Summary
Sugarcane is the world's most efficient feedstock for commercial production of bioethanol due to its superior biomass production and accumulation of sucrose in stems. Integrating first‐ and second‐generation ethanol conversion processes will enhance the biofuel yield per unit area by utilizing both sucrose and cell wall‐bound sugars for fermentation. RNAi suppression of the lignin biosynthetic gene caffeic acid O‐methyltransferase (COMT) has been demonstrated to improve bioethanol production from lignocellulosic biomass. Genome editing has been used in a number of crops for creation of loss of function phenotypes but is very challenging in sugarcane due to its highly polyploid genome. In this study, a conserved region of COMT was targeted with a single‐transcription activator‐like effector nuclease (TALEN) pair for multi‐allelic mutagenesis to modify lignin biosynthesis in sugarcane. Field‐grown TALEN‐mediated COMT mutants showed up to 19.7% lignin reduction and significantly decreased syringyl to guaiacyl (S/G) ratio resulting in an up to 43.8% improved saccharification efficiency. Biomass production of COMT mutant lines with superior saccharification efficiency did not differ significantly from the original cultivar under replicated field conditions. Sanger sequencing of cloned COMT amplicons (1351–1657 bp) revealed co‐editing of 107 of the 109 unique COMT copies/alleles in vegetative progeny of line CB6 using a single TALEN pair. Line CB6 combined altered cell wall composition and drastically improved saccharification efficiency with good agronomic performance. These findings confirm the feasibility of co‐mutagenesis of a very large number of target alleles/copies for improvement in crops with complex genomes.
Keywords: TALEN, field performance, genome editing, sugarcane, COMT, lignin, biofuel
Introduction
Sugarcane is considered a prime feedstock for sustainable biofuel production due to its high biomass yield and ratooning ability (Byrt et al., 2011). The first‐generation biofuel conversion process utilizes sucrose, extracted from sugarcane stems with roller mills, for bioethanol production by a direct fermentation process. This process uses the abundant lignocellulosic sugarcane biomass residue, also called bagasse, for generation of electricity (Tew and Cobill, 2008). More value can be captured from this abundant resource by second‐generation conversion processes, which use pretreatment of biomass and enzymatic hydrolysis for the saccharification of the cell wall‐bound cellulose and hemicellulose (Naik et al., 2010; Vermerris et al., 2007). Thus, the integration of first‐ and second‐generation conversion technology should have environmental and economic benefits (Joelsson et al., 2016; Somerville et al., 2010) and is currently explored for commercial production of sugarcane‐derived bioethanol in Brazil (Dias et al., 2014).
Lignocellulosic sugarcane biomass consists of cellulose, hemicellulose, and lignin (Karp and Shield, 2008). Lignin is a recalcitrance factor for biofuel production from lignocellulosic biomass. Lignin prevents the accessibility of cellulose microfibrils by cellulase enzyme as well as adsorbs hydrolytic enzymes; thereby, it inhibits the release of cell wall‐bound sugars (Chen and Dixon, 2007; Weng et al., 2008). The lignin polymer consists of p‐hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monomers (Li et al., 2008).
Conversion of 5‐hydroxyconiferyl alcohol to sinapyl alcohol, which is a component of the S subunit, is mediated by caffeic acid O‐methyl transferase (COMT) via the phenylpropanoid pathway (Hisano et al., 2009; Humphreys and Chapple, 2002). RNAi suppression or a knockout mutation of COMT resulted in reduction in both total lignin content and S/G ratio. Altered lignin content and composition following COMT suppression/mutagenesis were demonstrated in biofuel and/or forage crops like sugarcane (Jung and Altpeter, 2016; Jung et al., 2012, 2013), switchgrass (Baxter et al., 2014; Fu et al., 2011; Samuel et al., 2014), corn (Piquemal et al., 2002), sorghum (Saballos et al., 2008; Sattler et al., 2012), tall fescue (Chen et al., 2004), and alfalfa (Chen and Dixon, 2007; Guo et al., 2001).
Although lignin reduction by targeted genome editing may reduce costs associated with regulatory approval (Wolt et al., 2016), the polyploid nature of sugarcane genome creates challenges for this approach. Modern sugarcane cultivars have a genome size of approximately 10 Gb, large number (100–130) of chromosomes and ~12 homo(eo)logs at each locus resulting in a high level of genetic redundancy (Le Cunff et al., 2008; Piperidis et al., 2010; de Setta et al., 2014). The recent assembly of the draft sugarcane genome (Riaño‐Pachón and Mattiello, 2017) will accelerate targeted crop improvement. However, the complex architecture of its polyploid genome will remain a challenge for these efforts (Okura et al., 2016; Souza et al., 2011).
Transcription activator‐like effector nuclease (TALEN) is a genome‐editing tool enabling precise genome modifications, such as targeted mutagenesis, gene replacement, or insertion (Gurushidze et al., 2014; Li et al., 2012; Zhang et al., 2013). Targeted mutagenesis with TALEN has been successful in number of crops for creation of loss of function genotypes (reviewed in Baltes and Voytas, 2015; Weeks et al., 2016). Recently, TALEN‐mediated targeted mutagenesis in a highly polyploid sugarcane was reported (Jung and Altpeter, 2016), including generation of events with integration and expression of TALEN, evidence of targeted mutagenesis by capillary electrophoresis and sequence analysis of 89–148 nt PCR amplicons encompassing the TALEN target site and phenotypes, which displayed reduced lignin and altered lignin and cell wall composition under glasshouse conditions. Here, we report for the first time evaluation of TALEN‐mediated COMT mutant sugarcane lines in replicated field plots for agronomic performance, lignin content, lignin, and cell wall composition as well as saccharification efficiency. We also carried out Sanger sequencing of cloned 1351‐ to 1657‐bp‐long PCR amplicons. This allows now to report the number of co‐mutated COMT targets, resulting in improved saccharification efficiency in field‐grown sugarcane plants without compromising agronomic performance.
Results
Generation of sugarcane with integration of COMT‐TALEN was reported by Jung and Altpeter (2016) and is briefly summarized in the Data S1.
Number of COMT copies/alleles, mutation frequency, and types
Sequencing of 1351‐ to 1657‐bp‐long PCR amplicons of genomic DNA from COMT mutant line CB6 was used to determine number of COMT copies/alleles in the sugarcane genome. A total of 440 COMT amplicon containing plasmid colonies were sequenced by the Sanger method, and chromatograms were visually inspected to select 389 high‐quality Sanger read pairs with overlapping ends. Of these 389 high‐quality reads, 109 unique reads representing different COMT copies/alleles were identified (Table 1). Of the 109 unique reads, two reads displayed WT sequence in the targeted mutation site, while 107 reads displayed nucleotide insertions or deletions (InDels) in the TALEN target site, which indicates a mutation frequency of 98%. Five different exon variants, outside of the targeted mutation site, and 104 different intron variants were identified (Table S1). Identical target site mutations were confirmed in many different COMT copies/alleles. Different COMT copies/alleles were associated with intron sequence variations like InDels resulting in variations of intron length from 865 nucleotides to 1171 nucleotides, as well as sequence variations (single‐nucleotide polymorphisms, SNPs) in introns of the same size. Sequence reads representing different target site mutations within the same COMT copy/allele were also identified. A total of six unique reads, representing 5.6% of all unique reads, displayed more than one mutation type (up to 3; Table S1). Four of them had 7‐ and 48‐bp deletions, one of them had 4‐, 36‐, and 48‐bp deletions, and one of them displayed 3‐ and 10‐bp deletions.
Table 1.
