Abstract
Bacillithiol is a low‐molecular weight thiol produced by many gram‐positive organisms, including Staphylococcus aureus and Bacillus anthracis. It is the major thiol responsible for maintaining redox homeostasis and cellular detoxification, including inactivation of the antibiotic fosfomycin. The metal‐dependent bacillithiol transferase BstA is likely involved in these sorts of detoxification processes, but the exact substrates and enzyme mechanism have not been identified. Here we report the 1.34 Å resolution X‐ray crystallographic structure of BstA from S. aureus. Our structure confirms that BstA belongs to the YfiT‐like metal‐dependent hydrolase superfamily. Like YfiT, our structure contains nickel within its active site, but our functional data suggest that BstA utilizes zinc for activity. Although BstA and YfiT both contain a core four helix bundle and coordinate their metal ions in the same fashion, significant differences between the protein structures are described here.
Keywords: bacillithiol, Staphylococcus aureus, transferase, X‐ray crystallography, low‐molecular‐weight thiol, Gram‐positive, detoxification
Short abstract
PDB Code(s): 5WK0
Introduction
Most organisms employ low‐molecular weight thiols (LMWTs) to serve as redox buffers and deactivate toxic compounds. Bacillithiol (Supporting Information Scheme S1) is the predominant LMWT in many Gram‐positive bacteria such as Staphylococcus aureus and Bacillus anthracis.1, 2, 3 It is the preferred thiol cosubstrate for FosB, a bacillithiol‐S‐transferase responsible for resistance to the antibiotic fosfomycin.4, 5 BstA is another bacillithiol‐S‐transferase from S. aureus that plays a role in detoxifying electrophilic xenobiotic compounds within the cell by producing bacillithiol‐S‐conjugated products, which are further processed and removed from the cell.6 BstA is active against the model electrophilic compound chloro‐2,4‐dinitrobenzene (CDNB), but the exact electrophilic substrate, or group of substrates, upon which BstA acts has not been identified. BstA belongs to the DinB/YfiT‐like superfamily of metal‐dependent enzymes.6, 7 Although the structure of the namesake bacillithiol transferase YfiT from B. subtilis has been determined,8 the study of bacillithiol transferases is still in its infancy, and additional structures are needed to characterize this group of enzymes. Here we present the 1.34 Å resolution crystal structure of BstA from S. aureus. The structure clearly demonstrates how the enzyme binds its metal cofactor and gives insight into the types of substrates BstA might bind. Furthermore, we demonstrate that BstA is likely zinc‐dependent.
Results and Discussion
Overall structure of BstA
The crystals of BstA used in this study contained one polypeptide comprising 156 residues in the asymmetric unit, including Ala 155 and Leu 156 of the C‐terminal hexahistidine tag. The Ramachandran statistics determined by the PDB Validation Report are excellent (Table 1). BstA adopts the four‐helix bundle fold that is typical of YfiT‐like metal‐dependent enzymes [Fig. 1(A)]. Examination of the unit cell contents in our study reveals a likely dimeric interface between a symmetry‐equivalent peptide involving helices 1 and 4 of each subunit [Fig. 1(B)]. This interface buries ∼1600 Å2 of surface area with a ΔG of −29 kcal/mol as calculated by PDBePISA.9 Gel filtration analysis also indicated BstA migrates as a dimer.
Table 1.
X‐ray Data Collection and Refinement Statistics
| Resolution Limits (Å) | 50.4–1.335 (1.340–1.335) |
| Unit cell dimensions (Å) | 66.0 × 42.6 × 50.4 |
| Space group | P21212 |
| Number of independent reflections | 32,630 (319) |
| Completeness (%) | 98.9 (96.1) |
| Redundancy | 7.7 (7.5) |
| Avg I/σ(I) | 13.3 (2.3) |
| R merge (%) | 7.9 (77.0) |
| CC1/2 | 1.00 (0.82) |
| R work (%) | 15.1 |
| R free (%) | 19.0 |
| No. protein atoms | 1278 |
| No. heteroatomsa | 127 |
| Average B values | |
| Protein atoms (Å2) | 22.8 |
| Solvent (Å2) | 39.7 |
| Weighted root‐mean‐square deviations from ideality | |
| Bond lengths (Å) | 0.006 |
| Bond angles (deg) | 0.79 |
| General planes (Å) | 0.005 |
| Ramachandran angles | |
| Favored (%) | 98.2 |
| Allowed (%) | 1.8 |
| Outliers (%) | 0 |
Including one nickel (II) ion and 126 water molecules.
