Summary
Lipid rafts are microdomains present in the membrane of eukaryotic organisms and bacterial pathogens. They are characterized by having tightly packed lipids and a subset of specific proteins. Lipid rafts are associated with a variety of important biological processes including signaling and lateral sorting of proteins. To determine whether lipid rafts exist in the inner membrane of B. burgdorferi, we separated the inner and outer membranes and analyzed the lipid constituents present in each membrane fraction. We found that both the inner and outer membranes have cholesterol and cholesterol glycolipids. Fluorescence anisotropy and FRET showed that lipids from both membranes can form rafts but have different abilities to do so. The analysis of the biochemically-defined proteome of lipid rafts from the inner membrane revealed a diverse set of proteins, different from those associated with the outer membrane, with functions in protein trafficking, chemotaxis and signaling.
Keywords: Lipid rafts, microdomains, Lyme disease, Borrelia
Introduction
Cholesterol and other sterols are essential structural components of eukaryotic cell membranes. One important characteristic of sterols is that they promote the formation of liquid ordered domains, often referred to as lipid rafts (Brown & London, 1998). Lipid rafts in eukaryotes are characterized by having membrane lipids that are more tightly packed than in the surrounding liquid disordered bilayer, by having a high concentration of cholesterol and sphingolipids, and by their association with a subset of specific proteins with important biological functions (Brown & London, 2000). Since prokaryotes cannot synthetize cholesterol, for a long time it was thought that lipid rafts were present only in eukaryotes. This notion has now been revised with the discovery of lipid rafts in Borrelia burgdorferi, the agent of Lyme disease (LaRocca et al., 2010, LaRocca et al., 2013, Burgdorfer et al., 1982, Benach et al., 1983), and functional microdomains in other bacteria (Lopez & Kolter, 2010). Rafts may be present in even more bacteria, as several have cholesterol in their membranes. Intracellular bacteria transmitted by ticks require incorporation of cholesterol onto their membranes (Lin & Rikihisa, 2003). Anaplasma phagocytophilum incorporates cholesterol through the low-density-lipoprotein (LDL) pathway of the host (Xiong et al., 2009). Mycoplasma spp. also incorporate cholesterol onto their membranes (Smith, 1971, Razin, 1978, Razin et al., 1982). B. burgdorferi and H. pylori incorporate host cholesterol (Crowley et al., 2013, Correia et al., 2014) and through the action of a galactose and a glucose transferase, respectively (Lebrun et al., 2006, Ostberg et al., 2007), make cholesterol glycolipids (Hirai et al., 1995). In B. burgdorferi, one of these cholesterol glycolipids, cholesteryl 6-O-palmitoyl-β-D-galactopyranoside (ACGal), is essential in the formation of lipid rafts (Huang et al., 2016), and a recent study detected a flotillin homolog enriched in detergent resistant membranes, suggesting the presence of lipid rafts in H. pylori (Hutton et al., 2017).
Functional microdomains in Bacillus subtilis lack cholesterol but functionally, and at least to some extent structurally, resemble eukaryotic lipid rafts (Lopez & Kolter, 2010). These domains require the biosynthesis of polyisoprenoid lipids and co-localize with flotillin (Lopez & Kolter, 2010), a scaffold protein that is important for the recruitment of raft-associated proteins into membrane microdomains in eukaryotes (Langhorst et al., 2005).
Another basis for bacterial membrane raft formation may involve hopanoids, a family of lipids commonly found in bacteria (Ourisson & Rohmer, 1992). Hopanoids have a ring structure similar to that of sterols, and are important for regulation of membrane fluidity (Welander et al., 2009). Hopanoids act as functional analogues of cholesterol (Saenz et al., 2015), and can interact with glycolipids in bacterial outer membranes to form a liquid ordered bilayer, similar to the way that cholesterol interacts with sphingolipids to form rafts in eukaryotic cells (Saenz et al., 2015). Recently, small lateral features in the membranes of Bacillus subtilis have been documented that share properties with lipid rafts (Nickels et al., 2017).
Despite having different lipid constituents, bacterial microdomains have characteristics shared with eukaryotic lipid rafts. For example, they are resistant to solubilization by non-ionic detergents, are enriched in proteins, which like flotillin, have SPFH (Stomatin, Prohibitin, Flotillin, HflC/HflK) domains, and have specific membrane-associated functions (Toledo et al., 2015, Saenz et al., 2015, Lopez & Kolter, 2010, Lopez & Koch, 2017, Huang & London, 2016). Overall, although membrane microdomains in prokaryotes are novel from structural, physiological and evolutionary perspectives, they establish a tantalizing structural and evolutionary link between prokaryotes and eukaryotes.
Although this new field has advanced rapidly, there are still fundamental questions that are unanswered. Gram-negative bacteria have two membranes, a cytoplasmic or inner membrane (IM) and an outer membrane (OM), which increases the complexity of processes such as signaling and trafficking. So far, membrane lipid domains have been identified in OMs. This leaves unanswered the question as to whether membrane domains exist in inner cytoplasmic membranes, and if so, what are the similarities and differences with domains in the OM?
To address these questions we studied B. burgdorferi, a diderm bacterium that lacks LPS but has a large number of outer surface proteins as well as cholesterol and cholesterol glycolipids (Radolf et al., 1994, Radolf et al., 1995, Brandt et al., 1990, Livermore et al., 1978). In previous studies on B. burgdorferi, we demonstrated the presence of lipid rafts by biophysical and biochemical means, and we visualized them in the OM through electron microscopy (LaRocca et al., 2013). Whether lipid rafts form in the IM was not studied. However, a proteomic study of B. burgdorferi lipid rafts (derived from whole cells) revealed the presence of predicted IM proteins in lipid rafts (Toledo et al., 2015). Moreover, SPFH domain-containing proteins (HflC and HflK) that are found in eukaryotic lipid rafts (Browman et al., 2007), were predicted to be in the IM of B. burgdorferi.
In this study we isolated the IM and OMs of B. burgdoferi and then demonstrated using biophysical, biochemical and proteomic approaches that lipid rafts can form in both membranes, but have different properties. Lipids from each membrane fraction had different abilities to form domains. In addition, as judged by biochemical methods (detergent resistance), lipid raft-associated proteins in the IM and OMs were different, with the IM rafts being enriched in transporters and signaling proteins, linking these domains with protein trafficking and sensing and signaling pathways. In contrast, the OM rafts were enriched in lipoproteins associated with tick/host adaptation which is important for the biology of B. burgdorferi. The presence of lipid rafts in the IM establishes another layer of biological complexity in fundamental biological processes that could exist in many Gram-negative and prokaryotes with double-membranes.
Results
Separation of the inner and outer membrane
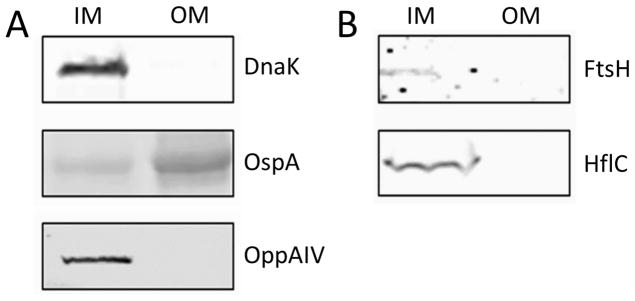
To determine whether lipid rafts were present in both membranes, we separated B. burgdorferi into IM and OM fractions and carried out a western blot using known markers to cytoplasmic, IM and OM proteins. The IM fraction contained: DnaK, a cytoplasmic chaperone; OppAIV, an IM protein; and traces of OspA, an OM protein (Figure 1A), indicating that the IM fraction includes the protoplasmic cylinder, which contains the cytoplasm and the peptidoglycan-cytoplasmic membrane complex, and traces of the OM (Figure 1A). Of these proteins, the OM fraction only showed the presence of OspA indicating that there was no detectable contamination from the cytoplasm or components from the IM (Figure 1A). Several membrane separations were carried out throughout this project with consistent results. For quality control purposes, fractions were probed with a battery of monoclonal and polyclonal antibodies to antigens with known locations in the cell (Figure S1). These included HtrA that is found in both IM and OM fractions; OspB, P66, and Lp6.6 which are enriched in the OM; and Lon protease, DnaK, and Flab which partition on the IM only.
Figure 1.
Subcellular localization of B. burgdorferi proteins DnaK, OspA, OppA4, HflC and FtsH analyzed by SDS-PAGE and Western blot. A. DnaK, OspA and OppA4 antibodies were used to assay the separation of the OM and the IM fractions. B. HflC and FtsH proteins were associated exclusively with the IM fraction, supporting their association with the inner membrane. OM: outer membrane; IM: Inner membrane. Controls using secondary antibodies alone were non-reactive (not shown).
