Abstract
In 2014, we established a pharmacogenetics unit with the intention of facilitating the integration of pharmacogenetic testing into clinical practice. This unit was centered around two main ideas: i) individualization of clinical recommendations, and ii) preemptive genotyping in risk populations. Our unit is based on the design and validation of a single nucleotide polymorphism (SNP) microarray, which has allowed testing of 180 SNPs associated with drug response (PharmArray), and clinical consultation regarding the results. Herein, we report our experience in integrating pharmacogenetic testing into our hospital and we present the results of the 2,539 pharmacogenetic consultation requests received over the past 3 years in our unit. The results demonstrate the feasibility of implementing pharmacogenetic testing in clinical practice within a national health system.
Keywords: individualization, pharmacogenetics, precision medicine
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
✓ The integration of pharmacogenetics into clinical practice has been challenging over the years, primarily due to economic reasons, certain prescribers’ skepticism and discomfort, and the difficulty of fitting case‐by‐case molecular analyses into the clinical routine.
WHAT QUESTION DID THIS STUDY ADDRESS?
✓ The feasibility of the implementation of a pharmacogenetic program in a university hospital supported by the Spanish NHS.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
✓ Here we present a pharmacogenetic implementation strategy centered around a multidisciplinary clinical pharmacogenetics consultation and a custom SNP microarray, based on a preemptive genotyping strategy in risk populations and individualization of the clinical recommendations.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE
✓ This strategy could be considered a model for the clinical implementation of pharmacogenetics in other public hospitals, mostly in our country, but also in other countries with a similar NHS.
The high variability in drug response between individuals has been of serious concern to health professionals. Studying the influence of genetic variations in drug response phenotypes can provide a more individualized and effective design for drug treatment.1
The rapid development of molecular techniques and bioinformatics in recent years has permitted the identification and characterization of many relevant genes and their association with undesired events, such as toxicity and therapeutic failure. Over the past decade, both the US Food and Drug Administration and the European Medicines Agency have incorporated pharmacogenetics information into drug labels available for prescribers and patients.2 However, the integration of this approach into clinical practice has been challenging, due primarily to economic reasons, certain prescribers’ skepticism and discomfort,3, 4 and the difficulty of fitting case‐by‐case molecular analyses into the clinical routine. Despite these limitations, the increasing evidence supporting the integration of pharmacogenetics information into a clinical setting and the development of pharmacoeconomic studies reflecting the cost‐effectiveness of a preemptive approach have to some extent fostered the transition to personalized medicine.
The difficulties of integrating pharmacogenetics into clinical practice are well recognized in the literature. A survey conducted in 2012 among members of the Spanish Societies of Pharmacology and Clinical Pharmacology classified the perceived barriers in Spain into three major groups related to low institutional promotion, the lack of clinical guidelines and protocols, and economic and institutional issues as well as ethical, legal, and social implications.5 These reported hurdles are similar to those found in other countries.6, 7 In 2011, the Translational Pharmacogenetics Program (TPP) of the National Institutes of Health (NIH) Pharmacogenomics Research Network was created to design solutions for the barriers to clinical pharmacogenetics implementation. Eight United States healthcare systems participated in this project by implementing custom pharmacogenetics implementation strategies. These institutions developed diverse solutions and workflows for pharmacogenetics implementation; however, they were all based on Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, which provided some consistency among the various institutions.8 Similarly, the Ubiquitous Pharmacogenomics Consortium and the Preemptive Pharmacogenomic Testing for Prevention of Adverse Drug Reactions (PREPARE) study were developed in Europe to design an implementation strategy and evaluate the impact of pharmacogenetics integration programs on clinical practice.9
In this context, in 2014 our group created a Pharmacogenetics Unit with the intention of facilitating the implementation of a strategy for the preemptive genotyping of pharmacogenetics biomarkers associated with drug response in the clinical practice of our hospital. This unit is based on a clinical pharmacogenetics consultation and a custom single nucleotide polymorphism (SNP) microarray, which has allowed screening of 180 SNPs associated with drug response (PharmArray). In this study, we report our strategy for the integration of pharmacogenetic testing into the clinical practice of a tertiary level hospital. We also report our activity over the last 3 years.
MATERIALS AND METHODS
Setting description
La Paz University Hospital (LPUH) in Madrid is a 1,308‐bed tertiary‐care teaching hospital of the Spanish NHS serving a population of ∼600,000 people. It is also a national reference center for up to 22 specific diseases. Our clinical pharmacogenetics unit is a multidisciplinary unit integrated into both the Clinical Pharmacology Department and the Institute of Medical and Molecular Genetics Institute (INGEMM) of our hospital.
The Clinical Pharmacology Department is linked to the Pharmacology and Therapeutics Department of the School of Medicine of the Autonomous University of Madrid. Therapeutic individualization has always been an important objective for our department, primarily through our therapeutic drug monitoring, therapeutic consultation, pharmacovigilance,10 and clinical toxicology11 programs. Therefore, adding a pharmacogenetics approach to our clinical practice appeared an essential tool for achieving a more personalized approach for the optimization of therapeutic strategies.
INGEMM was created in 2008 and belongs to the LPUH Health Research Institute. Over 40,000 genetic studies are performed every year in our institution as well as several competitive research projects in various areas related to omics.
