Summary
Leptospires and other members of the evolutionarily ancient phylum of Spirochaetes are bacteria often characterized by long, highly motile spiral- or wave-shaped cells. Morphology and motility are critical factors in spirochete physiology, contributing to the ability of these bacteria to successfully colonize diverse environments. However, the mechanisms conferring the helical structure of Leptospira spp. have yet to be fully elucidated. We have identified 5 L. biflexa bactofilin proteins, a recently characterized protein family with cytoskeletal properties. These 5 bactofilins are conserved in all species of the Leptospiraceae, indicating that these proteins arose early in the evolution of this family. One member of this protein family, LbbD, confers the optimal pitch distance in the helical structure of L. biflexa. Mutants lacking lbbD display a unique compressed helical morphology, a reduced motility, and a decreased ability to tolerate cell wall stressors. The change in the helical spacing, combined with the motility and cell wall integrity defects, showcases the intimate relationship and coevolution between shape and motility in these spirochetes.
Summary
Spirochetes are a diverse group of pathogenic and free-living bacteria that share an unusual spiral- or wave-form morphology, but relatively few components of cell shape have been identified. Here, we demonstrate that one protein influences both morphology and motility, indicating that both characteristics have evolved together to optimize spirochete movement.
Introduction
Spirochaetes are a diverse and evolutionarily ancient branch of eubacteria (Paster et al., 1991), with many having medical and veterinary relevance, including Treponema pallidum (syphilis), Borrelia spp. (Lyme disease and relapsing fever), Brachyspira hyodysenteriae (swine dysentery), and pathogenic Leptospira spp. (leptospirosis). Other members of this phylum are free living saprophytes, which can inhabit environments as disparate as Antarctica (Franzmann & Dobson, 1992) and hot springs (Patel et al., 1985). Traditionally, the spiral or wave-shaped morphology and internal periplasmic flagella were characteristic of the phylum Spirochaetes and used for classification.
Spirochete motility is driven by the periplasmic flagella, which function both in motility and as cytoskeletal elements conferring shape on the cells (Wolgemuth et al., 2006). Motility is particularly important to the life cycle of both pathogenic and free-living spirochetes, allowing these bacteria to bore though dense media, be it sediment or tissue, to rapidly move throughout their environment. Their quick dispersion is an important virulence factor in the pathogenic spirochetes, and many become avirulent with the loss of their flagella (Rosey et al., 1996, Sultan et al., 2013, Wunder et al., 2016). The flagella also contribute to cell shape, conferring the flat wave morphology in Borrelia burgdorferi (Motaleb et al., 2000), the irregular helix in Treponema denticola (Ruby et al., 1997), and the characteristic hooked ends in Leptospira (Picardeau et al., 2001).
Because the flagella play such a significant role in spirochete morphology, shape and motility in spirochetes are very closely related. Yet despite the significance of morphology to various aspects of spirochete physiology and life cycle, relatively little is known about how shape is determined in Leptospira cells. Peptidoglycan and the actin homolog MreB both contribute to the helical shape of leptospires (Slamti et al., 2011). Cytoplasmic and periplasmic filaments have been visualized in L. interrogans, but their function remains unknown (Malmstrom et al., 2009, Raddi et al., 2012). While some contributing factors in Leptospira architecture have been identified, other components, and the nature of the interactions between components, remain to be defined.
L. biflexa is a free living, saprophytic species of the Leptospiraceae family with a helical structure and hooked ends, typical of the genus. Relative to many spirochetes, L. biflexa cells are fast growing and comparatively easy to genetically manipulate, thus making this organism a good model for structural studies. Previously, we generated a proteomic map of L. biflexa (Stewart et al., 2015) and identified a bactofilin homolog, a family of proteins demonstrated to contribute to cell shape in other bacteria (Kuhn et al., 2010). Bactofilins are cytoskeletal proteins, defined as containing the conserved DUF583 domain (synonymous with DUF342) (Kuhn et al., 2010). Widely dispersed in bacteria and cytoskeletal in nature, the functions of bactofilins have not been characterized in spirochetes, but in bacteria they have consistent roles in morphology and are often associated with cell curvature. In some bacteria, including Proteus mirabilis and Myxococcus xanthus, cells lacking bactofilins become curled (Hay et al., 1999) or kinked (Koch et al., 2011), respectively. In Helicobacter pylori, however, bactofilin-deficient cells straighten (Sycuro et al., 2012). Many bacteria have more than one bactofilin, ranging from one to six per species (Kuhn et al., 2010). Interestingly, despite being highly conserved and widely distributed, bactofilins do not appear to be essential. In many cases, the observed phenotype is conferred by a single, predominant bactofilin (BacA in Caulobacter crescentus and BacM in M. xanthus, e.g.), while the loss of the other bactofilins does not appear to have an obvious affect.
In 2014, Vasa et al. determined that bactofilins have an unusual β-helical architecture (Vasa et al., 2015). A combination of solid-state NMR and scanning electron microscopy demonstrated that bactofilins form filamentous polymers through the interactions of the DUF583 domains, producing a rigid β-helical core. Though this structure has not been reported in other bacterial cytoskeletal filaments, it is similar to that of the fungal prion protein HET-s (Wasmer et al., 2008). This three-faced structure allows bactofilins to form the triangular polymeric fibers that act as cytoskeletal elements.
Due to their consistent roles in bacterial morphology, and our previous proteomic work identifying a leptospiral homolog, we sought to characterize the contribution of bactofilins to the morphology and physiology of L. biflexa. Here, we identified five bactofilin genes in L. biflexa and characterized LEPBI_I1431, which we termed LbbD (for Leptospira biflexa bactofilin D). We show that LbbD contributes to the periodic spacing of the cell helix, the integrity of the cell wall, and to motility, suggesting that L. biflexa morphology and motility evolved together to confer optimal shape and movement.