In a previous study, in which we characterized the biochemically-defined lipid raft proteome of B. burgdorferi whole cells, we found proteins in the lipid raft fraction that were predicted to be associated with the IM, most notably HflC, HflK and FtsH (Toledo et al., 2015). Western blots confirmed that they partitioned in the IM fraction (Figure 1B), suggesting that lipid rafts could also exist in the IM.
Cholesterol and cholesterol glycolipids are present in the inner membrane fraction
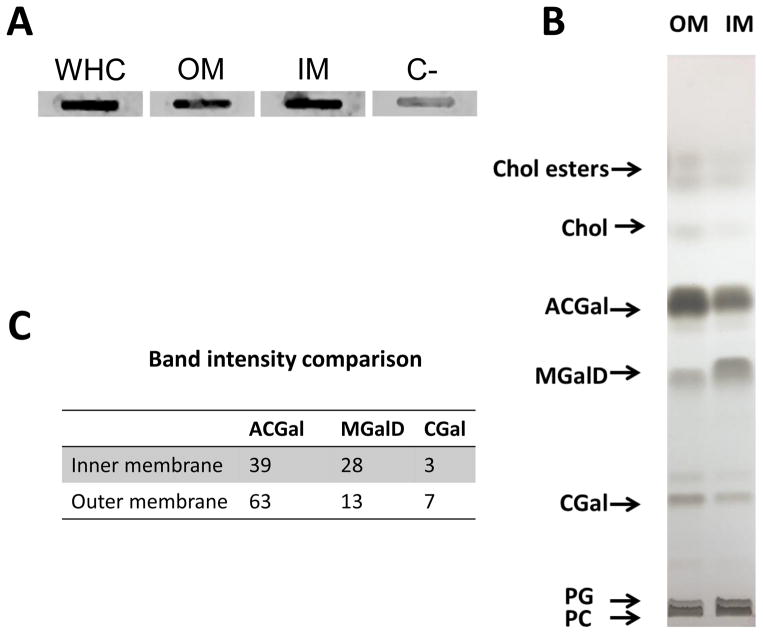
The presence of cholesterol glycolipids was assayed in both IM and OM fractions using an antibody to asialo GM1 (Figure 2A) (Garcia Monco et al., 1993, Garcia-Monco et al., 1995). In addition, total lipid extracts from both IM and OM fractions were subjected to TLC to separate the main lipids. Both the IM and OM fractions had the same lipids: including phosphatidylcholine (PC), phosphatidylglycerol (PG), mono-α-galactosyl-diacylglycerol (MGalD) cholesteryl-β-D-galactopyranoside (AGal) and cholesteryl 6-O-palmitoyl-β-D-galactopyranoside (ACGal), as well as free cholesterol and cholesterol esters (Figure 2B). However, the relative quantity of cholesterol glycolipids (ACGal and CGal) was higher in the OM whereas the IM was richer in MGalD (Figure 2C).
Figure 2.
Cholesterol and cholesterol glycolipids are present in both the inner and the outer membrane of B. burgdorferi. A. Detection of cholesterol glycolipids in the inner and outer membrane by slot blot using an antibody to asialo GM1. B. Thin liquid chromatography of total lipid extracts from the outer and the inner membrane of B. burgdorferi. C. Relative amount of ACGal, CGal and MGalD measured by densitometry. Numbers represent arbitrary units. Controls using secondary antibodies alone were non-reactive (not shown); C− = negative control.
Borrelia lipid extracts from the inner and outer membrane fractions do not form bilayers with equal levels of membrane order
Previous studies have shown the presence of cholesterol-lipid containing microdomains in B.burgdorferi in vitro and in vivo (LaRocca et al., 2010, LaRocca et al., 2013), and have evaluated the contribution of different Borrelia lipid components to domain formation (Huang et al., 2016). The finding that there are raft-associated proteins as well as cholesterol and cholesterol glycolipids in the IM led us to study whether rafts can also be formed from their lipids. To study the ordered state formation in bilayers formed from the IM and OM lipids, multilamellar vesicles (MLVs) subjected to mild sonication were prepared from total lipid extracts from both membranes. For comparison, DOPC was used to prepare vesicles that exist in a fully disordered state.
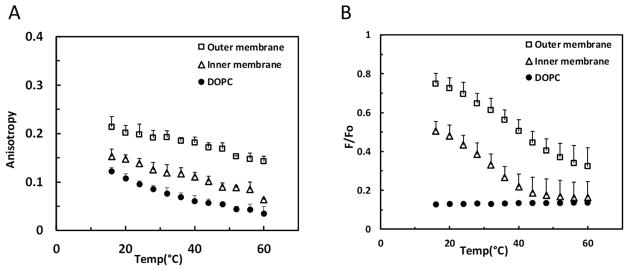
Membrane order was measured by DPH fluorescence anisotropy. The values for anisotropy are high (~0.3) in the solid-like gel and in liquid-ordered states, whereas the values for anisotropy are much lower in the liquid-disordered state (~0.05–0.15) (Gidwani et al., 2001). Figure 3A shows that vesicles made with IM and OM lipids showed a higher value of fluorescence anisotropy than the disordered state forming control vesicles formed from DOPC. The anisotropy values for the OM lipids were higher than those of the IM (Figure 3A) indicating that they have a higher degree of membrane order.
Figure 3.
Anisotropy and FRET analysis of membrane order and domain formation by inner and outer membrane lipids of B. burgdorferi. A. Anisotropy values of MLVs made of total lipids from the OM and IM. DOPC vesicles were used as a negative control. B. FRET values of MLVs made of total lipids from the OM and IM. DOPC vesicles were used as a negative (low membrane order) control. Error bars denote standard deviation from three biological replicates.
The higher degree of order in the vesicles formed from Borrelia membrane lipids could reflect the formation of ordered membrane domains, with the level of ordered domain formation being higher in the OM lipid vesicles than those formed from IM lipids. Alternately, the membranes could lack domains, and simply have different levels of order due to the presence of different levels of cholesterol lipids. The temperature dependence of anisotropy did not distinguish between these possibilities. Anisotropy values decrease as temperature increases because membrane disorder increases at elevated temperature (Gennis, 1989). Sometimes transitions between ordered and disordered states can be detected by a cooperative transition between a high and low anisotropy state as temperature increases, but that was not the case for vesicles formed from either IM or OM lipids.
Borrelia lipid extracts from the inner and outer membrane fractions have different abilities to form ordered domains
Because fluorescence anisotropy did not differentiate between homogenous membranes with a high degree of order and heterogeneous membranes where ordered and disordered domains co-exist, FRET was used to detect whether lipids from IM and OM fractions could segregate into co-existing ordered and disordered domains. Domains were detected by FRET using fluorescence probes that partition differentially into ordered and disordered domains, pyrene-DPPE and rhod-DOPE (Pathak & London, 2011). Because of partial segregation of pyrene-DPPE into ordered domains and partition of rhod-DOPE into disordered domains, donor fluorescence ratio in the presence of acceptor to that in its absence (F/Fo) is higher (i.e. FRET is weaker) in segregated bilayers than in homogenous bilayers. As temperature increases, domains melt and disappear, and the bilayer becomes homogeneous, increasing FRET (i.e. decreasing F/Fo). Figure 3B shows that vesicles composed of lipids from the IM and those composed of lipids from the OM both showed high values of F/Fo (weak FRET) relative to that in DOPC vesicles which do not form domains. In addition, donor fluorescence showed a sigmoidal temperature dependence characteristic of a thermal melting transition from a state in which ordered and disordered domains co-exist to one at high temperature in which the bilayer is in a homogenous disordered state. As judged from higher F/Fo values, the vesicles formed from the OM lipids appeared to have a greater degree of segregation into ordered and disordered domains than those formed from the IM lipids, i.e. exist with a higher fraction of the membrane in the form of ordered domains over the experimental temperature range, in agreement with the anisotropy results. It is also possible that the difference in FRET reflects larger domains in the OM samples or a greater difference in donor and acceptor partitioning into OM lipid domains. The transition temperature was higher for the vesicles prepared from OM lipids relative to those formed from IM lipids. This indicates that ordered domains were more thermally stable in vesicles formed from OM lipids. The influence of temperature on the physical state of the lipid rafts may have an important in vivo corollary on the ability of B. burgdorferi to persist in diverse environments.