Number of COMT variants in sugarcane line CB6
| Genotype | Number of quality reads | Number of quality reads with targeted mutation | Number of unique reads with WT sequence | Total number of unique reads with variation in exon and intron outside of targeted mutation site |
|---|---|---|---|---|
| COMT mutant | 389 | 387 | 2 | 109 |
Growth performance of TALEN‐mediated COMT mutant lines in the replicated field plots
The events evaluated in replicated field plots for biomass yield and agronomic traits included eight TALEN‐mediated COMT mutant lines including CB3‐8 derived from biolistic gene transfer and direct embryogenesis and CA4 and CA17 derived from Agrobacterium mediated gene transfer and indirect embryogenesis. In addition, control plants including the original cultivar (wild type; WT), callus‐derived control harboring the nptII gene (TC1), a callus‐derived non‐transgenic control (TC2), a direct embryogenesis‐derived transgenic control with no mutation (TC3), and a line with COMT suppression by RNAi (B401) were evaluated in these field plots (Table 2 and Figure 1). The plant height of COMT mutant lines varied from 174.4 cm (CB3) to 195.3 cm (CB5). However, these variations were not significantly different from WT (181.8 cm). Number of tillers produced by COMT mutant lines was non‐significantly higher than WT (8.2) except CA4 and CA17, which displayed significantly more tillers. Stalk diameter varied from 19.3 mm (TC1 and TC2) to 22.7 mm (CB5). Among the COMT mutant lines, CA17 had significantly thinner stalks (15.9 mm) than WT (21.4 mm). Dry biomass yield of COMT mutant lines, transgenic, and tissue culture controls was not significantly different from WT (36.0 t/ha) with the exception of line CA17 and control TC2, which displayed a reduced biomass yield. Both, CA17 and TC2, were regenerated from callus while all CB lines as well as control TC3 were derived from direct embryogenesis. There was a trend to elevated biomass production in several of the COMT mutants with CB5 producing 43.7 t/ha, followed by CB7 with 40.4 t/ha and CB8 with 36.5 t/ha. Lines with a trend to elevated biomass displayed a reduction in soluble solids/Brix units by 17%–19% compared to WT with no significant difference in juice volume. Line CB6 resembled WT with no significant difference in any of the phenotypic data.
Table 2.
Phenotypic performance of COMT mutants and controls under field conditions
| Line | Plant height (cm) | No. of tillers per plant | Stalk diameter (mm) | Biomass yield (t/ha) | Juice volume (mL/100 g of fresh stalks) | Soluble solids (° Brix) |
|---|---|---|---|---|---|---|
| WT | 181.8 | 8.2 | 21.4 | 36.0 | 37.1 | 21.0 |
| TC1 | 153.0* | 11.9* | 19.3 | 29.7 | 40.6 | 20.2 |
| TC2 | 177.0 | 9.2 | 19.3 | 23.6* | 40.1 | 20.5 |
| TC3 | 181.2 | 8.2 | 22.2 | 32.0 | 39.3 | 21.1 |
| CB3 | 174.4 | 9.2 | 19.5 | 32.0 | 32.0 | 18.7 |
| CB4 | 182.3 | 8.8 | 21.3 | 35.3 | 34.0 | 20.0 |
| CB5 | 195.3 | 9.2 | 22.7 | 43.7 | 32.6 | 17.5* |
| CB6 | 186.8 | 9.0 | 20.0 | 35.4 | 29.7 | 20.4 |
| CB7 | 190.8 | 8.8 | 21.4 | 40.4 | 31.1 | 17.4* |
| CB8 | 189.7 | 9.1 | 21.5 | 36.5 | 32.7 | 17.0* |
| CA4 | 186.3 | 10.4* | 19.5 | 31.7 | 30.4 | 16.6* |
| CA17 | 191.9 | 10.8* | 15.9* | 24.0* | 30.5 | 16.4* |
| B401 | 173.8 | 11.8* | 21.7 | 36.8 | 42.9 | 21.3 |
WT, wild‐type sugarcane; TC1, callus‐derived control harboring the nptII gene; TC2, callus‐derived non‐transgenic control; TC3, direct embryogenesis‐derived transgenic control with no mutation; CB3‐CB8, transgenic lines derived from biolistic transformation; CA4‐CA17, transgenic lines derived from Agrobacterium‐mediated transformation; B401, COMT RNAi line.
Values with asterisk in the same column indicate significant difference compared to WT (n = 3, P < 0.05) as determined by t‐test.
Figure 1.

(a) Field trial of TALEN‐mediated COMT mutants. (b) Field performance of COMT mutant lines and wild type (WT). CB6 and CB7 are mutant lines derived through direct embryogenesis and biolistic transformation. (c) Stems of field‐grown TALEN‐mediated COMT mutant (CB6) in comparison with wild type (WT), immediately after juice extraction with roller mills.
COMT expression in COMT RNAi and TALEN‐mediated COMT mutant lines
Quantitative real‐time RT‐PCR analysis was performed to quantify COMT gene expression in COMT mutant or RNAi‐suppressed plants compared to WT plants (Figure S1). COMT expression in RNAi line was 41.8% for B401, which corresponds to 58% COMT gene suppression (Figure S1). The COMT expression in TALEN‐mediated COMT mutant lines varied from 26.7% (CA4) to 91.4% (CB3) of WT transcripts.
Mutation frequency and lignin modification due to TALEN‐mediated COMT mutation
Mutation frequencies of field‐grown COMT mutant lines were determined by capillary electrophoresis (CE) analysis (Table 3). Mutation frequency ranged from 51.4% (CB4) to 92.5% (CB6). Replicated field samples of mutant lines displayed uniform CE mutation pattern despite variations in the relative fluorescence (Figure 2). WT plants displayed no mutation and its estimated lignin content was 241.8 mg/g of DW. The amount of lignin reduction with respect to WT varied from 10.9% reduction in CB5 to 19.7% in CB7. Lignin reduction did not correlate with mutation frequencies of field‐grown plants. COMT mutant lines with high biomass production were used for the estimation of lignin monomers such as syringyl (S) and guaiacyl (G). The S/G ratio of WT was 0.91, whereas CB5, CB6, and CB7 displayed reduced S/G ratios of 0.80, 0.63, and 0.71, respectively (Table 3).
Table 3.
Mutation frequency determination of TALEN‐mediated lignin reduction lines under field conditions
| Line | Mutation frequency (%)a | AcBr Lignin content (mg/g DW)b | Lignin reduction (%) | S/G molar ratioc |
|---|---|---|---|---|
| WT | 0.0 | 241.8 ± 1.3 | – | 0.91 |
| TC3 | 0.0 | 241.2 ± 4.2 | 0.2 | 0.89 |
| CB3 | 70.0 | 200.6 ± 2.6* | 17.0 | n.a. |
| CB4 | 51.4 | 209.4 ± 5.3* | 13.4 | n.a. |
| CB5 | 69.9 | 215.5 ± 4.5* | 10.9 | 0.80* |
| CB6 | 92.5 | 194.6 ± 6.1* | 19.5 | 0.63* |
| CB7 | 68.1 | 194.1 ± 2.0* | 19.7 | 0.71* |
| CB8 | 71.7 | 197.7 ± 5.9* | 18.2 | n.a. |
| CA4 | 80.0 | 198.9 ± 5.8* | 17.7 | n.a. |
| CA17 | 89.3 | 195.9 ± 4.3* | 19.0 | 0.47* |
| B401 | n.a. | 219.0 ± 6.3* | 9.4 | n.a. |
WT, wild‐type sugarcane; TC3, direct embryogenesis‐derived transgenic control with no mutation; CB3‐CB8, transgenic lines derived from biolistic transformation; CA4‐CA17, transgenic lines derived from Agrobacterium‐mediated transformation; B401, COMT RNAi line; DW, dry weight; n.a., not analyzed.
Values with asterisk in the same column indicate significant difference compared to WT (n = 3, P < 0.05) as determined by t‐test.
Mutation frequency estimated by relative fluorescent quantitation based on capillary electrophoresis electropherogram. Mutation frequency (%) = (Sum of peak height of all mutant peaks over sum of peak height of all peaks including wild‐type peak) × 100.
Acetyl Bromide (AcBr) method was used to determine total lignin content.
Ratio of monolignol compositions of S (syringyl) and G (guaiacyl) subunits and expressed as mg/g DW.
Figure 2.
