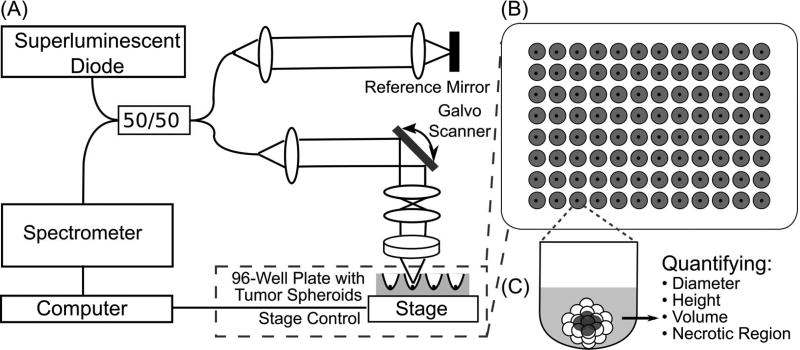
A customized spectral domain OCT (SD-OCT) system was developed (Figure 1A) to acquire 3D images of tumor spheroids. A superluminescent diode (SLD) (Thorlabs, SLD1325) was used to provide broadband light with a central wavelength of 1320 nm and spectral range of 110 nm. Output of the SLD was split 50/50 with a fiber coupler and transmitted to the sample and reference arms of the SD-OCT system. A 2D galvanometer (Thorlabs, GVS002) was used to scan the optical beam in transverse directions on the sample. A 5× objective lens with an effective focal length of 40 mm was employed in the sample arm. The beam diameter at the objective was measured to be ~5 mm, yielding an effective numerical aperture (NA) of ~0.062 in air. This leads to a calculated depth of field (DOF) of ~340 µm in air. The DOF of the OCT system was measured to be ~400 µm in air. During experiments, efforts were made to consistently maintain the focal position at ~100 – 200 µm below the top surface of each tissue sample. As a result, the effect of non-uniform depth-wise focal profile would be relatively minimum in our experiments. The transverse resolution was measured to be ~7.8 µm using a USAF target. Interference signals between the reference arm and the sample arm were detected by a spectrometer, which consists of a diffraction grating, a F-theta lens (Thorlabs, FTH100-1064) and a 1024-pixel InGaAs lines can camera (Sensors Unlimited, SU1024LDH2) operating at 20 kHz A-scan rate. The maximum depth detection range of the SD-OCT system was ~3.4 mm in air or ~2.5 mm in tissue. The axial resolution of the system was measured to be ~4.9 µm in tissue (refractive index of tumor spheroids was assumed to be 1.37 (45,46)). Sensitivity of the OCT system was measured as ~104 dB. In addition, a 2D motorized stage (Aerotech, Pittsburgh, PA) was used to provide horizontal translation of the 96-well plate in order to image tumor spheroids in different wells (Figure 1B).
Figure 1.
Illustration of a custom spectral domain optical coherence imaging (SD-OCT) platform for 3D tumor spheroids imaging. (A) A schematic diagram of the SD-OCT platform. (B) 3D tumor spheroids were cultured in a round-bottom, ultra-low-attachment 96 wells plate. (C) From depth-resolved images of a single 3D tumor spheroid, the diameter, height, volume and distribution of necrotic tissue can be measured.
OCT Image Acquisition and Processing
OCT images of tumor spheroids were acquired at 4, 7, 11, 14, 18 and 21 days after spheroid initiation for both cell lines, Numbers of tumor spheroids from U-87 MG and HCT16 cell lines imaged at each time point are shown in Table 1. OCT scanning range was adjusted to cover the whole tumor spheroid as it developed. Actual scanning range varied from 0.46×0.42 mm2 to 2.0×1.7 mm2. Each 3D dataset of a tumor spheroid was acquired in 10 – 15 seconds. Obtained OCT datasets of tumor spheroids were first processed to generate OCT structural images (in 8-bit) with corrected scales. OCT intensity was normalized by dividing the OCT image value by 255, the maximum value of 8-bit data prior to next steps of image analysis (Figure 1C).
Table 1.
Numbers of U-87 MG and HCT 116 tumor spheroids imaged by OCT.
| Day | 4 | 7 | 11 | 14 | 18 | 21 |
| U-87 MG | 12 | 11 | 9 | 6 | 5 | 4 |
| HCT 116 | 24 | 21 | 18 | 15 | 6 | 3 |
Viability Tests and Fluorescent Imaging
After OCT imaging of all tumor spheroids at each time point, 1–2 tumor spheroids were randomly selected for viability tests to evaluate the distribution of live and dead cells. These spheroids were treated with calcein AM (ThermoFisher Scientific) and propidium iodide (Biotium, Hayward, CA) for live and dead staining, respectively. The whole plate was incubated for 30 minutes prior to fluorescent imaging. Fluorescent images were obtained using an Olympus IX51 inverted microscope with a 10× objective for small tumor spheroids and a 4× objective for large tumor spheroids. A manual image registration was performed to correlate fluorescent images with 3D OCT images based on landmark features of tumor spheroids, such as surface irregularity or cell sprouting. Due to the destructive effect of fluorescent dyes, these 1–2 selected tumor spheroids were no longer used in later stages.
Histology and Immunohistochemistry
At selected time points, 1–2 tumor spheroids were processed for histology and immunohistochemistry (IHC) to compare with corresponding OCT results. Each tumor spheroid was collected in a single 1.5 mL micro-centrifuge tube filled with 10% formaldehyde and fixed for 48 hours. Histology and immunohistochemistry processes of each tumor spheroid were performed by HistoWiz (New York, USA) using standard paraffin embedding techniques. Sections of tumor spheroids (5 µm in thickness) were stained for hematoxylin and eosin (H&E) and TUNEL apoptosis detection (Promega), respectively. A counter staining of hematoxylin was applied to TUNEL. Images were obtained using a Leica AT2 scanner.