Double bond content of inner and outer membrane lipids
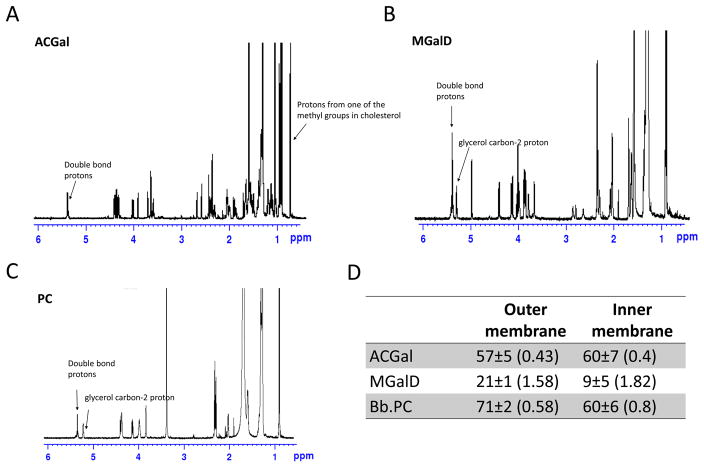
FRET and anisotropy analysis showed that the OM lipids formed vesicles with a higher level of order than the IM as well as a greater ability to form ordered domains. These properties are consistent with the higher level of cholesterol lipids in the vesicles prepared from OM lipids. However, this difference could also reflect a different level of acyl chain saturation in IM and OM lipids. Lipids having unsaturated acyl chains (acyl chains containing double bonds) have a decreased ability to form ordered domains relative to those with saturated acyl chains (London, 2002). The saturation of acyl chains is important for the formation of tightly packed ordered domains in eukaryotic cells (London, 2002), and in B. burgdorferi (Huang et al., 2016). Therefore, using 1H NMR we measured the number of double bonds in the three major Borrelia lipid species: PC, ACGal and MGalD.
The percentage of saturation for each lipid is showed in Figures 4A–C. There was no significant difference in saturation levels between ACGal from the IM and OM (Figure 4D). In contrast, the level of saturation of the acyl chains of MGalD and PC were significantly lower (p-value< 0.001) in the IM, reflecting a higher double bond ratio for these lipids (Figure 4D). This suggests these IM lipids have a decreased ability to form ordered domains relative to the corresponding lipids in the OM.
Figure 4.
Double bond content of outer and inner membrane major lipids. A. 1H-NMR spectrum of B. burgdorferi PC. B. 1H-NMR spectrum of B. burgdorferi ACGal. C. 1H-NMR spectrum of B. burgdorferi MGalD. D. Lower limit percent of saturated acyl chains in the outer and inner membranes; in parenthesis is shown the average double bonds per acyl chain. Error bars denote standard deviation from three replicates.
For outer membrane PC, the 71% estimate for lower limit to the percentage of saturated acyl chains gives an upper limit of 58% of PC molecules having one double bond. The lower limit is calculated by assuming that no acyl chains have more than one double bond. The actual percent of saturated acyl chain is slightly higher due to the presence of acyl chain with multiple double bonds. Prior studies indicate that only a small fraction of unsaturated acyl chains in Borrelia have multiple double bonds (Hossain et al., 2001). This means that 42% or more of the PC molecules have two fully saturated acyl chains. In contrast, there is an upper limit of 80% of PC molecules with one double bond in the IM, which could mean as little as 20% of inner membrane PC have two fully saturated acyl chains. For MGalD, there is a high enough level of double bonds (~1.6 double bonds per MGalD in the OM and ~1.8 in the IM MGalD) so that every MGalD could contain one or more double bonds.
The lipid rafts from the inner membrane and outer membrane have different associated proteins
In a previous study we characterized the lipid raft proteome of B. burgdorferi whole cells (Toledo et al., 2015). One of the most interesting findings was the detection of proteins predicted to be associated with the IM in the lipid raft fractions, which suggested that these could also exist in the IM. However, in our first proteome of lipid rafts using whole cells, we could not differentiate between proteins associated with the IM or OM.
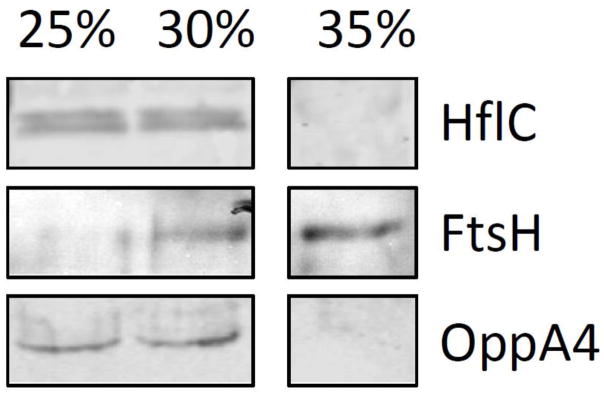
After purifying the IM and OMs, we treated them with Triton-X100 and separated the soluble and detergent resistant membrane fractions by ultracentrifugation in an iodixanol density gradient. The insoluble and soluble fractions from the IM were probed with antibodies against HflC, FtsH and OppAIV (Figure 5) demonstrating that HflC and OppAIV were associated with rafts. However, FtsH appeared to be present in insoluble and soluble fraction. To get an overall picture of the proteins present in both fractions, we compared them by mass spectrometry to find those that were enriched in the detergent resistant fraction (Table 1). The lipid rafts from the IM were rich in proteins associated with binding/transport (38.2%) (Figure S2A), especially members of the transport system superfamily of ATP-binding cassette (ABC) transporters (Table 1). Also proteins associated with metabolic (20%) and catabolic (10.9%) processes were found and to a lesser extend proteins associated with sensing and signaling (7.3%) and scaffold proteins (3.6%) (Figure S2A). Among these proteins were, HflC, HflK, FtsH, LysM, HtrA, Mcp-2 and Lon-1, proteins previously found highly enriched in lipid rafts in B. burgdorferi derived from whole cells. In addition, we analyzed in silico the predicted localization of the proteins found in lipid rafts in the IM fraction, finding that 89.1% of these were associated with this membrane or the periplasm (Figure S2B).
Figure 5.
Western blot showing the association of OppAIV and HflC with the detergent resistant membrane (DRMs) fractions. FtsH is only partially associated with DRM fractions by western blot. The 25%–30% of iodixanol represent the DRM fractions and the 35% of iodixanol represents the soluble fraction.
Table 1.
Proteome of inner membrane lipid rafts. Normalized spectra count of proteins in both the inner membrane lipid raft (DRM) and the soluble fractions for three technical replicates (TR).
| Name | Symbol | Gene | Lipid rafts inner membrane | Soluble fractions | T-test (p-value) | ||||
|---|---|---|---|---|---|---|---|---|---|
| TR1 | TR2 | TR3 | TR1 | TR2 | TR3 | ||||
| Uncharacterized protein | BB_0039 | 13.008 | 10.339 | 11.632 | 0 | 0 | 0 | 0.00011 | |
| Uncharacterized protein | BB_0058 | 6.0705 | 6.8924 | 4.9852 | 0 | 0 | 0 | 0.00041 | |
| V-type ATP synthase subunit B | atpB | BB_0093 | 18.211 | 17.231 | 16.617 | 0 | 0 | 0 | < 0.00010 |
| ATP synthase subunit A | BB_0094 | 11.274 | 9.477 | 11.632 | 3.6696 | 2.3942 | 3.6156 | 0.00065 | |
| Uncharacterized protein | BB_0103 | 5.2033 | 4.3077 | 4.9852 | 1.2232 | 0 | 0 | 0.00083 | |
| Periplasmic serine protease | HtrA | BB_0104 | 22.547 | 21.539 | 21.603 | 6.1159 | 5.9856 | 4.8208 | < 0.00010 |
| membrane fusion protein | BB_0141 | 6.0705 | 5.1693 | 6.6469 | 0 | 1.1971 | 0 | 0.00069 | |
| Protein translocase subunit SecA | BB_0154 | 9.5393 | 10.339 | 9.9704 | 0 | 0 | 0 | < 0.00010 | |
| guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase | spoT | BB_0198 | 4.336 | 5.1693 | 4.9852 | 0 | 0 | 0 | < 0.00010 |
| Protein HflK | hflk | BB_0203 | 4.336 | 6.8924 | 6.6469 | 0 | 0 | 0 | 0.0019 |
| protein HflC | HflC | BB_0204 | 6.9377 | 5.1693 | 6.6469 | 0 | 0 | 0 | 0.00034 |
| phosphate ABC transporter substrate-binding protein | pstS | BB_0215 | 10.407 | 10.339 | 9.1396 | 0 | 0 | 0 | < 0.00010 |
| Uncharacterized protein | BB_0228 | 15.61 | 16.369 | 15.786 | 0 | 0 | 0 | < 0.00010 | |
