Abstract
Evolving understanding of structural and biological complexity of tumors has stimulated development of physiologically relevant tumor models for cancer research and drug discovery. A major motivation for developing new tumor models is to recreate the three-dimensional environment of tumors and context-mediated functional regulation of cancer cells. Such models overcome many limitations of standard monolayer cancer cell cultures. Under defined culture conditions, cancer cells self-assemble into three-dimensional constructs known as spheroids. Additionally, cancer cells may recapitulate steps in embryonic development to self-organize into three-dimensional cultures known as organoids. Importantly, spheroids and organoids reproduce morphology and biologic properties of tumors, providing valuable new tools for research, drug discovery, and precision medicine in cancer. This Progress Report discusses uses of both natural and synthetic biomaterials to culture cancer cells as spheroids or organoids, specifically highlighting studies that demonstrate how these models recapitulate key properties of native tumors. The report concludes with our perspectives on the utility of these models and areas of need for future developments to more closely mimic pathologic events in tumors.
Keywords: three-dimensional tumor models, spheroids, organoids, natural materials, synthetic materials
The table of contents entry
This Progress Report presents the state-of-the-art approaches using natural, synthetic, and composite biomaterials for three-dimensional tumor modeling, and discusses how these models uniquely reproduce key properties of native tumors to facilitate basic and applied cancer research and cancer drug discovery efforts.
1. Introduction
Basic and applied cancer research has historically relied on various in vitro cell-based models to investigate signaling pathways and mechanisms underlying different phenotypes and functions of cancer cells including metabolism, growth, migration, matrix invasion, and drug resistance.[1, 2] Additionally, cancer drug discovery efforts in academia and pharmaceutical industries have long benefited from cell-based disease models to evaluate toxicity profiles and biological activities of compounds against cancer cells, mechanisms of drug effects, and off-target interactions.[3, 4] Importantly, the adaptability of cell-based models to miniaturized culture platforms enables automated, high throughput screening of libraries of chemical compounds to expedite identification of lead compounds for subsequent tests in animal models and clinical trials.
Monolayer cultures of adherent cancer cells have traditionally been used for these applications.[5] The ease of forming and maintaining two-dimensional (2D) cultures of cells and their compatibility with various culture vessels and a broad range of biochemical assays have made 2D cultures indispensable to cancer research.[6] Despite these benefits, evolving understanding of the complexity of cancer clearly establishes that 2D cultures fail to recapitulate pathophysiological features of human tumors. Adhesion of cells to non-physiologic stiff substrates such as plastic and glass, lack of a compact morphology and close cell-cell and cell-matrix contacts, exposure of cells to an environment of uniform nutrients and oxygen content, and absence of matrix proteins all are major shortcomings of 2D cancer cell cultures. Although 2D models allow co-cultures of cancer and stromal cells to study heterotypic cellular interactions, disparities between 2D cultures and native tumors necessitate conducting these studies with more relevant in vitro tumor models to ensure reliability of resulting data.
Limitations of 2D culture systems for chemical compounds library screening and drug discovery also contribute to well-documented inefficiencies in identifying compounds that translate successfully to clinical oncology.[7] For example, several promising drug candidates developed for aggressive pancreatic, brain, and lung cancers based on success in initial screening with standard cell assays ultimately failed clinically.[8] Despite significant time and resource investment to develop new cancer drugs, currently up to 95% of candidate drugs effective in preclinical tests fail in clinical trials.[9, 10] This low productivity significantly increases costs of cancer drug discovery to ~$2B for a single drug.[11–13] More widespread utilization of in vitro models that more closely model actual human tumors will help identify safe and effective compounds, reducing costs and time invested in compounds that fail later in drug development.
The need for better in vitro cancer models has fueled intense research both in academia and industry, leading to development of three-dimensional (3D) models as major tools both for basic cancer research and drug discovery applications.[14] These models are generated using different sets of technologies and offer various degrees of complexity including self-assembled and freestanding spherical aggregates of cancer cells as cellular spheroids, tumorspheres, organotypic spheroids, matrix-mediated assembled cellular aggregates, multilayered cultures of cancer cells or tumor slices, organoids, and microfluidics- and microfabricated-mediated cultures of cancer cells.[15–21] Importantly, inclusion of various stromal cells (such as carcinoma-associated fibroblasts, immune cells, and vascular cells), addition of matrices of generic or defined compositions, modulation of mechanical and biochemical properties of the stroma, and generation of physiologic levels of fluid flow have all been demonstrated in a broad range of studies. We will focus this Progress Report only on two popular 3D tumor modeling approaches based on spheroids and organoids developed using natural or synthetic biomaterials. We highlight and discuss studies that demonstrate using 3D models and reproducing key biologic properties of tumors. In addition, we provide perspectives on the utility of biomaterials-based approaches to tumor modeling and discuss areas of need and potential opportunities that can be addressed with these models.
2. Biomaterials-based 3D cancer models
Advances in materials science and engineering have led to development and use of synthetic and natural materials in tissue engineering for a variety of applications, including the rapidly growing area of engineering 3D models of cancer.[22–25] These materials are used to construct scaffolds of defined mechanical and/or biochemical properties to physically support cell adhesion and growth and facilitate self-assembly of cells into 3D clusters [Figure 1]. Tissue-engineered models of cancer also enable cellular interactions with specific biochemical factors conjugated to scaffolds, homotypic interactions of cancer cells, and signaling among cancer cells, stromal cells, and matrix proteins. Natural materials such as collagen, laminin rich extracellular matrix (lrECM), hyaluronic acid (HA), alginate, and chitosan have been used alone or in a variety of combinations to provide biomimetic supportive structures for cancer and stromal cells and allow the cells to remodel the matrix. However, natural matrix proteins are subject to certain limitations, such as batch-to-batch compositional variations and changes in cross-linking and assembly density, tissue sources from which materials are derived, and the expense to derive and purify them. On the other hand, synthetic materials such as polyethylene glycol (PEG), poly-lactic-glycolic-acid (PLGA), and polycaprolactone (PCL) can be conveniently engineered with defined properties such as stiffness, porosity, and presentation of specific signaling molecules present in tumor microenvironments. The ability to engineer and control these properties provides novel approaches to elucidate effects of defined mechanical and biochemical cues on cancer cells. We will present the use of a variety of both natural and synthetic materials used to construct 3D cancer models; discuss how these in vitro models help recapitulate certain properties of native tumors; and present how these models advance understanding of tumor biology and facilitate testing for new cancer therapies. Supplementary Table S1 provides a summary of some of these studies in terms of type of biomaterials, cancer cells, and major outcomes.
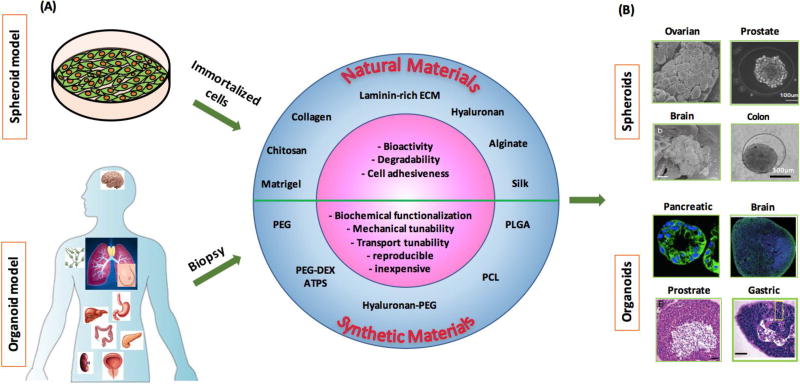
Figure 1.
Biomaterial based spheroid and organoid modeling. (A) Approaches to develop 3D tumor models from immortalized cell lines as spheroids and biopsies as organoids. Various natural and synthetic materials offer unique advantages to facilitate self-assembly or directed assembly of cells into these models. (B) Representative images of spheroids and organoids formed using cells of different cancers. Images in (B) were reproduced with permission. [74] Copyright 2010, Elsevier. [92] Copyright 2014, Elsevier. [101] Copyright 2016, John Wiley and Sons, [163] Copyright 2015, Nature Publishing Group. [165] Copyright 2017, Nature Publishing Group. [158] Copyright 2014, Elsevier. [157] Copyright 2014, Nature Publishing Group.
3. Spheroid models
Cancer cell spheroids, also known as multicellular spheroids, are the simplest in vitro model of solid tumors. Spheroids are generated due to the inherent property of epithelial cancer cells to form intercellular adhesions and self-assemble into a compact aggregate on a non-adherent surface or within a 3D matrix. Spheroids of different sizes ranging from few tens of micrometers to a millimeter scale can conveniently be made. The 3D morphology of spheroids mimics avascular tumors in terms of close cell-cell and cell-ECM adhesions, exposure of cells within a spheroid to non-uniform concentrations of soluble factors, low oxygen tension in the core of a spheroid resulting in hypoxic, slow cycling, and dormant cells, and an acidic extracellular environment. These properties are implicated in a wide range of biological processes in cancer as highlighted with the following examples: loss of cell-cell contacts through downregulation of cadherin junctions and catenins allows detachment of cancer cells from a tumor mass, enabling transition of the cells to a migratory, mesenchymal-like state to facilitate metastasis;[26, 27] cancer cell-ECM signaling mediated by adhesion complexes promotes cell proliferation and survival;[28] dynamic cell-ECM adhesion and detachment through integrins leads to traction forces connecting the matrix to actomyosin filaments to facilitate cancer cell migration;[29, 30] a hypoxic tumor environment promotes evolution of cancer stem cells with the ability to repopulate a tumor mass and resist drug treatments;[31, 32] and low pH in the acidic extracellular environment reduces uptake of weakly basic drugs, such as doxorubicin, conferring resistance to chemotherapy.[33] Additionally, spheroids offer flexibility of incorporating different stromal components to accommodate studies on how physical interactions between cancer cells and tumor stroma and intercellular signaling regulate tumor growth, angiogenesis, invasion, and drug resistance. Therefore, despite being a relatively simple model, spheroids are inherently suited for a broad range of tumor biological studies.