Results
Bactofilins are well conserved in the Leptospiraceae family
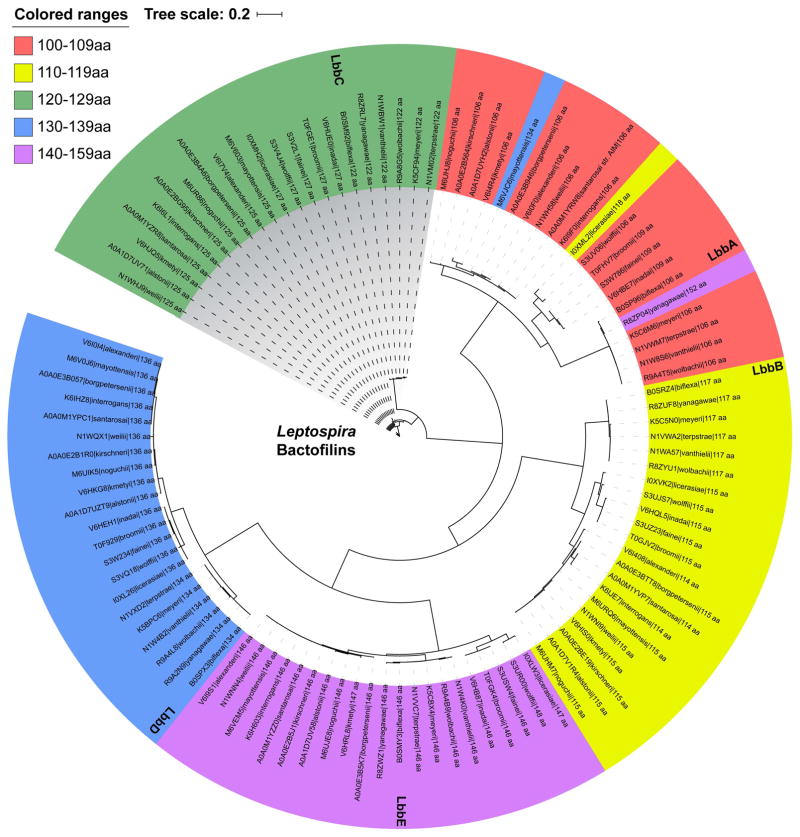
Five bactofilins were identified in the L. biflexa genome based on sequence similarity using NCBI BLAST (Altschul et al., 1990). The bactofilins range in size from 106–146 amino acids (Table 1), with relatively low amino acid identity (Fig. S1) but maintaining a conserved tertiary structure of beta-helices (DUF342). Further BLAST searching revealed that all sequenced Leptospira spp. have five genes encoding bactofilins (Fig. 1), which can be subdivided into protein families based on amino acid similarity and size. The closely related bacteria Turneriella parva and Leptonema illini have 5 and 6 bactofilin genes, respectively. Leptospiraceae bactofilins cluster more closely by size and amino acid sequence across the family than they do within individual species, suggesting that bactofilins arose early in the Leptospira lineage and are highly conserved. We termed the bactofilins of L. biflexa LbbA-E, for Leptospira biflexa bactofilins A-E, ranked by increasing number of amino acids (Table 1). LbbD was the only bactofilin identified from a previous proteomic mapping experiment (Stewart et al., 2015) and we selected this protein for further characterization of its contribution to L. biflexa morphology and physiology.
Table 1.
L. biflexa bactofilins
| UniProt Identifier | Name | Number of Amino Acids | kDa | % Similarity |
|---|---|---|---|---|
| LEPBI_I1310 | LbbA | 106 | 11.4 | 51.1 |
| LEPBI_I3470 | LbbB | 117 | 12.7 | 49.1 |
| LEPBI_I2643 | LbbC | 122 | 12.8 | 42.1 |
| LEPBI_I1431 | LbbD | 134 | 14.2 | 100 |
| LEPBI_I1047 | LbbE | 146 | 15.9 | 55.1 |
Percent similarity to LbbD (bold) was determined by the Jotun Hein alignment method.
Fig. 1. Bactofilins are conserved throughout the Leptospira.
Bactofilins from each sequenced Leptospira spp. were compared by amino acid similarity. Phylogenetic distances were estimated with the BLOSUM45 matrix and is an indication of amino acid changes per site. The InterPro database accession number, species name, and the number of amino acids of each Leptospira bactofilin is provided.
Generation of L. biflexa strains
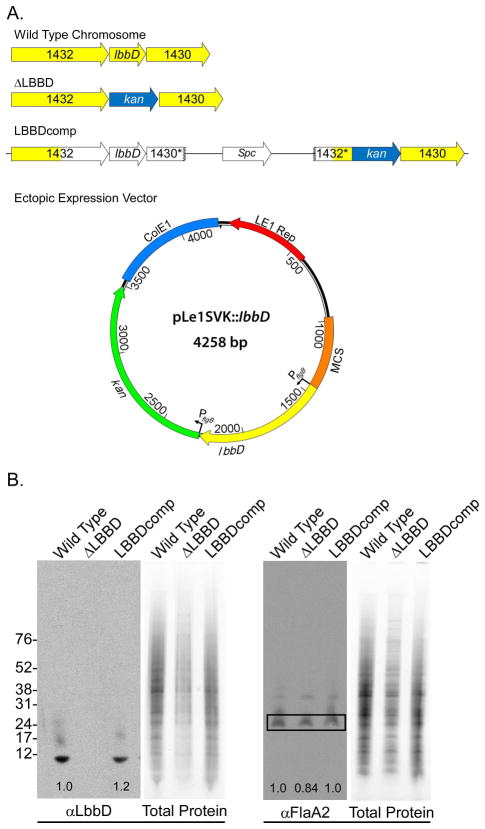
An lbbD deletion mutant was engineered using allelic replacement with a kanamycin-resistance cassette and designated ΔLBBD (Fig. 2A). The presence of the kanamycin-resistance gene and absence of lbbD were confirmed by PCR and by Southern blot analysis (data not shown). The lack of protein production was also confirmed by immunoblot with an αLbbD antibody (Fig. 2B). Given the low sequence conservation between L. biflexa bactofilin proteins (Fig. S1), the αLbbD antibody appears to be mono-specific. Genetic complementation of lbbD was engineered by integration of the E. coli plasmid vector, containing lbbD and a spectinomycin-resistance cassette, at the native locus (Fig. 2A). The complemented strain, LBBDcomp, was confirmed by PCR to contain the restored lbbD gene, and production of LbbD was demonstrated by immunoblot (Fig. 2B). Quantitative immunoblots demonstrated that the levels of the flagellar protein FlaA2 were similar in all strains (Fig. 2B). Ectopic expression of the bactofilin was achieved by cloning lbbD under the control of the constitutive B. burgdorferi flgB promoter on the shuttle vector pLe1SVK (described in the Experimental Procedures section), and the resulting strain was named SV1LBBD. The presence of pLe1SVK:: lbbD in L. biflexa transformants was confirmed by Southern blot analysis (data not shown).
Fig. 2. Genetic structure and protein production of bactofilins from wild type and mutant strains.
A. Genetic structure of the wild type, lbbD mutant, complement, and SV1LBBD (ectopic expression of lbbD). In the complementation strain, LBBDcomp, the yellow-colored arrows represent chromosomal loci and open arrows denote DNA derived from integration of the E. coli complementation vector. * indicates partial genes formed by the insertion of the complementing plasmid. The shuttle vector expressing lbbD contains both a leptospiral (LE1 Rep) and an E. coli (ColE1) origins of replication, both lbbD (yellow arrow) and the kanamycin-resistance cassette (green arrow) are fused to borrelial flgB promoters. MCS, multiple cloning site
B. Quantitation of LbbD and FlaA2 levels in L. biflexa strains. Antiserum against purified LbbD and FlaA2 was used in quantitative immunoblots standardized to the total protein load of each lane and the results are shown as numbers (relative to wild type levels) at the base of each blot.
