Abstract
Hemerythrin‐like proteins have generally been studied for their ability to reversibly bind oxygen through their binuclear nonheme iron centers. However, in recent years, it has become increasingly evident that some members of the hemerythrin‐like superfamily also participate in many other biological processes. For instance, the binuclear nonheme iron site of YtfE, a hemerythrin‐like protein involved in the repair of iron centers in Escherichia coli, catalyzes the reduction of nitric oxide to nitrous oxide, and the human F‐box/LRR‐repeat protein 5, which contains a hemerythrin‐like domain, is involved in intracellular iron homeostasis. Furthermore, structural data on hemerythrin‐like domains from two proteins of unknown function, PF0695 from Pyrococcus furiosus and NMB1532 from Neisseria meningitidis, show that the cation‐binding sites, typical of hemerythrin, can be absent or be occupied by metal ions other than iron. To systematically investigate this functional and structural diversity of the hemerythrin‐like superfamily, we have collected hemerythrin‐like sequences from a database comprising fully sequenced proteomes and generated a cluster map based on their all‐against‐all pairwise sequence similarity. Our results show that the hemerythrin‐like superfamily comprises a large number of protein families which can be classified into three broad groups on the basis of their cation‐coordinating residues: (a) signal‐transduction and oxygen‐carrier hemerythrins (H‐HxxxE‐HxxxH‐HxxxxD); (b) hemerythrin‐like (H‐HxxxE‐H‐HxxxE); and, (c) metazoan F‐box proteins (H‐HExxE‐H‐HxxxE). Interestingly, all but two hemerythrin‐like families exhibit internal sequence and structural symmetry, suggesting that a duplication event may have led to the origin of the hemerythrin domain.
Keywords: up‐and‐down bundle, nonheme iron protein, hemerythrin‐like superfamily subgroups, oxygen‐binding protein
Abbreviations
- Fqo
F420H(2)‐dependent quinone reductase
- HMM
hidden Markov model
- IRP2
iron regulatory protein 2
- LLM
luciferase‐like flavin monooxygenase
- MCP
methyl‐accepting chemotaxis protein
- PDB
protein data bank
- PNPOx
pyridoxamine 5'‐phosphate oxidase
- RIC
repair iron‐center
Introduction
Hemerythrin was initially described as a multimeric O2 carrier‐protein with a binuclear nonheme iron center and with a distribution that, at first, appeared to be limited to three phyla of marine invertebrates, Brachiopoda, Priapulida, and Annelida.1 Over the past decade, however, hemerythrin‐like proteins have been identified in many taxonomically distant groups including humans,2, 3, 4 plants,5 bacteria,6, 7, 8, 9, 10 and archaea (PDB code: 2P0N). The function of these binuclear nonheme Fe‐containing proteins (e.g., hemerythrin, myohemerytrin, bacteriohemerythrin) is tightly related to O2‐binding and activation,11, 12, 13 a trait indicative of the major evolutionary pressure exerted by atmospheric and oceanic oxygenation since the late Archaean Eon on the biosphere.14 It has been proposed that hemerythrin homologs that reversibly bind O2 (hemerythrin, myohemerytrin,15 and bacteriohemerythrin16) belong to a monophyletic group that appeared during this period of increasingly oxidizing conditions.14
The basic structural fold of hemerythrin‐like proteins consists of an up‐and‐down four helix bundle with an overall right‐handed path.17, 18, 19 Most structures exhibit a binuclear cation‐binding site in which each helix contributes to the metal‐ion coordination with at least one residue. The first coordination sphere of the binuclear five‐coordinate/six‐coordinate nonheme Fe(II) site of O2‐binding hemerythrins includes five histidines, two bridging carboxylates from a glutamic and an aspartic acid residues, and a µ‐oxo/hydroxo bridge.20 However, the cation‐binding residues are not conserved in all solved hemerythrin structures (Table 1). For instance, the catalytic site of hemerythrin in YtfE,24 a repair iron centers protein from Escherichia coli, has been shown to involve four instead of five histidine residues, and two carboxylate bridges from two glutamic residues instead of one glutamic and one aspartic acid residues. This arrangement results in a five‐coordinate/five‐coordinate binuclear Fe(II)‐binding site with nitric oxide‐reductase activity24 that is likely to be part of a defense mechanism against DNA damage associated with NO. A survey of the literature reveals that many other functionally characterized hemerythrin‐containing proteins have a cation‐binding motif that is different from the H‐HxxxE‐HxxxH‐HxxxxD motif of signal‐transduction and oxygen‐carrier hemerythrins. For instance, the primary structure of the Rv2633c hemerythrin‐like catalase from Mycobacterium tuberculosis (Uniprot ID: P9WL59),25 and hemerythrin homologs in Mycobacterium smegmatis (Uniprot IDs: A0QXI3, A0QV17, and A0R5J3);26 Anabaena sp. strain PCC7120 (Uniprot ID: Q8YS92);27 Aeromonas hydrophila (Uniprot ID: A0KMZ0);10 Acidothermus cellulolyticus (Uniprot ID: A0LQU2);28 Oryza sativa subsp. japonica (Uniprot IDs: V9G2Z0; and Q6AUD8) and Arabidopsis thaliana (Uniprot ID: Q8LPQ5),5 suggests that these proteins also have differences in the first coordination‐sphere of the iron centers, but there is no available confirmation from structural data.
Table 1.
Structural Characteristics of Cation‐coordination in Solved Hemerythrin‐like Domain Structures
| Protein | Organism | Function | Coordination number | Ligation | Source [PDB code] |
|---|---|---|---|---|---|
| Hemerythrin (subunit A) | Themiste dyscritum | Reversible O2 binding | 6C/5C | 5H/1E/1D | [1HMD]20 |
| Myohemerythrin | Themiste hennahi | Reversible O2 binding | 6C/5C | 5H/1E/1D | [2MHR]21 |
| Bacteriohemerythrin (McHr) | Methylococcus capsulatus | Reversible O2 binding | 6C/5C | 5H/1E/1D | [4XPX]16 |
| Hemerythrin‐like domain (DcrH) | Desulfovibrio vulgaris | O2 sensing | 6C/5C | 5H/1E/1D | [2AWC]22 |
| Hemerythrin‐like domain (FBXL5) | Homo sapiens | Iron sensing | 6C/5C | 4H/3E | [3V5X]23 |
| Hemerythrin‐like‐domain (YtfE) | Escherichia coli | 2NO + 2H+ + 2e‐ → N2O + H2O | 5C/5C | 4H/2E | [5FNN]24 |
| Hemerythrin domain (hypothetical protein NMB1532) | Neisseria meningitidis | Unknown | 5C/5C | 4H/2E | [2P0N] |
| Hemerythrin domain (uncharacterized protein PF0695) | Pyrococcus furiosus | Unknown | – | – | [3CAX] |
Although the evolutionary relationships among a group of protein sequences closely related to signal‐transduction and oxygen‐carrier hemerythrins have been described elsewhere,14, 29, 30, 31 a comprehensive evolutionary analysis and classification of hemerythrin‐like proteins is lacking. In this study, we report the outcome of an extensive bioinformatic analysis of hemerythrin‐like proteins and present their classification into three major groups based on the conservation of cation‐coordinating residues.