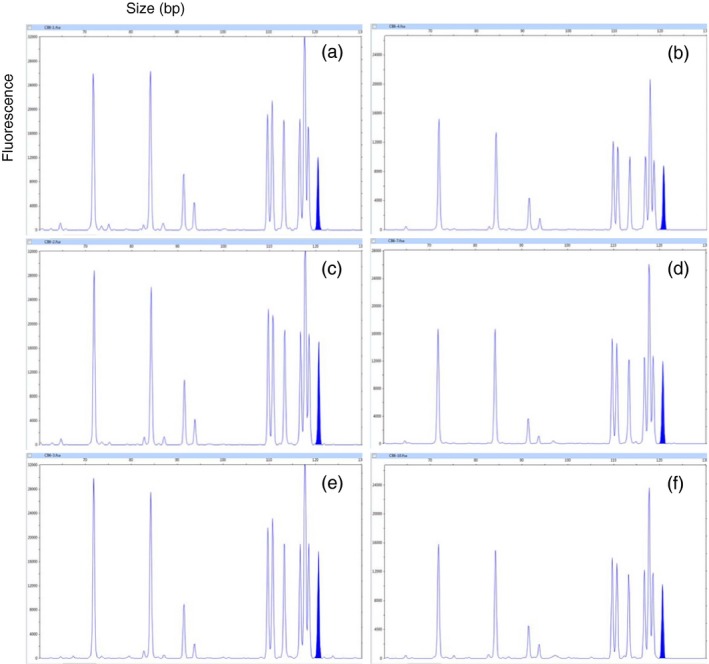
Representative electropherogram for different tillers and different plants of COMT mutant line CB6. (a,b) Two different tillers of the same plant in replication 1. (c,d) Two different tillers of the same plant in replication 2. (e,f) Two different tillers of the same plant in replication 3. Peak representing unmodified COMT is highlighted in blue.
Effect of TALEN‐mediated mutation on cell wall carbohydrates
The composition of cell wall carbohydrates was evaluated in COMT mutants, RNAi‐suppressed COMT line, and WT sugarcane plants (Table 4). The amount of glucose in the plant cell wall of COMT mutant or RNAi‐suppressed COMT sugarcane did not differ from that of WT. TALEN‐mediated COMT mutant lines CA17 and CB6 displayed a significant increase in both xylose and arabinose contents as compared to WT by 3.4%–7.6% and 19.2%–22.5%, respectively (Table 4).
Table 4.
Cell wall carbohydrates in COMT suppressed or mutant sugarcane lines
| Line | Cell wall carbohydrates (mg/g DW) | Total sugar | ||
|---|---|---|---|---|
| Glucose | Xylose | Arabinose | ||
| WT | 437.3 ± 3.0 | 193.2 ± 1.1 | 24.5 ± 0.5 | 655.0 |
| TC3 | 433.0 ± 6.9 | 189.1 ± 11.3 | 26.0 ± 0.7 | 648.1 |
| B401 | 427.9 ± 7.7 | 194.0 ± 0.3 | 29.5 ± 0.1* | 651.4 |
| CA17 | 434.2 ± 5.9 | 199.8 ± 2.4* | 30.0 ± 0.9* | 664.0 |
| CB5 | 437.6 ± 6.8 | 195.8 ± 3.6 | 22.7 ± 1.7 * | 656.1 |
| CB6 | 434.7 ± 6.1 | 208.5 ± 3.4* | 29.2 ± 0.5* | 672.4 |
| CB7 | 435.2 ± 0.2 | 185.4 ± 1.7* | 27.3 ± 0.6* | 647.9 |
WT, wild‐type sugarcane; TC3, direct embryogenesis‐derived transgenic control with no mutation; CB5, CB6, and CB7, transgenic lines derived from biolistic transformation; CA17, transgenic lines derived from Agrobacterium‐mediated transformation; B401, COMT RNAi line; DW, dry weight.
Values with asterisk in the same column indicate significant difference compared to WT (n = 3, P < 0.05) as determined by t‐test.
Improvement in saccharification efficiency by COMT mutation
Glucose yield from lignocellulosic biomass of COMT mutant or RNAi‐suppressed COMT lines was determined by enzymatic hydrolysis with dilute acid pretreatment. COMT mutant sugarcane displayed significantly improved glucose yield from lignocellulosic biomass by up to 44% compared to WT. The saccharification efficiencies were 39%, 50%, 57%, and 54% for WT, B401, CB5, and CB6, respectively (Figure 3).
Figure 3.
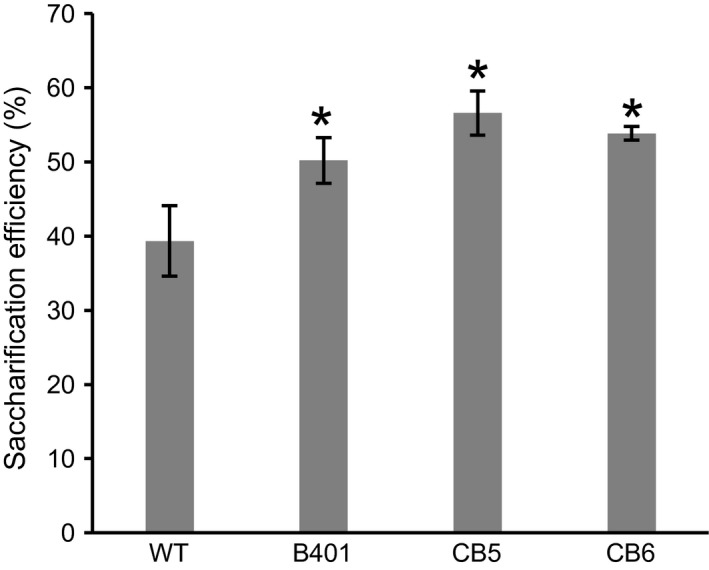
Saccharification efficiency of lignocellulosic biomass into directly fermentable glucose. Wild‐type sugarcane (WT); sugarcane with RNAi suppression of COMT (B401), TALEN‐mediated COMT mutant sugarcane lines (CB5, CB6). Error bars indicate standard error (n = 2). Asterisk above the bars indicates significant difference from WT at P < 0.05 in t‐test.
Effect of lignin modification on resistance to disease and insects and lodging
No significant differences were observed between COMT mutated/suppressed lines and WT for orange rust or red lesion on midrib (Table S2). qRT‐PCR analysis indicated the absence of sugarcane yellow virus (SCYLV) infection in COMT mutant or WT plants (Table S2). Lodging was absent from all entries at the time of harvest (Figure 1a), and therefore, data are not shown.
Discussion
Designer nucleases like TALEN, zinc finger nucleases, meganucleases, CRISPR/Cas, or Cpf are key to enabling site‐directed genome modifications by targeting double‐stranded DNA breaks in genes of interest. Refinement of these genome‐editing tools have enabled unprecedented precision in genetic modifications, revolutionizing functional genomics and crop improvement (Altpeter et al., 2016; Hilscher et al., 2017; Voytas and Gao, 2014). Data from field experiments revealing the agronomic performance of genome‐edited crops are just emerging (Shi et al., 2017). Here, we present the first report of agronomic and/or conversion performance of a crop, which underwent genome editing with TALEN. Sugarcane is highly polyploid (x = 10–13), interspecific hybrid with chromosome number 2n = 100–130 and the genome size of ~10 Gb (Parthasarathy, 1948; Brandes and Artschwager, 1958; Piperidis et al., 2010; de Setta et al., 2014). This highly redundant genomic context requires extensive co‐editing of a large number of targets for creation of a ‘loss of function’ phenotype. Our most remarkable finding is that the extensive co‐editing of more than 100 copies/alleles of the lignin biosynthetic gene caffeic acid O‐methyltransferase (COMT) did not compromise agronomic performance under field conditions. However, the targeted mutagenesis of the vast majority of the more than 100 COMT copies/alleles improved saccharification of cell wall‐bound sugars by 39%–44% for biofuel production. This was accompanied by significantly reduced lignin content and altered lignin monomer ratio.