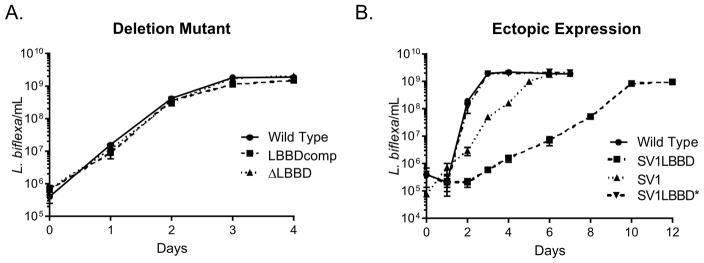
Ectopic-expression of LbbD reduces growth rate
The ΔLBBD strain grew at the same rate as wild type and complemented strains in liquid culture (Fig. 3A). In contrast, the ectopic-expression strain grew significantly slower than the control strain SV1, which contains only the shuttle vector (Fig. 3B). However, the shuttle vector lacks any active partitioning system and is itself unstable, requiring cultivation in the presence of antibiotic selection, which appears to impart a minor growth defect. When grown without antibiotic, both strains lose the shuttle vector, as confirmed by Southern blot analysis (data not shown). An isolate in which the shuttle vector expressing lbbD was lost (strain SV1LBBD*) had a wild type growth rate (Fig 3B), further supporting that ectopic expression of lbbD reduced the growth rate. The SV1LBBD strain does not survive long-term storage at −80° C, likely due to cell lysis, while SV1LBBD* and SV1 strains survive the same storage conditions. Over-expression of C. crescentus bactofilins also resulted in cell lysis (Kuhn et al., 2010).
Fig. 3. Ectopic expression of lbbD affects L. biflexa growth rate.
A. Growth rates of the wild type, ΔLBBD mutant and LBBDcomp strains. The growth rate slopes of each strain are not statistically different from wild type as determined by linear regression analysis.
B. Growth rates of the the lbbD expressing strain SV1LBBD, the control strain SV1 containing pLe1SVK only, and cells that had lost the expression plasmid (SV1LBBD*) compared to wild type. Data represent the mean ± SD calculated from triplicate independent cultures. Wild type and SV1LBBD* strains were grown in the absence of antibiotic selection, whereas the other strains were grown in the presence of kanamycin. The growth rate slopes of wild type and SV1LBBD* are not statistically different from one another, but those of SV1 and SV1LBBD are different from wild type (p=0.0016), as determined by linear regression analysis.
LbbD contributes to cell morphology and motility
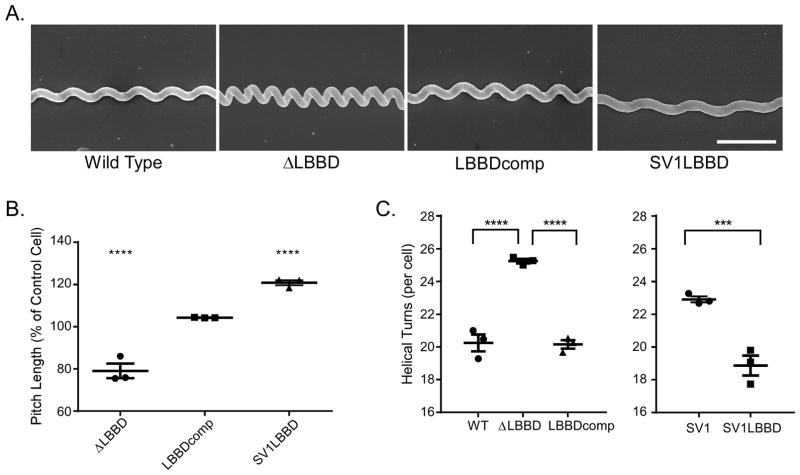
Because bactofilins influence cell morphology, we assessed the contribution of LbbD to various aspects of Leptospira morphology including cell length and helical pitch distance. Although cell length was similar in the wild type, ΔLBBD, and LBBDcomp strains (Fig. 4, left panel), the distance between consecutive helices (helical pitch) revealed a significant shortening of the helical pitch in lbbD mutant cells relative to wild type, and this distance was restored in the complemented strain (Fig. 5A). The helical pitch distance in spirochetes lacking LbbD decreased by an average of 20% compared to wild type (Fig. 5B), giving the cells a distinctive compressed morphology. However, the ΔLBBD spirochetes have an average of 5 more helices per cell than wild type (Fig. 5C); these additional coils in the deletion mutant thus confer the appearance of similar cell lengths between strains (Fig. 4, left panel).
Fig. 4. Ectopic expression but not loss of LbbD affects cell length.
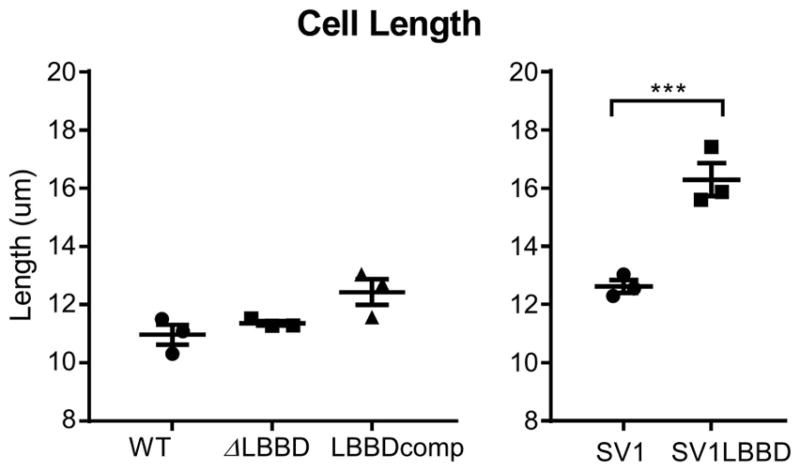
Individual spirochete lengths were measured by live cell imaging under darkfield microscopy with a 40x objective. SV1LBBD and shuttle vector control strains were grown in the presence of kanamycin to ensure retention of the plasmid and are not comparable to the wild type, mutant or complemented strains, which were grown without antibiotic selection. Each data point represents the average length determined from 15 cells and calculated from three biological replicates; shown are the mean ± the standard error of the mean. Ordinary one-way ANOVA was applied to determine whether differences were significant (*** denotes P value < 0.001).
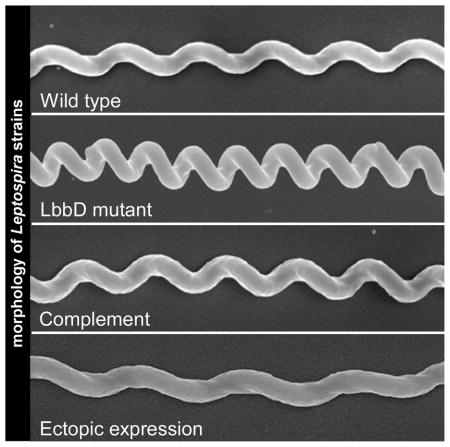
Fig. 5. Cells lacking LbbD display an altered helical morphology.
A. Scanning electron micrographs exemplifying the different helical morphologies of various L. biflexa strains. All micrographs are shown to the same scale, and the scale bar in the SV1LBBD panel represents 1 μm.
B. LbbD affects average pitch length between helices in L. biflexa. The average pitch lengths of each strain are displayed relative to control strains. The percent of pitch distance of ΔLBBD and LBBDcomp are compared to that of wild type, and the SV1LBBD pitch length is compared to that of the SV1 strain (shuttle vector only). The pitch lengths were measured by determining the distance between peak-to-peak and trough-to-trough of each helix. Six pitch measurements were taken per cell and averaged, and each point on the graph represents the averaged measurements of 23–64 cells. Three biological replicate samples were measured per strain. Mean and standard error of the mean are shown.