Results and Discussion
Structural analysis
The structures of hemerythrin‐like homologs in the Protein Data Bank (PDB) were identified using the HHpred webserver.32 We found 27 structures corresponding to eight different proteins from different source organisms: YtfE (PDB IDs: 5FNN, 5FNP, 5FNY) from Escherichia coli, DcrH (PDB IDs: 2AVK, 2AWC, 2AWY, 3AGT, 3AGU, 3WAQ, 3WHN) from Desulfovibrio vulgaris; bacteriohemerythrin (PDB IDs: 4XPW, 4XPX, 4XPY) from Methylococcus capsulatus; NMB1532 (PDB ID: 2P0N) from Neisseria meningitidis; PF0695 (PDB ID: 3CAX) from Pyrococcus furiosus; myohemerythrin (PDB IDs: 1A7D, 1A7E, 2MHR) from Themiste hennahi; hemerythrin (PDB IDs: 1HMD, 2HMQ, 2HMZ, 1HMO) from Themiste dyscritum; and FBXL5 (PDB IDs: 3U9M, 3V5Y, 3U9J, 3V5X, 3V5Z) from Homo sapiens. To avoid redundancy, only one representative per protein was analyzed. Distinctive traits in the tertiary structure of these proteins were evaluated by pairwise comparisons of X‐ray crystal structures (Table 2). While seven of the analyzed eight structures showed a right‐handed four‐alpha‐helix bundle, characteristic of hemerythrin, a distinctive two‐helix swap was identified in the hemerythrin‐like structure of YtfE. This rearrangement preserves the up‐and‐down topology of the fold, but results in a left‐handed four‐helix bundle (Fig. 1). Because of this topological difference, the alignment of full protein structures produced suboptimal results (Supporting Information Table SI). We therefore performed further structural analysis of hemerythrin‐like proteins based on the comparison of their metal‐binding sites using MetalS2.33 The local similarity between pairs of proteins was evaluated by their MetalS2 score and by the percent identity of the superposition‐derived sequence alignment (Table 2). Scores lower than 2.25 indicate a high level of structural similarity.33
Table 2.
Metal S 2 Score and Percent Identity of Pairwise Structural Alignments of Hemerythrin Homologs and the Uncharacterized Protein Q4MWP8
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
|---|---|---|---|---|---|---|---|---|---|
| Hemerythrin, subunit A (Uniprot ID: P02246) | 1 | 0.000 | 0.637 (55%) | 0.759 (50%) | 0.797 (44%) | 1.907 (14%)a | 1.623 (31%) | 1.611 (18%)a | 1.814 (16%)a |
| Myohemerythrin (Uniprot ID: P02247) | 2 | 0.000 | 0.832 (45%) | 0.723 (42%) | 1.937 (17%)a | 1.514 (14%)a | 1.514 (20%)a | 1.748 (14%)a | |
| Bacteriohemerythrin, McHr (Uniprot ID: Q60AX2) | 3 | 0.000 | 0.740 (50%) | 1.926 (20%)a | 1.560 (21%)a | 1.569 (18%)a | 1.813 (18%)a | ||
| Hemerythrin‐like domain of DcrH (Uniprot ID: Q726F3) | 4 | 0.000 | 1.853 (20%)a | 1.559 (28%)a | 1.870 (24%)a | 1.946 (20%)a | |||
| Hemerythrin‐like domain of FBXL5 (Uniprot ID: Q9UKA1) | 5 | 0.000 | 1.286 (25%)a | 1.583 (29%)a | 2.233b (26%)a | ||||
| Hemerythrin‐like‐domain of YtfE (Uniprot ID: P69506) | 6 | 0.000 | 1.061 (23%)a | 1.489 (18%)a | |||||
| Hemerythrin‐like domain of NMB1532 (Uniprot ID: Q9JYL1) | 7 | 0.000 | 1.521 (18%)a | ||||||
| Uncharacterized protein Q4MWP8 | 8 | 0.000 |
The numbers in parenthesis refer to the percent identity of the superposition‐derived sequence alignment.
Percent identity <30%.
MetalS2 score >2.
Figure 1.
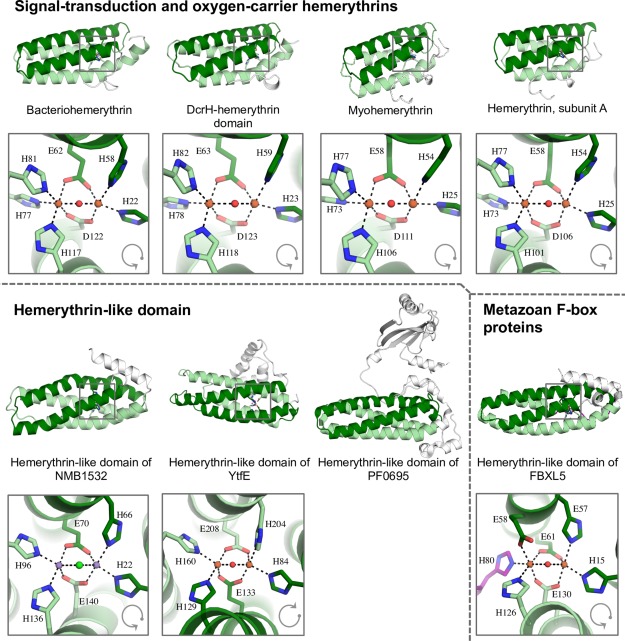
Ribbon diagram of distant hemerythrin homologues. In all structures, helices 1 and 2 are colored in dark green; helices 3 and 4 are colored in light green. Non‐homologous regions are shown in white. A disordered region in the tertiary structure of hemerythrin in FBXL5 is highlighted in purple. Gray arrows indicate the sequential arrangement of the helices in the hemerythrin fold.
The entire set of hemerythrin homologs aligned with MetalS2 scores of below 2. Hemerythrin, myohemerythrin, bacteriohemerythrin, and the hemerythrin‐like domain of DcrH from Desulfovibrio vulgaris are closely related (sequence identity >40%) and aligned with a MetalS2 score below 1. These hemerythrin homologs have a characteristically conserved 5H/1E/1D ligation of the binuclear iron site (Table 1).
The hemerythrin‐like domains of NMB1532 (PDB ID: 2P0N) and YtfE (PDB ID: 5FNN) are distantly related (sequence identity <25%), but they have a very similar tertiary structure, as shown by their structure alignment score (Table 2). The two‐helix swap in YtfE maintains the local structure of the binuclear site, in which helix α1 and helix α3 each donates one histidine residue, and both helix α2 and helix α4 donate a histidine and a glutamate. An important difference between these two structures is the presence of two manganese ions coordinated to the hemerythrin domain of NMB1532 from Neisseria meningitidis (PDB code: 2P0N). Because presently no functional data is available for NMB1532, it is unknown whether manganese ions are naturally present in hemerythrins.