Developing a strategy for the implementation of a pharmacogenetics unit
In the process of developing a strategy for the implementation of a pharmacogenetics unit in our hospital, we identified three main issues to address:
To obtain a minimum infrastructure to develop this activity, including setting up a pharmacogenetics unit in association with the INGEMM as well as the development of affordable and quick genotyping strategies.
To increase the knowledge and acceptance of pharmacogenetics among physicians through clinical sessions and development of clinical protocols and research projects.
To gain a wide acceptance and the necessary funding by the management of the hospital and the regional health system, demonstrating the cost‐effectiveness and utility of adding a pharmacogenetics approach to clinical practice.
Developing an affordable and easy‐to‐use genotyping method
SNP selection
For the SNP selection process we decided to compose an in‐house multidisciplinary team of clinical pharmacologists, molecular geneticists, and technicians to achieve good coverage of relevant pharmacogenetics associations in drug metabolism and transporter genes. Information about the pharmacogenetic testing variants and their impact on drug response was gathered mostly from the variant and clinical annotations in PharmGKB.12 We relied mostly on level 1 through 3 Levels of Evidence based on PharmGKB Clinical Annotations. We also included some lower‐evidence associations recently described but not yet implemented in the clinical routine that we found of local interest for further studies and research.
Array design and technology
We used the OpenArray technology first on the ABI Biotrove OpenArray NT image cycler system (Applied Biosystems, Foster City, CA) and recently updated to the QuantStudio 12K Flex Real‐Time PCR System (Thermo Fisher Scientific, Pittsburgh, PA). TaqMan SNP Genotyping Assays were selected in the catalog of PreDesigned and DME SNP Genotyping Assays for optimal typing of the polymorphic targets (Pub no: MAN0009593). The assays were then preloaded in the plates and configured onto OpenArrays.
The loading process is automatic, using the QuantStudio 12K Flex OpenArray AccuFill system. The arrays are then inserted into glass cases and sealed for polymerase chain reaction (PCR) and subsequent imaging. Genotypes are then determined using the TaqMan Genotyper software available from Thermo Fisher Scientific and the data are analyzed by a custom script (PharmHulp v. 1.0).
Array validation
Twenty commercially available DNA samples with previously reported CYP2D6, CYP2C19, and CYP2C9 genotypes were acquired from the Coriell Biorepository (Camden, NJ) in order to test the specificity and accuracy of the included assays and validate our platform. All samples were tested blindly.
Evaluating patient and physician's satisfaction
Since 2015 our unit is ISO9001:2008‐certified and patient and physician's satisfaction is evaluated through surveys that include dichotomous (Yes/No), multiple choice, and numeric questions. Index of satisfaction (IS) was calculated using the following formula:
Estimation of the costs associated with our activity
We made an approximation of the cost associated with our activity. For this calculation, we included: i) the price of health services (Consult price ‐115€‐ and clinical inform price ‐71€‐) provided by the NHS, and ii) the cost of pharmacogenetics determination (PharmArray and Sanger sequencing/INNO‐LIPA) including DNA extraction, consumables, and personnel costs for the hospital.
RESULTS
Developing a strategy for the implementation of a pharmacogenetics unit at La Paz University Hospital
In the process of implementing pharmacogenetics in clinical practice, our main objective was to evolve from the usual ad hoc (case‐by‐case) genotyping strategy, in which the decision to perform a pharmacogenetics test is individualized and always linked to prescription, to a preemptive strategy in which genetic information would be obtained ab initio in risk populations (those who are susceptible to receive a drug from which pharmacogenetics information could be available) and would therefore be available at the moment of prescription.13
To this end, we developed protocols in collaboration with the petitionary clinical services including pharmacogenetics markers relevant for drugs used for specific diseases, and therefore adopting an approach between a case‐by‐case strategy and a full preemptive one (Table 1).
Table 1.
Established drug–gene protocols for preemptive genotyping of PhGx markers in specific diseases in LPUH
| Drug/gene pairs | Diseases | Requesting department |
|---|---|---|
| Thiopurines/TPMT | Inflammatory bowel disease, psoriasis | Gastroenterology, Dermatology |
| Immunosuppressants/CYP3A5, CYP3A4, ABCB1, POR | Pediatric kidney transplantation, Psoriasis, | Pediatric nephrology, Dermatology |
| Voriconazole/CYP2C19 | Aspergillosis in child bone marrow transplantation patients | Pediatric hemato‐oncology |
| Anticoagulant agents/CYP2C9, VKORC1, CYP4F2, APOE | Thromboembolic disease and atrial fibrillation | Internal Medicine, Hematology |
| Simvastatin/ SLCO1B1, ABCG2 | High cardiovascular risk | Cardiology |
| Methotrexate/ MTHFR | Leukemia, psoriasis | Pediatric hemato‐oncology, Rheumatology, Dermatology |
| Irinotecan/UGT1A1 | Colorectal cancer | Oncology |
| Fluoropyrimidines/ DPYD, TP53 | Colorectal cancer | Oncology |
Finally, our strategy is centered around the personalized medicine idea, in which an individualized interpretation of pharmacogenetics results is essential. Thus, unlike strategies based on alerts included in prescribing systems, each patient's clinical background, individual interactions, and other factors are taken into account, in addition to genetic information, to deliver an individual clinical recommendation.