Due to these advantageous features of spheroids, there has been a major push to incorporate spheroids as surrogate tumor models in cancer drug discovery. Original platforms for 3D cell cultures were cumbersome, expensive, and not amenable to high throughput screening operations that are critical to the workflow in the pharmaceutical industry. However, recent technological developments generate large numbers of consistently-sized spheroids in standard microplates compatible with standard robotic instruments such as liquid handlers, high content imagers, and plate readers. These capabilities simplify and expedite formation and drug treatment of spheroids, as well as analysis of cellular responses to chemical compounds.[34–37] Importantly, several commercially available biochemical assays originally developed for 2D cultures, such as PrestoBlue and CellTiter Glo, have been optimized for 3D cultures to provide quantitative, end-point measures of drug effects on cancer cells.[38, 39] Collectively, these advances help streamline the use of spheroids as physiologically-relevant tumor models in cancer drug discovery.
3.1 Natural Materials
A. Collagen
Collagen is an abundant structural protein in the body and serves as a major substrate for cell adhesion. Collagen is also the main matrix protein of desmoplastic tumors of pancreas, breast, lung, colorectal, and skin. Cancer cells binding to the integrin binding motif Arg-Gly-Asp of collagen fibrils is mediated by α1 β1 and α2 β1 integrins.[40, 41] Adhesion of cancer cells to collagen facilitates multiple key processes including cell proliferation, survival, migration, and invasion.[42–44] Considering the importance of collagen in biology of tumors, various collagen-based 3D cancer models have been developed to study these events. These models primarily use dispersed single cells or spheroids of cancer cells in a collagen matrix, as well as co-cultures of cancer cells with stromal cells such as fibroblasts, endothelial cells, and adipose derived cells. Collagen used for in vitro studies is often derived from bovine skin, rat tail, and human placenta.
Cancer cells cultured in a 3D collagen scaffold showed morphological and gene expression similarities to tumors. MCF-7 breast cancer cells cultured alone in collagen showed elongated mesenchymal-like morphology, upregulation of transcription factors SLUG, SNAIL, ZEB1/2, TWIST1/2, LEF1 involved in epithelial-mesenchymal transition (EMT) of cancer cells, and concurrent downregulation of the epithelial cell marker E-cadherin and gain of vimentin, β-catenin, and osteonectin expression. [45] Collagen protected migrating HT1080 sarcoma cancer cells against doxorubicin by modulating cell motility proteins focal adhesion kinase (FAK), RhoA, and β1-integrin. Doxorubicin at a non-toxic concentration inhibited migration of HT1080 cells cultured in 2D by 70% and significantly decreased FAK, RhoA, and β1-integrin levels. However, cells cultured in a 3D collagen matrix showed significant resistance against doxorubicin-mediated inhibition of migration by maintaining activity of cell motility proteins, resulting in only 8% inhibition of migration. This finding was supported by a study that demonstrated the genes involved in focal adhesion pathway were upregulated in tumor cells by doxorubicin treatment of breast cancer patients.[46] Biomechanical properties of collagen also influence cancer cell functions. For example, spheroids of MDA-MB-231 breast cancer cells cultured in collagen matrices showed stiffness dependent apoptosis and migration.[47] Increasing the matrix stiffness from 0.3 kPa, to 1.2 kPa, and to 6.0 kPa resulted in greater resistance to paclitaxel treatment. Additionally, increasing the matrix stiffness to 1.2 kPa and 6.0 kPa caused a reduction of over 20% and 50% in cells invading the collagen matrix. This finding was consistent with data from ex vivo measurements of bulk modulus of freshly excised tumors from a mouse model of breast cancer.[48] Tests showed a significant direct correlation between collagen content of each tumor and its bulk modulus and stiffer tumors were associated with less frequent, smaller local recurrences and less extensive metastases.
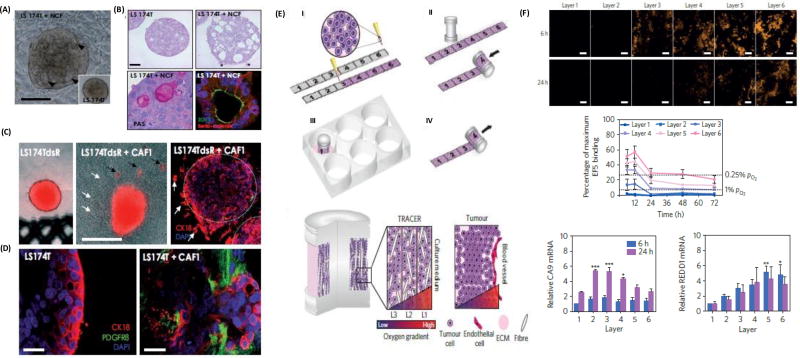
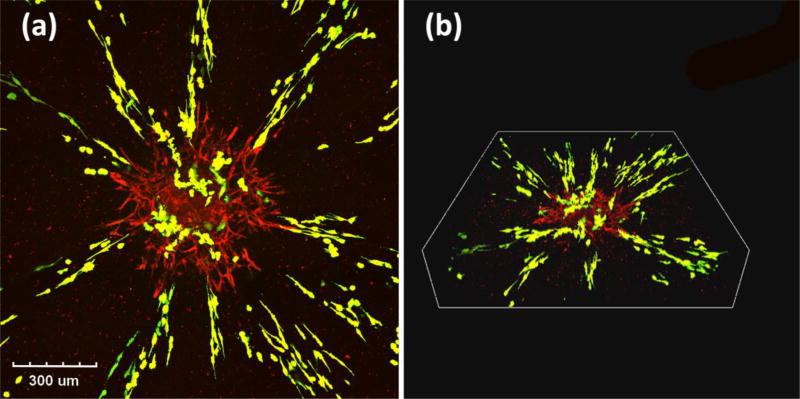
Dynamic interactions among cancer cells, stromal cells, and the extracellular matrix in tumors regulate morphology, proliferation, motility, and drug responses of cancer cells. 3D models that facilitate tumor-stromal interactions provide a unique tool to understand tumor biology. Co-culturing breast cancer cells with stromal fibroblasts in 3D collagen matrices restored the epithelial phenotype of cancer cells. When MCF7 cells were cultured with normal mammary fibroblasts in a collagen matrix, they formed tight clusters with a distinct border and showed apical-basal polarity with a lumen. By comparison, mono-cultured MCF7 cells in collagen formed loose clusters without polarity or lumen.[49] A similar finding was reported with co-cultures of LS174T colon cancer spheroids and normal colon fibroblast (NCF) cells in a 3D collagen matrix.[50] Cancer cells displayed well organized glandular epithelial structures and established desmosomes, adherence junctions, and tight luminal junctions, whereas mono-cultures of LS174T spheroids only showed rudimentary desmosomes [Figure 2A,B]. Co-culture LS174T-NCF spheroids showed close intercellular contacts with staining of NCF cells for fibroblast activation protein (FAP). Collagen fibers showed contraction in the presence of NCF cells, which nested around the epithelial cells as observed in the human colon adenocarcinoma. Co-cultures with NCF cells reduced mitotic potential of LS174T cells and enhanced effectiveness of a PI3K inhibitor, LY294002, to induce apoptosis in cancer cells and reduce the volume of the spheroids. Importantly, co-culturing LS174T cell spheroids with cancer-associated fibroblasts (CAFs) derived from colon adenocarcinoma promoted cancer cell invasion into the collagen matrix [Figure 2C]. Confocal imaging revealed invasion of LS174T into the collagen gel both as single cells and collectively in areas with high CAFs density [Figure 2D]. Spheroids of LS174T cells alone in collagen retained a compact morphology and did not invade the matrix.
Figure 2.
Collagen-based modeling of cancers. (A–B) Mono-cultured LS174T cells only show rudimentary desmosomes, whereas co-culture of LS174T with normal colon fibroblasts (NCF) results in established desmosomes, adherence junctions, and tight luminal junction. (C–D) Invasion of LS174T cells from the spheroid into the collagen matrix in the co-culture model. (E) Schematic representation of a six-layered rollable collagen-cell composite to develop physiologic oxygen concentrations. (F) Induction of hypoxia due to decreasing oxygen levels into the core of the model and upregulation of hypoxic genes. (A–D) were reproduced with permission. [50] Copyright 2011, Elsevier. (E–F) were reproduced with permission. [54] Copyright 2016, Nature Publishing Group.
A consequence of fibroblasts-mediated collagen contraction is reduced diffusion of nutrients and oxygen to the spheroid embedded in a collagen matrix. This may result in hypoxia and necrosis in the central zone of a spheroid and reduced proliferative capacity of cancer cells.[51, 52] Both limited proliferation of cancer cells and diffusive transport may reduce effectiveness of chemotherapeutics. For example, co-culture spheroids of HepG2 liver cancer cells and NIH 3T3 normal fibroblasts embedded in a collagen matrix were highly resistant to doxorubicin treatment compared to freestanding spheroids.[53]
Reduced oxygen diffusion to spheroids of HT-29 cancer cells in a dense collagen hydrogel was observed through positive staining of cells for pimonidazole during a two-week culture.[51] Hypoxia led to production of VEGF by HT-29 cells and release into the matrix. Earlier and higher levels of VEGF production resulted when 3T3 fibroblasts or endothelial cells were incorporated in the model. This indicated a major role for signaling between stromal and cancer cells to promote angiogenesis. A similar finding was reported with cultures of MDA-MB-231 breast cancer cells in type I collagen hydrogels that showed a significant upregulation of HIF-1α and VEGF-A within three days of culture.[40] Decreasing cancer cell density to 25% of the initial tests delayed the upregulation of both markers by 2–4 days due to reduced competition for available oxygen. Decreasing the hydrogel thickness by 50% mitigated diffusion limitations of oxygen and nutrients and changes in expression of both markers remained insignificant.
A novel model to study hypoxia-mediated changes in cell metabolism was developed using a complex rollable composite design known as TRACER.[54] A suspension of breast cancer cells in type I collagen was loaded onto a ~35 µm thick cellulose scaffold strip, which was sufficiently thin for free access of cells to oxygen and nutrients. Rolling the composite onto an oxygen permeable metallic core gave a six-layered configuration to generate the length scale over which oxygen gradients develop in tumors [Figure 2E]. When placed in culture media, oxygen and nutrients diffusion to cells progressively reduced moving from the outer to inner layers, mimicking oxygen gradients in tumors at progressively increasing distances from nearby blood capillaries. Importantly, the 3D spatial locations of cells in the model was mapped to their positions along the unrolled strip to facilitate collection and characterization of populations of cells from different layers. Those cells located in the middle layers (2–4) were under mild hypoxia and showed upregulation of the HIF-1α regulated gene, carbonic anhydrase 9, whereas cells in the inner layers (4–6) were under severe hypoxia (O2<0.1%) within 6 hrs and showed upregulation of the HIF-1α target gene, REDD1 [Figure 2F]. Interestingly, a slight increase in oxygen concentration in layers deep in the construct was observed at 12 hours, indicating an adaptive hypoxia response and reduced oxygen consumption by cells. This 3D model helped identify various known and unknown mediators of metabolic adaptation of cancer cells to hypoxia.