COMT catalyzes O‐methylation of 5‐hydroxyconiferaldehyde and 5‐hydroxyconiferyl alcohol, diverting metabolic flux to formation of the syringyl (S) lignin monomer (Callazo et al., 1992; Vignols et al., 1995; Bout and Vermerris, 2003). Altered lignin composition creates brown vascular tissue in the stem and leaves. Brown midrib mutants (bmr) were among the earliest described and characterized mutants in maize (Jorgenson, 1931; Kuc and Nelson, 1964; Vignols et al., 1995). In diploid crops like maize and sorghum, bmr mutants arise naturally or following chemical mutagenesis (Jorgenson, 1931; Porter et al., 1978). However, the genetic redundancy of the highly polyploid sugarcane genome prevents the generation of natural or chemically induced bmr mutants in this crop. RNAi targeted toward a highly conserved catalytic domain in the first exon of COMT reduced S/G monomer ratio and lignin content and resulted in improved saccharification efficiency in sugarcane (Jung et al., 2012, 2013). This result informed a strategy for TALEN‐mediated targeted mutagenesis of this highly conserved catalytic COMT domain (Jung and Altpeter, 2016). Based on short 454 amplicons of the COMT target region and capillary electrophoresis, an almost complete knockout of the target COMT alleles was reported in sugarcane along with reduction in lignin and S/G lignin monomer ratio (Jung and Altpeter, 2016). However, the total number of the mutated COMT copies could not be assessed with short amplicon sequencing, as the genome of the parental sugarcane cultivar CP88‐1762 has not been fully sequenced and due to the high conservation of the targeted first COMT exon. Here, we confirm that only four SNPs exist in the entire first exon. To reveal different COMT copies/alleles and account for the number of targeted COMT modifications, we included the first COMT intron as indicator of sequence variation in different alleles/copies in the sequence analysis. Therefore, 1351‐ to 1657‐bp‐long, cloned COMT amplicons spanning exon 1 encompassing the mutation target site, intron 1 and the 5′ region of exon 2, were sequenced using the Sanger method (Figures S2 and S3). The sequence alignment of 389 paired quality reads from cloned COMT amplicons of bmr line CB6 with good agronomic performance revealed 109 unique COMT copies/alleles with five exon variants outside the targeted mutation site and 104 intron variants (Tables 1 and S1). In sharp contrast, the haploid genome of sorghum (diploid species) a close relative of sugarcane contains only seven copies of the COMT gene. The drastically lower redundancy of COMT in sorghum allowed successful chemical mutagenesis for generation of bmr phenotypes in this crop (Bout and Vermerris, 2003; Porter et al., 1978).
A single TALEN pair targeted to the highly conserved catalytic domain in the first exon of COMT was able to co‐mutate 107 of the 109 identified COMT copies/alleles in sugarcane bmr line CB6 without compromising its agronomic performance. To our knowledge, the maximum number of genomic sites previously reported to be simultaneously edited has been 62 (Yang et al., 2015) using two CRISPR‐Cas9 gRNAs targeted to the highly conserved catalytic domain of porcine endogenous retrovirus polymerase genes (PERVs) in PK15 cells of pigs. However, the successful application of this cellular approach to animals is still pending. TALEN has been widely used for the improvement in agronomic traits in a variety of diploid and polyploid crops (Baltes and Voytas, 2015; Hilscher et al., 2017; Weeks et al., 2016). However, so far typically only a small number of co‐mutated target genes were reported in plants. For instance, in potato, Clasen et al. (2016) mutated four alleles/copies of the vacuolar invertase gene (VInv) using TALEN. Both FAD2‐1A and FAD2‐1B genes were co‐mutated by TALEN in soya bean (Haun et al., 2014). Wang et al. (2014) co‐mutated three copies of the TaMLO gene in hexaploid bread wheat with a single TALEN pair targeted to a conserved region in exon 2. Multiplexing of up to eight gRNAs in CRISPR/Cas9‐mediated targeted co‐mutagenesis of seven genes of the OsFTL gene family in rice (Ma et al., 2015).
The effective spacer length for cleavage by a TALEN pair is 12–21 nucleotides (Miller et al., 2011) or 13–16 bp for TALEN pairs with C‐terminal truncation (Christian et al., 2012). Both repeated cleavage by constitutively expressed TALENs and incomplete editing before the first division of the embryogenic cell that received the genome‐editing tool may result in a cellular mosaic pattern or chimerism (Jung and Altpeter, 2016; Zhang et al., 2014). Typical for such mosaic pattern is the observation that many of the same copies/alleles in different plant cells or vegetative progenies have different mutation frequencies or different mutation types. Six unique COMT reads from bmr mutant CB6 displayed more than one mutation type (up to 3), which represents only 5.6% of all unique reads that contain mutations (Table S1). These mutation type variants most likely arose due to progression of deletions until cleavage was terminated by insufficient spacer length. However, 98% of all sequence reads in bmr mutant CB6 were co‐mutated (Table 1). Capillary electrophoresis (CE) in multiple replications and tillers displayed identical peak patterns suggesting that the vast majority of the targeted COMT mutations have occurred before the first division of the embryogenic cell that received the genome‐editing tool. Mutation frequencies observed in the replicated field samples corresponded well to glasshouse‐grown COMT mutant lines reported earlier (Jung and Altpeter, 2016). Brown coloration was observed immediately after crushing or cutting of stems of all COMT mutants with significantly reduced lignin content and in contrast to tissue culture and transgenic controls and WT. Plants from the entire field experiment were crushed with a roller mill and all the crushed stems from COMT mutants with significantly reduced lignin content were instantly brown, suggesting the absence of chimerism in the mutated events and stability of the trait in the vegetative progenies (Figure 1c). Earlier, brown coloration in freshly cut stem tissues of transgenic sugarcane or switchgrass with RNAi suppression of COMT and lignin reduction was also reported (Fu et al., 2011; Jung et al., 2012).
Targeting the TALEN to the highly conserved catalytic domain of COMT may have contributed to a loss of function phenotype even in the case of silent mutations by deleting one or several essential amino acids for catalytic activity. This may explain the absence of a correlation between COMT expression level and observed cell wall modifications in COMT mutants. CB7 displayed a similar (19.7%) lignin reduction as CB6 (19.5%) as well as a similar reduction in S/G lignin monomer ratio. However, both lines displayed a different mutation frequency (68% vs 93%) according to CE analysis. This suggests that a large proportion of the COMT copies/alleles are either non‐functional, are expressed at a low level or differ in the mutation type. The abundant allelic COMT variation in sugarcane can now be further explored for targeted mutation of specific copies/alleles similar to the CRISPR/Cas9 approach reported for the 4CL gene family in Populus (Zhou et al., 2015).
Identification of off‐target mutations is a daunting task in sugarcane, even with the recent assembly of the draft sugarcane genome (Riaño‐Pachón and Mattiello, 2017). TALEN appears to be superior to CRISPR/Cas9 in regard to on‐target mutagenesis (Shan et al., 2015). In the absence of genomewide analysis of off‐target mutagenesis, agronomic performance is the best indicator for unintended mutations that may arise by genome editing or somaclonal variation. All COMT mutant lines and transgenic controls, which underwent direct embryogenesis and biolistic gene transfer (CB3–CB8; TC3), resembled the original sugarcane cultivar (WT) in biomass yield and biomass related traits (Table 2). However, COMT mutant line CA17, which was derived from Agrobacterium‐mediated gene transfer and the tissue culture control TC2 which like CA17 regenerated plants following the longer tissue culture process of indirect embryogenesis, displayed significantly reduced biomass yields. This also suggests that minimizing somaclonal variation by reducing the time in tissue culture (Taparia et al., 2012) and/or by using biolistic instead of Agrobacterium‐mediated gene transfer (Joyce et al., 2014; Wu et al., 2015) may contribute to better agronomic performance. Callus‐derived sugarcane lines with RNAi suppression of COMT also displayed a moderate decrease in biomass yield compared to the original cultivar along with reduced stalk length and stalk diameter (Jung et al., 2013). Transgenic switchgrass with RNAi suppression of COMT and lignin reduction did not display any negative impact on biomass production or diseases susceptibility (Baxter et al., 2014).