Development of an affordable and easy‐to‐use genotyping method: Application of a custom SNP array (PharmArray) in a clinical pharmacogenetics unit
We designed a customized SNP microarray based on OpenArray Technology (Thermo Fisher Scientific) for the pharmacogenetics test (PharmArray, registration number 4571001). Human leukocyte antigen (HLA)‐B studies were performed by Sanger sequencing and INNO‐LIPA HLA‐B probe assays (Fujirebio, Malvern, PA) for accurate allele discrimination.
SNP selection
We selected the 16‐sample x 192 SNP format because it fulfilled our requirements and best fit the particular workflow and volume of patients in our hospital at that time. Our original version (PharmArray 2013) allowed testing of 192 SNPs in the same run. However, because of technical specifications and changes in manufacturer formatting, we recently had to update the number of probes to 180. The final compilation of SNPs included in our custom array and the specific treatments related to each gene are shown in Table 2.
Table 2.
Final design of the PharmArray 180 SNPs format
| Gene | Rs# | No. of SNPs | Treatment |
|---|---|---|---|
| ABCB1 | rs2032582*; rs1045642*; rs3213619; rs1128503 | 4 | Immunosuppressants and antiplatelet drugs |
| ABCC2 | rs717620; rs56296335**; rs3740066; rs56199535*; rs56220353* | 5 | Tenofovir (antiretroviral agent) |
| ABCG2 | rs2231142; rs2273697; rs72552713 | 3 | Statins, Methotrexate (cytotoxic agent), Imatinib (tyrosine‐kinase inhibitor) |
| APOE | rs7412 | 1 | Anticoagulants, Pravastatin (Statin) |
| COMT | rs4680 | 1 | Nicotine |
| CFTR | rs75527207; rs113993960; rs199826652; rs267606723*; rs193922525; rs80282562; rs121909013; rs74503330; rs121909041; rs121908755*; rs121909005; rs121908757 | 12 | Ivacaftor (CFTR potentiator) |
| CYP2C19 | rs4244285*; rs4986893; rs12248560*; rs28399504; rs56337013; rs72552267; rs72558186; rs41291556 | 8 | Voriconazole (triazole antifungal agent), antiplatelet and psychotropic therapy |
| CYP2C8 | rs11572080; rs10509681; rs1058930; rs11572103 | 4 | Paclitaxel (cytotoxic agent), psychotropic therapy and oral antidiabetic agents |
| CYP2C9 | rs1799853; rs1057910 | 2 | Anticoagulants, psychotropic therapy |
| CYP2D6 | rs1080985; rs28371725; rs35742686; rs3892097; rs5030655; rs5030865*; rs5030867; rs5030656; rs1065852; rs1058164; rs1135840; rs16947; rs28371706; rs61736512; rs769258 | 15 | Psychotropic therapy, opioids |
| CYP3A4 | rs55785340; rs4646438 | 2 | Immunosuppressants |
| CYP3A5 | rs776746; rs55965422*; rs10264272; rs41303343; rs41279854 | 5 | Tacrolimus (immunosuppressant) |
| CYP4F2 | rs2108622 | 1 | Anticoagulants |
| DPYD | rs3918290; rs55886062*; rs55886062*; rs67376798; rs1801159; rs1801265 | 6 | Fluoropyrimidines (Cytotoxic agents) |
| ERCC1 | rs11615; rs3212986 | 2 | Cisplatin (cytotoxic agent) |
| EPHX1 | rs1051740 | 1 | Cisplatin (cytotoxic agent) |
| FCGR2A | rs1801274 | 1 | Biological therapy |
| HTR2A | rs6311 | 1 | Psychotropic therapy |
| IL10 | rs1800896; rs1800872; rs1800871 | 3 | Biological therapy |
| IL23R | rs7517847; rs10489629; rs11465804; rs1343151 | 4 | Biological therapy |
| KCNJ6 | rs2070995 | 1 | Analgesics |
| MTHFR | rs1801133; rs4846051; rs1801131 | 3 | Methotrexate (cytotoxic agent) |
| POR | rs1057868; rs2868177 | 2 | Immunosuppressants |
| SLC15A2 | rs2293616; rs2257212; rs1143671; rs1143672 | 4 | Others |
| SLC22A1 | rs72552763; rs55918055*; rs36103319*; rs34059508*; rs628031; rs4646277; rs2282143; rs4646278*; rs12208357 | 9 | Tramadol (Opioid), Metformin (oral antidiabetic agent) |
| SLC22A2 | rs316019; rs8177516; rs8177517; rs8177507*; rs8177504 | 5 | Fampridine (potassium channel‐blocking agent), Metformin (oral antidiabetic agent) |
| SLC22A6 | rs11568626 | 1 | Others |
| SLCO1B1 | rs4149056; rs2306283; rs56101265; rs72559745; rs56061388; rs55901008*; rs59502379; rs56199088*; rs55737008; rs4149015 | 10 | Statins, Irinotecan (cytotoxic agent), oral antidiabetic agents, conjugated estrogens |
| TLR2 | rs4696480; rs11938228 | 2 | Biological therapy |
| TLR9 | rs352139 | 1 | Biological therapy |
| TNF | rs1800629 | 1 | Biological therapy |
| TP53 | rs1042522 | 1 | Cisplatin (cytotoxic agent) |
| TPMT | rs1800460; rs1800462; rs1142345; rs1800584 | 4 | Thioguanines |
| UGT1A1 | rs887829; rs4148323; rs34993780; rs35350960; rs55750087; rs4124874 | 6 | Irinotecan (cytotoxic agent) |
| UGT2B7 | rs7438135 | 1 | Morphine, micophenolate (immunosuppressant) |
| VKORC1 | rs9934438 | 1 | Anticoagulants |
| XPC | rs2228001 | 1 | Cisplatin (cytotoxic agent) |
| XRCC1 | rs25487 | 1 | Cisplatin (cytotoxic agent) |
The table shows the final selection of SNPs (dbSNP b146) and genes included in our custom array. We also included two sex markers (rs768983 and rs3913290) for internal quality control of the genotyping process and the SNPs‐related drugs. SNPs followed by * are reported to have more than two allelic variants.