Figure 1.
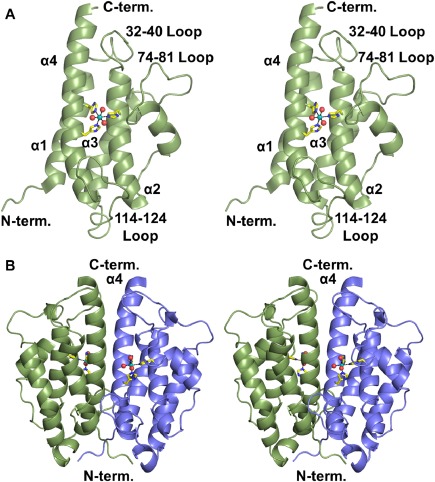
Overall structure of BstA. A: Stereo image of the BstA monomer. The dotted lines represent interactions with the bound metal. B: Stereo image of the BstA dimer colored by subunit.
Active site
The putative BstA active site faces the solvent adjacent to the dimeric interface. Helices 2 and 4 of the four helix bundle, along with three major loops (consisting of residues 32–40, 74–81, and 114–124) extend out from these helices and constitute two walls of the active site pocket. Helices 1 and 4 of the other subunit define the final wall. The structure contains a metal ion coordinated octahedrally by His 47, His 135, His 139, and three water molecules [Fig. 2(A)]. X‐ray fluorescence scans of crystals grown in the same crystallization drop as the crystal used in our structural study demonstrated the presence of zinc in very low abundance and nickel in high abundance [Fig. 2(B)]. As a result of this fluorescence scan, we have chosen to model nickel in the active site of the structure reported here. To identify which metal ion BstA uses preferentially, we conducted metal‐dependence assays. We dialyzed BstA in the presence of EDTA to remove bound metals, and then we removed the EDTA through another round of dialysis. We reacted BstA with bacillithiol, CDNB, and nickel or zinc and monitored the formation of the BSH‐CDNB adduct (Supporting Information Fig. S1). The reaction with zinc yielded a specific activity ten times greater than the reaction with nickel (55 ± 6 nmol min−1 mg−1 and 5.9 ± 0.7 nmol min−1 mg−1, respectively). Thus, we suspect BstA utilizes a zinc‐dependent mechanism, but its native zinc ion was exchanged for a nickel ion during purification via nickel‐affinity chromatography. Attempts to soak in or co‐crystallize with zinc have not yet met with success.
Figure 2.
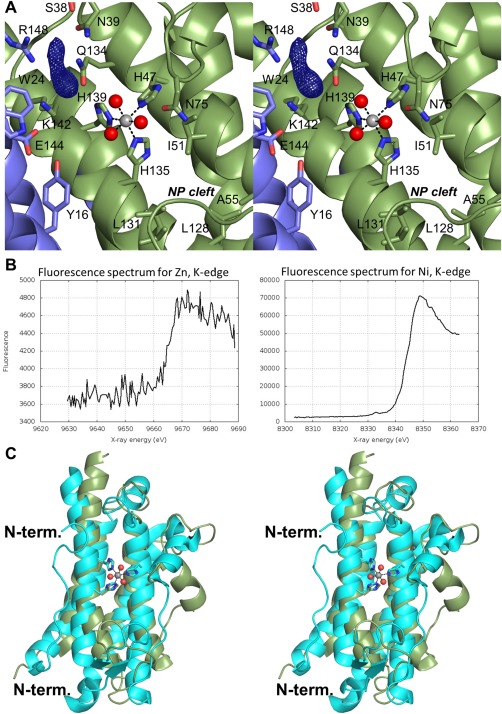
BstA active site and comparison with YfiT. A: Close‐up view of the putative BstA active site colored by subunit. The F o – F c electron density map is contoured at 2.5σ. The non‐polar (NP) cleft is labeled. B: X‐ray fluorescence scans for zinc (left) and nickel (right). C: Comparison of BstA (green) with YfiT from B. subtilis (cyan).