COMT mutant sugarcane lines and controls did not display any lodging prior harvest (Figure 1a) and disease scores did not differ significantly from control plants in this study (Table S2). For a final conclusion on disease response, large‐scale and multisite field testing is required. Alternative targets beside COMT have been explored for suppression of the lignin biosynthetic pathway, and most of them compromise plant growth or impair the plant defense system (Bonawitz and Chapple, 2013; Chen and Dixon, 2007; Eudes et al., 2014). However, examples of lignin‐modified plants with increased resistance to pathogens due to accumulation of lignin precursors and other phenolic compounds were also reported (McKeehen et al., 1999; Quentin et al., 2009; Sattler and Funnell‐Harris, 2013).
Lignin is a major recalcitrance factor for saccharification of cell walls (Chen and Dixon, 2007; Li et al., 2008). Field‐grown COMT mutant lines with reduced lignin and S/G ratio displayed 39%–44% improvement in saccharification efficiency over the original sugarcane cultivar (Figure 3). This moderately exceeds the reported improvements in saccharification efficiency following RNAi suppression of COMT in switchgrass (17%–22%, Fu et al., 2011; 9%–34%, Baxter et al., 2014) and sugarcane (19%–32%, Jung et al., 2013). Reduction in total lignin content and S/G ratio observed in this experiment were consistent with earlier reports from RNAi suppression of COMT in switchgrass (Baxter et al., 2014; Fu et al., 2011; Samuel et al., 2014), sugarcane (Jung et al., 2012, 2013), maize (Piquemal et al., 2002), tall fescue (Chen et al., 2004), alfalfa (Chen and Dixon, 2007), poplar (Studer et al., 2011), and bmr mutant sorghum (Dien et al., 2009). RNAi in contrast to genome editing depends on continued expression of transgenes. In contrast, targeted mutagenesis by sequence‐specific nuclease is heritable over generations in the absence of transgene expression (Char et al., 2015; Gao et al., 2010; Wang et al., 2014).
Sugarcane is vegetatively propagated for commercial production to maintain the original genotype while avoiding the extensive segregation of the highly polyploid genome. Therefore, removing the genome‐editing tool by Mendelian segregation of mutant and transgene alleles as described for seeded crops (Xu et al., 2015) would compromise the agronomic performance of sugarcane. Alternatively, non‐transgenic approaches to genome editing that have recently been demonstrated in other model and crop systems and are currently explored for generation of genome‐edited sugarcane without transgene footprint. These include transient delivery of DNA, or delivery as non‐integrative molecules like RNA, protein, or ribonucleoprotein complex (Luo et al., 2015; Svitashev et al., 2016; Zhang et al., 2016).
Conclusion
A single TALEN pair designed to target the highly conserved domain of COMT was able to mutate 98% of the more than 100 COMT copies/alleles as revealed by Sanger sequencing of cloned, long PCR amplicons. Field‐grown COMT mutants displayed improved saccharification efficiency of up to 44% along with reduced lignin content and S/G lignin monomer ratio with no significant difference in biomass production and agronomic performance compared to the original sugarcane cultivar (WT). These findings demonstrate the great potential of genome editing for crop improvement and will contribute to more efficient and sustainable biofuel production from sugarcane.
Experimental procedures
Generation of TALEN‐mediated COMT mutant lines from sugarcane is described in Jung and Altpeter (2016) and is briefly summarized in the Data S1.
Number of COMT copies/alleles in sugarcane genome
Line CB 6 was chosen for this analysis since it combined a high mutation frequency based on capillary electrophoresis and 454 sequencing (Jung and Altpeter, 2016) with excellent conversion and agronomic performance. Genomic DNA was isolated from leaf tissues of field‐grown bmr mutant line (CB6) using CTAB method (Murray and Thompson, 1980). Isolated DNA was quantified and 100 ng DNA was used as a template for PCR. A region of COMT, spanning exon 1 through exon 2 (~1600 bp), which encompasses mutation target site and intron (Figure S3), was amplified by Q5 High‐Fidelity DNA Polymerase (NEB, Ipswich, MA) under the following conditions: 98 °C for 30 s, 34 cycles of amplification at 98 °C for 10 s, 72 °C for 10 s, and 72 °C for 10 s, and final extension at 72 °C for 2 min using 4F and BK_RO7 primers (Table S3). PCR amplicons were separated in a 1.5% agarose gel electrophoresis and targeted gel fragment excised under UV light after ethidium bromide staining. Amplicons were purified from gel fragments using QIAquick kit (Qiagen) and ligated into pGEM®‐T easy vector using T4 DNA ligase (Promega, Madison, WI). The ligated plasmid containing insert was electroporated into competent E. coli cells (NEB) and grown at 37 °C for 16 h. Transformed colonies were harvested after blue‐white select screening, and DNA was prepared using GeneJET miniprep kit (Thermo Fisher Scientific Inc., Waltham, MA). Sanger sequencing was performed on both strands using M13F and M13R primers at the Eurofins Genomics (Huntsville, AL). Sequence chromatograms were visually checked for quality, and sequences were trimmed manually. Quality reads with overlapping ends following both M13F and M13R sequencing were aligned with the multiple sequence alignment tool CLUSTALW (Table S1).
Phenotypic evaluation of COMT mutants in replicated field plots
For field testing, mature nodes of glasshouse‐grown COMT mutant lines (eight lines) and control plants (transgenic controls, tissue culture control, RNAi control, and WT) were planted in 3‐L pots containing Fafard No. 2 mix (Sun Gro Horticulture, FL) irrigated once a day and fertilized biweekly with Miracle‐Gro Lawn Food (Scotts Miracle‐Gro, Marysville, OH). Three vegetative tillers were randomly selected for each of the eight TALEN‐mutated lines. Each of the three vegetative progeny was used for planting a separate replication of the field plots. Five plants were planted for each of the three replications per line. Therefore, 15 uniformly grown plants were selected and transplanted for each of the eight TALEN lines and for each of the controls to the field plots on Mar 25, 2015 at the University of Florida, Plant Science Research and Education Unit (PSREU), Citra, Florida, USA, under USDA/APHIS permit 13‐299‐101r‐a1. The field plots were laid out in a randomized complete block design (RCBD) of single row plots per accession with three replications in loamy sand soil. Each single row plot represents five plants per replication of mutant line or control plants, and each replication was surrounded by one row of the original cultivar (WT) as border plants. The spacing between rows and plants within each row was 120 and 60 cm, respectively. Weeds were removed by a minirototiller between rows and manually within rows during plant establishment. Established sugarcane plots were monitored for disease symptoms and insect damage (Table S2). Symptoms of sugarcane orange rust and elongated red lesion on midrib (red rot) were visually inspected and severity of symptoms scored using a rating scale of 0–4 as described in Glynn et al. (2013). 0 represents no disease; and 4 represents completely diseased plants. Sugarcane yellow leaf virus infection was also determined by the quantitative real‐time PCR analysis (Table S2) as described below using YLV‐F and YLV‐R primers (Table S3) and normalized against glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as reference gene. For the control of insects such as mealybugs, scales, and aphids, Bifenthrin (Brigade® 2EC), Imidacloprid (Admire® Pro) or Sulfoxaflor (Transform™) were applied at the labeled rate. For the control of orange rust, Pyraclostrobin (Headline®) was applied at the labeled rate. Plots were fertilized with 34 kg/ha N, 11 kg/ha P, and 34 kg/ha K at planting and 1 month after planting and irrigated daily with a rate of 10 mm for 15 days following transplanting. During the grand growth period, plots were fertilized twice in intervals of 6 weeks with 85 kg/ha N, 20 kg/ha P, and 85 kg/ha K. The established plots were irrigated up to three times a week depending on rainfall to provide at least 30 mm of irrigation per week.
One month before harvest COMT mutant lines and control plants were evaluated for plant height (length from base of the plant to shoot apical meristem), number of tillers per plant, and stalk diameter (middle of the stalk). Three plants per row were measured for each agronomic trait and means derived for statistical analysis. Plots were harvested on October 26, 2015 for determination of biomass, juice volume, and Brix (soluble solids). For biomass weight determination, all plants per row were harvested and the aboveground fresh biomass weight measured. To determine dry weight of the biomass, a subsample was taken from each row consisting of two mature stalks. These were chopped into small pieces, the fresh weight was determined and the biomass was dried at 60 °C for 4 weeks until constant weight was reached and the dry weight was determined. Biomass dry weight of mutant lines and controls was calculated by multiplication of the fresh biomass weight with the ratio of each subsample dry weight/fresh weight.