Volumetric quantification of 3D tumor spheroids
To calculate the tumor volume based on measured diameter of the tumor spheroid, three cross-sectional images in the XY, XZ, and YZ imaging planes across the centroid of the spheroid were selected. Diameter of the spheroid was measured in XY plane and its height was measured in XZ plane. Diameter-based tumor volume was calculated as:
where d was the average diameter of the spheroid measured in the X and Y directions.
To quantify actual tumor volume, we employed a voxel-based method: First, a 3D average filter was performed to remove the speckles within tumor spheroids in OCT images. Then, the tumor spheroid was segmented using a canny edge detection filter frame by frame. Connective voxels were grouped and the mean distance of grouped connective voxels to the manually chosen spheroid centroid was calculated. The group with the minimum mean distance was identified as the spheroid region. The voxels within this region were summed and then multiplied by the actual volume of an individual voxel (volume/voxel), yielding the total volume of the spheroid.
Necrotic tissue detection of 3D tumor spheroids
Necrotic region in 3D tumor spheroids can be identified with intrinsic optical attenuation measurement. Since OCT signals originate from back-scattering at different depths, OCT intensity detected as a function of depth in a homogeneous medium can be described by the Beer-Lambert Law (47):
where z represents the depth and μ is the optical attenuation coefficient, and I0 is the incident intensity to the sample. OCT images are typically presented in logarithmic scale to increase image contrast. Therefore, the optical attenuation coefficient can be obtained by finding the derivative of the log-scale OCT intensity:
To calculate the optical attenuation from the logarithmic scale OCT images, we first plotted the OCT intensity profile along each axial scan from cross-sectional OCT images. Next we obtained local optical attenuation coefficients by linear fitting the log-scale OCT intensity profile over a certain depth range, extracting its slope, and multiply the slope by −1/2. The aforementioned method was further applied to each axial scan across the entire spheroid. Note that we applied a 3D average filter over OCT data to reduce speckle noise in OCT images before optical attenuation calculation. The attenuation coefficient at each voxel within the segmented spheroid region was calculated based on the slope of OCT intensity profile in a 10-voxel depth window (~40 µm in depth), with the voxel located in the middle of the window.
Statistical analysis
Student’s paired t-tests were performed to compare diameter and height measurements, as well as diameter-based and voxel-based volumes at each time point. A p value of <0.05 was employed to indicate statistically significant differences between the paired measurements (diameter versus height, diameter-based volume versus voxel-based volume).
Results
Longitudinal Volumetric Imaging of Tumor Spheroids using OCT
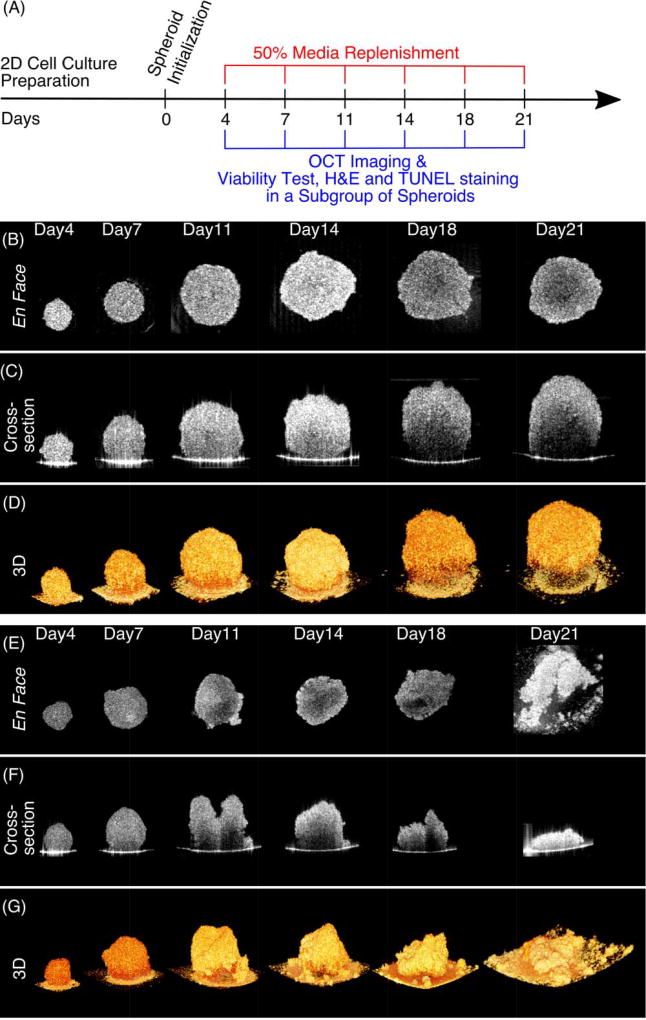
OCT was employed to perform volumetric imaging of tumor spheroids for both U-87 MG and HCT 116 cell lines at 6 different time points over 21 days (Figure 2A). Figure 2B–D and Supplementary Materials (Video-1) show the development of a U-87 MG tumor spheroid over 21 days. En face (Figure 2B), cross-sectional (Figure 2C), and 3D rendered images (Figure 2D) were shown to illustrate the growth dynamics of the same sample. We observed that U-87 MG cells gathered to form a spheroid with a diameter of ~250 µm on Day 4. The spheroid continued to grow and reached ~550 µm in diameter on Day 14, after which growth slowed down and no significant changes in diameter were observed. The U-87 MG tumor spheroid remained tightly-packed spherical shape throughout the 21-day development.