The hemerythrin‐like domain of FBXL5, in which a disordered region substitutes part of helix α3, exhibits the worst superimposition scores of the homologous set. The binuclear site of the hemerythrin domain in these proteins is asymmetrical: α2 donates an additional glutamic acid, and the disordered region that replaces part of helix α3 donates the third histidine residue of the cation‐coordination.
Cluster analysis
To map out sequence and evolutionary relationships between the members of the hemerythrin‐like superfamily in a comprehensive fashion, we searched for hemerythrin‐like sequences in 2580 fully sequenced genomes (Table 3) and identified a total of 6599 sequences, which were subsequently clustered in CLANS34 (Fig. 2) based on their all‐against‐all pairwise similarities as measured by BLAST P values. In addition to a large family of oxygen‐carrier and oxygen‐sensing hemerythrins, several independent clusters were identified. These clusters fall into three broadly defined groups based on the conservation of cation‐coordinating residues: signal‐transduction and oxygen‐carrier hemerythrins (H‐HxxxE‐HxxxH‐HxxxxD), hemerythrin‐like (H‐HxxxE‐H‐HxxxE), and metazoan F‐box (H‐HExxE‐H‐HxxxE) proteins. Their phylogenetic distribution indicates that the metazoan F‐box proteins set is the most recent one. Internal symmetry within most hemerythrin‐like and metazoan F‐box protein sequences (Fig. 3, Supporting Information Table SI) strongly suggests that the hemerythrin fold originated by duplication and fusion of an ancestral helix‐loop‐helix motif.
Table 3.
Presence/Absence of Hemerythrin Homologs
| Phylogenetic group | Presence | Absence |
|---|---|---|
| Archaea (n = 171) | 74 | 97 |
| Nanoarchaeota (n = 2) | – | 2 |
| Euryarchaeota (n = 104) | 41 | 63 |
| Candidatus korarchaeota (n = 1) | 1 | – |
| Crenarchaeota (n = 56) | 20 | 36 |
| Thaumarchaeota (n = 9) | 9 | – |
| Unclassified archaea (n = 15) | 3 | 12 |
| Uncultured archaea (n = 4) | – | 4 |
| Bacteria (n = 3998) | 2062 | 1936 |
| Aquificae (n = 11) | 9 | 2 |
| Thermodesulfobacteria (n = 2) | 2 | – |
| Thermotogae (n = 12) | 8 | 4 |
| Caldiserica (n = 1) | 1 | – |
| Chrysiogenetes (n = 1) | 1 | – |
| Deferribacteres (n = 6) | 6 | – |
| Dictyoglomi (n = 1) | 1 | – |
| Chloroflexi (n = 34) | 12 | 22 |
| Actinobacteria (n = 561) | 309 | 252 |
| Planctomycetes (n = 26) | 9 | 17 |
| Chlamydiae (n = 11) | 3 | 8 |
| Verrucomicrobia (n = 13) | 2 | 11 |
| Lentisphaerae (n =1) | – | 1 |
| Candidatus omnitrophica (n = 3) | – | 3 |
| Nitrospirae (n = 12) | 4 | 8 |
| Acidobacteria (n = 10) | 4 | 6 |
| Synergistetes (n = 15) | – | 15 |
| Cyanobacteria/melainabacteria group (n = 98) | 54 | 44 |
| Firmicutes (n = 907) | 431 | 476 |
| Tenericutes (n = 71) | 3 | 68 |
| Fibrobacteres (n = 3) | 2 | 1 |
| Elusimicrobia (n = 3) | 1 | 2 |
| Bacteroidetes/chlorobi group (n = 334) | 244 | 90 |
| Candidatus cloacimonetes (n = 2) | 1 | 1 |
| Candidatus latescibacteria (n = 1) | 1 | – |
| Candidatus marinimicrobia (n = 1) | – | 1 |
| Deinococcus‐thermus (n = 17) | 3 | 14 |
| Armatimonadetes (n = 3) | 1 | 2 |
| Spirochaetes (n = 41) | 30 | 11 |
| Fusobacteria (n = 15) | 5 | 10 |
| Gemmatimonadetes (n = 7) | 1 | 6 |
| Proteobacteria (n = 1327) | 857 | 470 |
| Nitrospinae/tectomicrobia group (n = 5) | 2 | 3 |
| Unclassified bacteria (n = 442) | 55 | 387 |
| Uncultured bacterium (n = 1) | – | 1 |
| Eukaryota (n = 770) | 392 | 378 |
| Alveolata (n = 45) | 3 | 42 |
| Amoebozoa (n = 7) | 4 | 3 |
| Apusozoa (n = 1) | 0 | 1 |
| Cryptophyta (n = 1) | 1 | 0 |
| Euglenozoa (n = 19) | 1 | 18 |
| Diplomonadida (n = 3) | 0 | 3 |
| Haptophyceae (n = 2) | 0 | 2 |
| Heterolobosea (n = 1) | 1 | 0 |
| Choanoflagellida (n = 2) | 1 | 1 |
| Fungi (n = 373) | 213 | 160 |
| Metazoa (n = 221) | 98 | 123 |
| Fonticula (n = 1) | 0 | 1 |
| Ichthyosporea (n = 2) | 2 | 0 |
| Parabasalia (n = 1) | 1 | 0 |
| Rhizaria (n = 2) | 2 | 0 |
| Rhodophyta (n = 2) | 2 | 0 |
| Stramenopiles (n = 23) | 7 | 16 |
| Viridiplantae (n = 64) | 56 | 8 |
Figure 2.
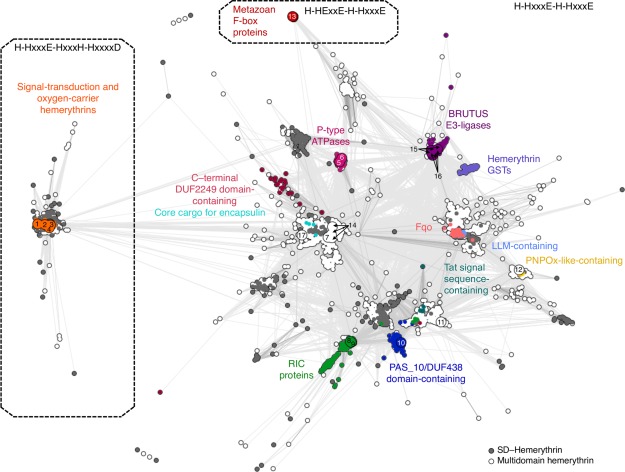
Cluster map of hemerythrin homologs. Cluster map of 6599 hemerythrin sequences in two‐dimensional space at a P value cutoff of 1e‐10. Dotted lines enclose three large groups formed at a P value cutoff of 1e‐13. Each dot represents a sequence; dots are colored by groups of sequences with known domain organization. (1) Q9PIQ3 and (2) Q0P932 from Campylobacter jejuni; (3) Q60AX2 from Methylococcus capsulatus, and Q726F3 from Desulfovibrio vulgaris; (4) Q9KSP0 from Vibrio cholerae; (5) Q9RJ01 from Streptomyces coelicolor; (6) Q92Z60 from Rhizobium meliloti; (7) Q8YS92 from Nostoc sp.; (8) P69506 from Escherichia coli; (9) Q7WX96 from Cupriavidus necator; (10) Q8U2Y3 from Pyrococcus furiosus; (11) A0KMZ0 from Aeromonas hydrophila; (12) Q9JYL1 from Neisseria meningitidis; (13) Q9UKA1 from Homo sapiens; (14) A0QXI3, A0R5J3, A0QV17 from Mycobacterium smegmatis; (15) Q8LPQ5 from Arabidopsis thaliana; (16) V9G2Z0 from Oryza sativa; and (17) Rv2633c from Mycobacterium tuberculosis.