Array validation
We genotyped 20 Coriell samples with previously reported genotypes for three relevant pharmacogenetics markers: CYP2C19, CYP2C9, and CYP2D6. Our design allowed an accurate determination of 95% of the genotypes; however, due to limitations of the technique, we found some discrepancies, especially among those samples showing copy number variants in CYP2D6 (*5 allele and xN alleles) and those expressing extremely rare haplotypes that were not included in our design (CYP2C19*10 and CYP2C9*9). These specific determinations are not included in our clinical routine.
Structure and functioning of the phamacogenetics unit at La Paz University Hospital
In‐house classification of pharmacogenetics tests
The clinical pharmacogenetics unit of La Paz University Hospital evaluates both the internal and external inquiries for which a pharmacogenetics test might be suited: i) drug response prediction; ii) the optimization of dosing requirements; and iii) the identification of therapy failure, adverse reactions, or interactions related to genetic variation. In order to handle the requests received, we classified pharmacogenetics tests into three groups (Figure 1):
Preemptive molecular screening of actionable genetic markers required for treatment selection (HLA‐B*57:01/abacavir; IL28B‐PEG‐interferon‐α). The pharmacogenetics result is directly referred to the petitioner because a complementary specialized clinical counseling is not deemed necessary and standard information is provided along with the results of the test. Presently, IL28B studies before the administration of interferon in hepatitis C virus (HCV) are no longer performed due to the implementation of alternative pharmacological treatments.
Drugs with a well‐defined protocol for pharmacogenetics treatment recommendations in a particular disease. Included here are those cases in which treatment based on pharmacogenetics testing has been agreed upon by clinical service(s) and are included in clinical protocols (Table 1). In this case, pharmacogenetics determination is requested along with other complementary tests performed in the initial stages of diagnosis and/or treatment.
Drugs without a well‐defined protocol. Ad hoc phenotyping of pharmacogenetics markers is available but not included in clinical protocols. In this case, a consultation is made to the pharmacogenetics unit in order to evaluate a specific therapeutic problem and determine whether the pharmacogenetics test is recommended.
Figure 1.
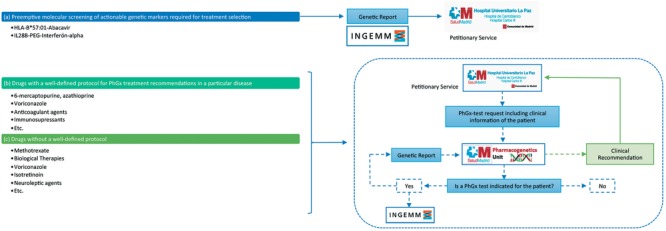
Classification of pharmacogenetics tests and pharmacogenetics unit workflow. We have divided pharmacogenetics tests into three main groups: For drugs belonging to group (a) in which the pharmacogenetics test is required before treatment prescription, requests and samples are directly forwarded to the genetics department (INGEMM) for sample processing and analysis. The final molecular report is directly sent to the petitionary service for treatment selection. Groups (b) and (c) share a different workflow: Petitionary services refer their request, including all clinical information, to the pharmacogenetics unit that will decide whether a pharmacogenetics test is indicated in each case. If the pharmacogenetics test is recommended, molecular analysis is performed in INGEMM and a genetic report is generated. Taking into account the molecular result and the clinical information of the patient, a final clinical recommendation is given to the original petitionary service. Clinical recommendations are specific for each patient because they are generated relying on a multifactorial basis. It is important to notice that groups (b) and (c) might share some particular drugs (e.g., voriconazole), given these might have a protocol when prescribed for a particular disease but not for a different pathology.
It is important to consider that this distribution is not static: one drug might fit into two different groups, depending on its clinical indication and the protocol established in agreement with the petitionary clinical service. Furthermore, one particular drug can change from group 3 to group 2 when a consistent protocol has been defined and a preemptive genotyping strategy is designed. This process is dynamic and continuously updated.
It is important to note that our implementation plan is aimed toward a preemptive pharmacogenetics approach; thus, the pharmacogenetics test is performed on risk populations in which patients might or might not receive treatment. Nevertheless, genetic information would be available should prescription be necessary; for example, prior to bone marrow transplantation procedures in which a preemptive pharmacogenetics study for voriconazole response is performed in case the patient develops aspergillosis after transplantation.