Figure 2.
Growth dynamics of 3D tumor spheroids measured with OCT. (A) Timeline for 3D tumor spheroid preparation and imaging. At each time point, OCT imaging was performed on all spheroids, prior to the viability test, H&E and TUNEL staining on 1–2 randomly selected spheroids. (B–D) Sequential en face, cross-section and 3D rendered OCT images to illustrate the growth dynamics of a U-87 MG tumor spheroids over 21 days. The U-87 MG spheroid remained tightly clustered throughout the 21-day development. (E–G) Growth dynamics of an HCT 116 tumor spheroid over 21 days. Distinct evolution of its geometric shape was observed after Day 11, where the spheroid became disrupted and flattened. Scale Bars: 100 µm.
The growth dynamics of another cell line (HCT 116) over the same duration were shown in Figure 2E–G and Supplementary Materials (Video-2). We found that HCT 116 cells formed a spheroid with a size of ~350 µm on Day 4, and reached ~500 µm on Day 7. As shown in en face images (Figure 2E), the HCT 116 cell cluster maintained a smooth and near-spherical shape and continued to grow, albeit rather slowly, before Day 18. However, from cross-section images (Figure 2F), the HCT 116 cluster was observed as a spheroid only on Days 4 and 7. On day 11, an irregular “two-peak” shape of the HCT 116 cluster was seen. In addition, tumor height was observed to reach its maximum on day 11 and reduced after that day. In 3D rendered images (Figure 2G), we clearly observed that the shape of the HCT 116 tumor spheroid was disrupted and flattened after day 11. Similar results were observed in all HCT 116 tumor spheroids investigated in this study. Therefore, the morphology of HCT 116 underwent distinctive evolution in their geometric shapes at late growth stage when compared with U-87 MG tumor spheroids.
Morphological Quantification of Tumor Spheroids
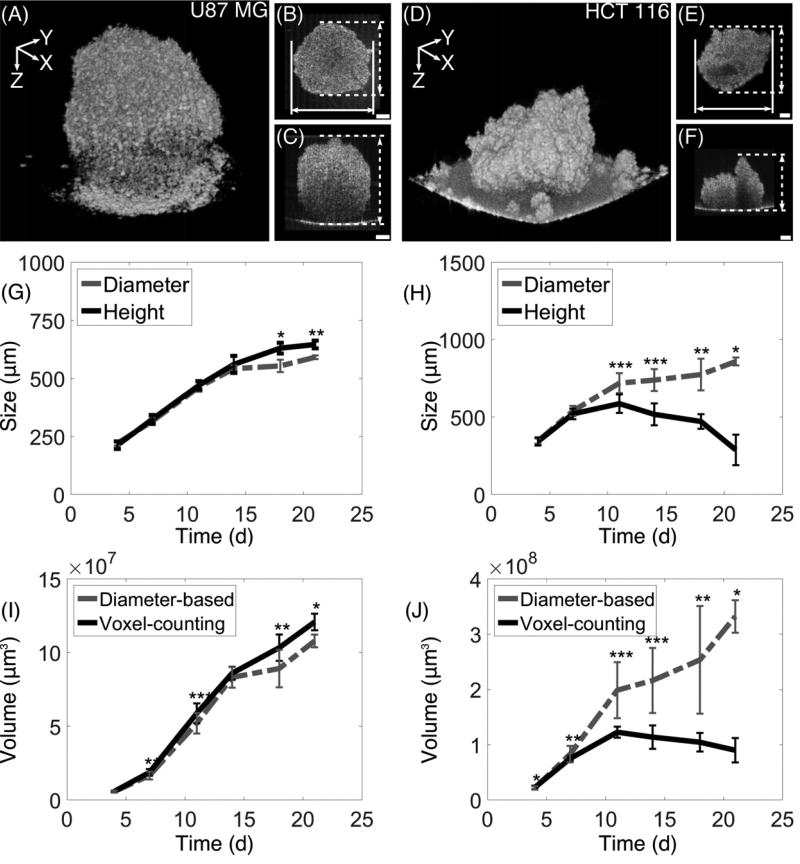
The growth kinetics of U-87 MG and HCT 116 tumor spheroids were quantitatively analyzed and compared in terms of diameter, height and volume (Figure 3). Figure 3A–F exhibits two tumor spheroids (Day 18) used in our determination of average diameter and height based on en face XY and cross-sectional XZ OCT images. The average diameter was calculated from cross-sectional measurements in the X and Y directions in the en face OCT image (Figure 3B, E). Similarly, the average height was measured from both XZ and YZ cross-sectional images (Figure 3C, F).
Figure 3.
Quantitative analysis of the growth kinetics of 3D tumor spheroids in terms of size (diameter and height) and volume. (A–F) Two tumor spheroids from U-87 MG and HCT 116 cell lines at Day 18 were shown as examples to illustrate how average diameter and height were measured from en face and cross-sectional OCT images. Growth curves of U-87 MG (G) and HCT 116 (H) tumor spheroids were shown. While similar growth trends of average diameter and height were revealed for U-87 MG spheroids in (G), discrepancy between average diameter and height of HCT 116 tumor spheroids after Day 11 was observed in (H), in consistence with our finding of heterogeneous geometry in OCT images (D–F). Comparisons of growth curves of the volume of U-87 MG (I) and HCT 116 (J) tumor spheroids between voxel-based measurements and diameter-based measurements were shown. While volumes obtained from both methods were similar for U-87 MG tumor spheroids throughout 21 days, volumes of HCT 116 tumor spheroids were significantly over-estimated with diameter-based measurements compared to voxel-based measurements, especially after Day 11. Scale Bars: 100 µm.