Highlight: Use of collagen for 3D tumor modeling in these studies reproduced a wide range of events critical for tumor progression: EMT of cancer cells; migration and invasion into the collagen matrix; effects of matrix stiffness; drug resistance; tumor-stromal interactions; metabolic adaptations to hypoxia; and angiogenesis. Understanding biological mechanisms underlying these events will facilitate discovery and development of new, molecularly targeted drugs to improve cancer therapy.
B. Laminin rich extracellular matrix (lrECM)
IrECM is a solubilized extract derived from Engelbreth-Holm-Swarm mouse sarcoma cells. Laminin is an essential component of the basement membrane for polarized epithelial morphogenesis. 3D lrECM preserves cancer tissue architecture and biology by restoring their biochemical and biomechanical properties.[55] Interestingly, malignant subtypes of cancer cells can be distinguished from their non-malignant counterparts based on their distinct morphological differences in 3D lrECM cultures. These distinct morphologies are also reflected in gene expression of the cells and correlate with drug responses in vivo. 3D lrECM promotes integrin-mediated signaling between cancer cells and ECM proteins such as laminin and fibronectin that drive cancer cell malignancy in vivo. As such, 3D lrECM provides a valuable tool to identify integrin targets in cancer cells.
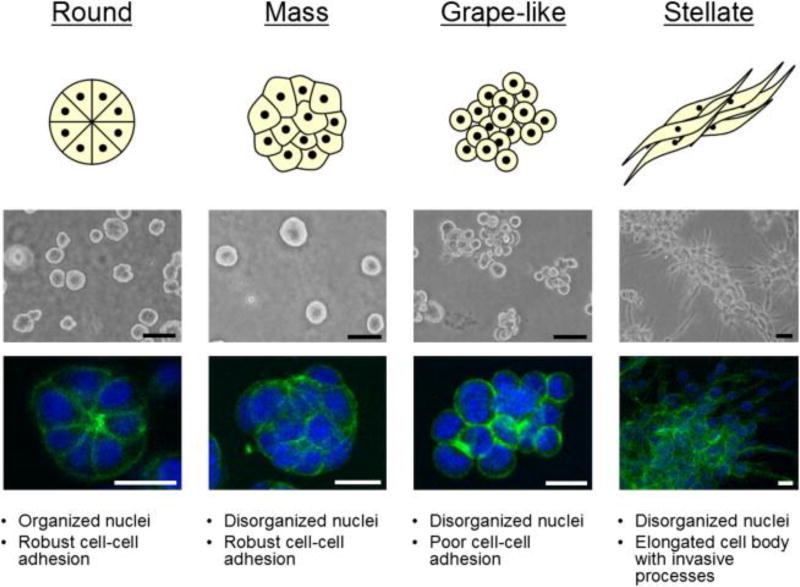
Non-malignant breast and prostate cancer subtypes cultured in 3D lrECM displayed distinct, polarized, growth arrested, acinus like structures, while malignant cancer subtypes formed disorganized, proliferative, and non-polar colonies.[56–58] Malignant breast and prostate cancer cells displayed four distinct morphologies in 3D lrECM based on their close cell-cell contacts and invasive potentials: round, mass, grape-like, and stellate [Figure 3].[57] Gene expression patterns of breast and prostate cancer cells with similar morphologies frequently clustered together, suggesting that the gene expression pattern strongly correlates with colony morphology in the 3D lrECM cultures. In another study, colorectal cancer cells cultured in 3D IrECM scaffolds gained round, mass, and stellate morphologies. [59] DLD-1, CaCO2, HT-29, SW-480 cells exhibited solid tumor formation capacity, whereas LOVO, COLO-205, COLO-206F formed grape-like structures. Regardless of morphology, cells showed reduced responses to EGFR inhibition in 3D IrECM compared to 2D cultures. Lewis lung carcinoma cells (LLC1) cultured in 3D IrECM formed clusters and showed significant cytoskeleton rearrangement without stress fibers. Relative to a 2D culture, LLC1 cells in 3D lrECM exhibited marked differences in expression of microRNAs, metabolic pathways, MAP kinase pathway, cell adhesion, and immune response genes. Comparison of expression levels of selected genes and miRNAs between LLC1 cells grown as 3D cultures and LLC1 tumors implanted in mouse indicated close correlation between the two model systems.[60] Non-malignant human mammary epithelial (HMEC) cultured in 3D lrECM showed downregulation of 22 genes during cells growth arrest and acini formation. These 22 genes were used as signatures to predict the prognosis of breast cancer patients in three large independent breast cancer microarray datasets. This gene signature based on 3D lrECM cultures accurately predicted breast cancer outcomes in estrogen receptor positive (ER+) and negative (ER−) tumors.[61]
Figure 3.
Laminin-rich ECM (lrECM) modeling of breast cancer. Breast cancer cells cultured in lrECM show four distinct morphologies that can be used to distinguish malignant and their non-malignant cells. Reproduced under the terms of the CC-BY-X license. [57] Copyright 2007, FEBS Press and Wiley.
Interactions of ECM proteins with integrin receptors of cancer cells modulate different functions of cancer cells. lrECM induces production of high levels of endogenous fibronectin in cancer cells. Fibronectin interacts with integrin α5β1 on cancer cells through its Arg-Gly-Asp and Pro-His-Ser-Arg-Asn synergy sequences. This interaction promotes proliferation, survival, and invasion of cancer cells.[62–64] The level of endogenous fibronectin secreted by malignant T4-2 breast cancer cells was 9.4-fold higher than non-malignant S1 cells, even though both sublines originated from the same parental cells. Inhibition of integrin α5β1 induced apoptosis in T4-2 cells by suppressing AKT signaling.[65] Treatment with a peptide that disrupts interactions of α5β1 integrin with fibronectin promoted apoptosis and enhanced the effect of radiation treatment on malignant cells (T-42 and MDA-MB-231). This was consistent with a study of gene expression data from breast cancer patients that revealed an association of high levels of α5-integrin with decreased survival.[65] A different study showed that the microenvironment provided by 3D IrECM promotes preferential enrichment of α5β1 integrin and endogenous fibronectin in breast cancer cell lines of the basal subtype as compared with luminal cells. Blocking β1-integrin in a panel of breast cancer cells (T4-2, MDA-MB-231, BT-474, MCF-7, and SKBR3) cultured as pre-formed 3D clusters successfully inhibited growth of these malignant cells.[66] Reducing α6- and β-integrins or vimentin levels reverted metastatic prostate cancer cells into a non-malignant type and reduced tumor growth in vivo.[67] Furthermore, reversion of malignant T4-2 breast cancer cells to non-malignant cells by inhibiting β1-integrin significantly reduced malignancy in vivo.[68] Inhibiting β1-integrin in HER2-amplified breast cancer cells (AU565, SKBR3, and HCC1569) significantly increased sensitivity of the cells to the HER2-targeting agents trastuzumab, pertuzumab, and lapatinib.[69] In a more complex 3D co-culture model of PC3 prostate cancer cells with HS5 bone stromal cells, α6- and/or β1-integrin expression in cancer cells increased compared to mono-culture of PC3 cells.[70]
Highlight: Using IrECM for tumor modeling distinguished malignant from non-malignant cells from their 3D morphologies and gene expression profiles; maintained close correlations of expression of selected genes and miRNAs between 3D cultures and tumors implanted in mice; accurately predicted outcomes in specific subtypes of breast cancer; and revealed the therapeutic value of targeting specific integrins in cancer cells to block signaling driven by ECM proteins, rendering metastatic cancer cells non-malignant, promoting apoptosis of cancer cells, reducing tumor burden in vivo, and enhancing drug responses of cancer cells.
C. Alginate, chitosan, hyaluronic acid, and their combinations
C.1 Alginate
Alginate is a family of naturally occurring polysaccharides extracted from brown seaweeds. It supports attachment and integration of cancer cells and promotes their growth. EpCAM-positive hepatocellular carcinoma cells cultured in an alginate matrix formed 3D clusters that recapitulated major features of glandular epithelium in vivo, such as acini, apical morphogenesis, and expression of stem cell-associated proteins with β-catenin signaling.[71] Using an alginate-based 3D model, the study identified that Wnt/β-catenin signaling pathway activation was essential for maintaining the hepatocellular carcinoma stemness, formation of spheroids, and maintaining acinous structures. EpCAM-positive cells cultured in the 3D matrix and treated with TGF-β showed EMT signaling, high tumorigenic activity, and resistance to doxorubicin and 5-fluorouracil in mouse models compared with cancer cells harvested from the same treatment in 2D cultures and used in animal tests.