| Uncharacterized protein | BB_0238 | 13.875 | 13.785 | 13.294 | 2.4464 | 2.3942 | 2.4104 | < 0.00010 | |
| M23 peptidase domain-containing protein | BB_0246 | 3.4688 | 3.4462 | 3.3235 | 0 | 0 | 0 | < 0.00010 | |
| leucine--tRNA ligase | leuS | BB_0251 | 5.2033 | 6.8924 | 4.9852 | 0 | 0 | 0 | 0.0007 |
| ATP-dependent protease La | lon-1 | BB_0253 | 8.6721 | 9.477 | 9.9704 | 0 | 0 | 0 | < 0.00010 |
| transglycosylase SLT domain-containing protein | BB_0259 | 4.336 | 5.1693 | 4.9852 | 0 | 0 | 0 | < 0.00010 | |
| Uncharacterized protein | BB_0261 | 5.2033 | 4.3077 | 3.3235 | 0 | 0 | 0 | 0.0014 | |
| Uncharacterized protein | BB_0298 | 6.0705 | 6.8924 | 5.8161 | 1.2232 | 1.1971 | 2.4104 | 0.00084 | |
| Uncharacterized protein | BB_0319 | 11.274 | 9.477 | 11.632 | 0 | 0 | 1.2052 | 0.00018 | |
| LysM domain protein | LysM | BB_0323 | 16.477 | 17.231 | 15.786 | 2.4464 | 3.5914 | 3.6156 | < 0.00010 |
| peptide ABC transporter substrate-binding protein | BB_0330 | 39.892 | 42.216 | 44.867 | 11.009 | 11.971 | 8.4364 | < 0.00010 | |
| peptide ABC transporter ATP-binding protein | BB_0334 | 7.8049 | 8.6154 | 5.8161 | 1.2232 | 0 | 0 | 0.0016 | |
| peptide ABC transporter ATP-binding protein | OppF | BB_0335 | 6.0705 | 7.7539 | 9.1396 | 0 | 0 | 0 | 0.00099 |
| glutamyl-tRNA(Gln) amidotransferase subunit A | gatA | BB_0342 | 3.4688 | 4.3077 | 4.9852 | 0 | 0 | 0 | 0.00063 |
| proline--tRNA ligase | proS | BB_0402 | 3.4688 | 4.3077 | 4.1543 | 0 | 0 | 1.2052 | 0.0017 |
| membrane protein insertase | YidC | BB_0442 | 4.336 | 3.4462 | 3.3235 | 0 | 0 | 0 | 0.00032 |
| ABC transporter ATP-binding protein | BB_0466 | 3.4688 | 4.3077 | 3.3235 | 0 | 0 | 0 | 0.00027 | |
| Aldose reductase | BB_0528 | 6.0705 | 6.0308 | 5.8161 | 0 | 0 | 0 | < 0.00010 | |
| Long-chain-fatty-acid CoA ligase PE=4 SV=1 | BB_0593 | 31.22 | 31.016 | 30.742 | 11.009 | 7.1827 | 10.847 | < 0.00010 | |
| methyl-accepting chemotaxis protein | BB_0596 | 7.8049 | 5.1693 | 5.8161 | 0 | 0 | 0 | 0.0014 | |
| methyl-accepting chemotaxis protein | BB_0597 | 8.6721 | 8.6154 | 5.8161 | 0 | 0 | 0 | 0.0012 | |
| aminopeptidase | apeB | BB_0627 | 13.875 | 12.923 | 14.125 | 0 | 0 | 0 | < 0.00010 |
| PTS system glucose-specific transporter subunit IIBC | BB_0645 | 3.4688 | 3.4462 | 2.4926 | 0 | 0 | 0 | 0.00062 | |
| protein translocase subunit | SecD | BB_0652 | 6.0705 | 4.3077 | 6.6469 | 0 | 0 | 0 | 0.0013 |
| Lipoprotein | BB_0664 | 2.6016 | 2.5846 | 3.3235 | 0 | 0 | 0 | 0.00031 | |
| signal recognition particle protein | ffh | BB_0694 | 9.5393 | 6.8924 | 8.3087 | 0 | 0 | 0 | 0.00042 |
| Uncharacterized protein | BB_0751 | 5.2033 | 4.3077 | 4.1543 | 1.2232 | 0 | 0 | 0.0014 | |
| Uncharacterized protein | BB_0752 | 18.211 | 12.062 | 17.448 | 0 | 0 | 0 | 0.0012 | |
| ABC transporter ATP-binding protein | BB_0754 | 8.6721 | 9.477 | 9.1396 | 0 | 0 | 0 | < 0.00010 | |
| pyridoxal kinase | BB_0768 | 6.0705 | 6.8924 | 5.8161 | 0 | 0 | 0 | < 0.00010 | |
| ATP-dependent zinc metalloprotease FtsH | FtsH | BB_0789 | 9.5393 | 8.6154 | 9.9704 | 0 | 0 | 0 | < 0.00010 |
| excinuclease ABC subunit A | uvrA | BB_0837 | 6.0705 | 4.3077 | 5.8161 | 0 | 0 | 0 | 0.0006 |
| ornithine carbamoyltransferase | argF | BB_0842 | 6.0705 | 6.0308 | 5.8161 | 0 | 0 | 0 | < 0.00010 |
| arginine-ornithine antiporter | BB_0843 | 3.4688 | 3.4462 | 3.3235 | 0 | 0 | 0 | < 0.00010 | |
| PF-49 protein | BB_A21 | 4.336 | 3.4462 | 4.1543 | 0 | 0 | 0 | 0.00013 | |
| Outer membrane protein | BB_A52 | 6.9377 | 5.1693 | 4.9852 | 0 | 0 | 0 | 0.00079 | |
| oligopeptide ABC transporter OppAIV | OppAIV | BB_B16 | 16.477 | 16.369 | 19.11 | 0 | 0 | 0 | < 0.00010 |
| Protein BptA | bptA | BB_E16 | 4.336 | 5.1693 | 4.9852 | 0 | 0 | 0 | < 0.00010 |
| pyrazinamidase/nicotinamidase PncA | PncA | BB_E22 | 8.6721 | 8.6154 | 9.1396 | 0 | 0 | 0 | < 0.00010 |
| protein coding | PF32 | BB_F24 | 3.4688 | 3.4462 | 2.4926 | 0 | 0 | 0 | 0.00062 |
| antigen P35 | P35 | BB_J41 | 9.5393 | 10.339 | 7.4778 | 1.2232 | 1.1971 | 2.4104 | 0.0013 |
| Uncharacterized protein | BB_K13 | 15.61 | 17.231 | 18.279 | 1.2232 | 1.1971 | 2.4104 | < 0.00010 | |
| PF-49 protein | BB_N33 | 2.6016 | 2.5846 | 3.3235 | 0 | 0 | 0 | 0.00031 | |
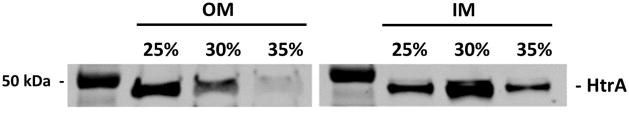
Similarly, we identified the proteins enriched in lipid rafts in the OM fraction by mass spectrometry and we compared them with those found in lipid rafts in the IM (Table 2). The OM was enriched with known proteins including OspA, OspB, P66 among others (Table 2). Proteins such as GroEL and HtrA were found in both the IM and OM fractions, but while HtrA was found associated with lipid rafts in both fractions (Figure 6), GroEL was only found enriched in lipid rafts in the OM fraction. Likewise, components of the flagella apparatus were found in both fractions but enriched only in the DRMs of the OM fraction.
Table 2.
Proteome of outer membrane lipid rafts. Normalized spectra count of proteins of the outer membrane DRM and soluble fractions for three technical replicates (TR).
| Name | Symbol | Gene | DRM | Soluble fraction | T-test (p-value) | ||||
|---|---|---|---|---|---|---|---|---|---|
| TR1 | TR2 | TR3 | TR1 | TR2 | TR3 | ||||
| Outer membrane protein P13 | P13 | BB_0034 | 62.712 | 57.696 | 59.003 | 13.052 | 13.451 | 12.824 | <0.0001 |
| Periplasmic serine protease | HtrA1 | BB_0104 | 58.853 | 64.427 | 68.988 | 0 | 0 | 0 | <0.0001 |
| Flagellar filament 41 kDa | flaB | BB_0147 | 198.75 | 212.51 | 214.23 | 67.118 | 72.941 | 67.997 | <0.0001 |
| Uncharacterized protein | BB_0227 | 9.648 | 7.6928 | 8.1696 | 0 | 0 | 0 | <0.0001 | |
| Integral outer membrane protein P66 | p66 | BB_0603 | 163.05 | 157.7 | 155.22 | 74.09 | 74.175 | 73.564 | <0.0001 |
| Pts system, fructose-specific iiabc component | BB_0629 | 3.8592 | 1.9232 | 5.4464 | 0 | 0 | 0 | <0.0001 | |
| 60 kDa chaperonin | GroEL2 | BB_0649 | 212.26 | 204.82 | 195.16 | 40.81 | 42.597 | 47.972 | <0.0001 |
| Flagellar filament | flaA | BB_0668 | 46.31 | 46.157 | 44.479 | 7.3872 | 6.282 | 7.9031 | <0.0001 |
| Antigen P83/100 | P83/P100 | BB_0744 | 29.909 | 30.771 | 31.771 | 0 | 0 | 0 | <0.0001 |
| Uncharacterized protein | BB_0840 | 5.7888 | 7.6928 | 5.4464 | 0 | 0 | 0 | 0.0008 | |
| Outer surface protein A | ospA | BB_A15 | 267.25 | 281.75 | 285.94 | 125.81 | 120.91 | 115.91 | <0.0001 |
| Outer surface protein B | ospB | BB_A16 | 716.85 | 712.54 | 682.62 | 143.08 | 132.21 | 137.67 | <0.0001 |
| Uncharacterized protein | BB_A41 | 12.542 | 10.578 | 9.9851 | 0 | 0 | 0 | 0.0001 | |
| Uncharacterized protein | BB_A46 | 9.648 | 10.578 | 10.893 | 0 | 0 | 0 | <0.0001 | |
| Complement regulator-acquiring surface protein 1 | cspA | BB_A68 | 13.507 | 12.501 | 14.524 | 1.4621 | 1.3432 | 1.1447 | <0.0001 |
| ErpA protein | erpA | BB_P38 | 4.824 | 4.808 | 4.5387 | 0 | 0 | 0 | <0.0001 |
HtrA been found associated with the inner and outer membrane lipid rafts.