As shown in Figure 3G, U-87 MG tumor spheroids showed a linear increase in the average diameter from Day 4 (Dia. 220 ± 20 µm) to Day 14 (Dia. 530 ± 30 µm), and then exhibited a slower growth rate until Day 21 (Dia. 580 ± 20 µm). This growth trend for spheroid diameter was consistent with previous studies (9,22). Similarly, the height of U-87 MG tumor spheroids linearly increased from Day 4 (Ht. 230 ± 10 µm) to Day 14 (Ht. 550 ± 40 µm), and then continued to increase at a slower rate until Day 21 (Ht. 640 ± 20 µm). The average height of U-87 MG tumor spheroids was slightly but significantly larger than the average diameter of the spheroids only after Day 14, indicating non-perfect sphericity of the spheroids at a late stage of culture. On the other hand, the average diameter of HCT 116 tumor spheroids exhibited a linear increase from Day 4 (Dia. 350 ± 30 µm) to Day 11 (Dia. 670 ± 90 µm), but then only slightly increased from Day 11 to Day 21 (Figure 3H). A different growth curve pattern was observed for the average height of the HCT 116 tumor spheroids. The spheroid height reached the maximum value on Day 11 (Ht. 560 ± 70 µm) and then rapidly dropped until Day 21 (Ht. 270 ± 100 µm). The diameter and height of HCT 116 increased at the same pace only on Day 4 and Day 7. Significant differences between height and diameter were observed at all other measured time points (p<0.001 on Days 11, 14; p<0.01 on Day 18, and p<0.05 on Day 21; Figure 3H). The discrepancy between average diameter and height of HCT 116 tumor spheroids was consistent with our observation of the heterogeneous geometry development and collapse of the spheroids at a late stage shown in reconstructed OCT 3D images (Figure 3D).
The volume growth curves of U-87 MG and HCT 116 tumor spheroids were measured and compared using a voxel-based method (Figure 3I, J). The actual average spheroid volume of U-87 MG tumor spheroids steadily increased by over 20 times from Day 4 to Day 21 (Figure 3I). The U-87 MG volumes derived from the traditional diameter-based measurements were comparable but slightly smaller than the volume obtained using voxel-based method. In comparison, Figure 3J shows the voxel-based volume measurements of HCT 116 spheroids. The average volume of HCT 116 tumor spheroids first increased from Day 4 to Day 11, but gradually decreased at subsequent time points. In contrast, the growth curve for diameter-based volumes of HCT 116 tumor spheroids displayed a steadily increasing trend throughout the entire period, showing a large deviation from the growth curve of the voxel-based measurements. Therefore, the diameter-based volumes of HCT 116 tumor spheroids were significantly over-estimated compared to the volume measured from the voxel-based method after Day 11. The diameter-based volume measurements were as high as ~276% compared to the voxel-based measurements on Day 21, which could be attributed to the significant reduction of the height of the HCT 116 spheroid (Figure 3H). Our results suggest that voxel-based volume measurements from 3D OCT data yield a more robust and accurate quantification of the 3D tumor spheroids compared to diameter-based volume measurements. OCT provides complimentary depth information of the samples, which is not readily observable with traditional imaging modalities but critical for the actual volume quantification of 3D tumor spheroid models.
Identification of Necrotic Regions Based on Intrinsic Optical Contrast
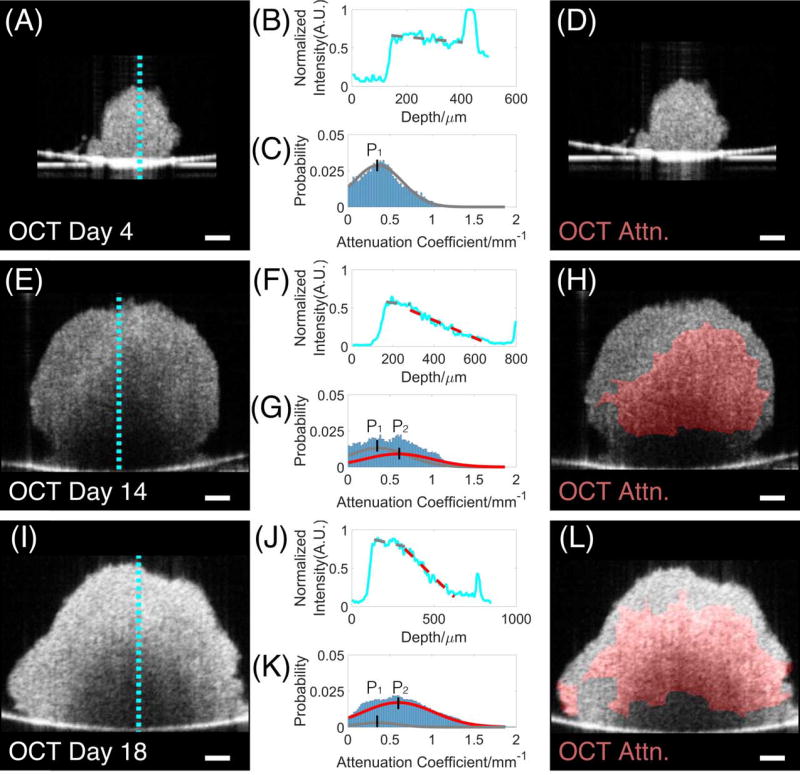
We detected necrotic regions in 3D tumor spheroids based on intrinsic optical attenuation contrast. Figure 4A–L showed representative analyses of optical attenuation of tumor spheroids on Days 4, 14 and 18, respectively. The backscattered signals within tumor spheroids (Figure 4A, E, I) were used to derive intensity profiles along each axial scan line (Figure 4B, F, J). We found that the intensity profile of the HCT 116 tumor spheroid on Day 4 (Figure 4B), although noisy, showed no significant drops (fitted gray line). On Days 14 and 18 (Figure 4F, J), two distinct slopes in the intensity profile were clearly observed: a slow decay of intensity at the top portion of the spheroid marked with a gray line and a fast decay marked with a red line. Therefore, high attenuation regions (indicated in red lines) could be observed from the intensity profiles of the tumor spheroids on Days 14 and 18 (Figure 4F, J), but not from the tumor spheroid on Day 4 (Figure 4B).