C. The ΔLBBD cells have 5 more helical turns per cell than wild type, while the SV1LBBD strain (ectopically-expressed lbbD) has ~4 helical turns less than control cells containing the shuttle vector (SV1). Each point represents the number of helical turns averaged from 15 cells. Three biological replicates were measured. Mean and standard error of the mean are shown. Ordinary one-way ANOVA was applied to determine the statistical analysis in all graphs (*** denotes P value < 0.001, **** denotes P value < 0.0001).
Ectopic expression of lbbD resulted in a significant (p<0.001) increase in cell length compared to control cells (Fig. 4, right panel) and produced cells with greater helical pitch distances, increasing the distance between helices by an average of 20% relative to cells containing only the shuttle vector (Fig. 5B). Furthermore, even though SV1LBBD cells are longer (Fig. 4, right panel), they have an average of 4 less helices than the shuttle vector-only cells (Fig. 5C). Although ectopic expression of lbbD affects multiple phenotypes, we were unable to demonstrate higher levels of the bactofilin protein by quantitative immunoblot.
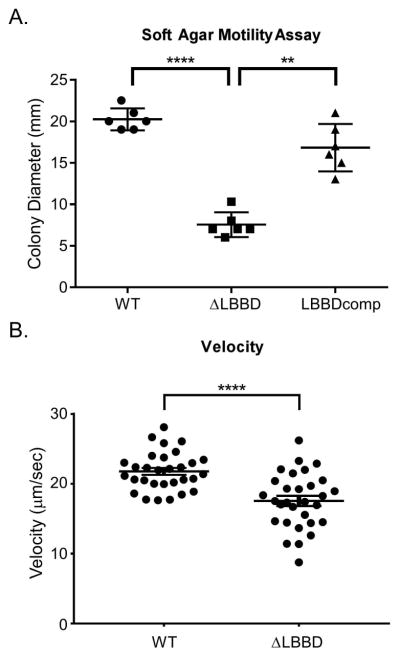
In spirochetes, structure and motility are intimately linked. Because we noticed such a distinctive change in morphology, we assessed the impact of the mutation on spirochete motility. Soft agar motility assays were conducted with wild type, ΔLBBD, and ΔLBBDcomp cells. In six biological replicate plates, the ΔLBBD cells formed significantly smaller colonies than did the wild type and ΔLBBDcomp (Fig. 6A), suggesting that deletion of lbbD results in impaired motility. However, it is possible that the motility phenotype in soft agar is due to an impaired chemotactic response and therefore does not migrate far in the soft agar motility assays. We further assessed the motility of the ΔLBBD strain by measuring the velocity of individual WT and mutant cells in 0.5 % methyl cellulose (Fig. 6B). The ΔLBBD cells were significantly slower than WT cells, further supporting the contribution of LbbD to optimal motility. Visualization of ΔLBBD cells in the presence of the viscosity agent methyl cellulose indicated they were motile and indistinguishable from the movement of wild type cells (Figs. S2 and S3). Combined with the FlaA2 quantitative immunoblot, the data indicate that the impaired motility phenotype does not appear to be due to a defect in the assembly of the flagellar machinery, as was observed in Bacillus subtilis (El Andari et al., 2015).
Fig. 6. Loss of LbbD reduces spirochete motility.
A. Colony size in soft agar plates was measured in the wild type, ΔLBBD, and LBBDcomp strains following a 7-day incubation period. Each point represents a biological replicate and mean and standard deviation bars are shown. Statistical significance was determined using Repeated Measurements one-way ANOVA. (** denotes P value < 0.01, **** denotes P value < 0.0001).
B. Velocity of spirochete strains in 0.5% methyl cellulose. Maximum speeds of individual cells were measured in three independent experiments and statistical significance was calculated using an unpaired t test with Welch’s correction (P < 0.0001). Mean and standard error of the mean are shown.
Cell wall integrity is weakened in spirochetes lacking LbbD
The deletion of BacM in M. xanthus led to an increased sensitivity to cell wall-targeting antibiotics (Koch et al., 2011). Therefore, we compared the 3 L. biflexa strains to both cell wall-targeting antibiotics and osmotic stress. We determined the minimum inhibitory concentrations to Amdinocillin, a drug that specifically targets Penicillin Binding Protein-2 (PBP-2), and to A22, which inhibits ATP binding to MreB (Bean et al., 2009). The MICs were determined in quadruplicate (Table 2). In all cases, ΔLBBD was almost twice as sensitive to cell wall-targeting antibiotics than wild type, indicating that LbbD contributes to cell wall stability. Surprisingly, the complemented strain did not restore resistance to amdinocillin to full wild type levels, which may relate to the large insertion of plasmid DNA at the lbbD locus (Fig. 2A) that might subtly alter the transcription rate.
Table 2.
Susceptibility of wild type, ΔLBBD, and LBBDcomp cells to cell wall targeting antibiotics
| A22 | Amdinocillin | |
|---|---|---|
| Wild Type | 10 μg/ml | 12.5 ± 1.0 μg/ml |
| ΔLBBD | 6.3 ± 0.5 μg/ml (p<0.0001) | 7 μg/ml (p<0.0001) |
| LBBDcomp | 9.0 μg/ml (ns) | 10 μg/ml ± 2.0 (p=0.0037) |
Susceptibility was calculated from four biological MIC replicates ± the standard deviation. Statistics were determined with 2-way ANOVA with Tukey’s multiple comparisons test with all strains compared to wild type. ns = not significantly different.
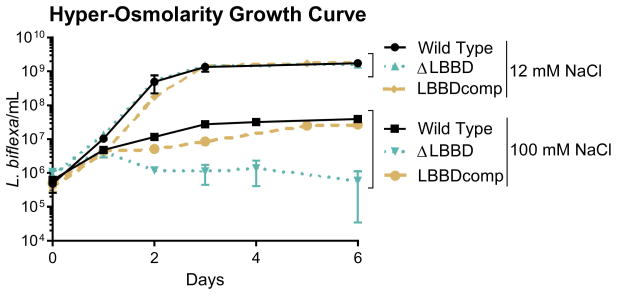
Cell wall integrity was further assessed by comparing growth of L. biflexa strains in hyper-osmotic conditions (100 mM NaCl) to the standard EMJH formulation containing 12 mM NaCl. Although all strains grew to similar high densities in standard EMJH media (>109 cells/mL), growth in hyper-osmotic EMJH medium significantly reduced growth of all 3 strains. Wild type and complemented strains attained densities of ~4 x107 L. biflexa/mL, while the ΔLBBD mutant was affected more severely, reaching only ~5x106 L. biflexa/mL before declining (Fig. 7). Together, the antibiotic and osmotic sensitivies of the LbbD mutant indicate that this bactofilin is an important component contributing to cell wall stability.
Fig. 7. The ΔLBBD mutant is more sensitive to osmotic stress than the wild type or complemented strains.