Quantitative real‐time RT‐PCR analysis
Top visible dewlap leaf was collected for analysis of the eight COMT mutant lines, one RNAi control and WT from each of the three replicated plots. Total RNA was isolated using TRIzol reagent (Invitrogen, Grand Island, NY) and treated with RNase‐Free RQ1 DNase (Promega, San Luis Obispo, CA) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of RNA using iScript cDNA synthesis kit (Bio‐Rad, Hercules, CA). The sugarcane glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) primers were used to amplify a GAPDH gene fragment as a reference for normalization of transcripts as described by Iskandar et al. (2004). COMT target site was amplified using COMT_EF1 and COMT_ER1 primers (Table S3). Quantitative real‐time PCR of the transcripts was performed in the CFX Connect Real‐Time PCR (Bio‐Rad, Hercules, CA) with SsoAdvanced SYBR Green Supermix (Bio‐Rad) under the following conditions: 95 °C for 3 min denaturation, 40 cycles at 95 °C for 10 s, and 58 °C for 45 s. Amplification specificity was verified by melt curve analysis from 55 to 95 °C. COMT expression levels in mutant and RNAi plants relative to WT were calculated using the method (Livak and Schmittgen, 2001).
Capillary electrophoresis
Genomic DNA was extracted from 150 mg leaf tissues of field‐grown COMT mutant lines and WT using CTAB method (Murray and Thompson, 1980). After DNA quantification, 100 ng of DNA was used as a template for PCR. PCR fragment encompassing the TALEN target site was amplified using 4F and 6‐FAM dye‐labeled 128R primers (Table S3). PCR was performed with Q5 High‐Fidelity DNA Polymerase (NEB, Ipswich, MA) under the following conditions: 98 °C for 30 s, 34 cycles of amplification at 98 °C for 10 s, 72 °C for 10 s, and 72 °C for 10 s, and final extension at 72 °C for 2 min. Capillary electrophoresis of the PCR amplicon was performed by GENEWIZ (South Plainfield, NJ) using Applied Biosystems 3730xl Genetic Analyzer (Life Technologies, Grand Island, NY). Electropherogram was analyzed using the Peak Scanner Software v2.0 (Life Technologies). A peak at 125 bp was considered as WT COMT, and peaks other than 125 bp were analyzed as mutant COMTs. Mutation frequency in the COMT amplicon population was estimated by quantifying relative fluorescence of the electropherogram peaks. Mutation frequency is the ratio of sum of peak height of all mutant peaks to sum of peak height of all mutant peaks plus WT peak and expressed as percentage.
Lignin and cell wall carbohydrates
Field‐grown matured sugarcane stalks were harvested from each plot and crushed with a roller mill to extract juice. The bagasse of each accession was dried at 45 °C until constant weight and ground using a Wiley mill (Thomas Scientific) with 2.0‐mm sieve. Two grams of ground sample was washed three times with 50% ethanol and one time with distilled water while incubated at 45 °C for 1 h during each washing step. The washed samples were dried at 45 °C for 1 day and passed through a 0.42‐mm sieve. The sample was further dried at 45 °C until constant weight. Total lignin contents were determined using the modified acetyl bromide method as described by Jung et al. (2012). The lignin content was calculated using sugarcane vascular bundle lignin molar extinction coefficient of 21.5 L/g/cm as reported in He and Terashima (1990).
Lignin compositions were determined using a pyrolysis molecular beam mass spectrometer. Each sample was prepared in triplicate by weighing about 1.0–2.5 mg into a stainless metal cup and pyrolyzed at 500 °C to produce volatile compounds. The volatile compounds were analyzed using a molecular beam mass spectrometer (Extrel Pittsburgh, PA). NIST 8492 (National Institute of Standards and Technology) lignin content 26.2% was used as a lignin standard (Jung et al., 2016).
Cell wall carbohydrates were analyzed using the National Renewable Energy Laboratory (NREL) protocol by Sluiter et al. (2008). Liberated monomeric sugars were identified and quantified with an Agilent/HP 1200 HPLC equipped with an RI detector (Agilent Technologies, Santa Clara, CA). The HPLC analysis was carried out using a Hi‐Plex H column (Agilent Technologies), operating at a flow rate of 0.6 mL/min using 5 mm H2SO4 as a mobile phase.
Enzymatic hydrolysis
Dilute acid pretreatment and enzymatic hydrolysis were performed as previously described (Jung et al., 2013). The enzymatic hydrolysis was performed for 72 h. Glucose from enzymatic hydrolysis was measured by an Agilent 1200 series modular HPLC with RI detector (Agilent Technologies). The HPLC analysis was carried out using a HPX‐87H column (Bio‐Rad), operating at a flow rate of 0.6 mL/min using 5 mm H2SO4 as a mobile phase. Saccharification efficiency was calculated as the ratio of glucose released following the enzymatic hydrolysis to the amount of glucose present in the cell wall before the hydrolysis.
Statistical analysis
Gene expression, total lignin content, cell wall carbohydrate content, saccharification efficiency, agronomic trait components and biomass yield data were statistically analyzed. T‐tests were performed using SAS™ version 9.3 (SAS Institute Inc., Cary, CA) to determine whether the means were significantly different between mutant lines and control plants (n = 3, P < 0.05).
Supporting information
Figure S1 Quantitative RT‐PCR analysis of COMT expression in field grown, TALEN mediated COMT mutants and control plants
Figure S2 (a) DNA sequences of TALEN binding and target sites in the first exon of the sugarcane COMT. P1‐P2 and P1‐P3 are primer binding sites for amplification of TALEN target site or long PCR amplicons for COMT copy/allele identification, respectively. (b) Sequence confirmation of TALEN mediated COMT mutation in PCR amplicons. Deletions ranging from 2 to 48 bp in one of the mutants (CB6) considered as different mutation types.
Figure S3 Schematic representation of the region of COMT that was PCR amplified, cloned and sequenced for identification of mutations in different COMT copies/alleles by the Sanger method.
Table S1 Number of copies/alleles of COMT and types of TALEN mediated target mutation in sugarcane.
Table S2 Disease symptoms and insect damage on COMT mutant, RNAi/suppressed sugarcane, WT and transgenic control plants.
Table S3 A list of primer pairs used in this study.
Data S1 Supplementary Experimental Procedures and Results.
Acknowledgements
The information, data, or work presented herein was funded in part by Syngenta. This research was also supported in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF) Funded by the Ministry of Education (grant number: 2015R1D1A1A01061365). The authors would like to thank Dr. Hardev Sandhu (Everglades Research and Educational Center, UF‐IFAS, Belle Glade, FL) for providing tops of sugarcane cultivar CP 88‐1762 and Sun Gro Horticulture, Apopka, FL, for donation of the Fafard #2 potting mix. In accordance with University of Florida policies and procedures, we report that corresponding author Fredy Altpeter is a consultant to a company that may be affected by the research reported in the enclosed paper. Those interests are fully disclosed to University of Florida, and an approved plan for managing any potential conflicts arising from this involvement is in place.