Figure 3.
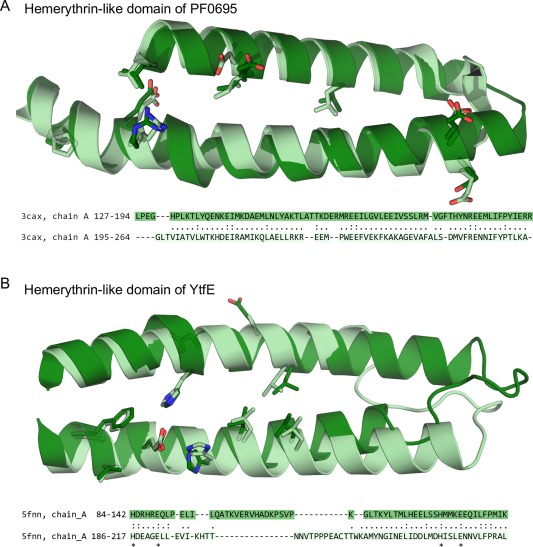
Repeat units of the hemerythrin‐like domain. Internal sequence symmetry was identified by HHpredID with a probability >90. Structural superimposition of the repeat units in (A) PF0695 from Pyrococcus furiosus (PDB ID: 3CAX), and in (B) YtfE from E. coli (PDB ID: 5FNN). Sequence alignments are based on the structural superimposition of the repeat units. Aligned residues are connected by a dot. Exact matches are connected by a colon. Amino acids that participate in cation‐coordination are marked with an asterisk.
O2‐carrier hemerythrins and closely related sequences
Signal‐transduction and O2‐carrier hemerythrins form a tightly connected group in the map containing 1424 hemerythrin sequences from bacterial, archaeal, and eukaryotic species, in which the motif H‐HxxxE‐HxxxH‐HxxxxD is conserved. These conserved positions, which represent a variation from the H‐HxxxE duplicate, contribute to the 5H/1E/1D ligation to iron (Table 1) in hemerythrin from Themiste dyscritum, myohemerythrin from Themiste hennahi, bacteriohemerythrin from M. capsulatus, and the hemerythrin‐like domain of DcrH from D. vulgaris. These hemerythrin homologs are all involved in O2 transport and storage through their ability to bind O2 reversibly,16, 20, 22, 23, 24 and O2 sensing in signal‐transduction proteins by autoxidation of the binuclear iron site upon O2 binding.35 Sequences with an H‐HxxxE‐HxxxH‐HxxxxD motif are highly conserved as shown by the low number of divergent projections in the cluster map, which may indicate a recent divergence of this group (Fig. 2, Supporting Information Table SI).
Hemerythrin‐like proteins containing signal transduction and chemotaxis domains were conspicuously identified in proteobacteria, where they also have been functionally characterized. For instance, the oxygen‐sensing protein DcrH (Uniprot ID: Q726F3) from D. vulgaris comprises an N‐terminal double sensory domain dCache_3 (Pfam accession: PF14827), followed by a domain of unknown function (Pfam accession: PF07889) and a methyl‐accepting chemotaxis‐protein signaling domain (Pfam accession: PF00015). Hemerythrin is located at the C‐terminal end of this protein. A different protein domain organization occurs in VC1216, a signal transduction protein from Vibrio cholerae in which hemerythrin is followed by a GGDEF diguanylate cyclase domain (Pfam accession: PF00990). Different arrangements of sensory and chemotaxis domains (such as TadZ_N, HPTransfase, MEKHLA, MCPsignal, TarH, and HAMP) were also present in sequences from Spirochaetes and Firmicutes species (Supporting Information Fig. S2), suggesting that in these cases, hemerythrin may be involved in a wide range of cellular responses to O2 (Fig. 2, Table 4).
Table 4.
Putative Functions of the Most Frequently Identified Nonhomologous Domains in Hemerythrin‐like Proteins
|
Pfam ID (Accession) |
Domain description | Putative functiona | Amino acid motif of the associated hemerythrin‐like domain |
|---|---|---|---|
| TadZ_N (PF16968) | N‐terminal domain of the pilus assembly protein TadZ | Signal‐transduction‐response receiver | H‐HxxxE‐HxxxH‐HxxxxD |
| HPTransfase (PF10090) | Histidine phosphotransferase | Signal transduction | H‐HxxxE‐HxxxH‐HxxxxD |
| MEKHLA (PF08670) | Shares similarity with the PAS domain | Signal sensor | H‐HxxxE‐HxxxH‐HxxxxD |
| GGDEF (PF00990) | GGDEF domain | Diguanylate cyclase, signal transduction | H‐HxxxE‐HxxxH‐HxxxxD |
| MCPsignal (PF00015) | Methyl‐accepting chemotaxis protein (MCP) signaling domain | Signal transduction | H‐HxxxE‐HxxxH‐HxxxxD |
| TarH (PF02203) | Tar (taxis towards aspartate and maltose, away from nickel and cobalt) ligand binding domain homolog | Signal transduction, chemotaxis | H‐HxxxE‐HxxxH‐HxxxxD |
| HAMP (PF00672) | HAMP domain (present in Histidine kinases, Adenyl cyclases, Methyl‐accepting proteins and Phosphatases) | Signal transduction | H‐HxxxE‐HxxxH‐HxxxxD |
| PDEase_II (PF02112) | cAMP phosphodiesterases class‐II | cAMP catabolic process | H‐HxxxE‐HxxxH‐HxxxxD |
| sCache_2 (PF17200) | Single cache (CAlcium channels and CHEmotaxis receptors) domain 2 | Signal transduction, small‐molecule recognition | H‐HxxxE‐HxxxH‐HxxxxD |
| Popeye (PF04831) | Member of the conserved barrel domain of the cupin superfamily | Unknown | H‐HxxxE‐HxxxH‐HxxxxD |
| SprT‐like (PF10263) | Domain of unknown function | Unknown | H‐HxxxE‐HxxxH‐HxxxxD |
| DUF1858 (PF08984) | Domain of unknown function | Unknown | H‐HxxxE‐HxxxH‐HxxxxD |
| Prok‐RING_2 (PF14445) | Prokaryotic RING finger family 2 | Associated with components of the ubiquitin‐based signaling and degradation system | H‐HxxxE‐H‐HxxxE |
| zf‐CHY (PF05495) | Zinc fingers motif | Unknown | H‐HxxxE‐H‐HxxxE |
| zf‐rbx1 (PF12678) | Zinc fingers motif | Unknown | H‐HxxxE‐H‐HxxxE |
| zinc_ribbon_6 (PF14599) | Zinc fingers motif | Unknown | H‐HxxxE‐H‐HxxxE |