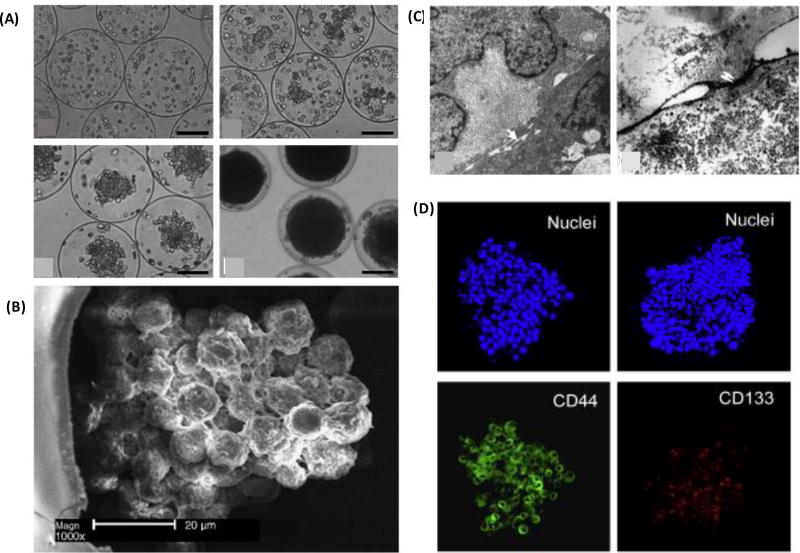
Alginate lacks major integrin binding sites and does not interact with integrins on cancer cells. However, alginate can be functionalized with an RGD (Arg-Gly-Asp) peptide sequence present in major ECM proteins, such as fibronectin, to allow ECM interactions and signaling. Constructing a 3D tumor angiogenesis model using RGD-conjugated alginate disks containing dispersed OSCC-3 oral cancer cells showed an increase in a proangiogenic marker, IL-8, due to cell-ECM interactions and independent of oxygen levels.[72] However, cell-ECM interactions only moderately altered secretion of VEGF. Increased pro-angiogenic molecules promoted invasion of endothelial cells into the matrix. Alginate microcapsules were also used to confine cancer cells and facilitate spontaneous formation of spheroids reproducing solid tumor properties. SMMC-7721 human hepatocarcinoma cells encapsulated in alginate-poly-l-lysine-alginate (APA) microcapsules of 200–300 µm diameter showed actin reorganization into networks to direct cells to form tumor-like clusters [Figure 4A]. Glucose consumption and lactate production of cells correlated well with the cellular proliferation kinetics. Cells in spheroids were arranged into trabecular structures morphologically similar to hepatocarcinoma in vivo [Figure 4B]. Cells expressed tight junctions, showed microvilli on their surface, and developed canaliculi-like structures essential for the integrity of tissues in vivo [Figure 4C].[73] A similar approach was used to encapsulate PC3 human prostate cancer cells in a miniaturized aqueous liquid core of microcapsules with an alginate hydrogel shell to form 3D cellular aggregates. This strategy effectively enriched expression of cancer stem cell genes NANOG, OCT4, CD44, and CD133 [Figure 4D].[74] It was suggested that this enrichment was due to retention of autocrine factors of cells in close proximity when cells were encapsulated in the microcapsules. Harvesting the PC3 aggregates and implanting them in mouse resulted in significantly larger tumors than when disperse PC3 cells or aggregates formed by liquid overlay cultures were injected. This was potentially due to the enhanced content of cancer stem cells in aggregates harvested from the core-shell microcapsules that mimic the structure of early embryos, the native home of totipotent-pluripotent stem cells. [75–77]
Figure 4.
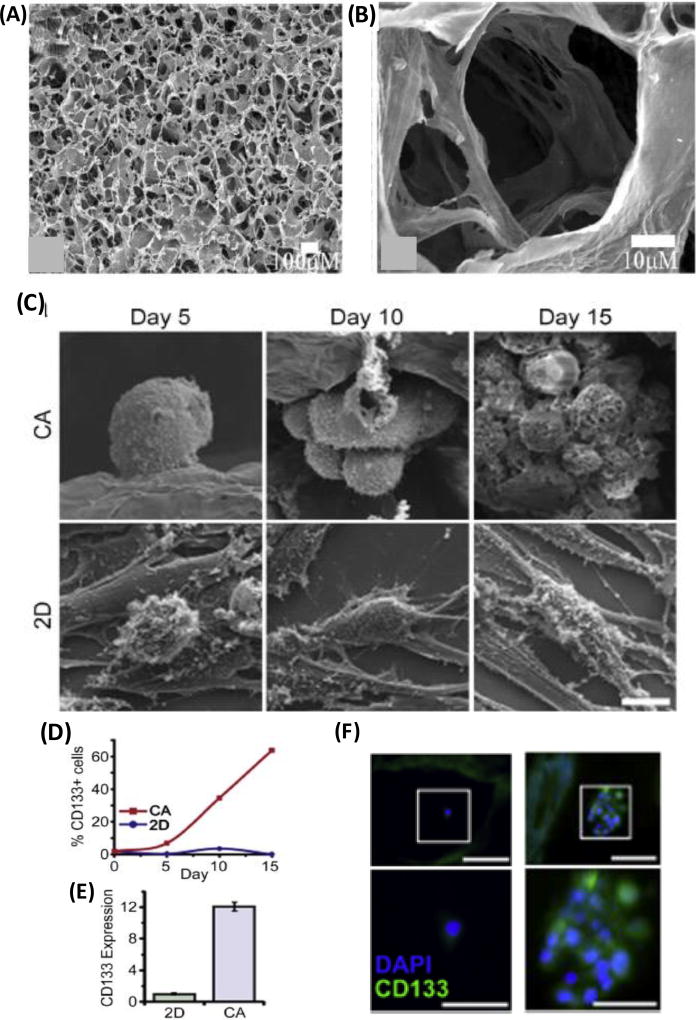
Alginate microcapsules to model hepatocellular carcinoma. Liver cancer cells contained in alginate microcapsule and form spheroids. (B) Cells show actin reorganization and arrange into trabecular structures, (C) express tight junctions and microvilli on their surface, and developed canaliculi-like structures. (D) prostate cancer spheroids encapsulated in the alginate core-shell microcapsule display cancer stem cell marker genes CD44 and CD133. (A–C) were reproduced with permission [73] Copyright 2006, John Wiley and Sons. (D) was reproduced with permission. [74] Copyright 2014, Elsevier.
C.2 Chitosan
Chitosan is a linear polysaccharide obtained from partial deacetylation of chitins of crustaceans. It shares structural similarities with glycosaminoglycans present in native ECM. In contrast to alginate (an anionic polymer), chitosan is a cationic polymer. It provides functional amino groups and surface charge to promote cell attachment.[78] Colon cancer and hepatocellular carcinoma cells cultured on chitosan membranes showed increased cell motility, drug resistance, self-renewal capacity, and cancer stem cell-like gene expression. Cancer stem cell genes OCT4, NANOG, CD133, CD44, and the epithelial maker EpCAM were highly upregulated in these cancer cells. Culturing cells on chitosan membranes led to activation of canonical Wnt/β-catenin-CD44 signaling in CD44+ colon cancer cells and non-canonical Wnt-STAT3 signaling in CD44− hepatocellular carcinoma cells.[79] Compared to 2D cultures, MCF-7 cells cultured in 3D scaffolds of chitosan, derived from prawn chitin, produced more lactate from glucose, showed 35% slower growth with 1 nM tamoxifen treatment, and required higher tamoxifen concentrations to show a comparable toxicity. Decreased activity of tamoxifen with 3D cultures in the chitosan matrix was mediated by the reduced uptake of an autocrine growth factor in breast cancer cells, cathepsin D.[80] Interestingly, chitosan was used as a targeting moiety on nanoparticles to eliminate CD44+ breast cancer cells.[81]
C.3 Chitosan-Alginate (CA)
A composite matrix of chitosan and alginate provides superior cell adhesion properties. A chitosan-alginate (CA) natural scaffold was synthesized by lyophilizing and cross-linking a mixture of chitosan and alginate [Figure 5A,B].[82] The resulting CA scaffold promoted enhanced attachment, integration, and proliferation of osteoblasts compared to a scaffold purely made of chitosan. Hepatocarcinoma cells in CA scaffolds showed slower proliferation than in 2D and Matrigel cultures.[83] Glypican-3 (GPC-3), a histochemical marker that is used to distinguish hepatocarcinoma from benign hepatocellular mass lesions,[84] increased by 5.5 folds in HepG2 cells in CA scaffolds compared to 2D culture. Hepatocarcinoma cells in a CA scaffold were more tumorigenic in animal models. Tumors generated from CA pre-cultured hepatocarcinoma cells were nearly twice as large than those generated using cells harvested from 2D or Matrigel cultures. Additionally, tumors from CA pre-cultured cells upregulated levels of pro-angiogenic growth factors IL-8, bFGF, and VEGF, and induced formation of large, well-rounded blood vessels with well-defined endothelial lining (retained features of normal blood vessels). Similar results were observed with cultures of U-87 MG brain cancer cells in CA scaffolds. Compared to 2D cultures, cells in CA showed slower growth, secreted higher levels of ECM proteins including fibronectin and laminin, exhibited a more rounded and interconnected morphology similar to tumor cells in vivo, and upregulated VEGF and MMP-2.[85] When CA pre-cultured U-87 MG glioblastoma cells were implanted in mice, they facilitated significantly higher recruitment of CD31+ endothelial cells compared to tumors seeded with U-87 MG cells harvested from 2D or Matrigel cultures, indicating improved ability for angiogenesis. In another study, U-87 MG and U-118 MG cells cultured in CA scaffolds showed enrichment for cancer stem cells [Figure 5C–F].[86] Various stem cell related genes including Nestin, GFAP, frizzled 4, GLI, HES, CD44, and CD133 were upregulated in the glioblastoma cells. CD44 induced overexpression of EMT transcription factors SNAIL1, SNAIL2, and Twist2. CD133+ cells from CA cultures formed tumors in mice, while cells lacking this marker failed to induce tumors. Chitosan and alginate were used to form a nanoscale matrix around T cells for co-delivery with bone marrow cells to treat leukemia in a mouse model. This chitosan-alginate matrix reduced the side effect of graft-versus-host disease (GVHD) without compromising the anti-leukemia capacity of T cells.[87]
Figure 5.
Chitosan-alginate (CA) composite scaffolds to model cancer. (A–B) SEM images of CA scaffold synthesized by lyophilizing and cross-linking a mixture of chitosan and alginate. (C) Growth and morphology of glioblastoma cells in CA scaffolds compared to monolayer culture of cells. (D–F) Unlike in monolayer cultures, cells in 3D culture in the CA scaffolds express high levels of cancer stem cell marker CD133. (A–B) were reproduced with permission. [82] Copyright 2005, Elsevier. (C–F) were reproduced with permission. [86] Copyright 2014, Elsevier.