GroEl is only associated with lipid rafts in the outer membrane but is found in both membrane fractions.
Figure 6.
Western blot showing the association of HtrA with the detergent resistant membrane (DRMs) and soluble fractions in the OM and IM.
Overall the diversity of proteins in lipid rafts in the IM is greater and are mainly associated with transport (Table 1), while in the OM are found the most abundant lipid raft proteins including OspA, OspB and GroEL (Table 2).
Discussion
In this study, we present biochemical and biophysical evidence that lipid rafts exist in the IM of B. burgdoferi. The lipid composition of the IM and OMs was similar and both had cholesterol glycolipids ACGal and CGal. Despite the differences in the quantity of ACGal and CGal between both membranes, the high level of cholesterol lipids was by itself a strong indicator for the potential to form lipid rafts in the IM of B. burgdorferi. The biophysical data obtained by anisotropy and FRET was consistent with the presence of lipid rafts in both membrane fractions. Nonetheless, the vesicles made from OM lipids had consistently higher values of fluorescence anisotropy, indicating that the OM is, in comparison, more ordered than the IM. Similarly, vesicles from the OM lipids had a higher degree of segregation into domains as assayed by FRET than those from the IM lipids, indicating that lipids from the OM have a higher ability to promote raft formation. This could be due, at least in part, to the higher quantity of ACGal, in the OM, because it appears to be the main lipid component inducing ordered-domain formation in B. burgdorferi (Huang et al., 2016). The higher degree of saturation level of the acyl chains of lipids from the OM could also contribute to their ability to support raft formation to a greater degree than IM lipids, since tight acyl chain packing promoted by lipid saturation is a key feature of lipid raft organization (Brown & London, 1998, Ahmed et al., 1997). Sphingolipids, which are key components of lipid rafts in eukaryotes, have largely saturated acyl chains that allow them to pack tightly together. In B. burgdorferi it has been shown that PC, at least partly, takes the place of sphingolipids in lipid rafts (Huang et al., 2016), probably due to its relative high level of acyl chain saturation (Huang et al., 2016, Hossain et al., 2001). Thus, the higher level of PC saturation in the outer membrane could contribute to the higher ability of outer membrane lipids to form lipid rafts.
The proteomes of lipid rafts from both membranes were characterized separately. The lipid rafts of the IM had a greater diversity of associated proteins, especially proteins with transport and signaling functions. Of these, ABC transporters were prominent in the IM. The presence of ABC transporters in the IM lipid raft fraction may not be coincidental as these transporters co-localize with cholesterol-rich microdomains in eukaryotes (Orlowski et al., 2007). A number of biological functions have been examined at the ABC transporter-lipid raft interface in eukaryotic cells (Neumann et al., 2017). Importantly, in bacteria with two membranes, ABC transporters are associated with the IM because they transport molecules from periplasm to cytosol. Despite finding several ABC transporters we did not find permeases, which are intregral membrane proteins, in the proteome of the IM (neither in the rafts nor in the soluble fraction); probably due to sensitivity limitations.
In contrast, the lipid rafts of the OM had more abundant proteins, notably lipoproteins OspA and OspB and GroEL. Some proteins were found in the lipid rafts from both membranes. This included, HtrA, which is not a surprise since HtrA is present in different subcellular locations including the IM, the periplasmic space and the OM (Russell et al., 2013, Coleman et al., 2013), and also has been found in vesicles as well as soluble extracellularly. Another interesting example was GroEL, which is primarily a cytoplasmic chaperone but is also found associated with the OM (Nowalk et al., 2006, Scopio et al., 1994). The role that GroEL plays in the OM is not known but our results indicate that is associated with rafts in the OM. We also found by mass spectrometry and western blot that HflC and HflK were in lipid rafts in the IM. HflC and HflK are known to modulate the activity of FtsH, which is required to infect the tick vector and the mammalian host as well as for in vitro growth (Chu et al., 2016), and was also found in lipid rafts. FtsH seems to be associated with rafts through the binding to HflC and HflK since it can also be found in soluble fraction by western blot. This observation is consistent with HflC and HflK having a role in regulating associated-proteins in rafts by either recruiting them (such as FtsH) or by facilitating the degradation of raft-associated proteins through the action of FtsH. Both HflC and HflK as well as other proteins with SPFH domains share several features, including subcellular sorting of proteins (Browman et al., 2007). Although the mechanism or the motifs by which proteins are recruited into rafts are unclear, these membrane domains seems to provide a platform for protein recruitment (Langhorst et al., 2005). DRMs are not synonymous with lipid rafts, but this approach is universally used to define the proteome of lipid rafts in eukaryotic cells (Kim et al., 2009, Insenser et al., 2006) and are useful to determine the protein composition of these structures (Brown, 2006).
In bacteria, it has been shown that flotillin homologs play an important role in microdomain formation (Hutton et al., 2017, Langhorst et al., 2005, Lopez & Kolter, 2010). Nonetheless, B. burgdorferi lacks a homolog for flotillin, thus it is possible that other SPFH proteins could be playing this role. Based on this observation and the fact that HflC and HflK are the only SPFH proteins is B. burgdorferi, they are good potential candidates to do so.
Based on these protein profiles, it seems likely that, lipid rafts in the OM play a role in environmental adaptation, whereas lipid rafts in the IM are associated with protein trafficking and signaling. As B. burgdorferi persist in diverse environments, tick mid gut and vertebrate host, the variation of lipid rafts in different milieus may be important to understand signaling processes that facilitate the transition of the spirochete from the tick vector to the vertebrate host and vice versa. Along these lines, this study reveals novel structural complexity in the lipid rafts of the IM and OM of B. burgdorferi. This complexity also extends to possible and different functional roles for this spirochete. The existence of cholesterol-based lipid rafts in this prokaryote suggests an evolutionary continuity for these structures.
Experimental procedures
Bacteria, cultures, antibodies, lipids and fluorescent probes
B. burgdorferi B31-A3 was grown in BSK-II media supplemented with 6% rabbit serum (Sigma-Aldrich), at 33°C. Escherichia coli was grown in Luria-Bertani broth at 37°C in a shaking incubator. A murine IgG1 monoclonal antibody to OspA (Coleman & Benach, 1992), a rabbit antibody to asialo GM1 ganglioside (Abcam, Cambridge UK), a rat monoclonal antibody to OppAIV (Mulay et al., 2007), a rabbit polyclonal antibody to FtsH (Chu et al., 2016) and a rabbit polyclonal antibody to HflC (Chu et al., 2016) were used for western blot and slot blot applications. Additional markers for quality control of fractions used murine monoclonal antibodies to OspB (Coleman et al., 1992); to FlaB (Coleman & Benach, 1989); to Lp6.6 (Katona et al., 1992); and DnaK (Coleman & Benach, 1987) were used to detect proteins in western blots of IM and OM fractions. Likewise, rabbit polyclonal antibodies to P66, Lon protease (Coleman et al., 2009); and HtrA (Coleman et al., 2013) were used for the same purpose.
For Förster resonance energy transfer (FRET) experiments, the following probes were used: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (rhod-DOPE) from Avanti Polar Lipids; and 2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(1-pyrenesulfonyl) from Molecular Probes, Inc., For anisotropy experiments 1,6-diphenyl-1,3,5-hexatriene (DPH) from Sigma-Aldrich was used. Borrelia lipids extracted from the IM and OMs, and 1,2-dioleoyl-phosphatidylcholine (DOPC) (Avanti Polar) were used to make vesicles as described below. Lipids and probes stock solutions were dissolved in chloroform and stored at −20°C. Concentration of fluorescent lipids were quantified from absorbance in methanol using εpyrene-DPPE=35,000 M−1cm−1 at 350 nm, εrhod-DOPE =95,000 M−1cm−1 at 560 nm, εDPH=84,800 M−1cm−1 at 352 nm (Haugland, 2002).