Pharmacogenetics unit workflow
Figure 1 shows the organization of our clinical pharmacogenetics unit, which includes six main steps (Figure 2). Petitionary services (treating physicians) send their request, including all clinical information, to the pharmacogenetics unit:
For drugs belonging to the first group, samples are directly remitted to the INGEMM at LPUH for molecular analysis. The molecular report and its interpretation is then generated and sent back to the petitionary service through the LPUH electronic health record (EHR).
In those cases in which a protocol has been agreed upon for the specific indication, clinical department samples are sent to the INGEMM and a consult with the pharmacogenetics unit is automatically created. A molecular report is generated by a molecular geneticist and remitted to the clinical pharmacologists for the elaboration of an individualized report, taking into account each patient's clinical record. This clinical report is usually sent to the treating physician through the EHR; however, a personal interview with the patient can be appointed if deemed necessary.
If an agreed‐upon protocol does not exist, the patient is directly referred to the pharmacogenetics unit. Then it is decided if a pharmacogenetic test is recommended; if this is the case the process would be the same as described for the second group of pharmacogenetics tests.
Figure 2.
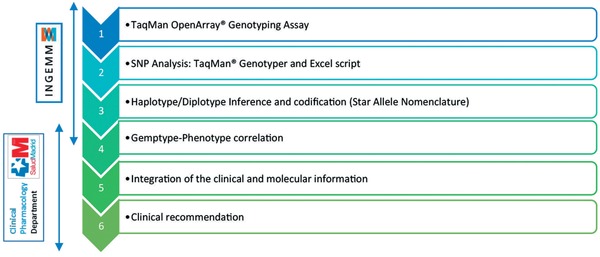
Analysis workflow. Our custom analysis workflow includes six main steps performed by both the Clinical Pharmacology Department and the INGEMM pharmacogenetics specialists. First, genomic DNA from the patients is automatically extracted from peripheral blood cells using Chemagen technology (Perkin‐Elmer, Boston, MA). However, in particular patients, DNA can be obtained from other biological samples such as saliva or tissue. It is necessary that all patients give informed consent to genetic analysis. Subsequently, a TaqMan OpenArray Genotyping Assay is performed using our custom design (PharmArray; Reg. no. 4571001). An individualized analysis of each SNP included in the pharmacogenetics protocol for each specific drug and disease is then performed. We then proceed to haplotype and diplotype inference using population databases and codification using the star‐allele nomenclature (*). Once genotypes are codified, phenotypes are also inferred. Finally, an integration of both the clinical and molecular information is performed for a more individualized clinical recommendation.
Activity of the pharmacogenetics unit from 2014 to 2016
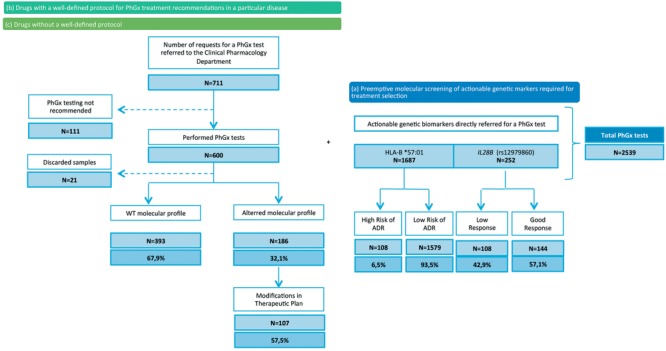
Between the implementation of the pharmacogenetics unit at LPUH in January 2014 and December 2016 (3 years), we received 2,539 requests (Figure 3); 1,939 were for actionable genetic marker testing required for treatment selection (87% for HLA‐ B*57:01 and 13% for IL28B) and were therefore directly sent for molecular screening because no previous clinical evaluation was needed. Approximately 6.5% of the patients tested for HLA‐B*57:01 showed a molecular profile related to a high risk of developing a hypersensitivity reaction to abacavir and, therefore, an alternative pharmacological treatment was recommended. Some 42.9% of the patients tested for IL28B before the administration of PEG‐ interferon‐alpha‐containing regimens in HCV genotype 1 patients showed a molecular profile related to low response to treatment.
Figure 3.
Patient Enrollment (2014–2016). Distribution of the 2,539 pharmacogenetics tests performed in our Pharmacogenetics Unit. PhGx: Pharmacogentic.
A total of 711 enquiries belonging to tests groups 2 and 3 were requested. After clinical evaluation, we found that 84% of the patients (600) met the specific inclusion criteria for the requested pharmacogenetics test. We found that 32.1% of the patients showed a molecular profile that could be related to the specific requested drug's pharmacokinetics and pharmacodynamics. Among these patients, a clinical recommendation for dose adjustment was performed in 107 (57.5%).
TPMT preemptive testing before the administration of thioguanines is the most common request (60.7%). Genetic variants in the MTHFR gene are also frequently studied as predictors of methotrexate‐induced liver toxicity (19.8%). The growing evidence regarding pharmacogenetics associations with the oral antifungal voriconazole and immunosuppressant agents’ pharmacokinetics has led to an increase in the number of requests over the past year, mainly for patients needing transplantation (2.3% for voriconazole and 6.7% for immunosuppressant). We also performed pharmacogenetics tests before the administration of acenocoumarol (2.7%), fluoropyrimidines (2.8%), and other drugs, such as psychotropic agents; however, these studies were performed on a less frequent basis (6%, all).