C.4 Hyaluronic acid (HA)
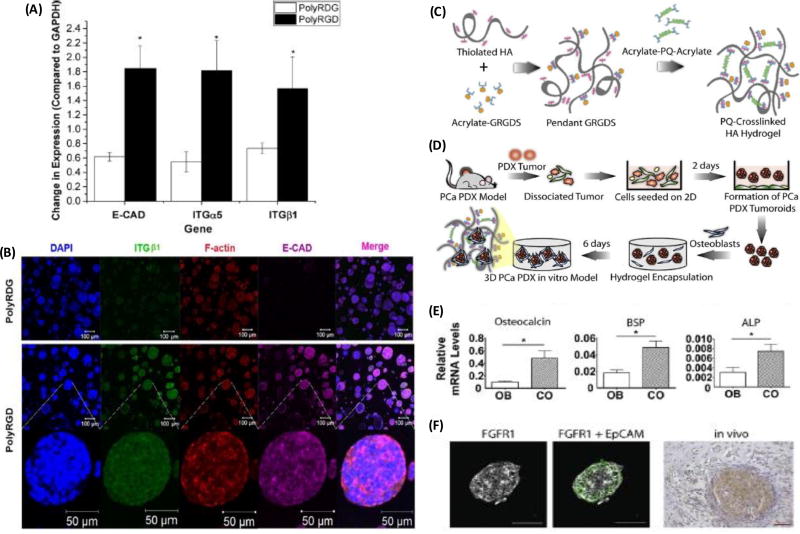
Hyaluronic acid (HA) is a natural anionic polymer and a rich ECM component often overexpressed in tumors. Its accumulation around tumors correlates with enhanced invasion of cancer cells, cancer cell malignancy, and poor patient outcomes. [88, 89] HA interacts with cell surface receptors (e.g., CD44 and RHAMM) and HA-binding proteins to mediate processes such as cell adhesion, migration, and proliferation.[90] The significance of HA in cancer and the ease of its production and chemical modification make it an attractive biomaterial for cancer research. Nanoparticles were decorated with HA for gene and drug delivery to target cancer stem cells.[91–93] HA scaffold hydrogels were successfully used to culture prostate cancer PDX cells that remained viable with continued expression of the androgen receptor, resisted docetaxel treatment in a 0–1 µM concentration range, but did not show significant growth in the HA-hydrogels.[94] Unmodified HA hydrogels do not support integrin-mediated cell engagement. A strategy to overcome this problem is to chemically conjugate cell adhesive RGD peptides to the HA matrix. A biomimetic hydrogel was constructed by mixing thiolated HA and acrylated co-polymer carrying multiple copies of cell adhesive cysteine containing peptide (PolyRGD-AC).[95] LNCaP prostate cancer cells encapsulated in HA-PolyRGD as dispersed single cells formed multicellular spheroids that expressed higher mRNA levels of E-cadherin, α5-integrin, and β1-integrin compared to cells in a negative control PolyRDG gels [Figure 6A,B].
Figure 6.
Functionalized hyaluronic acid (HA) to model prostate cancer. (A–B) Higher mRNA levels of E-cadherin, α5-integrin, and β1-integrin in HA-PolyRGD compared to the PolyRGD control. (C–D) Modification and crosslinking of HA with GRGDS and MMP-degradable peptides to co-culture prostate PDX cells with osteoblast cells, to (E) enable bone forming capability of PDX (E) and (F) preserve in vivo expression of FGFR1. (A–B) were reproduced with permission. [95] Copyright 2016, American Chemical Society. (C–F) were reproduced with permission. [96] Copyright 2016, Elsevier.
Bone is the most common site of prostate cancer metastasis. Co-cultures of prostate cancer and bone cells in HA hydrogels mimic the bone microenvironment of prostate cancer metastasis. HA was specifically modified with integrin-binding peptides GRGDS and cross-linked matrix MMP-degradable peptides to enable co-culturing MDA PCa 118b prostate cancer PDX cells with MC 3T3-E1 osteoblastic precursor cells [Figure 6C,D].[96] The co-culture increased transcript levels of osteoblast-enriched markers osteocalcin, bone sialoprotein, and alkaline phosphatase (ALP) in MC 3T3-E1 osteoblastic cells, indicating the 3D HA hydrogel model helped retain the inherent ability of the PDX cells to induce bone formation [Figure 6E]. The study identified that cross-talk between PDX and osteoblastic cells in the hydrogel was mediated by autocrine signaling through fibroblast growth factor receptor 1 (FGFR1) on the PDX cells. Similarly, these receptors were highly expressed by MDA PCa 118b cells in vivo [Figure 6F]. This indicated that the co-culture model reliably recapitulated in vivo properties of prostate cancer-bone metastasis. Inhibition of FGFR1 using dovitinib decreased interactions of PDX-derived prostate cancer cells and osteoblastic cells. This inhibition reduced cellularity of co-cultures and increased osteogenic activity of MC 3T3-E1 cells. This was consistent with a clinical study where dovitinib showed efficacy in patients with advanced metastatic castrate resistant prostate cancer (CRPC) and bone metastasis by reducing lesion size and intensity on bone scans, lymph node size, and tumor specific symptoms without proportional declines in prostate specific antigen.[97] HA hydrogels are attractive substrates for binding various cytokines. A cytokine releasing HA-based bilayer hydrogel system was constructed to allow sustained release of a heparin-binding epidermal growth factor-like growth factor from the heparin decorated hydrogel particle in the top layer into the bottom layer containing LNCaP prostate cancer cells.[98] This bioengineered growth factor signaling in the HA hydrogel allowed formation of large spheroids that showed close cell-cell contacts, cortically organized F-actin, and increased protein and mRNA expression of pro-angiongenic factors VEGEF165 and IL-8.
C.5 Chitosan-hyaluronan (CH)
Chitosan-hyaluronan (CH) is formed by grafting the amine group of chitosan with the carboxyl group of HA to form a stable covalent bond. A549 and H1299 small cell lung cancer cells cultured in CH scaffolds formed compact spheroids and displayed slower proliferation compared to the 2D cultures of cells.[99] Furthermore, cells in spheroids showed strong upregulation of N-cadherin, vimentin, and fibronectin. Similarly, anti-apoptotic genes such as BCRC5 and BCL2, EMT-related transcription factor TWIST1, and cancer stem cell genes CD44, CD133, SOX2, NANOG, POU5F1 were significantly upregulated in spheroids formed in CH scaffolds. Glioblastoma (GBM6) cultured in CH scaffold showed enhanced stem cell marker expression resisted treatment of alkylating agents.[100]
D. Silk
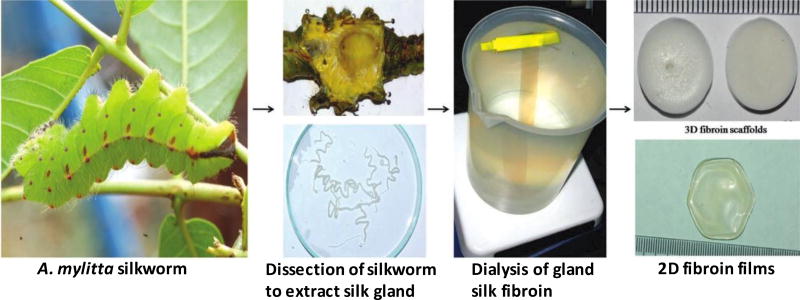
Silk is a natural polymer and widely used clinically as sutures. It is composed of fibroin, a filament core protein, and a glue-like coating of sericin proteins.[101, 102] Oxygen and water permeability, slow degradability, cell adhesiveness, relatively low thrombogenicity, and amenability to convenient surface modification make silk an attractive biomaterial for tissue engineering applications including tumor modeling.[103] Osteosarcoma cells cultured in 3D silk scaffolds proliferated slower than 2D cultures, and showed similar levels of proliferation maker genes such as Cyclin B, E2F1, Ki67, and PcNA as observed in a SCID mouse model.[104] Immunocytochemistry showed increased levels of pro-angiogenic markers HIF-1α, VEGF-A, and VEGF receptor in cancer cells comparable to the native tissue in mouse xenografts. In 3D scaffolds made using fibroin from the silk gland of the tropical silkworm Antheraea mylitta [Figure 7], growth of MDA-MB-231 breast cancer cells cultured for 60 days followed the Gompertz law, similar to the growth of avascular solid tumors.[105] Cell proliferation in the construct showed spatial variations and a larger number of proliferating cells localized toward the periphery of the scaffold. There was a marked increase in levels of pro-angiogenic markers VEGF and IL-8 receptors in the cancer cells that was suppressed by combination treatments of paclitaxel with either celecoxib (a COX-2 inhibitor) or ZD6474 (a VEGFR2 inhibitor). Interestingly, the source from which the silk fibroin is derived to construct the 3D silk scaffold influences proliferation, viability, and metabolism of cancer cells. Silk derived from A. mylitta provided superior cell adhesion and improved viability and proliferation compared to that from B. mori. Similarly, glycolysis of MDA-MB-231 cells in silk scaffolds of A. mylitta-derived fibronin was similar to that in vivo.[106]
Figure 7.
Stepwise process of 3D silk fibroin generation from tropical silkworm A. mylitta used to model breast cancer. Silkworms were dissected to extract silk glands. Fibroin was isolated from the glands and dialyzed to obtain silk fibroin solutions. The solution was used to fabricate scaffolds and films. Reproduced with permission. [105] Copyright 2012, John Wiley and Sons.
Heterotypic cultures of cancer and stromal cells in silk scaffolds are also used to study tumor-stromal interactions such as breast cancer-bone metastasis and evaluate efficacy of targeted therapies.[107] Folate receptors are highly expressed in various cancer cells and are attractive drug targets.[108] Nanoparticles of silk fibroin, derived from A. mylitta, were used to conjugate folic acid and loaded with doxorubicin, to target breast cancer-bone metastasis in a co-culture model of osteoblasts (MG63) and MDA-MB-231 human breast cancer cells. This targeted delivery through folate receptors on cancer cells decreased the population of cancer cells, and the invasiveness and angiogenic capabilities of the cells in terms of VEGF secretion particularly. Interestingly, there was minimal effects on the proliferation and function of bone cells, indicating increased specificity of the treatment toward the cancer cells while protecting normal bone cells.
Highlight: The above studies utilized matrices made of alginate, chitosan, hyaluronic acid, silk, or combinations of these materials and demonstrated the ability to reproduce key properties of tumors in vivo. These included arrangement into structures morphologically similar to native tumors; slower cell proliferation compared to 2D cultures but similar to tumors in mouse models; glucose consumption and lactate production by cancer cells; production of angiogenic molecules by cancer cells including HIF-1α, IL-8, and VEGF to promote recruitment of endothelial cells, consistent with mouse xenografts; enrichment of cancer stem cells; and activation of stem cell signaling pathways. Additionally, harvesting cancer cells from 3D cultures in these matrices and implanting them in mice enhanced tumor formation and drug resistance relative to cells from 2D cultures. Thus, the 3D environment of these matrices maintained malignant properties of cancer cells much more effectively than standard 2D cultures.