Figure 4.
Determination of necrotic regions of HCT 116 tumor spheroids on Days 4, 14, and 18 based on intrinsic optical attenuation contrast. The backscattered signals in cross-sectional OCT images (A, E, I) were used to derive intensity profiles along each axial scan line (B, F, J). High attenuation regions (indicated in red lines in F, J) could be clearly observed in the intensity profiles of the tumor spheroids on Days 14 and 18 (F, J), but not on the tumor spheroid on Day 4 (B). Further analyses of optical attenuation coefficient histograms (C, G, K) were performed to determine the threshold to separate low and high attenuation regions (i.e. 0.48 mm−1), which is calculated as the median of the two peak values (P1=0.36 mm−1, P2=0.60 mm−1). High-attenuation regions above the threshold highlighted in red (H, L) were detected as the necrotic cores in the tumor spheroids. The region of necrotic tissue clearly increased as the spheroid developed. Scale Bars: 100 µm.
The quantitative determination of necrotic region is based on attenuation coefficients instead of the intensity profiles. To determine the threshold to separate low and high attenuation regions, we first plotted histograms of attenuation coefficients from OCT images of specimens from different time point (Figure 4C, G, and K). It can be clearly seen that live and necrotic tissue have distinct peak locations (Figure 4C, G). The histograms were then fit as the combination of two Gaussian curves, where the peak locations (P1, P2) were determined from the curve-fitting. The best fitting results were observed at Peak 1 (P1 = 0.36 mm−1) and Peak 2 (P2 = 0.60 mm−1) for all tumor spheroids tested in this study with R2 > 0.95. The threshold of high attenuation region was determined as the mean of the two peak values (i.e. 0.48 mm−1). Binarized maps were generated by highlighting high attenuation regions above the threshold (indicated in red) (Figure 4D, H, L). Since the OCT imaging technology was completely non-destructive, we further monitored the progression of tumor necrotic regions from the same sample throughout the growing period (Supplementary Video-3). Increase of high-attenuation regions was clearly visible as the spheroid developed. Similar results were also found in the progression of necrotic regions of U-87 MG tumor spheroids.
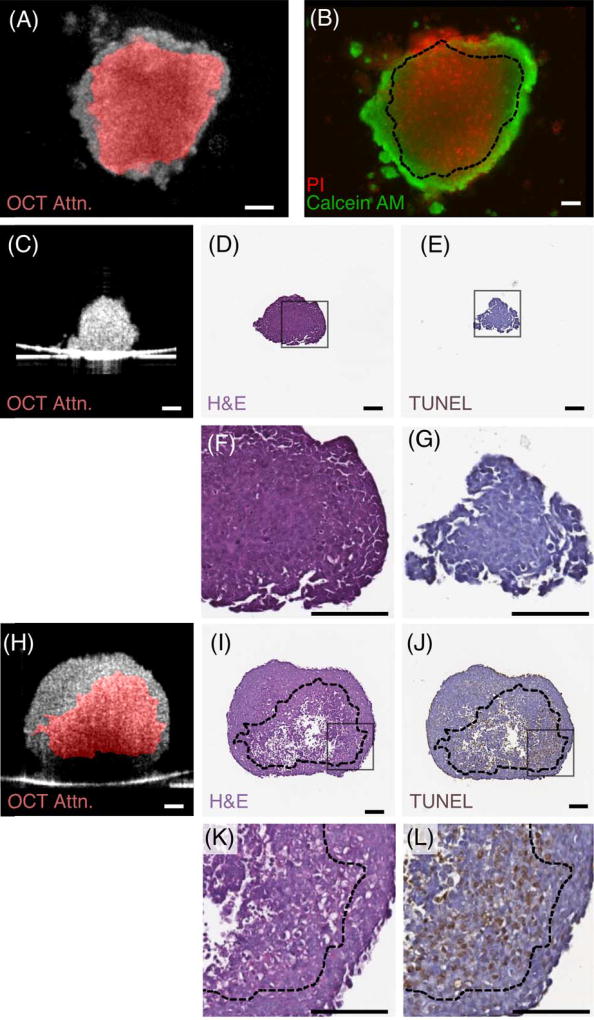
This non-destructive necrotic region detection technique was confirmed by comparing spheroid images obtained using fluorescence, histology and immunohistochemistry (IHC) with corresponding OCT optical attenuation images (Figure 5). Figure 5A–B shows the correlation between en face OCT attenuation image of an HCT 116 tumor spheroid (Figure 5A) at Day 14 and the corresponding fluorescent image (Figure 5B). The necrotic region within the tumor spheroid indicated by the viability test (PI stained, red region in Figure 5B) was consistent with the distribution of the high-attenuation region derived from the OCT image (red region, Figure 5A). Figure 5C–L further showed the correlation between OCT attenuation image of two HCT 116 tumor spheroids at Days 4 and 14 and the corresponding histology and IHC spheroid slices stained with H&E and TUNEL, respectively. On Day 4, no necrotic region was observed in OCT attenuation image, H&E and TUNEL stained slices (Figure 5C–G). On Day 14, OCT attenuation image (Figure 5H) showed a good match with H&E and TUNEL slices (Figure 5I–L), as indicated by analysing the features within the regions in H&E and TUNEL slices marked by dash lines derived from the contour of OCT high attenuation regions. In H&E slices, the necrotic regions were indicated by less dense and aggregated structure located within the dashed line region (Figure 5I, K). In TUNEL slices, a good match was observed between high attenuation region and TUNEL labelled apoptotic cellular region (Figure 5J, L).