References
- Altpeter, F. , Springer, N.M. , Bartley, L.E. , Blechl, A. , Brutnell, T.P. , Citovsky, V. , Conrad, L. et al (2016) Advancing crop transformation in the era of genome editing. Plant Cell, 28, 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artschwager, E. and Brandes, E.W. (1958) Sugarcane (Saccharum officinarum L.): Origin, classification, characteristics and descriptions of representative clones. USDA Handbook No. 122. Washington, D.C. [Google Scholar]
- Baltes, N.J. and Voytas, D.F. (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33, 120–131. [DOI] [PubMed] [Google Scholar]
- Baxter, H.L. , Mazare, I.M. , Labbe, N. , Kline, L.M. , Cheng, Q. , Windham, M.T. , Mann, D.G.J. et al (2014) Two‐year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol. J. 12, 914–924. [DOI] [PubMed] [Google Scholar]
- Bonawitz, N.D. and Chapple, C. (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24, 336–343. [DOI] [PubMed] [Google Scholar]
- Bout, S. and Vermerris, W. (2003) A candidate‐gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O‐methyltransferase. Mol. Genet. Genomics, 269, 205–214. [DOI] [PubMed] [Google Scholar]
- Byrt, C.S. , Grof, C.P.L. and Furbank, R.T. (2011) C4 Plants as biofuel feedstocks, optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 53, 120–135. [DOI] [PubMed] [Google Scholar]
- Char, S.N. , Unger‐Wallace, E. , Frame, B. , Briggs, S.A. , Main, M. , Spalding, M.H. , Vollbrecht, E. et al (2015) Heritable site‐specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 13, 1002–1010. [DOI] [PubMed] [Google Scholar]
- Chen, F. and Dixon, R.A. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Auh, C.K. , Dowling, P. , Bell, J. , Lehmann, D. and Wang, Z.Y. (2004) Transgenic down‐regulation of caffeic acid O‐methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea). Funct. Plant Biol. 31, 235–245. [DOI] [PubMed] [Google Scholar]
- Christian, M.L. , Demores, T.Z.L. , Starker, C.G. , Osborn, M.J. , Nyquist, M.D. , Zhang, Y. , Carlson, D.F. et al (2012) Targeting G with TAL effectors, a comparison of activities of TALENs constructed with NN and NK repeat variable di‐residues. PLoS ONE, 7, e45383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen, B.M. , Stoddard, T.J. , Luo, S. , Demorest, Z.L. , Li, J. , Cedrone, F. , Tibebu, R. et al (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo, P. , Montoliu, L. , Puigdomènech, P. and Rigau, J. (1992) Structure and expression of the lignin O‐methyltransferase gene from Zea mays L. Plant Mol. Biol. 20, 857–867. [DOI] [PubMed] [Google Scholar]
- Dias, M.O.S. , Cavalett, O. , Filhob, R.M. and Bonomi, A. (2014) Integrated first and second generation ethanol production from sugarcane. Chem. Eng. Trans. 37, 445–450. [Google Scholar]
- Dien, B.S. , Sarath, G. , Pedersen, J.F. , Sattler, S.E. , Chen, H. , Funnell‐Harris, D.L. , Nichols, N.N. et al (2009) Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenergy Res. 2, 153–164. [Google Scholar]
- Eudes, A. , Liang, Y. , Mitra, P. and Loqué, D. (2014) Lignin bioengineering. Curr. Opin. Biotechnol. 26, 189–198. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Mielenz, J.R. , Xiao, X. , Ge, Y. , Hamilton, C.Y. , Rodriguez, M. , Chen, F. et al (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl Acad. Sci. USA, 108, 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Smith, J. , Yang, M. , Jones, S. , Djukanovic, V. , Nicholson, M.G. , West, A. et al (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187. [DOI] [PubMed] [Google Scholar]
- Glynn, N.C. , Laborde, C. , Davidson, R.W. , Irey, M.S. , Glaz, B. , D'Hont, A. and Comstock, J.C. (2013) Utilization of a major brown rust resistance gene in sugarcane breeding. Mol. Breed. 31, 323–331. [Google Scholar]
- Guo, D. , Chen, F. , Wheeler, J. , Winder, J. , Selman, S. , Peterson, M. and Dixon, R.A. (2001) Improvement of in‐rumen digestibility of alfalfa forage by genetic manipulation of lignin O‐methyltransferases. Transgenic Res. 10, 457–464. [DOI] [PubMed] [Google Scholar]
- Gurushidze, M. , Hensel, G. , Hiekel, S. , Schedel, S. , Valkov, V. and Kumlehn, J. (2014) True‐breeding targeted gene knock‐out in barley using designer TALE‐nuclease in haploid cells. PLoS ONE, 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun, W. , Coffman, A. , Clasen, B.M. , Demorest, Z.L. , Lowy, A. , Ray, E. , Retterath, A. et al (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. [DOI] [PubMed] [Google Scholar]
- He, L. and Terashima, N. (1990) Formation and structure of lignin in monocotyledons. III. heterogeneity of sugarcane (Saccharum officinarum l.) lignin with respect to the composition of structural units in different morphological regions. J. Wood Chem. Technol. 10, 435–459. [Google Scholar]
- Hilscher, J. , Burstmayr, H. and Stoger, E. (2017) Targeted modification of plant genomes for precision crop breeding. Biotechnol. J. 12, 1–14. [DOI] [PubMed] [Google Scholar]
- Hisano, H. , Nandakumar, R. and Wang, Z.Y. (2009) Genetic modification of lignin biosynthesis for improved biofuel production. In Vitro Cell. Dev. Biol. 45, 306–313. [Google Scholar]
- Humphreys, J.M. and Chapple, C. (2002) Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229. [DOI] [PubMed] [Google Scholar]
- Iskandar, H.M. , Simpson, R.S. , Casu, R.E. , Bonnett, G.D. , Maclean, D.J. and Manners, J.M. (2004) Comparison of reference genes for quantitative real‐time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol. Biol. Rep. 22, 325–337. [Google Scholar]
- Joelsson, E. , Erdei, B. , Galbe, M. and Wallberg, O. (2016) Techno‐economic evaluation of integrated first‐ and second‐generation ethanol production from grain and straw. Biotechnol. Biofuels, 9, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson, L.R. (1931) Brown midrib in maize and its linkage relations. J. Am. Soc. Agron. 23, 549–557. [Google Scholar]
- Joyce, P. , Hermann, S. , O'Connell, A. , Dinh, Q. , Shumbe, L. and Lakshmanan, P. (2014) Field performance of transgenic sugarcane produced using Agrobacterium and biolistics methods. Plant Biotechnol. J. 12, 411–424. [DOI] [PubMed] [Google Scholar]
- Jung, J.H. and Altpeter, F. (2016) TALEN mediated targeted mutagenesis of the caffeic acid O‐methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 92, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J.H. , Fouad, W.M. , Vermerris, W. , Gallo, M. and Altpeter, F. (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 10, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Jung, J.H. , Vermerris, W. , Gallo, M. , Fedenko, J.R. , Erickson, J.E. and Altpeter, F. (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field‐grown sugarcane. Plant Biotechnol. J. 11, 709–716. [DOI] [PubMed] [Google Scholar]
- Jung, J.H. , Kannan, B. , Dermawan, H. , Moxley, G.W. and Altpeter, F. (2016) Precision breeding for RNAi suppression of a major 4‐coumarate:coenzyme A ligase gene improves cell wall saccharification from field grown sugarcane. Plant Mol. Biol. 92, 505–517. [DOI] [PubMed] [Google Scholar]
- Karp, A. and Shield, I. (2008) Bioenergy from plants and the sustainable yield challenge. New Phytol. 179, 15–32. [DOI] [PubMed] [Google Scholar]
- Kuc, J. and Nelson, O.E. (1964) The abnormal lignins produced by the brown‐midrib mutants of maize. Arch. Biochem. Biophys. 105, 103–113. [DOI] [PubMed] [Google Scholar]
- Le Cunff, L. , Garsmeur, O. , Raboin, L.M. , Pauquet, J. , Telismart, H. , Selvi, A. , Grivet, L. et al (2008) Diploid/polyploid syntenic shuttle mapping and haplotype‐specific chromosome walking toward a rust resistance gene (Bru1) in highly polyploid sugarcane (2n–12x~115). Genetics, 180, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Weng, J.K. and Chapple, C. (2008) Improvement of biomass through lignin modification. Plant J. 54, 569–581. [DOI] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, S. , Li, J. , Stoddard, T.J. , Baltes, N.J. , Demorest, Z.L. , Clasen, B.M. , Coffman, A. et al (2015) Non‐transgenic plant genome editing using purified sequence‐specific nucleases. Mol. Plant, 8, 1425–1427. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- McKeehen, J.D. , Busch, R.H. and Fulcher, R.G. (1999) Evaluation of wheat (Triticum aestivum L.) phenolic acids during grain development and their contribution to Fusarium resistance. J. Agric. Food Chem. 47, 1476–1482. [DOI] [PubMed] [Google Scholar]