| TAT_signal (PF10518) | TAT (twin‐arginine translocation) pathway signal sequence. Transport of folded proteins across energy‐transducing membranes | Transport across membranes | H‐HxxxE‐H‐HxxxE |
| E1‐E2_ATPase (PF00122) | Proton‐ATPase | Transport across membranes | H‐HxxxE‐H‐HxxxE |
| GST_N_3 (PF13417) | Glutathione S‐transferase, N‐terminal domain | Glutathione metabolism. | H‐HxxxE‐H‐HxxxE |
| Pyrid_ox_like (PF16242) | Pyridoxamine 5'‐phosphate oxidase like | Putative a defense mechanism against oxidative stress | H‐HxxxE‐H‐HxxxE |
| Hydrolase (PF00702) | Haloacid dehalogenase‐like hydrolase | Hydrolase activity | H‐HxxxE‐H‐HxxxE |
| UMPH‐1 (PF05822) | Pyrimidine 5'‐nucleotidase | Hydrolase activity | H‐HxxxE‐H‐HxxxE |
| Bac_luciferase (PF00296) | Luciferase‐like flavin monooxygenase (LLM) domain | Oxygenase activity | H‐HxxxE‐H‐HxxxE |
| MetRS‐N (PF09635) | N‐terminal domain of methionyl‐tRNA synthetase which adopts a glutathione S‐transferase (GST)‐like fold | Unknown | H‐HxxxE‐H‐HxxxE |
| ScdA_N (PF04405) | Repair of iron centers domain | Unknown | H‐HxxxE‐H‐HxxxE |
| DUF2249 (PF10006) | Domain of unknown function | Unknown | H‐HxxxE‐H‐HxxxE |
| F420H2_quin_red (PF04075) | F420H(2)‐dependent quinone (Fqo) reductase | Unknown | H‐HxxxE‐H‐HxxxE |
| DUF2481 (PF10654) | Domain of unknown function | Unknown | H‐HxxxE‐H‐HxxxE |
| DUF1858 (PF08984) | Domain of unknown function | Unknown | H‐HxxxE‐H‐HxxxE |
| PAS_10 (PF13596) | PAS: Per‐Arnt‐Sim domain (present in period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single‐minded protein) | Involved in oxidative stress protection | Not conserved |
| DUF438 (PF04282) | Domain of unknown function | Unknown | Not conserved |
| F‐box_4 (PF15966) | F‐box protein. | Protein‐protein interactions | H‐HExxE‐H‐HxxxE |
| LRR_9 (PF14580) | Leucine‐rich repeat. | Protein‐protein interactions | H‐HExxE‐H‐HxxxE |
Information about putative domain functions was gathered from InterPro.36
H‐HxxxE‐H‐HxxxE hemerythrins
At a P value cutoff of 1e‐13, a total of 4957 bacterial, archaeal, and eukaryotic sequences exhibiting a characteristic conservation of the H‐HxxxE repeat formed many distinct, but profusely connected clusters. These hemerythrin‐like sequences comprise more than a dozen groups, the majority of which are poorly studied. As shown in Table 4, some of the divergent sequences from this group could be annotated, as is the case for repair iron‐center (RIC) hemerythrins; PAS_10‐containing hemerythrins; hemerythrin‐like proteins with a C‐terminal DUF2249 domain (Pfam accession: PF10006); hemerythrin‐like ATPases; Bac_luciferase‐containing hemerythrins (Pfam accession: PF00296); F420H(2)‐dependent quinone reductases (Pfam accession: PF04075); pyridoxamine 5'‐phosphate oxidases (Pfam accession: PF16242); BRUTUS E3‐ligases; and hemerythrin‐containing glutathione S‐transferases (GST). Recently reported hemerythrin‐like cargo proteins detected by the encapsulin system37 were identified and are indicated on the cluster map (Fig. 2). This system is involved in iron mineralization and oxidative stress protection through encapsulation in Firmicutes.37
For the most part, linear combinations of domains in hemerythrin‐like proteins are both cluster‐ and phylum‐specific, with the clear exception of ScdA_N‐ and PAS_10‐containing hemerythrins, which were identified in taxonomically distant species.
Hemerythrin‐like sequences of repair iron‐center proteins form a large sub‐cluster (colored dark green in the map). These hemerythrin‐like proteins contain a domain of unknown function termed ScdA_N (Pfam accession: PF04405). Two characterized hemerythrin‐like RIC proteins, NorA from the denitrifier species Ralstonia eutropha and YtfE from Escherichia coli, have the ability to bind nitric oxide. Nitric oxide and reactive nitrogen species are deleterious products of denitrification and host immune system responses, particularly damaging to iron‐sulfur clusters38 and to DNA.39 The hemerythrin‐like domain of YtfE has been shown to catalyze the reduction of nitric oxide to nitrous oxide.24, 40
DUF438‐ and/or PAS‐containing hemerythrin‐like proteins (dark blue) often have an additional domain (DUF1858, Pfam accession: PF08984) similar to ScdA_N. This group is closely related to hemerythrin‐like RIC proteins. PF0695 is a PAS‐containing hemerythrin‐like protein from Pyrococcus furiosus that has been structurally determined by X‐ray crystallography (PDB code: 3CAX), but its biological function remains uncharacterized. Interestingly, multiple sequence alignments show that the first and the last iron‐coordinating histidine residues are not conserved in PF0695 and most DUF438‐ and/or PAS‐containing hemerythrin‐like proteins, suggesting that these may be naturally occurring metal‐free proteins. Internal structure and sequence symmetry was detected in the hemerythrin‐like domain of YtfE and PF0695 (HHrepID P values of 2.5e‐7 and 4.4e‐12, respectively). The N‐ and C‐ terminal halves of both hemerythrin‐like structures were superposed at RMSDs of less than 3Å (Fig. 3).