Evaluating patient and physicians satisfaction
Patient's satisfaction has been evaluated in about 12% of the patients attending our consult (years 2015–2016). Also, we evaluated physician's perception (19 physicians from 9 different departments) about pharmacogenetics, their expectations on the application of pharmacogenetics, and the perceived utility of our pharmacogenetics unit. Questions and results of both surveys are presented in Table 3.
Table 3.
Patient's and physician's survey
| Patient's s survey (n = 51) | |
|---|---|
| YES (%) | |
| Do the facilities seem to you decent, clean, and in accordance with the Clinical Genetic visit you have obtained? | 100.00% |
| The expert who has attended to you has taken the adequate time to explain to you all the necessary details in order to understand the scope of the Clinical Pharmacogenetic visit? | 100.00% |
| If studies were requested, have you understood what are they about, what it is expected from the result and how long it will take approximately? | 100.00% |
| Do you consider that the information was complete? | 100.00% |
| Regardless the number of experts assisting you in the visit, have you had enough privacy to comment with them the problems that afflict you? | 100.00% |
| IS (%) | |
|---|---|
| Transparency of the expert's information. | 89.57% |
| Time of response of the requested analysis with regard to your necessities. | 79.13% |
| Degree of confidence that you have in the results of the Clinical visit of this Institute. | 80.43% |
| Answer and/or possibility of new consultations if requested. | 83.48% |
| Telephone answering service of the administrative assistants when you call in order to make an appointment or clarify doubts. | 84.78% |
| GLOBAL IS | 83.47% |
| Physician's survey (n = 19) | |
|---|---|
| N (%) | |
| Service/Unit | |
| Internal Medicine | 4 (21.1) |
| Dermatology | 3 (15.8) |
| Pediatric nephrology (kidney transplantation) | 3 (15.8) |
| Gastroenterology | 2 (10.5) |
| Pediatric hemato‐oncology | 2 (10.5) |
| Psychiatry | 2 (10.5) |
| Oncology | 1 (5.3) |
| Clinical Immunology | 1 (5.3) |
| Thromboembolic unit | 1 (5.3) |
| Do you think that in the last 3 years have improved your understanding about the utility of pharmacogenetics in clinical practice? | |
| Yes | 19 (100) |
| If it has improved, which factors have contributed to it?* | |
| Contact with the Unit of Pharmacogenetics | 17 (89.5) |
| Development of protocol and guidelines | 4 (21.1) |
| Development of research projects related to pharmacogenetics | 11 (57.9) |
| Education, sessions or specific courses | 8 (42.1) |
| You make use of Pharmacogenetics according to:* | |
| Protocols agreed with the Unit of Pharmacogenetics | 8 (42.1) |
| Request of tests of individual cases | 14 (73.7) |
| In case of a recommendation of the Unit of Pharmacogenetics: | |
| I follow the indications reported by the Unit of Pharmacogenetics | 16 (84.2) |
| I do not follow the exact indications but I consider the genetic result | 3 (15.8) |
| I neither follow the indications nor I consider them | 0 (0) |
| IS (%) | |
|---|---|
| Do you consider appropriate the way of requesting a test and/or visit? | 79.0% |
| Do you consider appropriate the time of response? | 76.8% |
| Do you consider appropriate and useful the provided pharmacogenetic information? | 94.7% |
| Do you think that the use of pharmacogenetics has an impact on the management of your patient? | 85.3% |
| Do you think that in the following years the use of pharmacogenetics will increase in your specialty? | 91.6% |
| N (%) | |
|---|---|
| Which aspects do you think could increase the use of pharmacogenetics in your specialty?* | |
| Clearer guidelines about the use of pharmacogenetics | 11 (57.9) |
| Greater level of evidence about its clinical validity and utility | 11 (57.9) |
| Evaluation of the cost‐effectiveness of the use of pharmacogenetics | 10 (52.6) |
| Time of response more appropriate | 5 (26.3) |
IS: index of satisfaction.
*More than one choice question can be selected.
Estimation of the costs associated with our activity
Global cost of the activity for the NHS for the 3 years was 202,140 €, including all the costs for the NHS. The costs differ based on the level of consultation; the cost of a complete process, including pharmacogenetics determination and individual consultation, is 216 €. For the sake of comparison, the cost of attendance of a patient for an intravenous drug administration (i.e., anti‐TNF administration) is 260 € and for a determination of drug plasma level is 106 €.
DISCUSSION
The growing evidence supporting the contribution of genetic variability in genes coding drug‐metabolizing enzymes and transporters to interindividual heterogeneity in drug response has identified pharmacogenetics as a relevant tool for achieving the personalized medicine paradigm.3 However, several barriers have been perceived both in Spain and around the world for routine integration of pharmacogenetics into the clinical practice of various national health systems (NHSs).5, 6, 7
Here we have presented our experience in the implementation of a custom strategy for integrating pharmacogenetics into the clinical practice of a Spanish tertiary‐level hospital through the creation of a clinical pharmacogenetics unit covered by the Spanish NHS and accessible to all patients. This strategy is centered around two main ideas: individualization of clinical recommendations and the evolution to a preemptive genotyping strategy. Our strategy shows several similarities with other implementation programs previously proposed both in the United States and in Europe. The use of multidisciplinary teams and the common use of CPIC guidelines are two main points shared by most of the centers, including ours. In addition, our analysis workflow (Figure 2) is consistent with the stepwise process used across the TPP sites and advised by the CPIC guidelines.8 One critical point in our approach is related to the interpretation of results and the development of the final clinical recommendation. In most of the mentioned approaches, the final step is a general clinical recommendation based on the predicted phenotype from the patient diplotype, mostly provided through the inclusion of warnings and simple pharmacogenetics information into prescription systems. Although this approach has some advantages and might be appropriate in some cases (e.g., clear, actionable biomarkers), in our opinion it can be insufficient at the present stage of knowledge and general acceptance for many of the pharmacogenetics tests and could even be harmful if the interpretation is not appropriate for a particular patient. Some reasons for this include the following:
A relevant number of patients have two or more variants affecting their treatment (of one or several concomitant drugs).