- Miller, J.C. , Tan, S. , Qiao, G. , Barlow, K.A. , Wang, J. , Xia, D.F. , Meng, X. et al (2011) A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, S.N. , Goud, V.V. , Rout, P.K. and Dalai, A.K. (2010) Production of first and second generation biofuels, A comprehensive review. Renew. Sustain. Energy Rev. 14, 578–597. [Google Scholar]
- Okura, V.K. , de Souza, R.S.C. , de Siqueira Tada, S.F. and Arruda, P. (2016) BAC‐pool sequencing and assembly of 19 Mb of the complex sugarcane genome. Front. Plant Sci. 7, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy, N. (1948) Origin of noble sugar‐canes (Saccharum officinarum L.). Nature, 161, 608. [DOI] [PubMed] [Google Scholar]
- Piperidis, G. , Piperidis, N. and D'Hont, A. (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genomics, 284, 65–73. [DOI] [PubMed] [Google Scholar]
- Piquemal, J. , Chamayou, S. , Nadaud, I. , Beckert, M. , Barrie, Y. , Mila, I. , Lapierre, C. et al (2002) Down‐regulation of caffeic acid O‐methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 130, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, K.S. , Axtell, J.D. , Lechtenberg, V.L. and Colenbrander, V.F. (1978) Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci. 18, 205–208. [Google Scholar]
- Quentin, M. , Allasia, V. , Pegard, A. , Allais, F. , Ducrot, P.H. , Favery, B. , Levis, C. et al (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño‐Pachón, D.M. and Mattiello, L. (2017) Draft genome sequencing of the sugarcane hybrid SP80‐3280. F1000Res., 6, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos, A. , Vermerris, W. , Rivera, L. and Ejeta, G. (2008) Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenergy Res. 1, 193–204. [Google Scholar]
- Samuel, R. , Pu, Y. , Jiang, N. , Fu, C. , Wang, Z.‐Y. and Ragauskas, A. (2014) Structural characterization of lignin in wild‐type versus COMT down‐regulated switchgrass. Front. Energy Res. 1, 1–9. [Google Scholar]
- Sattler, S.E. and Funnell‐Harris, D.L. (2013) Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front. Plant Sci. 4, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, S.E. , Palmer, N.A. , Saballos, A. , Greene, A.M. , Xin, Z. , Sarath, G. , Vermerris, W. et al (2012) Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the Caffeic O‐Methyltranferase (COMT) of sorghum. Bioenergy Res. 5, 855–865. [Google Scholar]
- de Setta, N. , Monteiro‐Vitorello, C. , Metcalfe, C. , Cruz, G.M. , Del Bem, L. , Vicentini, R. , Nogueira, F.T. et al (2014) Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genom. 15, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Zhang, Y. , Chen, K. , Zhang, K. and Gao, C. (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Gao, H. , Wang, H. , Lafitte, H.R. , Archibald, R.L. , Yang, M. , Hakimi, S.M. et al (2017) ARGOS8 variants generated by CRISPR‐Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. https://doi.org/10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter, A. , Hames, B. , Ruiz, R.O. , Scarlata, C. , Sluiter, J. , Templeton, D. and Crocker, D. (2008) Determination of structural carbohydrates and lignin in biomass. NREL LAP TP‐510‐42618.
- Somerville, C. , Youngs, H. , Taylor, C. , Davis, S.C. and Long, S.P. (2010) Feedstocks for lignocellulosic biofuels. Science, 329, 790–792. [DOI] [PubMed] [Google Scholar]
- Souza, G.M. , Berges, H. , Bocs, S. , Casu, R. , D'Hont, A. , Ferreira, J.E. , Henry, R. et al (2011) The sugarcane genome challenge, strategies for sequencing a highly complex genome. Trop. Plant Biol. 4, 145–156. [Google Scholar]
- Studer, M.H. , Demartini, J.D. , Davis, M.F. , Sykes, R.W. , Davison, B. and Keller, M. (2011) Lignin content in natural Populus variants affects sugar release. Proc. Natl Acad. Sci. USA, 108, 6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev, S. , Schwartz, C. , Lenderts, B. , Young, J.K. and Mark Cigan, A. (2016) Genome editing in maize directed by CRISPR‐Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparia, Y. , Gallo, M. and Altpeter, F. (2012) Comparison of direct and indirect embryogenesis protocols, biolistic gene transfer and selection parameters for efficient genetic transformation of sugarcane. Plant Cell Tissue Organ Cult. 111, 131–141. [Google Scholar]
- Tew, T.L. and Cobill, R.M. (2008) Genetic improvement of sugarcane (Saccharum spp.) as an energy crop In Genetic Improvement of Bioenergy Crops (Vermerris W., ed.), pp. 273–294. Springer New York. [Google Scholar]
- Vermerris, W. , Saballos, A. , Ejeta, G. , Mosier, N.S. , Ladisch, M.R. and Carpita, N.C. (2007) Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci. 47, S142–S153. [Google Scholar]
- Vignols, F. , Rigau, J. , Torres, M.A. , Capellades, M. and Puigdomenech, P. (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O‐Methyltransferase. Plant Cell, 7, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D.F. and Gao, C. (2014) Precision genome engineering and agriculture, opportunities and regulatory challenges. PLoS Biol. 12, e1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.‐L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Weeks, D.P. , Spalding, M.H. and Yang, B. (2016) Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 14, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J.K. , Li, X. , Bonawitz, N.D. and Chapple, C. (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172. [DOI] [PubMed] [Google Scholar]
- Wolt, J.D. , Wang, K. and Yang, B. (2016) The regulatory status of genome‐edited crops. Plant Biotechnol. J. 14, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H. , Awan, F.S. , Vilarinho, A. , Zeng, Q. , Kannan, B. , Phipps, T. , McCuiston, J. et al (2015) Transgene integration complexity and expression stability following biolistic or Agrobacterium‐mediated transformation of sugarcane. In Vitro Cell. Dev. Biol. Plant, 51, 603–611. [Google Scholar]
- Xu, R.‐F. , Li, H. , Qin, R.‐Y. , Li, J. , Qiu, C.‐H. , Yang, Y.‐C. , Ma, H. et al (2015) Generation of inheritable and “transgene clean” targeted genome‐modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5, 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Guell, M. , Niu, D. , George, H. , Lesha, E. , Grishin, D. , Aach, J. et al (2015) Genome‐wide inactivation of porcine endogenous retroviruses (PERVs). Science, 350, 1101–1104. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, F. , Li, X. , Baller, J.A. , Qi, Y. , Starker, C.G. , Bogdanove, A.J. et al (2013) Transcription activator‐like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.‐L. et al (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Jacobs, T.B. , Xue, L.J. , Harding, S.A. and Tsai, C.J. (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4‐coumarate:CoA ligase specificity and redundancy. New Phytol. 208, 298–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Quantitative RT‐PCR analysis of COMT expression in field grown, TALEN mediated COMT mutants and control plants
Figure S2 (a) DNA sequences of TALEN binding and target sites in the first exon of the sugarcane COMT. P1‐P2 and P1‐P3 are primer binding sites for amplification of TALEN target site or long PCR amplicons for COMT copy/allele identification, respectively. (b) Sequence confirmation of TALEN mediated COMT mutation in PCR amplicons. Deletions ranging from 2 to 48 bp in one of the mutants (CB6) considered as different mutation types.
Figure S3 Schematic representation of the region of COMT that was PCR amplified, cloned and sequenced for identification of mutations in different COMT copies/alleles by the Sanger method.
Table S1 Number of copies/alleles of COMT and types of TALEN mediated target mutation in sugarcane.
Table S2 Disease symptoms and insect damage on COMT mutant, RNAi/suppressed sugarcane, WT and transgenic control plants.
Table S3 A list of primer pairs used in this study.
Data S1 Supplementary Experimental Procedures and Results.