The group of hemerythrin‐like ATPase transporters includes two forms with slightly different domain organization; a C‐terminal hemerythrin‐like domain and an N‐terminal E1‐E2_ATPase domain (Pfam accession: PF00122) flank either a UMPH‐1 domain (Pfam accession: PF05822) or a hydrolase domain (Pfam accession: PF00702). This group includes two functionally characterized proteins from Acidothermus cellulolyticus 28 (Uniprot ID: A0LQU2) and from Sinorhizobium meliloti 41 (Uniprot id: Q92Z60). It has been suggested that both proteins participate in Fe2+ (or Ni2+) transport across membranes, and that the C‐terminal hemerythrin‐like domain could function as an iron sensor to avoid harmful intracellular iron overload.28, 41
Plant‐specific hemerythrin‐like sequences form two distinct groups: hemerythrin‐containing BRUTUS proteins and hemerythrin GSTs. BRUTUS proteins were identified in species of Chlorophyceae, Trebouxiophyceae, Mamiellophyceae, Klebsormidiophyceae, and Streptophytina. These sequences contain several zinc fingers motifs (zf‐CHY, zf‐rbx1, zinc_ribbon_6), and a Prok‐RING_2 domain (Pfam accession: PF14445), which suggests that they are E3‐ligases that participate in iron regulation,5, 42 a process equivalent to that present in animals (see below). Internal sequence symmetry found in most hemerythrin homologs was not detected in hemerythrin‐like sequences from BRUTUS proteins. Hemerythrin GST sequences were found exclusively in species of Streptophytina. While most hemerythrin GSTs have a GST_N_3 (Pfam accession: PF13417) N‐terminal domain, seven sequences in this group contain a structurally similar43 MetRS‐N fold (Pfam accession: PF09635). Hemerythrin GSTs may be involved in heavy metal‐detoxification processes in plants.44, 45
Metazoan iron‐sensing hemerythrins
The most distant group of homologues we have detected consists of a set of F‐box‐like iron‐sensing proteins possessing a conserved H‐HExxE‐H‐HxxxE motif. Leucine‐rich repeats are present in most of these proteins (Fig. 2). In the case of the human protein FBXL5, the N‐terminal hemerythrin domain undergoes conformational changes depending on oxygen and iron availability.3, 4 Both leucine‐rich repeats (Pfam accession: PF14580) and F‐box domains (Pfam accession: PF15966) mediate protein–protein interactions; the central F‐box domain interacts with the E3 ubiquitin–ligase complex, and leucine‐rich repeats have been proposed to have role in binding to the iron regulatory protein 2 (IRP2).3, 4
A case of local structure convergence
A similarity search using MetalS346 gathered structures with equivalent metal coordination sites and surrounding chemical species within 5 Å from the metal‐ion. A significant result (total score < 2) was obtained for Q4MWP8, an uncharacterized protein from Bacillus cereus G9241. Unlike hemerythrins, in which the cation coordination internally crosslinks the four helices of the fold, the binuclear nonheme coordination site in Q4MWP8 is located at the interface of four discontinuous fragments of sequence from bromodomain‐like folds (Fig. 4). Bromodomains are putative protein–protein interaction domains47 with no apparent phylogenetic relationship to the hemerytrin‐like domain superfamily, and the local structural similarity around the binuclear iron‐binding site apparently constitutes a case of local structural convergence (Fig. 4).
Figure 4.
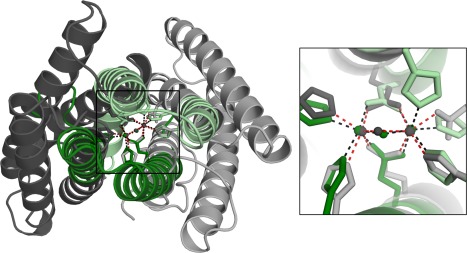
Local structural similarity between hemerythrin and Q4MWP8 from Bacillus cereus G9241. Ribbon diagram of two evolutionarily unrelated protein structures, Q4MWP8 from Bacillus cereus G9241 (shown in gray, PDB ID: 3DBY) and hemerythrin‐like domain of DcrH (shown in green, PDB ID: 2AWC), aligned with MetalS2. The inset shows the superposition of the metal‐binding sites.
Conclusions
Here we present a bioinformatic analysis of hemerythrin homologs, which compose a diverse multifunctional protein domain superfamily. We have identified at least three broad groups within the hemerythrin‐like superfamily by well‐defined sequence and structure similarity criteria. These are characterized by having a set of conserved residues at putative cation‐coordinating sites. Sequences in the hemerythrin‐like group exhibit symmetrical traits both in sequence and structure, suggesting a possible origin of hemerythrin through a duplication and fusion event involving a primordial two‐up‐and‐down helix motif containing a single H‐HxxxE cation‐coordination site (Fig. 5). This duplication resulted in an increase of the functional properties of the metal site, as the contemporary role of characterized hemerythrins relies on the presence of both iron‐binding sites. Moreover, binuclear non‐heme Fe enzymes essentially perform O2‐dependent reactions.13 This functional trait must have been incorporated to cellular metabolism as a response to free O2 conditions. Both the sequence cluster topology and the specialized function of signal‐transduction and oxygen‐carrier hemerythrins (H‐HxxxE‐HxxxH‐HxxxxD) as well as F‐box proteins (H‐HExxE‐H‐HxxxE) suggest a recent divergence of these families from the core cluster. Incorporation of hemerythrin domains into proteins by domain shuffling events and lateral gene transfer appears to be a recurrent trait during the complex evolutionary history of this fold superfamily.
Figure 5.
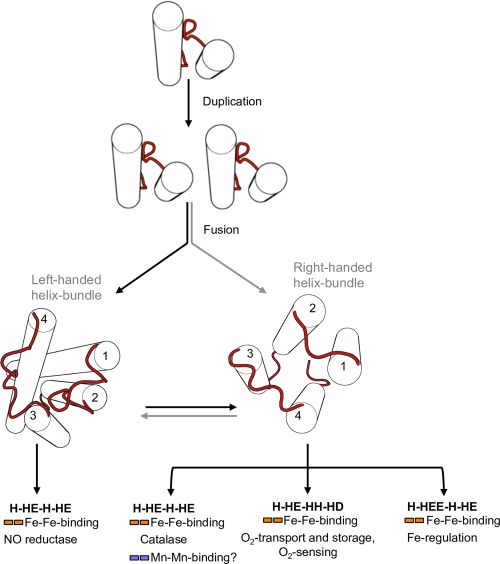
Fold change in the evolution of the hemerythrin‐like domain superfamily. Schematic representation of the evolution of hemerythrins depicting possible scenarios for the origination of different handedness of the hemerythrin fold.
Materials and Methods
Homology search
To identify evolutionarily distant members of the hemerythrin‐like domain superfamily, a database of profile HMMs, comprising Protein Data Bank (PDB) entries clustered down to a pairwise sequence identity of 70% (PDB70), was searched using HHpred with default parameters,48 using as query the sequence of bacteriohemerythrin (Uniprot ID: Q60AX2) from Methylococcus capsulatus. Protein sequences having an aligned region with HHpred probability higher than 90% were retrieved. We constructed multiple sequence alignments of these aligned regions with HHblits49 using default parameters. The multiple sequence alignments were then converted to Hidden Markov Models (HMMs) with HMMER3.50 We used hmmsearch50 to identify statistically significant matches (E value cutoff of 1e‐3 or lower) to the generated HMMs in the sequence database Uniprot.51
Sequence analysis
To delineate the domain composition of proteins gathered by hmmsearch, profile HMMs were built for every sequence following the same methodology as for the initial homology search, and were subsequently compared to the PfamA 30.0 profile HMM database52 using HHsearch. Only protein domains identified with a probability of 70% or higher were considered.