3.2 Synthetic materials
A. Polyethylene glycol (PEG) and its modified/functionalized forms
A major benefit of using PEG to create cell-based tissue engineered constructs is liquid-to-solid transition to form hydrogels containing cells. Although PEG in its native form is biologically inert and compatible with cell culture, its unique chemistry allows modifying or conjugating it with natural or synthetic bioactive molecules. This design strategy enables interactions of PEG with cancer cells to facilitate formation of 3D cultures. In addition to simple PEG-based hydrogels for culturing cancer cells, hybrid materials using PEG and synthetic or natural materials are also utilized for specific applications. For example, natural materials such as hyaluronic acid found in certain tumors may be used to better mimic tumor microenvironments. In addition, the use of PEG and dextran (DEX) as highly aqueous, immiscible polymeric solutions allows a scaffold-free approach to form mono-culture and co-culture spheroids. Below is a summary of studies using PEG-based materials for 3D cancer cell cultures.
PEG hydrogels
PEG hydrogels were synthesized from peptide functionalized multi-arm PEG macromolecules using enzymatic reactions.[109] The flexibility of PEG chemistry allowed functionalization of the hydrogels with RGD peptides to facilitate engagement of the matrix with integrins on cancer cells, and MMP-sensitive peptides to allow cell-secreted MMPs degrade the hydrogels. OV-MZ-6 and SKOV-3 ovarian epithelial cancer cells embedded in the synthesized hydrogels formed compact spheroids and secreted ECM proteins. Proliferation of cells, and size and number of spheroids in the hydrogel were dependent on the integrin binding capacity of the hydrogels and significantly enhanced with inclusion of RGD peptides. Ovarian cancer cell spheroids showed resistance to paclitaxel treatment, which correlated with a significant upregulation of several integrins (α3, α5, β1) and MMP-9 levels.
A composite hydrogel was synthesized by covalent attachment of fibrinogen fragments to PEG diacrylate (PEG-DA).[110] A Michael-type addition reaction was used to form the ester bond between the free thiols in the fibrinogen cysteines and acrylate end groups on the PEG-DA (PEGylation). The fibrinogen backbone in the PEG hydrogels presented cell adhesion motifs and allowed proteolytic degradation by cells. PEG-fibrinogen precursors were used to synthesize microspheres of adjustable stiffness and porosity for 3D culture of breast cancer cells MCF-7, SK-BR-3, and MDA-MB-231.[111] Cancer cells pre-mixed with the PEG-fibrinogen polymer precursor were suspended on a polydimethylsiloxane substrate and photo-crosslinked to form cancer cell-containing hydrogel microspheres. Hydrogels degraded over time and cells proliferated. Unlike MCF-7 and SK-BR-3 cells that formed compact spheroids in hydrogels of different Young’s moduli, MDA-MB-231 cells showed an elongated morphology in softer hydrogels but assumed a round morphology in hydrogels of ~10 kPa stiffness. Cancer cell-containing PEG-fibrinogen hydrogels were also made by suspending cancer cells in a PEG-fibrinogen pre-polymer solution, adding it to an oil phase to form cell-containing aqueous drops, and photo-crosslinking to form microspheres. Microspheres were then retrieved by vortexing and maintained in culture media.[112] Different breast and prostate cancer cells were cultured as spheroids for several weeks in microspheres. This approach resulted in a large number of spheroids, albeit with a wide range of size distributions, due to the size heterogeneity of the aqueous drops formed in the oil phase. Nevertheless, spheroids in PEG-fibrinogen microspheres displayed hallmarks of malignant cancer cells such as significant loss of apico-basal polarity, cellular and nuclear atypia, increased disorganization, elevated nuclear cytoplasmic ratio and nuclear volume density, and reduced length of cell-cell junctions.
A redox responsive PEG hydrogel was also reported for generation and recovery of cancer cell spheroids.[113] A cysteine (reducing agent) responsive PEG hydrogel was synthesized from octa-thiolated PEG derivative (8-arm PEG SH), horseradish peroxidase, and small phenolic compound (Glycyl-L-tyrosine). HepG2 hepatocarcinoma cells premixed with this precursor solution formed hydrogels containing cells that proliferated to form spheroids. Addition of a cysteine solution recovered HepG2 spheroids that produced significantly higher albumin and urea compared to the cells cultured in monolayer. Albumin secretion from the HepG2 spheroids was at 37–61 µg/106 cells per day, in close agreement with the secretion rate of ~61 µg/106 cells per day from hepatocytes in the body.
Hyaluronic acid (HA)-PEG
A 384-microplat format, multi-layered, 3D co-culture system was engineered using aqueous solutions of thiolated HA and thiol reactive PEG diacrylate (HA-PEGDA).[114] This composite formed a cushion layer preventing cellular interactions with the plate surface and overlaid with a layer of cancer cells, such as a bone metastatic prostate cancer cells and uterine cancer cells, suspended in HA-PEGDA. Both cancer cells proliferated and formed compact spheroids. A more complex model was also developed by including stromal cells, such ESS1 endometrial stromal sarcoma cells or HS27A bone marrow stroma cells, in HA-collagen and overlaying the suspension on the layer containing cancer cells. In co-cultures, spheroids of both cells showed their native phenotypes. Cells in prostate cancer spheroids expressed prostate-specific antigen (PSA) and epidermal growth factor receptor (EGFR), and the uterine cancer cells expressed mucin 1 and an estrogen-induced gene 121 protein. Morphologically, stromal cells in the co-culture model formed small aggregates rather than showing an elongated shape typical of fibroblast cells. Co-culture with cancer cells also led to both cytoplasmic and nuclear of expression HDAC2, in contrast to its nuclear localization in monocultures. It was also shown that stromal cells poorly adhered to cancer cells, indicating that communication between cancer and stromal cells is mediated by paracrine signaling. The utility of the model was demonstrated for high throughput screening of 232 chemical compounds that generated data reflecting the performance of the drugs in vivo.
The utility of thiolated hyaluronic acid (HA-SH) and PEG-DA to form HA-SH/PEG-DA hydrogels was shown by culturing patient derived prostate cancer cells.[94] The tumoroids cultured in the hydrogels retained close cell-cell contacts and the epithelial phenotype of the native tumors. Androgen receptor was mainly localized in the nucleus of the MDA PCa 183 cells (androgen dependent prostate carcinoma) in tumoroids compared to MDA PCa118b cells (androgen-receptor negative castrate-resistant prostate carcinoma), consistent with in vivo models. Primary cells in 3D hydrogel cultures were resistant to a chemotherapy drug, docetaxel, compared to spheroids generated form a bone metastatic prostate cancer cell line (C4-2B) that showed dose-response to the drug.
Highlight: These studies benefited from the flexible chemistry of PEG to either functionalize PEG hydrogels with ligands for integrins on cancer cells or create composite PEG-based hydrogels. Morphology of 3D cultures of metastatic cancer cells in PEG-based hydrogels depended on matrix stiffness. Cancer cells, including primary patient-derived cells, in these hydrogels formed spheroid cultures that displayed hallmarks of malignant cancer cells, such as significant loss of polarity and organization, elevated nuclear cytoplasmic ratio and nuclear volume density, expression of tumor-specific antigens, gain of tissue-specific functions such as protein secretion at physiologic levels, and resistance to chemotherapy drugs.
B. Polyethylene glycol-dextran (PEG-DEX) aqueous two-phase systems (ATPS)
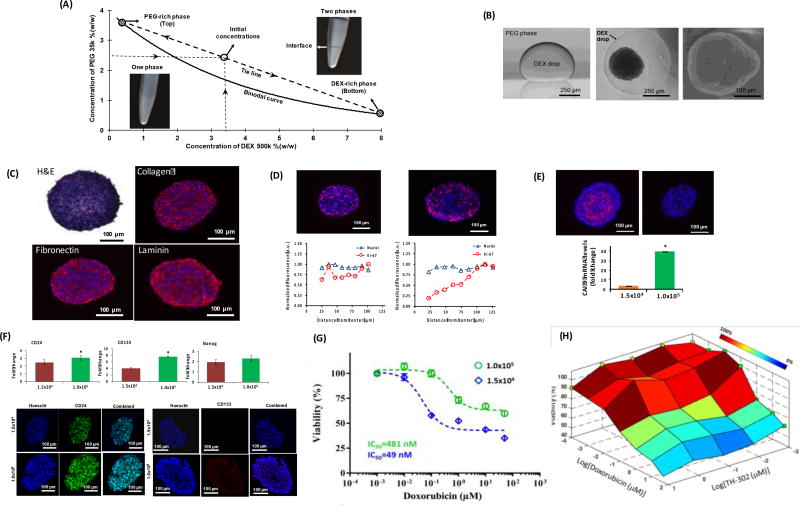
Aqueous solutions of PEG and DEX above certain concentrations result in two immiscible, highly aqueous phases [Figure 8A].[115] PEG-DEX ATPS provide a mild environment for various cells, including cancer cells, and have been widely used for cell and biomolecule micropatterning applications. [116–121]The PEG-DEX ATPS was used to develop a scaffold-free approach to conveniently generate spheroid cultures of cancer cells. The denser aqueous DEX phase solution containing cancer cells was dispensed as a submicroliter drop into a non-adherent microwell containing the immersion aqueous PEG phase.[38, 122] An ATPS was formulated with specific concentrations and molecular weights of PEG and DEX to result in an ultralow interfacial tension of 0.012 mJ/m2 between the two aqueous phases and effectively partition cancer cells to the DEX phase drop.[123] Confinement of cancer cells within a nanodrop promoted their self-assembly and aggregation into a single spheroid within 24 hours of incubation [Figure 8B]. Importantly, the PEG-DEX ATPS allows free diffusion of nutrients from the immersion PEG phase into the DEX phase drop containing the spheroid and diffusive removal of waste products of cells from the drop phase. After formation of spheroids, addition of media reduces concentrations of the polymers and reverts the ATPS to a single media phase. The trace amounts of PEG and DEX remaining in the media do not interfere with cell viability and growth, or diffusion of drug molecules to the spheroid.[38]
Figure 8.