Many patients also have concomitant diseases that can interact in various ways with the genotype of the patient.
Drug interactions can appear (or severity can be different), depending on the genotype of the patient.
Guidelines on these situations are rarely available and a more deep evaluation of evidence is needed.
Therefore, both clinical and molecular information should be individually integrated for each patient in order to develop a true personalized clinical recommendation based not only on genetic information but also taking into account medical history, other clinical factors, concomitant treatments, and the patient's preferences, provided by medical records and the patient encounter. This implies that our implementation strategy relies on the presence of a clinical pharmacogenetics specialist throughout the process, always in close agreement with other clinical specialists: selection of pharmacogenetics biomarkers and their integration in clinical protocols, selection of the patients for whom a pharmacogenetics test is indicated, and a final clinical recommendation and genetic counseling for patients when necessary.
In our experience, an individualized interpretation of both pharmacogenetics and clinical information is needed to achieve an accurate and optimized therapeutic plan. CPIC and other guidelines represent a useful framework for unification of the clinical recommendations among different centers, demonstrating that actionable recommendations for drugs can be implemented with minimal ambiguity.8 However, we found it was essential to adjust this genotype‐guided strategy to each patient's specific clinical background by a pharmacogenetics specialist. In addition, one of the challenges reported for the delivery of pharmacogenetics results is the identification of the right person to receive a recommendation as well as the various uses of information that might be made by each healthcare discipline.14, 15 Moreover, we found that the elaboration of an accurate clinical recommendation (with a clinical report and a posttest consultation when necessary), in addition to the molecular report, made the comprehension and application of the pharmacogenetics results easier for the specialists of the petitionary clinical departments, overcoming the already mentioned barriers to interpretation and application of the use of pharmacogenetics.
We also find that the stepped evolution from ad hoc genotyping strategies to a preemptive genotyping strategy is essential for bringing pharmacogenetics and clinics together. Our designed custom genomic tool PharmArray (based on the TaqMan OpenArray) was of great help to this aim. Although tests based on next‐generation sequencing (NGS) technologies16, 17, 18, 19 have been reported in the literature, we found that our platform results are much more suitable for the clinical approach in terms of simplicity of analysis, turnaround times, and affordable costs in our NHS. This genotyping tool allowed us the implementation of a semipreemptive strategy with eight preestablished protocols agreed upon with various clinical services in risk populations. In our view, this is a very appropriate intermediate step to a full preemptive strategy. In this context, the evolution of this intermediate approach to a more extensive or even complete preemptive pharmacogenetics strategy still needs to be supported by cost‐effectiveness studies.
Another key point for the implementation of pharmacogenetics in our hospital was bringing together research and clinical practice by promoting collaborative investigation20, 21 and decision making; several investigation projects are underway evaluating the clinical results of this implementation. This approach led to a direct impact on clinical practice and a more active interaction with other medical specialties through lectures and clinical meetings, allowing the creation of more efficient and optimized treatment guidelines and protocols, as shown in the physician survey performed. In this survey it is also remarkable that all physicians consider that, in the last 3 years, its comprehension of the utility of pharmacogenetics in the clinical practice has improved, mainly due to their interaction with our unit (88.2%). Another important result is that physicians consider that pharmacogenetic information is useful, with a global index of satisfaction of 94.74%. Moreover, most physicians follow the recommendations of our unit (84,2%).
As reported in the literature, our results show that 6.5% of the patients tested for HLA‐B*57:01 prior to the administration of abacavir showed a molecular profile associated with a high risk of developing severe hypersensitivity reactions to the drug; therefore, abacavir is not recommended in this subpopulation.22 Similarly, the number of patients with a low response genotype (IL28B; rs12979860; MAF = 0.31 T) for PEG‐interferon‐alpha‐containing regimes was 42.9%, as expected. This means 11.1% of the patients belonging to the first group of pharmacogenetics tests, i) actionable genetic markers required for treatment selection, showed a molecular profile in which a modification in the initial therapeutic plan was needed (Figure 3). For pharmacogenetic tests belonging to groups ii) drugs with a well‐defined protocol for a specific disease and iii) drugs without a predefined protocol for a particular disease, we found that 32.1% of the patients showed an altered molecular profile needing a reevaluation of the standard therapeutic strategies and dosing regimens. From these, we recommended modifications to the initial therapeutic plan for 57.5% of the patients (Figure 3).
The calculated costs are frequently lower than those described even for just genotyping in the United States and other countries.23 Despite that our costs seem cost‐effective and similar to usual procedures within our NHS, a formal cost‐efficiency study of our strategy would be necessary for stronger institutional support and expansion.