Cluster analysis
Sequence regions comprising only hemerythrin‐like domains were clustered in CLANS34 based on their BLAST P values. Clustering was performed to equilibrium in two‐dimensional space at a P value cutoff of 1e‐10, using default settings. Sequence clusters formed at a cutoff value of 1e‐13 were aligned with MAFFT.53
Finally, in order to detect internal symmetry in hemerythrin‐like proteins, sequence clusters formed at a P value cutoff of 1e‐18 were aligned, and the resulting multiple sequence alignments were analyzed with HHrepID,32 using default parameters. Noncluster forming sequences were analyzed individually. To corroborate the presence of internal symmetry at the structure level, the repeat fragments were superposed in hemerythrin‐like proteins of known structure with TMalign.54
Local structural analysis of the metal ion sites
Three‐dimensional models of hemerythrin homologs revealed a helix swap on one of the hemerythrin‐like domain families, which made it difficult to obtain a correct structural alignment with most algorithms (Supporting Information Table SI). To evaluate the structural similarity between cation‐coordinating sites of hemerythrin homologs we used MetaS2, which is a local structure alignment strategy that starts by superimposing the metal ions and the donor atoms of a pair of structures. The MetaS2 algorithm scores alignments considering sequence similarity, fractional coverage of the smallest site and fragmentation.33 Lower scores indicate more similar environments of a pair of metal‐binding sites.33 For comparison, we also included the structure of an uncharacterized protein (Q4MWP8 from Bacillus cereus G9241, PDB code: 3DBY) with a cation‐coordination site structurally similar to that of hemerythrin homologs (Fig. 1). The Metals2 score was used as a measure of the local structural pairwise superposition of the metal‐binding sites compared.33
Supporting information
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Figure S3
Supporting Information Figure S4
Supporting Information Table S1
Acknowledgemnts
Claudia Alvarez was a doctoral student in the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM). Part of this work was carried out at the Max Planck Institute for Developmental Biology, during a leave of absence of C. Alvarez with support from CONACYT. We are indebted to Ricardo Hernández for his help with the manuscript.
The hemerythrin‐like superfamily comprises a large number of protein families. We propose a classification of this superfamily into three broad groups on the basis of their cation‐coordinating residues. Furthermore, internal sequence and structural symmetry of hemerythrin‐like domains strongly suggests that the hemerythrin fold originated by duplication and fusion of an ancestral helix‐loop‐helix motif.
References
- 1. Terwilliger NB (1998) Functional adaptations of oxygen‐transport proteins. J Exp Biol 201:1085–1098. [DOI] [PubMed] [Google Scholar]
- 2. Rouault TA (2009) An ancient gauge for iron. Science 326:676–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salahudeen AA, Thompson JW, Ruiz JC, Ma H‐W, Kinch LN, Li Q, Grishin NV, Bruick RK (2009) An E3 ligase possessing an iron‐responsive hemerythrin domain is a regulator of iron homeostasis. Science 326:722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA (2009) Control of iron homeostasis by an iron‐regulated ubiquitin ligase. Science 326:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron‐binding E3 ligase mediates iron response in plants by targeting basic helix‐loop‐helix transcription factors. Plant Physiol 167:273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiong J, Kurtz DM, Ai J, Sanders‐Loehr J (2000) A hemerythrin‐like domain in a bacterial chemotaxis protein. Biochemistry 39:5117–5125. [DOI] [PubMed] [Google Scholar]
- 7. Karlsen OA, Ramsevik L, Bruseth LJ, Larsen Ø, Brenner A, Berven FS, Jensen HB, Lillehaug JR (2005) Characterization of a prokaryotic haemerythrin from the methanotrophic bacterium Methylococcus capsulatus (Bath). FEBS J 272:2428–2440. [DOI] [PubMed] [Google Scholar]
- 8. Kendall JJ, Barrero‐Tobon AM, Hendrixson DR, Kelly DJ (2014) Hemerythrins in the microaerophilic bacterium Campylobacter jejuni help protect key iron‐sulphur cluster enzymes from oxidative damage. Environ Microbiol 16:1105–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X, Tao J, Hu X, Chan J, Xiao J, Mi K (2014) A bacterial hemerythrin‐like protein MsmHr inhibits the SigF‐dependent hydrogen peroxide response in mycobacteria. Front Microbiol 5:800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zeng WB, Chen WB, Yan QP, Lin GF, Qin YX (2016) Hemerythrin is required for Aeromonas hydraphlia to survive in the macrophages of Anguilla japonica . Genet Mol Res 15. gmr.15028074. [DOI] [PubMed] [Google Scholar]
- 11. Solomon EI, Park K (2016) Structure/function correlations over binuclear non‐heme iron active sites. J Biol Inorg Chem 21:575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snyder RA, Betzu J, Butch SE, Reig AJ, DeGrado WF, Solomon EI (2015) Systematic perturbations of binuclear non‐heme iron sites: structure and dioxygen reactivity of de novo due ferri proteins. Biochemistry 54:4637–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nordlund P, Eklund H (1995) Di‐iron‐carboxylate proteins. Curr Opin Struct Biol 5:758–766. [DOI] [PubMed] [Google Scholar]
- 14. Alvarez‐Carreño C, Becerra A, Lazcano A (2016) Molecular evolution of the oxygen‐binding hemerythrin domain. PLoS One 11:e0157904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurtz DM (1999) Oxygen‐carrying proteins: three solutions to a common problem. Essays Biochem 34:85–100. [DOI] [PubMed] [Google Scholar]
- 16. Chen KH‐C, Chuankhayan P, Wu H‐H, Chen C‐J, Fukuda M, Yu SS‐F, Chan SI (2015) The bacteriohemerythrin from Methylococcus capsulatus (Bath): crystal structures reveal that Leu114 regulates a water tunnel. J Inorg Biochem 150:81–89. [DOI] [PubMed] [Google Scholar]
- 17. Hendrickson WA, Klippenstein GL, Ward KB (1975) Tertiary structure of myohemerythrin at low resolution. Proc Natl Acad Sci USA 72:2160–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber PC, Salemme FR (1980) Structural and functional diversity in 4‐alpha‐helical proteins. Nature 287:82–84. [DOI] [PubMed] [Google Scholar]
- 19. Presnell SR, Cohen FE (1989) Topological distribution of four‐alpha‐helix bundles. Proc Natl Acad Sci USA 86:6592–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stenkamp RE, Sieker LC, Jensen LH, McCallum JD, Sanders‐Loehr J (1985) Active site structures of deoxyhemerythrin and oxyhemerythrin. Proc Natl Acad Sci USA 82:713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheriff S, Hendrickson WA, Smith JL (1987) Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol 197:273–296. [DOI] [PubMed] [Google Scholar]