Figure 5.
Comparison of necrotic regions identified based on optical attenuation contrast with fluorescent, histology and immunohistochemistry (IHC) results. OCT attenuation image of a HCT 116 tumor spheroid on Day 14 (A) was compared with the corresponding fluorescent image (B). The spheroid in fluorescent image was stained with calcein AM (live cells, green) and propidium iodide (dead cells, red). A good match between necrotic regions identified from OCT attenuation image (A: high attenuation region in red) and fluorescent image (B: PI staining in red) was observed. OCT attenuation images of two HCT 116 tumor spheroids on Day 4 (C) and Day 14 (H) were compared with corresponding H&E (D, I) and TUNEL stained (E, J) slices, respectively. No necrotic core was observed in either OCT, H&E or TUNEL images on Day 4. In contrast, the tumor spheroid on Day 14 clearly exhibited a necrotic region in all images (H–L). The highlighted high attenuation region in OCT image (H) matched well with the necrotic region (I) and apoptotic region (J) identified by H&E and TUNEL staining, respectively. Zoom-in views of gray square-highlighted areas in H&E and TUNEL images (D, E, I, J) were shown in (F, G, K, L), respectively. The contours of high attenuation region in OCT image were indicated in black dash lines in H&E and TUNEL images (B, I–L). Scale Bars: 100 µm.
Discussion and Conclusions
Quantitative assessment of 3D tumor spheroids, including the size, shape, and growth curve is of great importance since tumor activity is highly relevant to its morphological structures. Currently, imaging platforms employing bright field, phase contrast or fluorescent imaging have been established for high-throughput, routine imaging and analysis of morphology or functions of the MCTS (8,9,18,22). However, they are unable to resolve the entire, large tumor structure due to limited depth penetration. In this study, we demonstrated the capability of OCT to acquire 3D label-free images of the entire MCTS as they develop over time. We observed different growth patterns between U-87 MG and HCT 116 cell lines: U-87 MG spheroids remained tightly-packed over time, while some HCT 116 spheroids became flattened and disrupted after Day 11. Nevertheless, from 2D images, both U-87 MG and HCT 116 tumor spheroids were observed to maintain their spherical shapes up to 18 days with diameters increasing gradually, yielding a false-positive observation of spheroid growth. In addition, quantitative analyses of HCT 116 spheroids showed that volume estimation based on 2D cross-sections led to large quantification errors when irregular shapes of tumor spheroids developed, while volume quantification based on 3D OCT data yielded an accurate result of the actual volume. Furthermore, 3D OCT images provide a unique view of MCTS, and offer complimentary depth information that is not available in 2D screening.
Viability tests remain a routine procedure used in functional analysis of the MCTS, especially for the purpose of drug screening (18). These tests require that spheroids be stained with fluorescent dyes, indicating live and dead cells, and be imaged with a fluorescent microscope. However, the destructive nature of fluorescent dyes staining indicates that these tests are only suitable for end-point studies. In this study, we demonstrated an alternative method that can characterize cell viability within the entire MCTS. Our results have indicated that OCT can clearly differentiate the necrotic region from the viable region in the spheroid based on intrinsic optical attenuation contrast. Additionally, OCT’s 3D imaging capability and non-destructive nature allowed us to quantitatively evaluate the distribution of necrotic tissues and monitor the progression of the necrotic core within the MCTS in situ over a period of 21 days, which is not feasible with traditional fluorescence viability tests.
One interesting observation is that the necrotic tissue within the tumor spheroid is not symmetrically distributed as expected from the concentric model for in vivo large tumor spheroids (4,48), but shifted towards the bottom of the tumor spheroid. This observation was clearly represented by OCT attenuation image (Figure 5H) and further confirmed by histology and immunohistochemistry results (Figure 5I–L). We suspect, though, that this asymmetric distribution is related to the surrounding environment of the MCTSs. In our experiment, the bottom surface of the MCTS is in close contact with the bottom of cell culture well. The heterogeneous distribution of oxygen and nutrients around the spheroid may partially explain the non-uniform distribution of necrotic tissue within the tumor spheroid. Early experimental evidence suggests that non-uniform distribution of oxygen tension exists within a stationary tumor spheroid (48). Our observations also indicated a thin layer of live tumor cells between the necrotic region and the bottom of the well (Figure 5I–L). In extra-large tumor spheroids (Dia. > 600 µm), we noticed that their bottom regions not highlighted in red in the OCT attenuation image. We suspected that it may be either due to insufficient penetration of light into these regions (Figure 4E, 4I, where intensity of these regions was close to the background intensity) or due to strong light reflection on top of tumor spheroid blocking incident lights.