Growth rates of wild type, ΔLBBD, and LBBDcomp strains cultivated in standard EMJH (12 mM NaCl) and in hyper-osmotic EMJH containing 100 mM NaCl. Data represent the mean ± the standard deviation calculated from triplicate independent cultures. Linear regression analysis indicates that only the hyper-osmotic growth rate slopes are significantly different from wild type (ΔLBBD, p=0.0319; LBBDcomp, p=0.0311).
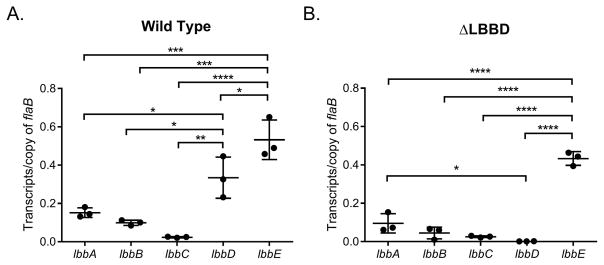
lbbD and lbbE are the most abundant bactofilin transcripts in L. biflexa
The transcript levels of the five bactofilin genes were determined by performing qRT-PCR on RNA isolated from log phase, in vitro-cultivated wild type L. biflexa. Gene lbbE was the most highly expressed bactofilin, followed by lbbD (Fig. 8A). The other three bactofilins were expressed at comparatively low levels. To determine if any of the bactofilins increased in expression to compensate for the deletion of lbbD, qRT-PCR was also performed on RNA isolated from the ΔLBBD mutant. As expected, lbbD dropped significantly, but the transcript levels of the other bactofilins did not change (Fig. 8B), indicating that bactofilin gene synthesis in L. biflexa does not compensate for the loss of an important component. Comparison of the transcript levels of the same genes between the strains did not indicate any significant differences, except for the expected change in lbbD, which was undetectable in the mutant (data not shown). We also examined the transcript levels of the genes flanking lbbD, lb1430 and lb1432, in both the wild type and ΔLBBD strains. Transcript levels from both genes bordered at the level of detection in each strain (data not shown), demonstrating that deletion of lbbD did not have polar effects on the surrounding genes.
Fig. 8. Bactofilin transcript levels do not change to compensate for the loss of lbbD transcripts.
Quantitative RT-PCR analysis of the transcript levels of the five bactofilin genes in wild type and mutant strains. Transcript from each gene was normalized to 1 copy of flaB transcript. Each point represents a biological replicate (averaged from 3 technical replicates) from independent RNA preparations ± the standard deviation. Ordinary one-way ANOVA was applied to determine statistical differences (* denotes P value < 0.05, ** denotes P value < 0.01,*** denotes P value < 0.001, **** denotes P value < 0.0001).
Discussion
Historically, it was assumed that a cytoskeleton was unique to eukaryotic cells and did not play a role in bacterial shape. However, it is now known that bacteria contain homologs or counterparts to each of the major cytoskeletal systems found in eukaryotes (Cabeen & Jacobs-Wagner, 2005). FtsZ, a homolog of tubulin, plays a role in cell division and shape determination (Bi & Lutkenhaus, 1991). Non-spherical bacteria were found to contain MreB, a filamentous protein similar to actin (Jones et al., 2001), that is responsible for rod morphology (Doi et al., 1988). While there is not a single widely conserved homolog to intermediate filaments, counterparts have been identified in various bacterial lineages (Ausmees et al., 2003). Bactofilins are a recently discovered class of cytoskeletal protein that do not have identified homologs in eukaryotic or archaean genomes. Polymeric and filamentous, they have been shown to play a role in cell wall integrity, motility, and shape determination (Hay et al., 1999, Kuhn et al., 2010, Koch et al., 2011, Sycuro et al., 2012, El Andari et al., 2015). We recently identified a bactofilin homolog while mapping the proteome of L. biflexa (Stewart et al., 2015). Previously identified as uncharacterized protein B0SGS8 but currently annotated as B0SPX3 in the Uniprot database, and here referred to as LbbD, the bactofilin partitioned with the membrane-associated protein fraction.
As bactofilins contribute to cell morphology and are widely distributed among bacterial species (Kuhn et al., 2010), we performed homology searches and identified multiple bactofilins in every genera of spirochetes (data not shown), with all leptospires encoding 5 bactofilins (Fig. 1). These Leptospiraceae bactofilin proteins phylogenetically segregate into 5 families discernible by amino acid similarity and length. This conservation across all leptospires indicates that these 5 protein families predate the speciation of the genera, and each bactofilin is important enough to the fitness of the cell that they remain highly conserved. As LbbD was the only bactofilin observed in the 2-dimensional gel proteome mapping project, with the caveat that the mapping was not exhaustive (Stewart et al., 2015), we deleted the gene (Figs. 2A and B) and characterized the phenotype of the mutant during in vitro cultivation.
The absence of LbbD did not impact growth rate (Fig. 3A) or the overall cell length (Fig. 4, left panel), but the helical pitch compressed by an average of 20% (Fig. 5B). The ΔLBBD mutant contains, on average, 5 more coils per cell than wild type (Fig. 5C). This suggests that the true linear length of the mutant is longer (if the cells could be straightened), but the increased number of compressed helices confers a similar apparent cell length to that of wild type. The purified peptidoglycan sacculus of L. biflexa is inherently helical (Slamti et al., 2011) and likely conforms the shape of the protoplasmic cylinder into a spiral. However, LbbD appears to exert an outward force on the helix, expanding its pitch by 20%. This is further supported by the LbbD ectopic expression strain, where cell length is increased, again by 20%, compared to controls (Fig. 4, right panel).
Bactofilin inactivation mutants produce structural changes that, in turn, often result in motility defects. P. mirabilis loses the ability to swarm in the absence of its bactofilin, although single cells retain motility (Hay et al., 1999). In B. subtilis, bactofilins contribute to the assembly of the flagellar apparatus and bactofilin mutants exhibit a complete loss of swimming motility (El Andari et al., 2015). L. biflexa cells lacking LbbD also display a motility defect, with a decreased ability to move through the soft agar medium relative to wild type (Fig. 6A) and measurements of spirochete velocity were significantly slower for the bactofilin mutant than for the wild type (Fig. 6B). The reduced motility and the altered helical pitch of the mutant suggest that there is an optimal spacing of the L. biflexa helix that allows for maximum translation. Unlike the mutants of B. subtilis, the flagellar machinery of L. biflexa appears intact as quantitative immunoblot analysis of FlaA2 indicate similar protein levels between WT, mutant and complemented cell lysates (Fig. 2B). Furthermore, qRT-PCR analysis of WT and ΔLBBD strains for flaB transcript normalized to putative outer membrane protein B0SQ62 (Uniprot) demonstrated similar levels between strains (data not shown). This outer membrane protein was previously identified as an abundant membrane-associated protein with Uniprot accession B0SGK2 (Stewart et al., 2015). Finally, mutant cell morphology during translation in viscous media was similar to that of WT (Figs. S2 and S3). Together, the data demonstrate that motility is impaired in cells lacking LbbD, but we do not observe any defect in the flagellar machinery. Our data does not exclude the possibility that chemotaxis is also affected in the mutant strain.