CONCLUSION
Based on our experience, the implementation of clinical pharmacogenetics programs in the clinical routine is feasible with the actual resources of the Spanish NHS. The clinical pharmacogenetics unit of La Paz University Hospital is centered around the two main ideas of i) individualization of clinical recommendations and ii) preemptive genotyping in risk populations. This is the first publication of a specific strategy for the implementation of pharmacogenetics in Spain that differs from genotype‐based clinical decision support tools integrated in prescription systems. Therefore, the multidisciplinary structure and workflow of this strategy could be considered a model for the clinical implementation of pharmacogenetic testing in other public hospitals of our country and in other countries with a similar NHS.
Conflict of Interest
The authors declare no competing interests for this work.
Author Contributions
A.M.B., I.D., and A.J.A.C. wrote the article; A.M.B., P.A., J.T., G.G., P.L., and A.J.A.C. designed the research; A.M.B., H.Y.T., M.M., R.H., and I.G. performed the research; I.D., P.A., J.T., G.G., E.R., J.F., and P.L. analyzed the data.
Funding
No funding was received for this work.
References
- 1. International Conference on Harmonisation; guidance on specifications: test procedures and acceptance criteria for biotechnological/biological products. Notice. Food and Drug Administration, HHS. Fed. Regist. 64, 44928–44935 (1999). [PubMed] [Google Scholar]
- 2. Ehmann, F. et al Pharmacogenomic information in drug labels: European Medicines Agency perspective. Pharmacogenomics J. 15, 201–210 (2015). [DOI] [PubMed] [Google Scholar]
- 3. Scott, S.A. Personalizing medicine with clinical pharmacogenetics. Genet. Med. 13, 987‐995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bielinski, S.J. et al Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time‐using genomic data to individualize treatment protocol. Mayo Clin. Proc. 89, 25–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agundez, J.A. et al Toward a clinical practice guide in pharmacogenomics testing for functional polymorphisms of drug‐metabolizing enzymes. Gene/drug pairs and barriers perceived in Spain. Front. Genet. 3, 273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunnenberger, H.M. et al Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horgan, D. et al An index of barriers for the implementation of personalised medicine and pharmacogenomics in Europe. Public Health Genomics 17 287–298 (2014). [DOI] [PubMed] [Google Scholar]
- 8. Luzum, J.A. et al The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin. Pharmacol. Ther. 102, 502‐510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manson, L.E. , van der Wouden, C.H. , Swen, J.J. & Guchelaar, H.J. The Ubiquitous Pharmacogenomics consortium: making effective treatment optimization accessible to every European citizen. Pharmacogenomics 18, 1041–1045 (2017). [DOI] [PubMed] [Google Scholar]
- 10. Ramirez, E. et al A pharmacovigilance program from laboratory signals for the detection and reporting of serious adverse drug reactions in hospitalized patients. Clin. Pharmacol. Ther. 87, 74–86 (2010). [DOI] [PubMed] [Google Scholar]
- 11. Muñoz, R. , Borobia, A.M. , Quintana, M. , Martínez‐Virto, A.M. , Frías, J. & Carcas, A.J. Development and validation of a poisoning surveillance program with automatic case detection in a tertiary care hospital (SAT‐HULP) Emergencias 25, 423–429 (2013). [Google Scholar]
- 12. Whirl‐Carrillo, M. et al Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borobia, A. Implementación de la farmacogenética en la práctica clínica: Hacia las estrategias de genotipado anticipado. IBJ Clin. Pharmacol. 1 (2016). [Google Scholar]
- 14. Freimuth, R.R. et al Implementing genomic clinical decision support for drug‐based precision medicine. CPT Pharmacometrics Syst. Pharmacol. 6, 153–155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. St Sauver, J.L. et al Integrating pharmacogenomics into clinical practice: promise vs reality. Am. J. Med. 129, 1093–1099 e1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lauschke, V.M. * Ingelman‐Sundberg, M. Requirements for comprehensive pharmacogenetic genotyping platforms. Pharmacogenomics 17, 917–924 (2016). [DOI] [PubMed] [Google Scholar]
- 17. Qiao, W. et al Long‐read single molecule real‐time full gene sequencing of cytochrome P450‐2D6. Hum. Mutat. 37, 315–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Twist, G.P. et al Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole‐genome sequences. npj Genomic Medicine 1, 15007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizzi, C. et al Personalized pharmacogenomics profiling using whole‐genome sequencing. Pharmacogenomics 15, 1223–1234 (2014). [DOI] [PubMed] [Google Scholar]
- 20. Borobia, A.M. et al An acenocoumarol dosing algorithm using clinical and pharmacogenetic data in Spanish patients with thromboembolic disease. PLoS One 7, e41360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tong, H.Y. et al A new pharmacogenetic algorithm to predict the most appropriate dosage of acenocoumarol for stable anticoagulation in a mixed Spanish population. PLoS One 11, e0150456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dean, L. Abacavir therapy and HLA‐B*57:01 genotype In: Pratt V., McLeod H., Dean L., Malheiro A., Rubinstein W. (eds). Medical Genetics Summaries. National Center for Biotechnology Information (US), Bethesda (MD) 2012. [PubMed] [Google Scholar]
- 23. Verbelen, M. et al Cost‐effectiveness of pharmacogenetic‐guided treatment: are we there yet? Pharmacogenomics J. 17, 395–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