- 22. Isaza CE, Silaghi‐Dumitrescu R, Iyer RB, Kurtz DM, Chan MK (2006) Structural basis for O2 sensing by the hemerythrin‐like domain of a bacterial chemotaxis protein: substrate tunnel and fluxional N terminus. Biochemistry 45:9023–9031. [DOI] [PubMed] [Google Scholar]
- 23. Ruiz JC, Bruick RK (2014) F‐box and leucine‐rich repeat protein 5 (FBXL5): sensing intracellular iron and oxygen. J Inorg Biochem 133:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lo F‐C, Hsieh C‐C, Maestre‐Reyna M, Chen C‐Y, Ko T‐P, Horng Y‐C, Lai Y‐C, Chiang Y‐W, Chou C‐M, Chiang C‐H, Huang WN, Lin YH, Bohle DS, Liaw WF (2016) Crystal structure analysis of the repair of iron centers protein YtfE and its interaction with NO. Chemistry 22:9768–9776. [DOI] [PubMed] [Google Scholar]
- 25. Ma Z, Strickland KT, Cherne MD, Sehanobish E, Rohde KH, Self WT, Davidson VL (2017) The Rv2633c protein of Mycobacterium tuberculosis is a non‐heme di‐iron catalase with a possible role in defenses against oxidative stress. J Biol Chem jbc.RA117.000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X, Li J, Hu X, Huang L, Xiao J, Chan J, Mi K (2015) Differential roles of the hemerythrin‐like proteins of Mycobacterium smegmatis in hydrogen peroxide and erythromycin susceptibility. Sci Rep 5:16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Padmaja N, Rajaram H, Apte SK (2011) A novel hemerythrin DNase from the nitrogen‐fixing cyanobacterium Anabaena sp. strain PCC7120. Arch Biochem Biophys 505:171–177. [DOI] [PubMed] [Google Scholar]
- 28. Traverso ME, Subramanian P, Davydov R, Hoffman BM, Stemmler TL, Rosenzweig AC (2010) Identification of a hemerythrin‐like domain in a P1B‐type transport ATPase. Biochemistry 49:7060–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martín‐Durán JM, De Mendoza A, Sebé‐Pedrós A, Ruiz‐Trillo I, Hejnol A (2013) A broad genomic survey reveals multiple origins and frequent losses in the evolution of respiratory hemerythrins and hemocyanins. Genome Biol Evol 5:1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. French CE, Bell JML, Ward FB (2008) Diversity and distribution of hemerythrin‐like proteins in prokaryotes. FEMS Microbiol Lett 279:131–145. [DOI] [PubMed] [Google Scholar]
- 31. Bailly X, Vanin S, Chabasse C, Mizuguchi K, Vinogradov SN (2008) A phylogenomic profile of hemerythrins, the nonheme diiron binding respiratory proteins. BMC Evol Biol 8:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alva V, Nam S‐Z, Söding J, Lupas AN (2016) The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andreini C, Cavallaro G, Rosato A, Valasatava Y (2013) MetalS2: a tool for the structural alignment of minimal functional sites in metal‐binding proteins and nucleic acids. J Chem Inf Model 53:3064–3075. [DOI] [PubMed] [Google Scholar]
- 34. Frickey T, Lupas A (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704. [DOI] [PubMed] [Google Scholar]
- 35. Onoda A, Okamoto Y, Sugimoto H, Shiro Y, Hayashi T (2011) Crystal structure and spectroscopic studies of a stable mixed‐valent state of the hemerythrin‐like domain of a bacterial chemotaxis protein. Inorg Chem 50:4892–4899. [DOI] [PubMed] [Google Scholar]
- 36. Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El‐Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler‐Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador‐Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios I, Yeh L‐S, Young S‐Y, Mitchell AL (2017) InterPro in 2017‐beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Giessen TW, Silver PA (2017) Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat Microbiol 2:17029. [DOI] [PubMed] [Google Scholar]
- 38. Justino MC, Almeida CC, Gonçalves VL, Teixeira M, Saraiva LM (2006) Escherichia coli YtfE is a di‐iron protein with an important function in assembly of iron‐sulphur clusters. FEMS Microbiol Lett 257:278–284. [DOI] [PubMed] [Google Scholar]
- 39. Caulfield JL, Wishnok JS, Tannenbaum SR (1998) Nitric oxide‐induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J Biol Chem 273:12689–12695. [DOI] [PubMed] [Google Scholar]
- 40. Strube K, De Vries S, Cramm R (2007) Formation of a dinitrosyl iron complex by NorA, a nitric oxide‐binding di‐iron protein from Ralstonia eutropha H16. J Biol Chem 282:20292–20300. [DOI] [PubMed] [Google Scholar]
- 41. Zielazinski EL, González‐Guerrero M, Subramanian P, Stemmler TL, Argüello JM, Rosenzweig AC (2013) Sinorhizobium meliloti Nia is a P1B‐5‐ATPase expressed in the nodule during plant symbiosis and is involved in Ni and Fe transport. Metallomics 5:1614–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthiadis A, Long TA (2016) Further insight into BRUTUS domain composition and functionality. Plant Signal Behav 11:e1204508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simader H, Hothorn M, Köhler C, Basquin J, Simos G, Suck D (2006) Structural basis of yeast aminoacyl‐tRNA synthetase complex formation revealed by crystal structures of two binary sub‐complexes. Nucleic Acids Res 34:3968–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y‐J, Han X‐M, Ren L‐L, Yang H‐L, Zeng Q‐Y (2013) Functional divergence of the glutathione S‐transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol 161:773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lallement PA, Brouwer B, Keech O, Hecker A, Rouhier N (2014) The still mysterious roles of cysteine‐containing glutathione transferases in plants. Front Pharmacol 5:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valasatava Y, Rosato A, Cavallaro G, Andreini C (2014) MetalS3, a database‐mining tool for the identification of structurally similar metal sites. J Biol Inorg Chem 19:937–945. [DOI] [PubMed] [Google Scholar]
- 47. Fujisawa T, Filippakopoulos P (2017) Functions of bromodomain‐containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol 18:246–262. [DOI] [PubMed] [Google Scholar]
- 48. Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Remmert M, Biegert A, Hauser A, Söding J (2011) HHblits: lightning‐fast iterative protein sequence searching by HMM‐HMM alignment. Nat Methods 9:173–175. [DOI] [PubMed] [Google Scholar]
- 50. Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. The Uniprot Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y, Skolnick J (2005) TM‐align: a protein structure alignment algorithm based on the TM‐score. Nucleic Acids Res 33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Figure S3
Supporting Information Figure S4
Supporting Information Table S1