The M. xanthus bactofilin BacM aids in maintaining the stability of the cell wall (Koch et al., 2011), while in C. crescentus two bactofilins mediate localization of peptidoglycan synthase to the stalked pole (Kuhn et al., 2010). H. pylori cells devoid of the bactofilin homolog lose the helical shape of wild type cells and increase the peptide crosslinking in the peptidoglycan sacculus (Sycuro et al., 2012). Interestingly, lbbD is flanked by genes encoding proteins with peptidoglycan-related functions: a multimodular transpeptidase-transglycosylase (penicillin binding protein 1A) and a peptidoglycan-specific endopeptidase. Although not definitive, previous cell fractionation experiments indicated that LbbD partitions with the membrane-associated proteins (Stewart et al., 2015), as do bactofilins in C. crescentus and P. mirabilis (Kuhn et al., 2010, Hay et al., 1999). Further experiments are required to confirm the spatial localization of LbbD. Cell wall integrity was weakened in the L. biflexa mutant, which was significantly impaired in its ability to cope with both osmotic stress and cell wall-targeting antibiotics (Fig. 7 and Table 2, respectively). These combined results for LbbD (contribution to cell wall integrity, genetic clustering with genes whose products are associated with cell wall synthesis and shape, and localization to the membrane fraction) further support the contribution of LbbD to cell wall stability in L. biflexa.
Finally, we determined the transcript levels of all 5 bactofilins during exponential growth of L. biflexa. Only lbbE was present in a greater amount than lbbD, while the other bactofilins were at comparatively low levels (Fig. 8). No changes in the relative transcript abundance of the other bactofilin genes was observed in the ΔLBBD strain, indicating that the other bactofilins do not increase to compensate for the loss of lbbD. Although we do not know the contribution of the other bactofilins to L. biflexa physiology, complementation experiments demonstrated that the observed phenotypes were due to the function of LbbD (Table 2, Figs. 5–7).
We propose a model in which LbbD acts as a structural support in L. biflexa, providing an outward force on the helical coils to counteract an inherent inward pressure, perhaps imposed by the peptidoglycan (Slamti et al., 2011) and/or other components of the cell wall. The intrinsic stability of bactofilin filaments may provide the rigidity necessary to maintain the proper spacing between helices in our model. The decrease in helical spacing in the ΔLBBD strain and the increase in the helical pitch of the ectopically-expressed lbbD cells supports this model. The opposing forces of LbbD and the peptidoglycan may produce the optimal spacing for the rotation of the periplasmic flagella or propagation of the wave generated by the flagella along the cell cylinder.
The motility and distinctive morphology of spirochetes allows for these microorganisms to quickly disseminate through viscous environments. We hypothesize that the L. biflexa bactofilin LbbD contributes to an optimized morphology that allows for more rapid motility. However, several intriguing avenues of investigation remain open: the role of the most highly transcribed bactofilin LbbE is unknown, as are the contributions of bactofilins to spirochetes with morphologies that differ from the leptospires, such as the planar wave-form of B. burgdorferi. The results presented here provide a platform for understanding the intimate co-evolution of morphology and motility in spirochetes.
Experimental procedures
Ethics statement
Animal experiments were conducted following guidelines from the National Institutes of Health with protocols approved by the Rocky Mountain Laboratories Animal Care and Use Committee. The Rocky Mountain Laboratories are accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Phylogenetic tree construction
The InterPro database (Hunter et al., 2009) was used to search for bacterial species possessing DUF583-containing proteins. Sequence information was taken from multiple strains and amino acid sequences were aligned with MAFFT version 7.245 (Katoh & Standley, 2013). The tree was inferred by approximately-maximum-likelihood methods implemented through FastTree version 2.1.8 (Price et al., 2010). This analysis utilized the Jones-Taylor-Thornton model of amino acid evolution (Jones et al., 1992) with the CAT model implemented to determine the evolutionary rate heterogeneity (Lartillot & Philippe, 2004), and local support values determined through the Shimodaira-Hasegawa test (Shimodaira & Hasegawa, 1999). The resulting tree was visualized with iTOL v3 (Letunic & Bork, 2016).
Bacterial strains
The sequenced organism L. biflexa serovar patoc strain Patoc I (Paris) was cultivated at 30°C in EMJH liquid media (Louvel & Picardeau, 2007) with shaking at 150 RPM. Plating media was made with 1.2% Noble Agar (final concentration) (BD Biosciences, San Jose, CA) and plates were incubated at 30°C, inverted. Kanamycin and spectinomycin, when needed, were added to a final concentration of 20 μg/mL. Plasmids were constructed in Escherichia coli strains TOP10 (ThermoFisher Scientific, Walthum, MA) or DE3-BL21 RIPL (Agilent Technologies, Santa Clara, CA).
Plasmid construction and L. biflexa transformations
L. biflexa cells were transformed using the electroporation protocol of Louvel and Picardeau (Louvel & Picardeau, 2007) and DNA was UV-treated (wavelength = 254nm) for 60 seconds prior to electroporation. GoTaq (Promega, Madison, WI), and proof-reading polymerases Vent (New England BioLabs, Ipswich, MA) and the Expand Long Template PCR System (Roche Applied Science, Indianapolis, IN) were used in fragment amplification. Fragments were confirmed by sequencing. All primers used in this study are listed in Supporting Information Table S1. The lbbD deletion mutant was constructed by allelic exchange with the kanamycin-resistance cassette (Fig. 2A) as follows: a 1,399 bp fragment consisting of lbbD and 441 base pairs upstream and 475 base pairs downstream was amplified from L. biflexa genomic DNA using primers Lb1431.Comp.NotI.F and Lb1431.Comp.NotI.Rev, cloned into pGEM (Promega), and designated pGEM::lbbD. Primers Lb1431.KO.SalI.F and Lb1431.KO.MluI.RC were used in an inverse PCR reaction using the Expand Long Template PCR System to delete lbbD. Primers Lb1431-Sal-pflgB.Fuse.F and Lb1431-Sal-Kan-fuse.RC were used to amplify the borrelial flgB promoter driving the kanamycin-resistance gene (Bono et al., 2000) from plasmid pBSV2 (Stewart et al., 2001) and also added 18 base pairs of sequence homology to the lbbD flanking regions. This PflgBkan fragment and the linearized lbbD deletion vector were combined using the Gibson Assembly Master Mix (New England BioLabs) following manufacturer’s instructions. The ampicillin-resistance marker in pGEM was inactivated by BspHI restriction enzyme digestion prior to transformation into L. biflexa. Spirochete transformants were confirmed by Southern blot analysis and PCR.
The complementation plasmid was constructed by ligating a cassette conferring spectinomycin-resistance into the BspHI site of pGEM::lbbD. Briefly, the spectinomycin resistance marker was amplified from the pGSBLe24 Leptospira shuttle vector (Bourhy et al., 2005) using primers Lb1431.Comp.Bsph.F and Lb1431.Comp.Bsph.Rev. Ligation into the BspHI site disrupts the ampicillin gene of the pGEM vector. The resulting plasmid, pflgSpec::lbbD-2-2, was electroporated into the ΔLBBD strain and transformants screened for integration of the entire vector. The genetic structure of the integration event into the L. biflexa chromosome is represented in Fig. 2A. Complementation of lbbD was confirmed by immunoblot, and the location of the recombination event was determined by PCR using the flanking primers Lb1432.Comp.Anchor.F and pGEM.Comp.Anchor.RC.