Isolation of B. burgdoferi outer and inner membranes
The IM and OMs of B. burgdoferi B31-A3 were separated by incubating Borrelia in the presence of 25 mM citrate buffer (pH 3,2) in agitation for 2 h at room temperature and isolated using a discontinuous sucrose gradient followed by a continuous sucrose density gradient protocol as previously described (Skare et al., 1995, Lenhart & Akins, 2010). The purity of the IM and OM fractions was assessed by western blot using antibodies to the outer membrane protein OspA (Barbour et al., 1983), the cytoplasmic protein DnaK (Tilly et al., 1993) and the inner membrane protein OppAIV (Brooks et al., 2006, Mulay et al., 2007). The IM and OM samples were used for thin-layer chromatography (TLC), slot blots, western blots, FRET and anisotropy applications or further treated, as described below, to isolate lipid rafts.
Detection of lipids in the outer and inner membranes
Lipids from the IM and OM were isolated following the Bligh and Dyer extraction protocol (Bligh & Dyer, 1959). The lipid extracts were dried under a nitrogen gas stream, quantified by dry weight, dissolved in chloroform and spotted on a high-performance TLC plate (EM Separations), multiple lanes with lipids from the outer and inner membranes were loaded onto each plate. After equilibrating the TLC chamber with a mobile phase of chloroform-methanol (85:15 v:v) for 15 min. the lipids were chromatographed and purified as previously described (Huang et al., 2016). The lipid mobility patterns of cholesterol, cholesterol glycolipids, and MGalD of B. burgdorferi have been studied and validated exhaustively in our previous publications (Huang et al., 2016, LaRocca et al., 2010, LaRocca et al., 2013). Lipid concentration was measured by dry weight and then, more accurately, by nuclear magnetic resonance (NMR) using anthracene as a reference standard as previously described (Huang et al., 2016) to prepare solutions with an estimated ~800 μM lipid. To do this, lipids and anthracene were dissolved in deuterated chloroform and then the 1H-NMR spectra of lipids were acquired on a Bruker Ascend™ 850 Mhz spectrometer with a 30° pulse, a delay time between acquisitions of 10 s to avoid saturation, and 32 acquisitions. The areas of lipid peaks corresponding to known numbers of protons per molecule were then compared to the area of anthracene proton peaks to calculate lipid concentration.
Model membrane vesicle preparation
Model membrane vesicles were made at a concentration of 50 μM total lipid by sonication. Briefly, lipids from the IM and OM were combined with fluorescent probes in a glass tube and dried for 10 min under nitrogen; samples were then transferred to a high vacuum and dried for 1 h. The dried lipids were heated to 70°C for 5 min and mixed with preheated (70°C) PBS (pH 7.8). The samples were sonicated in a bath sonicator (Laboratory Supplies) for 5 min or until the lipids appeared homogenously dispersed and then cooled to room temperature as described previously (Huang et al., 2016).
Temperature dependence of fluorescence anisotropy
IM or OM lipid vesicles were prepared at a concentration of 50 μM with 0.1 mol % DPH (mixed with the lipid in organic solvent) as described above. Fluorescence anisotropy was measured in a SPEX FluoroLog 3 spectrofluorometer (Jobin-Yvon), equipped with an automated polarization apparatus, using quartz semi-microcuvettes (excitation path length, 10 mm; emission path length, 4 mm). Anisotropy vs. temperature was measured by increasing temperature at 4°C intervals from 16°C to 60°C as described previously (Huang et al., 2016) in three separate experiments. Lipids from two different membrane preparations gave similar results.
Temperature dependence of FRET
Model membrane vesicles for FRET experiments were prepared by sonication as described above. The “F samples” were vesicles composed of a mixture IM lipids, OM lipids or other unlabeled lipids, plus a FRET donor and a FRET acceptor. “Fo samples” had the same unlabeled lipids and FRET donor but lacked the FRET acceptor. The F background samples were the same as F samples but lacked the FRET donor, while Fo background samples were the same as F samples except they lacked both the FRET donor and acceptor. The donor/acceptor pair used for FRET was 0.1 mol% pyrene-DPPE/5 mol % rhodamine-DOPE. The final samples contained unlabeled lipid at a concentration of 50μM dispersed in PBS. The fluorescence of the samples was measured as function of increasing temperature from 16°C to 60°C at 4°C intervals in a SPEX FluoroLog 3 spectrofluorometer as previously described (Huang et al., 2016) in three separate experiments. Lipids from two different membrane preparations gave similar results.
Lipid unsaturation level measurement by 1H-NMR
Dried lipids were mixed with deuterated chloroform, and then 1H-NMR spectra of lipids were acquired on the Bruker Ascend™ 850 MHz spectrometer at 850 MHz with a 30° pulse, using a delay time between acquisitions of 10s to avoid signal saturation, and 32 acquisitions. The areas of lipid peaks corresponding to known numbers of protons per molecule were used as the reference signal calculation (For PC and MGalD, a single proton attached to glycerol carbon 2, which has a chemical shift at about 5.2ppm; for ACGal, three protons from one of the methyl groups in cholesterol, which has a chemical shift close to 0.7ppm). The intensity (area) of the double bonds proton signal at chemical shift 5.5ppm were compared to the area of reference proton peaks in order to calculate unsaturation levels in terms of the number of double bonds per lipid molecule. Note that there is one double bond proton from sterol in ACGal in the double bond peak, this was subtracted to calculate the number of double bonds in acyl chains for ACGal.
Isolation and analysis of detergent resistant membranes (DRM) of B. burgdorferi
DRMs from the IM and OM were isolated by detergent insolubility using Triton X-100, followed by gradient separation using the caveola/raft isolation kit and following the manufacturer’s instructions (Sigma). The fractions were precipitated using trichloroacetic acid (TCA) as previously described (Toledo et al., 2015) and used for proteomic analysis or for western blot.
Immunoblots
Proteins from IM and OM samples were separated by electrophoresis (12.5% SDS-PAGE) and transferred to nitrocellulose membranes (GE Healthcare). Murine monoclonal antibodies were used to detect OspA and DnaK, a rat monoclonal was used to detect OppAIV, and FtsH and HflC were detected by a rabbit polyclonal antiserum. The immunoblots were resolved with goat anti-rabbit IgG IR800; goat anti-mouse IgG IR800; or anti-rat IgG IR800 and were read in an Odyssey scanner (Li-Cor Biosciences) in the 800-nm channel. For slot blots, whole cell B. burgdorferi lysate, its IM and OMs, and E. coli lysate were diluted in phosphate-buffered saline (PBS), applied to nitrocellulose membrane in a PR-648 slot blot apparatus (Hoefer), and blocked with 5% nonfat dry milk. Cholesterol glycolipids were detected using a rabbit antibody to asialo-GM1 that recognizes cholesterol glycolipids as described previously (Toledo et al., 2014). Slot blots were read in an Odyssey scanner in the 800-nm channel. Class-specific secondary antibodies were used alone as controls (no primary) to detect possible nonspecific reactivity in all immunoblot experiments.
Trypsin digestion
Samples were dissolved in 8 M urea and 0.1 M NH4HCO3, reduced with 4 mM DTT and then alkylated with 8.4 mM iodoacetamide. The proteins were subsequently digested with trypsin (Trypsin Gold, Mass Spectrometry Grade, Promega) at a 25:1 protein:trypsin mass ratio, incubating for 16 h at 37 °C. The digests were brought to 2% formic acid (FA) and desalted with Supel-Tips C18 Micropipette Tips (Sigma-Aldrich) using FA containing solutions with varied acetonitrile (ACN) essentially as described in vendor’s bulletin. The solvent was removed from the eluted peptides using a vacuum centrifuge and the resultant dried peptide was stored at −80 °C. The eluted peptides were dissolved in 2% ACN, 0.1% FA (buffer A) for analysis by automated microcapillary liquid chromatography-tandem mass spectrometry (LC/MS/MS).