The current OCT setup for this experiment takes 10–15 seconds to acquire one 3D dataset of a single tumor spheroid. Imaging speed can be further improved for high-throughput screening either by using state-of-the-art OCT systems equipped with a high speed tunable laser source (49), or by employing parallel OCT imaging with a space-division multiplexing setup (50). For example, a column of 16 wells in a 384-well plate could be scanned simultaneously with a 16-channel parallel OCT system, in order to achieve a 16x speed improvement for high throughput screening.
In summary, we have demonstrated that OCT is capable of visualizing and characterizing 3D structure of multicellular tumor spheroids over time. In addition to diameter measurements, OCT provides complementary depth information to better characterize tumor morphology, yielding an accurate and robust quantification of volumes and heights of MCTSs. Moreover, OCT can be reliably utilized to identify necrotic tissues within tumor spheroids based on intrinsic optical contrast, suggesting a promising alternative of viability tests in tumor spheroids. Further development of this OCT-based imaging platform with a parallel imaging scheme will facilitate high-throughput screening of MCTS for cancer drug discovery as well as other 3D tissue models for various biomedical applications.
Supplementary Material
Acknowledgments
Financial support: This work was supported by NSF IDBR grant 1455613, PFI:AIR-TT grant, 1640707, NIH grants R21EY026380 and R15EB019704, and Lehigh University startup fund.
The authors would like to acknowledge Mr. Wentao Shi for his assistance in cell culture and histological characterizations.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
References
- 1.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–6. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 2.Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18:240–9. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol J. 2014;9(9):1115–28. doi: 10.1002/biot.201300492. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240(4849):177–84. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 5.Mueller-Klieser W. Three-dimensional cell cultures: from molecular mechanisms to clinical applications. Am J Physiol Cell Physiol. 1997;273:C1109–C23. doi: 10.1152/ajpcell.1997.273.4.C1109. [DOI] [PubMed] [Google Scholar]
- 6.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3(9–10):1172–84. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4(3):309–24. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 8.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136(3):473–8. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaBarbera DV, Reid BG, Yoo BH. The multicellular tumor spheroid model for high-throughput cancer drug discovery. Expert Opin Drug Discov. 2012;7:819–30. doi: 10.1517/17460441.2012.708334. [DOI] [PubMed] [Google Scholar]
- 11.Pampaloni F, Ansari N, Stelzer EHK. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013;352:161–77. doi: 10.1007/s00441-013-1589-7. [DOI] [PubMed] [Google Scholar]
- 12.Lovitt CJ, Shelper TB, Avery VM. Miniaturized three-dimensional cancer model for drug evaluation. Assay Drug Dev Technol. 2013;11(7):435–48. doi: 10.1089/adt.2012.483. [DOI] [PubMed] [Google Scholar]
- 13.Wenzel C, Riefke B, Gründemann S, Krebs A, Christian S, Prinz F, et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323(1):131–43. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Astashkina A, Grainger DW. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv Drug Deliv Rev. 2014;69–70:1–18. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014;12(4):207–18. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong X, Lin C, Cheng J, Su J, Zhao H, Liu T, et al. Generation of multicellular tumor spheroids with microwell-based agarose scaffolds for drug testing. PLoS ONE. 2015;10(6):e0130348. doi: 10.1371/journal.pone.0130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann OI, Ilmberger C, Magosch S, Joka M, Jauch K-W, Mayer B. Impact of the spheroid model complexity on drug response. J Biotechnol. 2015;205:14–23. doi: 10.1016/j.jbiotec.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 18.Martinez NJ, Titus SA, Wagner AK, Simeonov A. High-throughput fluorescence imaging approaches for drug discovery using in vitro and in vivo three-dimensional models. Expert Opin Drug Discov. 2015;10:1347–61. doi: 10.1517/17460441.2015.1091814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacology & Therapeutics. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Zhou Q, Voss TC, Quick KL, LaBarbera DV. High-throughput imaging: Focusing in on drug discovery in 3D. Methods. 2016;96:97–102. doi: 10.1016/j.ymeth.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ham SL, Joshi R, Thakuri PS, Tavana H. Liquid-based three-dimensional tumor models for cancer research and drug discovery. Exp Biol Med. 2016;241(9):939–54. doi: 10.1177/1535370216643772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kessel S, Cribbes S, Dery O, Kuksin D, Sincoff E, Qiu J, et al. High-Throughput 3D tumor spheroid screening method for cancer drug discovery using Celigo image cytometry. SLAS Technol. 2017;22(4):454–65. doi: 10.1177/2211068216652846. [DOI] [PubMed] [Google Scholar]
- 23.Stock K, Estrada MF, Vidic S, Gjerde K, Rudisch A, Santo VE, et al. Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci Rep. 2016;6:28951. doi: 10.1038/srep28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakuri PS, Ham SL, Luker GD, Tavana H. Multiparametric analysis of oncology drug screening with aqueous two-phase tumor spheroids. Mol Pharm. 2016;13(11):3724–35. doi: 10.1021/acs.molpharmaceut.6b00527. [DOI] [PubMed] [Google Scholar]