The lbbD gene was ectopically-expressed on the shuttle vector pLe1SVK (Fig. 2A), a modified L. biflexa shuttle vector derived from pGSBLe24 (Bourhy et al., 2005), which incorporates the kanamycin-resistance cassette, ColE1 origin of replication, and the multiple cloning sites region from pBSV2 (Stewart et al., 2001). The plasmid pLe1SVK contains more cloning sites and is smaller than pGSBLe24. The shuttle vector-lbbD containing strain was engineered by constitutively expressing lbbD with the B. burgdorferi flgB promoter. Primers Lb1431.Expr.NdeI.F and Lb1431.Expr.HindIII.RC were used to amplify lbbD from genomic DNA using Vent Polymerase (New England BioLabs), and to add NdeI and HindIII restriction sites. The lbbD fragment was ligated into vector pBSV2ex, which contains NdeI and HindIII sites downstream of the cloned flgB promoter. The PflgB-lbbD fragment was excised using NotI restriction sites and ligated into pLe1SVK and electroporated into wild type L. biflexa and the resulting strain designated SV1LBBD. Transformants were confirmed by PCR, Southern blot, and immunoblot analysis. For comparative purposes, SV1LBBD was passaged without antibiotics to isolate strain SV1LBBD*, which had lost the expression vector.
Antibody production and quantitative immunoblots
Recombinant LbbD was expressed in E. coli using expression vector pET28a (EMD Millipore, Billerica, MA). Primers Lb1432.XhoI.T7.Rev and Lb1431.pet28.NcoI.F were used to amplify lbbD, and the resulting fragment was cloned into Zero Blunt TOPO PCR Vector (ThermoFisher Scientific). The lbbD gene and pET28a vector were digested with NdeI and XhoI and ligated together. After confirming expression of LbbD, the protein was purified from inclusion bodies. Purified protein was inoculated into New Zealand White Rabbits to generate LbbD antisera. All animal experiments and protocols were approved by the Animal Care and Use Committee of the Rocky Mountain Laboratories following the guidelines of the National Institutes of Health. The Rocky Mountain Laboratories are accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Quantitative immunoblots were used to compare protein levels of LbbD and FlaA2 among strains. Mini-protean TGX Stain Free protein gels (Bio-Rad, Hercules, CA) were used to separate and visualize protein loads using a ChemiDoc MP Imaging System with Image Lab software version 5.2.1 (Bio-Rad). Each lane contained lysates of approximately 5 x 108 cells. Immunoblots were imaged and quantified using the Image Lab software, according to the manufacturer’s recommendations.
Growth rate comparisons
Growth rate comparisons were performed in triplicate. Cultures were inoculated at 5x105 cells/mL from a starter culture grown to exponential phase and counted each day with a Petroff-Hauser counting chamber (Hausser Scientific, Horsham, PA) under dark-field illumination.
Scanning electron microscopy and cell measurements
Strains were grown to log phase (approximately 5x108 cells/mL), centrifuged at 5000x g for 10 minutes and resuspended in 1/10 the amount of media (final cell concentration ~5x109). Approximately 30 μl of cell suspension was placed on a poly-L-lysine coated silicon wafer chip and allowed to adhere for 30 min in a 30°C incubator. Chips were rinsed to remove non-adherent cells with 1 ml 0.1 M sodium cacodylate buffer (Ted Pella, Inc. Redding, CA) and then fixed with 2.5% glutaraldehyde in cacodylate buffer for 30 min at room temperature. Samples were processed using a Biowave Pro laboratory microwave equipped with a Coldspot water circulator (Ted Pella, Inc.; power output for rinses and dehydration was performed at 250 watts, all other steps used 167 watts) as follows: 45 sec rinse in 0.1 M sodium cacodylate buffer; post-fixation in 0.5% OsO4, 0.8% K4Fe(CN)6 in 0.1 M sodium cacodylate buffer for 2 cycles of 2 min on, 2 min off, 2 min on, 2 min off, 2 min on; 45 sec rinse in 0.1 M sodium cacodylate buffer. The cycle was repeated twice in the same fashion in 1% aqueous tannic acid with a 45 sec rinse in 0.1 M sodium cacodylate buffer. The cycle was repeated in the same fashion for 2 cycles in 0.5% OsO4, 0.8% K4Fe(CN)6 in 0.1 M sodium cacodylate buffer, followed by a 45 sec rinse with dH20. Ethanol dehydration was performed for 45 sec in varying concentrations: 1x in 25% EtOH, 1x in 50% EtOH, 1x in 95% EtOH, and 3x in 100% EtOH. The samples were critical point dried with a Bal-Tec CPD 030 (Leica Microsystems, Buffalo Grove, IL); placed on a stub using carbon sticky tape, and sputter coated with 75 Å of iridium (IBS/e; South Bay Technology, Inc., San Clemente, CA). Samples were placed in a Hitachi SU8000 scanning electron microscope operating at 2.0 kV, 10 μA, and a working distance of ~8 mm. All low mag images were taken with a secondary electron detector at the same nominal magnification of 25,000x.
Cell dimensions were measured using ImageJ 1.50i imaging software (Rasband, NIH) according to the protocol by Goldstein et al. (Goldstein et al., 1996). Briefly, the pitch was measured by determining the distance between crests and between troughs of the cell helix. Each cell’s pitch measurement was an average of 6 measurements, 23–65 cells were measured per strain. Measurements were performed on bacteria from 3 independent cultures. The number of helices per cell was determined by counting helical turns on 15 cells from three biological replicates. Cell length was measured with the 40x objective of a Nikon Eclipse 80i with Nikon NIS-Elements AR 4.20.02.
Soft Agar Motility Assays and Velocity Measurements
Soft agar motility assay plates were made with EMJH diluted 5x and Noble Agar diluted 0.5x. Each plate was made with 10 mL liquid EMJH, 31.75 mL water, and 8.25 mL of a 1.2% Noble Agar solution for 50 mL per plate. Holes were bored into the media with a sterile Pasteur pipette and a 5 μL suspension containing approximately 1x106 spirochetes in water was inoculated into each hole. Plates were incubated at 30°C for 7 days before the colony diameter was measured to the nearest millimeter.
Spirochete velocity was measured in 0.5 % methyl cellulose/EMJH solution. A 2 % methyl cellulose (Sigma Aldrich, 4,000 cP) stock solution was made in distilled water, autoclaved, and mixed at 4° C for approximately 3 days until solubilized. Methyl cellulose was added to L. biflexa cultures in mid-log phase (~5 x 108 spirochetes/mL) to achieve a 0.5 % final concentration and the suspension was incubated at 30° C with 360° rotation for 1–2 hours. Video recordings of spirochete motility were made on a Nikon Eclipse 80i and cells were tracked and velocities calculated using ImageJ FIJI version 1.0. Three independent cultures were used per strain and > 20 cells were measured per culture with the 10 fastest measurements used to calculate maximum spirochete velocity.