LC/MS/MS
Fused-silica capillaries (100 μm inner diameter - i.d.) were pulled using a P-2000 CO2 laser puller (Sutter Instruments,) to a 5 μm i.d. tip and packed with 10 cm of 5 μm ProntoSil 120-5-C18H (Bischoff Chromatography) using a pressure bomb. The samples were loaded via an Dionex WPS-3000 autosampler, part of an Dionex Ultimate 3000 system (Germering). The column was installed in-line with a Dionex LPG-3000 Chromatography HPLC pump running at 300 nL min−1. The peptides were eluted from the column by applying a 5 min linear gradient from 0% buffer B (98% ACN, 0.1% FA) to 10% buffer B, followed by a 120 min linear gradient from 10% buffer B to 45% buffer B. The gradient was switched from 45% to 80% buffer B over 10 min. Finally, the gradient was changed from 80 % buffer B to 0 % buffer B over 10 min, and then held constant at 0 % buffer B for 20 more minutes. The application of a 2.2 kV distal voltage electrosprayed the eluting peptides directly into an LTQ Orbitrap XL ion trap mass spectrometer (Thermo Fisher) equipped with a nano-liquid chromatography electrospray ionization source. Full mass spectra (MS) were recorded on the peptides over a 400 to 2000 m/z range at 60,000 resolution, followed by top-five MS/MS scans in the ion-trap. Charge state dependent screening was turned on, and peptides with a charge state of +2 or higher were analyzed. Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Fisher, San Jose, CA). MS/MS spectra were extracted from the RAW file with ReAdW.exe (http://sourceforge.net/projects/sashimi). The resulting mzXML data files were searched with GPM X!Tandem against a recent Unitprot Borrelia burgdorferi (strain ATCC 35210/B31/CIP 102532/DSM 4680) proteome database. The data was also analyzed and collated with Scaffold 4 (Proteome Software, Inc).
Database searching
Tandem mass spectra were extracted from the RAW file with ReAdW.exe (http://sourceforge.net/projects/sashimi). Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version X! Tandem Sledgehammer (2013.09.01.2)). X! Tandem was set up to search the BORBU_b31_cc database (2914 entries) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 20 PPM. Carbamidomethyl of cysteine was specified in X! Tandem as a fixed modification. Glu->pyro-Glu of the n-terminus, ammonia-loss of the n-terminus, gln->pyro-Glu of the n-terminus, deamidated asparagine and glutamine, oxidation of methionine and tryptophan and dioxidation of methionine and tryptophan were specified in X! Tandem as variable modifications.
Criteria for protein identification
Scaffold (version Scaffold_4.7.5, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm (Keller et al., 2002) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
The normalized spectra count was obtained by normalizing the total spectra counts at the MS level using Scaffold (version Scaffold_4.7.5, Proteome Software Inc., Portland, OR). The normalized spectral abundance factor (NSAF) value was obtained as previously described (Zybailov et al., 2006). NSAF values for each protein were compared in the insoluble fraction to those in the soluble fraction to determine which proteins were enriched in lipid rafts in the inner membrane using a t-test. Enriched proteins in the lipid raft fraction vs. non-raft fraction with a P-value less than 0.0015 were included in Table 1. To determine which proteins were enriched in the OM relative to the IM, the proteome of lipid rafts from both OM and IM were compared. Proteins enriched in the OM relative to the IM, and with a P-value less than 0.05 were included in Table 2.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD007904.
Database analysis
Uniprot identifiers were assigned based on mass spectroscopy. Gene ontology (GO) terms and protein FASTA sequences were derived from Uniprot (http://www.uniprot.org/). The GO was not available for many of the proteins identified, therefore CELLO (Yu et al., 2004) and Psort (Yu et al., 2010) were used to predict the protein subcellular localization.
Supplementary Material
Figure S1. Western blot showing the partitioning of antigens in the inner membrane (IM) and outer membrane (OM). Both IM and OM were probed with a battery of monoclonal and polyclonal antibodies to antigens with known locations in the cell. These included HtrA that is found in both IM and OM fractions; OspB, P66, and Lp6.6 which are enriched in the OM; and Lon protease, DnaK, and Flab which partition on the IM only. GM1 = antibody to asialo GM1 ganglioside to detect glycolipids.
Figure S2. Predicted biological processes (A) and subcellular localization (B) of raft-associated proteins in the inner membrane.
Acknowledgments
Antibody to OppAIV was generously provided by Drs. Justin Radolf and Melissa Caimano, University of Connecticut Health Center, Farmington. Antibodies to HflC and FtsH antibodies were generously provided by Drs Philip Stewart and Patricia Rosa from the Rocky Mountain Laboratories (NIH). This work was supported by NIH grants AI-125806-01 to A.T; GM-122493 to E.L; AI-027044 to J.L.B. The authors have no conflict of interest to declare.
References
- Ahmed SN, Brown DA, London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Tessier SL, Todd WJ. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Habicht GS, Bast TF, Cameron DJ, Ziegler JL, Barbour AG, Burgdorfer W, Edelman R, Kaslow RA. Spirochetes isolated from the blood of two patients with Lyme disease. N Engl J Med. 1983;308:740–742. doi: 10.1056/NEJM198303313081302. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brandt ME, Riley BS, Radolf JD, Norgard MV. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect Immun. 1990;58:983–991. doi: 10.1128/iai.58.4.983-991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Vuppala SR, Jett AM, Akins DR. Identification of Borrelia burgdorferi outer surface proteins. Infect Immun. 2006;74:296–304. doi: 10.1128/IAI.74.1.296-304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Chu CY, Stewart PE, Bestor A, Hansen B, Lin T, Gao L, Norris SJ, Rosa PA. Function of the Borrelia burgdorferi FtsH Homolog Is Essential for Viability both In Vitro and In Vivo and Independent of HflK/C. MBio. 2016;7:e00404–00416. doi: 10.1128/mBio.00404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JL, Benach JL. Isolation of antigenic components from the Lyme disease spirochete: their role in early diagnosis. J Infect Dis. 1987;155:756–765. doi: 10.1093/infdis/155.4.756. [DOI] [PubMed] [Google Scholar]
- Coleman JL, Benach JL. Identification and characterization of an endoflagellar antigen of Borrelia burgdorferi. J Clin Invest. 1989;84:322–330. doi: 10.1172/JCI114157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JL, Benach JL. Characterization of antigenic determinants of Borrelia burgdorferi shared by other bacteria. J Infect Dis. 1992;165:658–666. doi: 10.1093/infdis/165.4.658. [DOI] [PubMed] [Google Scholar]
- Coleman JL, Crowley JT, Toledo AM, Benach JL. The HtrA protease of Borrelia burgdorferi degrades outer membrane protein BmpD and chemotaxis phosphatase CheX. Mol Microbiol. 2013;88:619–633. doi: 10.1111/mmi.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JL, Katona LI, Kuhlow C, Toledo A, Okan NA, Tokarz R, Benach JL. Evidence that two ATP-dependent (Lon) proteases in Borrelia burgdorferi serve different functions. PLoS Pathog. 2009;5:e1000676. doi: 10.1371/journal.ppat.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JL, Rogers RC, Benach JL. Selection of an escape variant of Borrelia burgdorferi by use of bactericidal monoclonal antibodies to OspB. Infect Immun. 1992;60:3098–3104. doi: 10.1128/iai.60.8.3098-3104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia M, Casal S, Vinagre J, Seruca R, Figueiredo C, Touati E, Machado JC. Helicobacter pylori’s cholesterol uptake impacts resistance to docosahexaenoic acid. Int J Med Microbiol. 2014;304:314–320. doi: 10.1016/j.ijmm.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Crowley JT, Toledo AM, LaRocca TJ, Coleman JL, London E, Benach JL. Lipid exchange between Borrelia burgdorferi and host cells. PLoS Pathog. 2013;9:e1003109. doi: 10.1371/journal.ppat.1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Monco JC, Seidman RJ, Benach JL. Experimental immunization with Borrelia burgdorferi induces development of antibodies to gangliosides. Infect Immun. 1995;63:4130–4137. doi: 10.1128/iai.63.10.4130-4137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Monco JC, Wheeler CM, Benach JL, Furie RA, Lukehart SA, Stanek G, Steere AC. Reactivity of neuroborreliosis patients (Lyme disease) to cardiolipin and gangliosides. J Neurol Sci. 1993;117:206–214. doi: 10.1016/0022-510x(93)90175-x. [DOI] [PubMed] [Google Scholar]
- Gennis RB. Molecular Structure and Function. Springer-Verlag New York; New York: 1989. p. 533. [Google Scholar]
- Gidwani A, Holowka D, Baird B. Fluorescence anisotropy measurements of lipid order in plasma membranes and lipid rafts from RBL-2H3 mast cells. Biochemistry. 2001;40:12422–12429. doi: 10.1021/bi010496c. [DOI] [PubMed] [Google Scholar]