Several water molecules are present near the metal and are likely part of the substrate binding pocket. In addition, a peak greater than 8σ in the F o – F c map was observed within hydrogen bonding distance to Lys 142 and Arg 148 [Fig. 2(A)]. This peak constitutes part of a “tube” of density that is more substantial than a water molecule. It is possible that the density in question is buffer or another component of the crystallization solution, but we were unable to unambiguously identify it, and we therefore chose not to include it in the model. Nonetheless, the proximity of this unmodeled density to the metal ion indicates that it is present within the enzyme's active site. Many polar and positively‐charged residues, including Lys 142 and Arg 148, are in this region, suggesting this may be the pocket where the negatively‐charged bacillithiol binds. In contrast, the cleft on the other side of the metal ion consists of several non‐polar side chains (Ile 51, Ala 55, Leu 128, and Ile 131), which may be where an electrophilic substrate similar to CDNB binds. We attempted to soak in or co‐crystallize BstA with bacillithiol and/or CDNB, but neither was observed in electron density maps. Examination of the symmetry‐equivalent polypeptides within the unit cell demonstrates that the loop containing Asp 84 and Glu 85 interacts with or is very near the metal ion binding site, which may explain why our soaking and co‐crystallization attempts were not successful.
Structural comparison
The other related bacillithiol‐S‐transferase in the PDB is YfiT from B. subtilis (PDB ID 1RXQ).8 With 19% amino acid sequence identity, these two structures superimpose with a root‐mean square deviation of 3.3 Å for 123 structurally equivalent α‐carbons [Fig. 2(C)]. Both enzymes contain the core four‐helix bundle and three‐histidine metal binding site. However, YfiT contains a ∼20 residue N‐terminal extension and forms a dimer along helices 2 and 3 instead of 1 and 4 (Supporting Information Fig. S2). In addition, the loops that connect the helices differ significantly from BstA, and the putative active site of YfiT comprises residues from only one subunit. Furthermore, little conservation is evident between the active sites, but notable similarities include Arg 148 (BstA) and Arg 55 (YfiT) as well as Asn 75 (BstA) and Glu 95 (YfiT) (Supporting Information Fig. S3). Although both enzymes utilize bacillithiol, the lack of conservation suggests there may be significant differences in the manner these enzymes accommodate their substrates.
Understanding bacillithiol metabolism and other physiological mechanisms that confer resistance to antibiotics will aid in the development of new therapies. Ligand‐bound structures of bacillithiol transferases from several Gram‐positive organisms are needed to understand the role of this family of enzymes in cellular detoxification processes in these organisms. The structure of BstA described here provides insight into the structure and function of these enzymes.
Experimental Procedures
BstA from S. aureus (Strain TCH1516) was expressed recombinantly in E. coli and purified via nickel affinity chromatography. Crystals of BstA with a C‐terminal hexahistidine tag were grown in Hampton Research GRID Screen MPD condition A2. The crystals were flash frozen and X‐ray data were collected at LS‐CAT at Argonne National labs. The structure of BstA was solved via molecular replacement using a similar four‐helix bundle protein (PDB entry 3CEX, unpublished) as a search model. Data collection and refinement statistics are shown in Table 1. Additional experimental details can be found in the Supporting Information.
Supporting information
Supporting Information
Acknowledgement
Authors thank Dr. Joseph Brunzelle for collecting X‐ray fluorescence data.
X‐ray coordinates were deposited in the PDB (Accession number: 5WK0).
References
- 1. Helmann JD (2011) Bacillithiol, a new player in bacterial redox homeostasis. Antioxid Redox Signal 15:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC (2009) Bacillithiol is an antixodant thiol produced in Bacilli. Nat Chem Biol 5:625–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chandrangsu P, Loi VV, Antelmann H, Helmann JD (2018) The role of bacillithiol in gram‐positive Firmicutes . Antioxid Redox Signal 28:445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamers AP, Keithly ME, Kim K, Cook PD, Stec DF, Hines KM, Sulikowski GA, Armstrong RN (2012) Synthesis of bacillithiol and the catalytic selectivity of FosB‐type fosfomycin resistance proteins. Org Lett 14:5207–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thompson MK, Keithly ME, Harp J, Cook PD, Jagessar KL, Sulikowski GA, Armstrong RN (2013) Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus . Biochemistry 52:7350–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perera VR, Newton GL, Parnell JM, Komives EA, Pogliano K (2014) Purification and characterization of the Staphylococcus aureus bacillithiol transferase BstA. Biochim Biophys Acta 1840:2851–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Newton GL, Leung SS, Wakabayashi JI, Rawat M, Fahey RC (2011) The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S‐transferases. Biochemistry 50:10751–10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajan SS, Yang X, Shuvalova L, Collart F, Anderson WF (2004) YfiT from Bacillus subtilis is a probable metal‐dependent hydrolase with an unusual four‐helix bundle topology. Biochemistry 43:15472–15479. [DOI] [PubMed] [Google Scholar]
- 9. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