PEG-DEX ATPS to model different to study tumor biology and conduct drug testing. (A) A characteristic phase diagram for PEG-DEX ATPS shows the binodal curve and a concentrations of phase polymers used to generate a two-phase system. (B) Skin cancer cells spheroid formation inside the DEX drop immersed in the PEG phase. Triple negative breast cancer spheroids secrete endogenous ECM proteins, (D) contain actively proliferating cells (Ki67+, pink color) distributed in a size-dependent manner, (E) displaying size-dependent hypoxia (pimonidazole, pink color), (F), show elevation of cancer stem cell markers under hypoxia, (G) display resistance to doxorubicin under hypoxia (diamonds), and (H) become sensitive to doxorubicin by combination treatment with a hypoxia pro-drug TH-302. (A–B) were reproduced with permission. [122] Copyright 2014, John Wiley and Sons. (C–H) were reproduced with permission. [124] Copyright 2016, John Wiley and Sons.
This facile technology eliminated major difficulties with other spheroid formation techniques, such as formation of multiple spheroids in wells, inconsistency of size of spheroids, need for special plates, loss of spheroids during liquid handling, and incompatibility with standard liquid handling tools and screening instruments.[6] Spheroids of triple negative breast cancer cells generated with the ATPS technology reproduce major biological properties of solid breast tumors.[124] This included growth of spheroids over time, secretion and deposition of major ECM proteins such as collagen I, fibronectin, and laminin by cancer cells [Figure 8C], gradients of proliferative cells [Figure 8D], size- and density-dependent hypoxia [Figure 8E], expression of cancer stem cell markers [Figure 8F], and hypoxia-mediated resistance to doxorubicin and cisplatin [Figure 8G]. Hypoxia was modelled by varying cellular density. Spheroids formed using 1×105 MDA-MB-157 cells showed high expression of hypoxia-related genes and proteins (HIF-1α and carbonic anhydrase 9), and cancer stem cell markers (CD24, CD133, NANOG), compared to spheroids formed using 1.5×103 cells that were not hypoxic. Chemotherapy drug resistance of hypoxic spheroids was significantly and synergistically reduced by a combination treatment using a hypoxia activated pro-drug, TH-302, and doxorubicin [Figure 8H]. This technology has successfully been used to form spheroids of various breast, brain, skin, and colon cancer cells. Moreover, ATPS works with standard microwell plates for automated generation, drug treatment, and in situ analysis of spheroids using robotic liquid handling tools.[125] The potential of this approach for high throughput drug screening was demonstrated by single- and dual-agent testing of a collection of anti-cancer compounds against spheroids of brain, breast, and colon cancer cells to identify treatments that effectively induce cytotoxic or cytostatic effects.[35, 36]
C. Polycaprolactone (PCL)
Polycaprolactone can be produced through poly-condensation of a hydroxycarboxylic acid, 6-hydroxyhexanoic acid, and the ring-opening polymerisation (ROP) of a lactone, epsilon-caprolactone (epsilon-CL).[126] Electrospinning was used to form PCL nanofibers that promote cancer cell infiltration and attachment. Immunohistochemical analysis of TC-71 Ewing sarcoma cell spheroids cultured in 3D electrospun PCL scaffolds showed that spheroids preserved major markers (CD99+, keratin− and smooth muscle actin) routinely used for diagnosis in patients.[127] Compared to cells in 2D cultures, spheroids displayed slower growth but significantly upregulated phosphorylation of IGF-1R, similar to activation of the receptor in xenograft tumors. Inhibition of IGF-1R signaling in 3D cultures of TC-71 cells in PCL scaffolds using MK-0646 (a humanized IgG1 monoclonal antibody) caused expression of c-kit and human epidermal growth factor receptor 2 (HER2/neu), both of which are implicated in resistance to IGF-1R–targeted therapy, and constitutive phosphorylation of insulin receptor IR-β. Formation of hybrid IGF-1R/IR receptor heterodimers has been reported as a major mechanism of resistance to IGF-1R targeted therapy.[128] Importantly, data from the 3D models were consistent with those from the xenograft tumors of Ewing sarcoma cells, which could not be captured with 2D culture models.
MCF-7 cells cultured in 3D printed circular PCL scaffolds showed enrichment of cancer stem cells.[129] Similarly, using composite electrospun PCL/chitosan nanofiber scaffolds to culture MCF-7 and T47-D breast cancer cells led to enrichment of CD44−/CD24− cells highly capable of forming mammospheres.[130] These cells isolated from the scaffold and cultured in microplates showed greater resistance to treatment with docetaxel and doxorubicin. Although mechanisms of enrichment of breast cancer stem cells was not fully explored, the technique enabled enrichment of cancer stem cells for drug discovery against these inherently drug-resistant cells.
Prostate cancer most commonly metastasizes to bone, underscoring the need to understand tumor growth and response to therapy in this environment. A 3D tissue engineered bone construct (TEB) was formed by mineralizing human osteoblast (hOB) cells in a PCL-tricalcium phosphate (mPCL-TCP) scaffolds.[131] Interactions of metastasized prostate cancer cells with hOB were studied by culturing PC3 or LNCaP cells in the TEB construct. This promoted aggressiveness of prostate cancer cells that showed elevated levels of steroidogenic enzymes and prostate specific antigen (PSA), a biomarker of prostate cancer progression, in hOB-LNCaP co-culture relative to LNCaP control only.[132] MMP-9 activity in hOB and PC3 co-culture was highly upregulated but not in the tissue engineered bone construct (TEB) construct control groups. Similar enhancement of MMP-9 activity was observed in xenografts colonized with PC3 cells.[133] Furthermore, incorporating LNCaP in PEG hydrogels to avoid direct cellular contacts of LNCaP and hOB cells led to the identification of potential paracrine signaling molecules in bone metastasis. Expression of androgen-regulated genes in prostate cancer spheroids was induced by bone cells in co-culture, indicating the role of bone derived stromal soluble factors in growth of prostate tumors.[134] This phenomenon has been described in co-culture studies of LNCaP and osteoblast-like SaOs2 cells,[135] and upregulation of TGF-β1 signaling has been implicated in bone metastasis of prostate cancer cells.[136]
D. Poly(lactic-co-glycolic) acid (PLGA)
PLGA is a copolymer of poly lactic acid (PLA) and poly glycolic acid (PGA). PLGA hydrogels promote adhesion and proliferation of cancer cells. Highly porous PLGA microspheres were synthesized using an oil-water emulsion. The surface of the microspheres was amino-lysed and coated with type I collagen to facilitate culturing HO8910 ovarian cancer cells. Cells on microspheres proliferated and expressed the epithelial cell marker E-cadherin.[137] Collagen-coated porous PLGA scaffolds were also used to culture U251 glioblastoma spheroids.[138] Compared to 2D or freestanding spheroid cultures of the cells, spheroids in PLGA scaffolds showed more glycolysis, higher expression of angiogenic factors, and greater resistance to doxorubicin treatment. The U251 cells in PLGA scaffolds resisted apoptosis (low caspase activity) by upregulating apoptosis-resistance proteins such as survivin and BCL-2. Cells also upregulated angiogenic factors VEGF and bFGF.[139] Under hypoxia, spheroids showed increased resistance to doxorubicin but interestingly, the apoptosis-resistance proteins (survivin and BCL-2) were downregulated, indicating that drug resistance of hypoxic spheroids was independent of these anti-apoptotic proteins and potentially dependent on pathways that involve VEGF and bFGF signaling. A more complex 3D nanofibrous scaffold was fabricated by electrospinning a mixture of PLGA, PLA, and monopolyethylene glycol (mPEG), that was designated as 3P.[140] Spheroid formation of MCF-7 breast cancer, PC3 prostate cancer, B16 melanoma, BG1 ovarian cancer, and LLC1 Lewis lung cancer cells was demonstrated and depended on the surface topography and charge of the 3P scaffolds. Compared to 2D culture of cells, spheroids in the 3P scaffolds showed upregulated levels of an EMT marker, vimentin, over time, and reduced E-cadherin expression. Treating spheroids with a PI3K inhibitor, LY294002, and a MAPK inhibitor, U0126, abrogated the EMT phenotype and restored E-cadherin expression. The 3P scaffolds were shown to also facilitate 3D culture formation with LLC1 cell suspension collected by fine needle aspirates from tumor biopsies of C57BL/6 mice.
In another study, a highly porous poly(lactide-co-glycolide) (PLG) matrix was fabricated using a gas foaming-particulate leaching process to culture OSCC-3 oral squamous cell carcinoma cells.[141] Temporal production of VEGF and IL-8 in spheroid cultures in the PLG scaffolds was similar to that present in tumor tissues. Tumors formed in xenografts by implanting 3D PLG pre-cultured OSCC-3 cells contained more blood vessels relative to the density of blood vessels in tumors formed by implanting 2D pre-cultured cells. The tumors formed from 3D PLG pre-cultured spheroids expressed higher α5-integrin receptors that are associated with malignancy of cancer cells.[142] The stroma invaded by tumor cells in vivo contained myofibroblasts, which are known to promote a permissive environment for cancer cell invasion and growth.[143] Furthermore, OSCC-3 spheroids in 3D PLG scaffolds resisted treatment with a PI3K inhibitor, LY294002, targeting driver PI3K mutations in this oral squamous cell carcinoma cell line.
E. Thermo-responsive hydrogels
Temperature responsive biomaterials allow self-assembly of hydrogels by a temperature change, encapsulation of cancer cells upon incubation in 37°C and supporting cells to form spheroids, and isolation of spheroids after cooling the hydrogels to liquefy. For example, a thermo-reversible hydrogel poly (N-isopropylacrylamide-co-acrylic acid) microgel (PNIPAM-AA) was constructed for in situ generation and release of HepG2 spheroids. PNIPAM-AA exhibited less shrinkage for long-term cultures and maintained the scaffold structure. HepG2 cells proliferated best in the hydrogel with 1% AA in the copolymer.[144] Galactosylated PNIPAM-AA microgels enhanced liver-specific functions of HepG2 spheroids in terms of albumin secretion and urea synthesis over a three-week culture period.[145]
Highlight: The above studies showed successful use of PEG-DEX ATPS, PCL hydrogels, PLGA hydrogels and modified forms of these hydrogels for spheroid cultures with a wide variety of cancer cells. In contrast to 2D cultures, spheroids showed slower growth but enhanced activity of receptor tyrosine kinases (RTK), such as IGF-1R, and resistance to corresponding targeted treatments as observed in xenograft tumors. Enrichment of cancer stem cells, glycolysis, expression of angiogenic factors VEGF, bFGF, and IL-8, upregulation of apoptosis-resistance proteins such as survivin and BCL-2, chemotherapy drug resistance, and stromal cell-mediated aggressiveness of metastatic cancer cells are hallmark properties of tumors reproduced in spheroid cultures with these biomaterials.