- Haugland RP. Handbook of Fluorescent Probes and Research Products. Eugene, OR: Molecular Probes; 2002. [Google Scholar]
- Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J Bacteriol. 1995;177:5327–5333. doi: 10.1128/jb.177.18.5327-5333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain H, Wellensiek HJ, Geyer R, Lochnit G. Structural analysis of glycolipids from Borrelia burgdorferi. Biochimie. 2001;83:683–692. doi: 10.1016/s0300-9084(01)01296-2. [DOI] [PubMed] [Google Scholar]
- Huang Z, London E. Cholesterol lipids and cholesterol-containing lipid rafts in bacteria. Chem Phys Lipids. 2016;199:11–16. doi: 10.1016/j.chemphyslip.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Toledo AM, Benach JL, London E. Ordered Membrane Domain-Forming Properties of the Lipids of Borrelia burgdorferi. Biophys J. 2016;111:2666–2675. doi: 10.1016/j.bpj.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton ML, D’Costa K, Rossiter AE, Wang L, Turner L, Steer DL, Masters SL, Croker BA, Kaparakis-Liaskos M, Ferrero RL. A Helicobacter pylori Homolog of Eukaryotic Flotillin Is Involved in Cholesterol Accumulation, Epithelial Cell Responses and Host Colonization. Front Cell Infect Microbiol. 2017;7:219. doi: 10.3389/fcimb.2017.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insenser M, Nombela C, Molero G, Gil C. Proteomic analysis of detergent-resistant membranes from Candida albicans. Proteomics. 2006;6(Suppl 1):S74–81. doi: 10.1002/pmic.200500465. [DOI] [PubMed] [Google Scholar]
- Katona LI, Beck G, Habicht GS. Purification and immunological characterization of a major low-molecular-weight lipoprotein from Borrelia burgdorferi. Infect Immun. 1992;60:4995–5003. doi: 10.1128/iai.60.12.4995-5003.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim KB, Kim BW, Choo HJ, Kwon YC, Ahn BY, Choi JS, Lee JS, Ko YG. Proteome analysis of adipocyte lipid rafts reveals that gC1qR plays essential roles in adipogenesis and insulin signal transduction. Proteomics. 2009;9:2373–2382. doi: 10.1002/pmic.200800811. [DOI] [PubMed] [Google Scholar]
- Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca TJ, Crowley JT, Cusack BJ, Pathak P, Benach J, London E, Garcia-Monco JC, Benach JL. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe. 2010;8:331–342. doi: 10.1016/j.chom.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca TJ, Pathak P, Chiantia S, Toledo A, Silvius JR, Benach JL, London E. Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog. 2013;9:e1003353. doi: 10.1371/journal.ppat.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun AH, Wunder C, Hildebrand J, Churin Y, Zahringer U, Lindner B, Meyer TF, Heinz E, Warnecke D. Cloning of a cholesterol-alpha-glucosyltransferase from Helicobacter pylori. J Biol Chem. 2006;281:27765–27772. doi: 10.1074/jbc.M603345200. [DOI] [PubMed] [Google Scholar]
- Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol Microbiol. 2010;75:692–709. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore BP, Bey RF, Johnson RC. Lipid metabolism of Borrelia hermsi. Infect Immun. 1978;20:215–220. doi: 10.1128/iai.20.1.215-220.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London E. Insights into lipid raft structure and formation from experiments in model membranes. Curr Opin Struct Biol. 2002;12:480–486. doi: 10.1016/s0959-440x(02)00351-2. [DOI] [PubMed] [Google Scholar]
- Lopez D, Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr Opin Microbiol. 2017;36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010;24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay V, Caimano MJ, Liveris D, Desrosiers DC, Radolf JD, Schwartz I. Borrelia burgdorferi BBA74, a periplasmic protein associated with the outer membrane, lacks porin-like properties. J Bacteriol. 2007;189:2063–2068. doi: 10.1128/JB.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Neumann J, Rose-Sperling D, Hellmich UA. Diverse relations between ABC transporters and lipids: An overview. Biochim Biophys Acta. 2017;1859:605–618. doi: 10.1016/j.bbamem.2016.09.023. [DOI] [PubMed] [Google Scholar]
- Nickels JD, Chatterjee S, Stanley CB, Qian S, Cheng X, Myles DAA, Standaert RF, Elkins JG, Katsaras J. The in vivo structure of biological membranes and evidence for lipid domains. PLoS Biol. 2017;15:e2002214. doi: 10.1371/journal.pbio.2002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowalk AJ, Nolder C, Clifton DR, Carroll JA. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics. 2006;6:2121–2134. doi: 10.1002/pmic.200500187. [DOI] [PubMed] [Google Scholar]
- Orlowski S, Comera C, Terce F, Collet X. Lipid rafts: dream or reality for cholesterol transporters? Eur Biophys J. 2007;36:869–885. doi: 10.1007/s00249-007-0193-8. [DOI] [PubMed] [Google Scholar]
- Ostberg Y, Berg S, Comstedt P, Wieslander A, Bergstrom S. Functional analysis of a lipid galactosyltransferase synthesizing the major envelope lipid in the Lyme disease spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 2007;272:22–29. doi: 10.1111/j.1574-6968.2007.00728.x. [DOI] [PubMed] [Google Scholar]
- Ourisson G, Rohmer M. Hopanoids. 2. Biohopanoids: a novel class of bacterial lipids. Accounts of Chemical Research. 1992;25:403–408. [Google Scholar]
- Pathak P, London E. Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation. Biophys J. 2011;101:2417–2425. doi: 10.1016/j.bpj.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Bourell KW, Akins DR, Brusca JS, Norgard MV. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J Bacteriol. 1994;176:21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Goldberg MS, Bourell K, Baker SI, Jones JD, Norgard MV. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect Immun. 1995;63:2154–2163. doi: 10.1128/iai.63.6.2154-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S. Cholesterol uptake is dependent on membrane fluidity in mycoplasmas. Biochim Biophys Acta. 1978;513:401–404. doi: 10.1016/0005-2736(78)90208-0. [DOI] [PubMed] [Google Scholar]
- Razin S, Efrati H, Kutner S, Rottem S. Cholesterol and phospholipid uptake by mycoplasmas. Rev Infect Dis. 1982;4(Suppl):S85–92. doi: 10.1093/clinids/4.supplement_1.s85. [DOI] [PubMed] [Google Scholar]
- Russell TM, Delorey MJ, Johnson BJ. Borrelia burgdorferi BbHtrA degrades host ECM proteins and stimulates release of inflammatory cytokines in vitro. Mol Microbiol. 2013;90:241–251. doi: 10.1111/mmi.12377. [DOI] [PubMed] [Google Scholar]
- Saenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M, Simons K. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci U S A. 2015;112:11971–11976. doi: 10.1073/pnas.1515607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopio A, Johnson P, Laquerre A, Nelson DR. Subcellular localization and chaperone activities of Borrelia burgdorferi Hsp60 and Hsp70. J Bacteriol. 1994;176:6449–6456. doi: 10.1128/jb.176.21.6449-6456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare JT, Shang ES, Foley DM, Blanco DR, Champion CI, Mirzabekov T, Sokolov Y, Kagan BL, Miller JN, Lovett MA. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J Clin Invest. 1995;96:2380–2392. doi: 10.1172/JCI118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PF. Biosynthesis of cholesteryl glucoside by Mycoplasma gallinarum. J Bacteriol. 1971;108:986–991. doi: 10.1128/jb.108.3.986-991.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Hauser R, Campbell J, Ostheimer GJ. Isolation of dnaJ, dnaK, and grpE homologues from Borrelia burgdorferi and complementation of Escherichia coli mutants. Mol Microbiol. 1993;7:359–369. doi: 10.1111/j.1365-2958.1993.tb01128.x. [DOI] [PubMed] [Google Scholar]
- Toledo A, Crowley JT, Coleman JL, LaRocca TJ, Chiantia S, London E, Benach JL. Selective association of outer surface lipoproteins with the lipid rafts of Borrelia burgdorferi. MBio. 2014;5:e00899–00814. doi: 10.1128/mBio.00899-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo A, Perez A, Coleman JL, Benach JL. The lipid raft proteome of Borrelia burgdorferi. Proteomics. 2015;15:3662–3675. doi: 10.1002/pmic.201500093. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander PV, Hunter RC, Zhang L, Sessions AL, Summons RE, Newman DK. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol. 2009;191:6145–6156. doi: 10.1128/JB.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Lin M, Rikihisa Y. Cholesterol-dependent anaplasma phagocytophilum exploits the low-density lipoprotein uptake pathway. PLoS Pathog. 2009;5:e1000329. doi: 10.1371/journal.ppat.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Western blot showing the partitioning of antigens in the inner membrane (IM) and outer membrane (OM). Both IM and OM were probed with a battery of monoclonal and polyclonal antibodies to antigens with known locations in the cell. These included HtrA that is found in both IM and OM fractions; OspB, P66, and Lp6.6 which are enriched in the OM; and Lon protease, DnaK, and Flab which partition on the IM only. GM1 = antibody to asialo GM1 ganglioside to detect glycolipids.
Figure S2. Predicted biological processes (A) and subcellular localization (B) of raft-associated proteins in the inner membrane.