- 25.Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep. 2016;6:19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, et al. Optical coherence tomography. Science. 1991;254(5035):1178–81. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wojtkowski M. High-speed optical coherence tomography: basics and applications. Appl Opt. 2010;49(16):D30–61. doi: 10.1364/AO.49.000D30. [DOI] [PubMed] [Google Scholar]
- 28.Vakoc BJ, Fukumura D, Jain RK, Bouma BE. Cancer imaging by optical coherence tomography: preclinical progress and clinical potential. Nat Rev Cancer. 2012;12(5):363–8. doi: 10.1038/nrc3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drexler W, Liu M, Kumar A, Kamali T, Unterhuber A, Leitgeb RA. Optical coherence tomography today: speed, contrast, and multimodality. J Biomed Opt. 2014;19(7):071412. doi: 10.1117/1.JBO.19.7.071412. [DOI] [PubMed] [Google Scholar]
- 30.Fujimoto J, Swanson E. The Development, Commercialization, and Impact of Optical Coherence Tomography History of Optical Coherence Tomography. Investigative Ophthalmology & Visual Science. 2016;57(9):OCT1–OCT13. doi: 10.1167/iovs.16-19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boppart SA, Brezinski ME, Pitris C, Fujimoto JG. Optical coherence tomography for neurosurgical imaging of human intracortical melanoma. Neurosurgery. 1998;43(4):834–41. doi: 10.1097/00006123-199810000-00068. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen FT, Zysk AM, Chaney EJ, Kotynek JG, Oliphant UJ, Bellafiore FJ, et al. Intraoperative evaluation of breast tumor margins with optical coherence tomography. Cancer Res. 2009;69(22):8790–6. doi: 10.1158/0008-5472.CAN-08-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C, Cohen DW, Wang Y, Lee HC, Mondelblatt AE, Tsai TH, et al. Integrated optical coherence tomography and microscopy for ex vivo multiscale evaluation of human breast tissues. Cancer Res. 2010;70(24):10071–9. doi: 10.1158/0008-5472.CAN-10-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villiger M, Lorenser D, McLaughlin RA, Quirk BC, Kirk RW, Bouma BE, et al. Deep tissue volume imaging of birefringence through fibre-optic needle probes for the delineation of breast tumour. Sci Rep. 2016;6:28771. doi: 10.1038/srep28771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apelian C, Gastaud C, Boccara AC. Proc. SPIE 10053, Optical Coherence Tomography and Coherence Domain Optical Methods in Biomedicine XXI. San Francisco, USA. Bellingham: SPIE; 2017. Extracting relevant information for cancer diagnosis from dynamic full field OCT through image processing and learning; p. 100531H. [Google Scholar]
- 36.Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15(10):1219–23. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kut C, Chaichana KL, Xi J, Raza SM, Ye X, McVeigh ER, et al. Detection of human brain cancer infiltration ex vivo and in vivo using quantitative optical coherence tomography. Sci Transl Med. 2015;7:292ra100. doi: 10.1126/scitranslmed.3010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma M, Verma Y, Rao KD, Nair R, Gupta PK. Imaging growth dynamics of tumour spheroids using optical coherence tomography. Biotechnol Lett. 2006;29(2):273–8. doi: 10.1007/s10529-006-9232-2. [DOI] [PubMed] [Google Scholar]
- 39.Jung Y, Nichols AJ, Klein OJ, Roussakis E, Evans CL. Label-free, longitudinal visualization of PDT response in vitro with optical coherence tomography. Isr J Chem. 2012;52(8–9):728–44. doi: 10.1002/ijch.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piccinini F, Tesei A, Arienti C, Bevilacqua A. Cancer multicellular spheroids: Volume assessment from a single 2D projection. Comput Methods Programs Biomed. 2015;118(2):95–106. doi: 10.1016/j.cmpb.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Rizvi I, Celli JP, Evans CL, Abu-Yousif AO, Muzikansky A, Pogue BW, et al. Synergistic enhancement of Carboplatin efficacy with photodynamic therapy in a three-dimensional model for micrometastatic ovarian cancer. Cancer Res. 2010;70(22):9319–28. doi: 10.1158/0008-5472.CAN-10-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 43.Li F, Xu T, Nguyen DH, Huang X, Chen CS, Zhou C. Label-free evaluation of angiogenic sprouting in microengineered devices using ultrahigh-resolution optical coherence microscopy. J Biomed Opt. 2014;19(1):016006. doi: 10.1117/1.JBO.19.1.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Song Y, Dryer A, Cogguillo W, Berdichevsky Y, Zhou C. Nondestructive evaluation of progressive neuronal changes in organotypic rat hippocampal slice cultures using ultrahigh-resolution optical coherence microscopy. Neurophoton. 2014;1(2):025002. doi: 10.1117/1.NPh.1.2.025002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi WJ, Jeon DI, Ahn SG, Yoon JH, Kim S. Full-field optical coherence microscopy for identifying live cancer cells by quantitative measurement of refractive index distribution. Opt Express. 2010;18(22):23285–95. doi: 10.1364/OE.18.023285. [DOI] [PubMed] [Google Scholar]
- 46.Klein OJ, Jung YK, Evans CL. Longitudinal, quantitative monitoring of therapeutic response in 3D in vitro tumor models with OCT for high-content therapeutic screening. Methods. 2014;66(2):299–311. doi: 10.1016/j.ymeth.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermeer KA, Mo J, Weda JJA, Lemij HG, de Boer JF. Depth-resolved model-based reconstruction of attenuation coefficients in optical coherence tomography. Biomed Opt Express. 2014;5(1):322–37. doi: 10.1364/BOE.5.000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mueller-Klieser WF, Sutherland RM. Influence of convection in the growth medium on oxygen tensions in multicellular tumor spheroids. Cancer Res. 1982;42(1):237–42. [PubMed] [Google Scholar]
- 49.Klein T, Huber R. High-speed OCT light sources and systems [Invited] Biomed Opt Express. 2017;8(2):828–59. doi: 10.1364/BOE.8.000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou C, Alex A, Rasakanthan J, Ma Y. Space-division multiplexing optical coherence tomography. Opt Express. 2013;21:19219–27. doi: 10.1364/OE.21.019219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.