Hyper-Osmolarity
EMJH media was made without addition of NaCl salt at 90% volume. Hyper-osmolarity experiments were performed by comparing the growth rate of wild type versus ΔLBBD in triplicate cultures. Sodium Chloride was added to the medium at either the standard EMJH concentration (12 mM) or to a hyper-osmotic concentration of 100 mM. Cultures were then inoculated with: 1) wild type in standard EMJH (12 mM); 2) wild type in hyper-osmotic EMJH (100 mM NaCl); 3) ΔLBBD in standard EMJH; and 4) ΔLBBD in hyper-osmotic EMJH. Cultures were counted each day using a Petroff-Hauser counting chamber.
Minimal inhibitory concentration
MIC was determined by broth microdilution assays, as previously described (Slamti et al., 2011, Murray & Hospenthal, 2004). Strains were inoculated at 1 × 106 cells/mL into 24 well, round bottom microtiter plates. Antibiotics A22 and Amdinocillin (Sigma Aldrich, St. Louis, MO) were added in 2 fold-serial dilutions; the potential minimal inhibitory concentration (MIC) was described by Slamti et al. (Slamti et al., 2011). Positive controls (strains cultured in the absence of antibiotic) and negative controls (uninoculated medium) were included in each assay and were performed in quadruplicate independent assays. Plates were incubated at 30°C for 48 hours, at which time AlamarBlue (Biorad, Hercules, CA) was added to all wells according to manufacturer’s instructions. Plate were incubated for an additional 12 hours at 30°C, and growth was determined colormetrically by a change from blue to red.
RNA extraction and qPCR
Cells were grown to exponential phase, lysed using a FastPrep-24 Tissue and Cell Homogenizer in Lysing Matrix B tubes (MPbio, Santa Ana, CA) and RNA was extracted with the Qiagen (Germantown, MD) RNeasy Mini Kit. DNA was removed using Qiagen on-column DNase digestion and the RNA quality confirmed with the Agilent 2200 TapeStation (Agilent Technologies). Applied Biosystems (Foster City, CA) High Capacity cDNA kit was used to convert 1 μg RNA to cDNA, according to manufacturer’s instructions, and cDNA was diluted 1:25 for subsequent qPCR experiments. Transcript levels were determined for 3 biological replicates using 5μL of the diluted cDNA with 2x TaqMan Universal Master Mix (Applied Biosystems) using a ViiA 7 Dx qPCR machine (Life Technologies, Carlsbad, CA). The flaB standard curve was with L. biflexa cells serially diluted in water and containing from 101–106 organisms.
Supplementary Material
Acknowledgments
We thank Kelsi Sandoz and Olivia Steele-Mortimer for critical review of the manuscript, Hunter Stone for construction of pLe1SVK, Tregei Starr and Chad Hillman for technical expertise with microscopy, and M. Moteleb for providing helpful discussions and advice on the soft agar motility assay protocols. We are grateful to Ryan Kissinger and Anita Mora for assistance with graphics, Tyler Evans for performing cell measurements, and Rocky Riviera and the Rocky Mountain Laboratory Veterinary Branch for animal care and support. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, III, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P, Frangeul L, Couve E, Glaser P, Saint Girons I, Picardeau M. Complete nucleotide sequence of the LE1 prophage from the spirochete Leptospira biflexa and characterization of its replication and partition functions. J Bacteriol. 2005;187:3931–3940. doi: 10.1128/JB.187.12.3931-3940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nature Reviews Microbiology. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Doi M, Wachi M, Ishino F, Tomioka S, Ito M, Sakagami Y, Suzuki A, Matsuhashi M. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988;170:4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andari J, Altegoer F, Bange G, Graumann PL. Bacillus subtilis Bactofilins Are Essential for Flagellar Hook- and Filament Assembly and Dynamically Localize into Structures of Less than 100 nm Diameter underneath the Cell Membrane. PLoS One. 2015;10:e0141546. doi: 10.1371/journal.pone.0141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann PD, Dobson SJ. Cell wall-less, free-living spirochetes in Antarctica. FEMS Microbiol Lett. 1992;76:289–292. doi: 10.1016/0378-1097(92)90350-w. [DOI] [PubMed] [Google Scholar]
- Goldstein SF, Buttle KF, Charon NW. Structural analysis of the Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J Bacteriol. 1996;178:6539–6545. doi: 10.1128/jb.178.22.6539-6545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D224–228. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. Embo J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvel H, Picardeau M. Genetic manipulation of Leptospira biflexa. Curr Protoc Microbiol. 2007;Chapter 12(Unit 12E):14. doi: 10.1002/9780471729259.mc12e04s05. [DOI] [PubMed] [Google Scholar]
- Malmstrom J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CK, Hospenthal DR. Broth microdilution susceptibility testing for Leptospira spp. Antimicrob Agents Chemother. 2004;48:1548–1552. doi: 10.1128/AAC.48.5.1548-1552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, Stanton TB, Zablen L, Mandelco L, Woese CR. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BKC, Morgan HW, Daniel RM. Thermophilic Anaerobic Spirochetes in New-Zealand Hot Springs. Fems Microbiology Letters. 1985;26:101–106. [Google Scholar]
- Picardeau M, Brenot A, Saint Girons I. First evidence for gene replacement in Leptospira spp., inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol Microbiol. 2001;40:189–199. doi: 10.1046/j.1365-2958.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosey EL, Kennedy ML, Yancey RJ., Jr Dual flaA1 flaB1 mutant of Serpulina hyodysenteriae expressing periplasmic flagella is severely attenuated in a murine model of swine dysentery. Infect Immun. 1996;64:4154–4162. doi: 10.1128/iai.64.10.4154-4162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Slamti L, de Pedro MA, Guichet E, Picardeau M. Deciphering morphological determinants of the helix-shaped Leptospira. J Bacteriol. 2011;193:6266–6275. doi: 10.1128/JB.05695-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Carroll JA, Olano LR, Sturdevant DE, Rosa PA. Multiple Posttranslational Modifications of Leptospira biflexa Proteins as Revealed by Proteomic Analysis. Appl Environ Microbiol. 2015;82:1183–1195. doi: 10.1128/AEM.03056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW, Motaleb MA. Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect Immun. 2013;81:2012–2021. doi: 10.1128/IAI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasa S, Lin L, Shi C, Habenstein B, Riedel D, Kuhn J, Thanbichler M, Lange A. beta-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E127–E136. doi: 10.1073/pnas.1418450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Wolgemuth CW, Charon NW, Goldstein SF, Goldstein RE. The flagellar cytoskeleton of the spirochetes. J Mol Microbiol Biotechnol. 2006;11:221–227. doi: 10.1159/000094056. [DOI] [PubMed] [Google Scholar]
- Wunder EA, Jr, Figueira CP, Benaroudj N, Hu B, Tong BA, Trajtenberg F, Liu J, Reis MG, Charon NW, Buschiazzo A, Picardeau M, Ko AI. A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol Microbiol. 2016;101:457–470. doi: 10.1111/mmi.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.