4. Organoid models
While tumor spheroid models have been widely applied in studies of basic tumor growth, angiogenesis, and drug resistance as discussed above, their clinical value has been hindered by reliance on established cancer cell lines, which fail to capture the complexity and functionality of real tumors. To overcome this limitation, organoid models that provide a more complex and physiologic model than cancer cell lines and spheroids have recently been developed. Organoids have demonstrated excellent potential for disease modeling,[146] drug screening,[147] and tissue engineering for drug testing and organ replacement. Known as 3D ex vivo cellular cultures, organoids form either through self-organization or directed assembly under specific organogenesis cues.[148] Organoids not only physically resemble the architecture, cellular organization, and composition of the original tissue, [149] they also recapture genetic signatures of their in vivo counterparts. [150] As compared with spheroids, organoids contain several cell spatially-restricted lineages of committed cell types generated from either pluripotent stem cells (PSCs) or organ-specific adult stem cells (ASCs).[151] They can be efficiently established and stably propagated to model benign and malignant tissues including kidney,[152] breast,[153] lung,[154, 155] colon,[156, 157] prostate,[158] stomach,[159] liver,[160] thymus,[161] pancreas,[162, 163] brain,[164, 165] ureter,[152] and lymph.[166]
4.1 Organoids as disease models
While research with organoids mainly has focused on tissue engineering and regeneration, [148, 167] there is also a significant clinical need for biomimetic tumor models to bridge the technological gap between standard 2D cultures, 3D cultures such as spheroids, and in vivo models of cancer generated from established cell lines. However, compared to the large body of work using tumor spheroids, very few studies have attempted to engineer spatio-temporally organized organoid platforms to recapitulate complex tumor microenvironments. Recent developments in human patient-derived organoids has shed light on precise disease modeling. Directly generated from patient biopsies and resections, tumor organoids (tumoroids) recapitulate patient-specific histological features and physiological phenotypes in a very efficient and stable manner, therefore showing great potential in drug screening and precision medicine.[158, 168] A key advance in organoid culture started from intestinal organoids developed by Sato and Clevers, where human intestinal stem cells self-assembled into crypt-like structures in Matrigel.[169] Matrigel encapsulation has since become the most commonly used approach for all types of organoid cultures. Later, Ootani et al. established a different type of organoid culture system that better mimics intestinal stem cell niches, using a collagen gel-based air-liquid interface (ALI) method.[170] The ALI method has been applied to derive organoids from oncogene transgenic mouse gastrointestinal tissues,[157] and subsequently human colorectal tissues.[171] The resulting patient-derived ALI tumoroids closely recapitulated epithelium structures of the original tumor and demonstrated resistance to cancer therapies.
Besides derivation from patient tumor samples, tumor organoids can also be generated from normal tissue using gene manipulation technologies.[172–174] Matano and Sato introduced five gene mutations, including tumor suppressor genes APC, SMAD4, and TP53, as well as the oncogenes KRAS and/or PIK3CA, into normal human intestinal organoids. Engineered organoids formed tumors following implantation in mice. However, compared to organoids derived from chromosome-unstable human adenomas that formed macrometastatic colonies, the genetically engineered organoids failed to colonize in the liver, indicating that 'driver' pathway mutations alone are not adequate to induce invasive behavior.[172] However, in another study, transplanted colon organoids recapitulated not only tumor formation but also metastasis in vivo.[175] Organoids either assembled from human patient-derived samples or genetically engineered mouse models showed time-dependent progression from adenoma to metastasis in vivo. Moreover, this study also demonstrated the significant role of dysregulated Wnt signaling in progression of disseminated colorectal cancer cells. These studies highlight how transplantation of ex vivo engineered organoids provides a flexible platform to model all stages of colorectal cancer development.
Building on successes with intestinal organoids, investigators now are applying this culture method to cancer and other diseases. For example, 3D prostate tumoroids derived from prostate cancer circulating tumor cells provide an alternative, efficient approach to investigate intratumor heterogeneity as compared to tissue biopsy.[176] Kidney organoids with renal progenitors derived from iPSCs successfully repaired acute kidney injury, suggesting the possibility of using organoids in regenerative therapy for kidney diseases.[177] Recently, patient-derived endometrial organoids also have been developed and applied for endocrine-based and drug sensitivity testing with high success rates and reasonable reproducibility.[147]
4.2 Biomaterials for organoids culture
Since organoid formation highly depends on the self-assembling capacity of cells, it is essential to create a microenvironment with required niche factors. Unlike spheroid cultures that have employed a variety of materials, natural biomaterials, especially animal derived-ECM matrices, such as Matrigel and collagen, are the primary materials used for organoid culture.
A. Matrigel
Matrigel is a basement membrane (BM) extract composed of a complex mixture of over 1000 proteins. It is well-known as the most BM-like natural material, with type IV collagen, laminin and nidogen as major components. As the most commonly-used material for organoid culture, Matrigel ultimately augments the self-assembling capacity of PSCs.[178, 179] Since its initial development several decades ago, applications of Matrigel far exceed other biomaterials due to several major advantages: built-in complex distribution of nutrients and protein gradients, ease in handling and fast gelling kinetics, and the ready availability of a commercialized product with high quality control. However, Matrigel has several notable limitations for tissue engineering. First, the inherent compositional variability usually results in lack of control over individual specific microenvironmental parameters. More importantly, due to the cocktail of growth factors in Matrigel, the simultaneous occurrence of signaling cascades may confound signal transduction in cells undergoing organogenesis, leading to an incomplete understanding of self-assembly mechanisms.[180] Second, the fast gelling of Matrigel does not allow precise control over gelation kinetics, leading to uncertain microstructure of the final network.[181] The inability to manipulate mechanical properties also limits its application in studying mechanotransduction during organogenesis. Finally, although Matrigel is a widely supplied commercialized product, issues with reproducibility could still arise due to the inherently inconsistent composition and batch-to-batch variability. The lack of consistency can result in problems of genetic drift in organoid formation, which is particularly significant if investigators subculture and passage organoids.
B. Other biomaterials
As described above, most studies of organoids as disease models have focused on addition of specific exogenous signaling molecules to modulate the organoid development, cellular biology, and dysfunction. The role of the microenvironment, especially mechanical cues such as matrix stiffness and permeability, have rarely been taken into consideration. The use of Matrigel or collagen results in imprecise control of the mechanical environment, further complicating studies of mechanical cues driving organoid formation and differentiation. For example, intestinal organoid models require a 3D matrix to facilitate complex mechanical functions, including contraction needed for peristalsis.[182, 183] In order to contract, the mechanical properties of matrix must permit elastic deformation with a defined porosity, providing binding sites for cell adhesion and cellular migration. However, conventional natural materials cannot satisfy these requirements because manipulation of stiffness by varying components concentrations or crosslinking usually leads to the changes in matrix density as well as architecture and biochemical factors. [181]Therefore, biomaterials with adjustable mechanical and biochemical properties are in demand to replace Matrigel for organoid culture.
Beck et al. overcame the uncontrollable mechanical properties of Matrigel by incorporating PEG to build a PEG-Matrigel composite hydrogel.[184] The varied ratio of PEGDA and PEG-Monoacrylate (PEGMA) changed the extent of crosslinking of the PEG network, leading to changes in rigidity (from 50 to 4000 Pa) of the matrix with constant concentrations of ECM ligands. However, organoids derived from mammary carcinoma did not show protrusive migration or local dissemination in PEG-Matrigel composite hydrogels with varied stiffness. To induce cell migration, a second series of materials in which adhesive peptides were non-covalently incorporated into the PEG network were developed. Although the resultant adhesive PEG-Matrigel composite hydrogel induced some dissemination of epithelial cells at low rigidity, the limited cell migration observed in this study indicates failure to reproduce tumor invasion in vivo, partially due to the unfavorable hydrogel microstructure. Recently DiMarco et al. developed a recombinant engineered ECM containing an elastin-like structural backbone and extensions of cell-adhesive RGD peptides to precisely control biomechanical and biochemical cues for intestinal organoids. This structure enabled independent tuning of matrix stiffness decoupled from adhesion to understand the individual effect of matrix biomechanical and biochemical forces on intestinal organoids.[185]
5. Perspectives/Outlook
5.1 Advanced tumor modeling
Spheroid and organoid models have already accelerated understanding of organogenesis. In addition, tumor models using spheroids and organoids also offer the potential to improve patient-specific precision medicine. A primary limitation of current tumoroids models is lack of control over biochemical and mechanical signals that are crucial to tumor formation and metastasis. For future advanced tumor modeling, we believe that addition of spatiotemporal chemical/mechanical gradients in the matrix will bridge the gap between ex vivo tumoroids culture and in vivo tumor-stromal crosstalk. An ability to further sculpt the biophysical and biochemical microenvironment will help control and dissect intercellular signaling in cancer. With the incorporation of functional biomaterials and niche factors, tumor modeling will provide a more mechanistic understanding of how microenvironmental factors influence tumorigenesis and metastasis.
This is highlighted in a recent work from our group that developed a tumor model by incorporating multicellular spheroids in a new 3D hybrid hydrogel system composed of collagen and alginate.[186] Within this well-defined mechanical microenvironment, we showed that human mammary fibroblast (HMF) cells facilitated migration of MDA-MB-231 breast cancer cells out of spheroids and into the surrounding matrix in a sun-burst pattern. Based on these findings, we further designed a model using the same gel but containing dissociated cancer cells and HMF spheroids. As shown in Figure 9, the spheroid made of HMF cells expressing chemokine CXCL12 was embedded in the collagen-alginate hydrogel with suspended MDA-MB-231 cancer cells expressing CXCR4. After 5 days of culture in the 3D gel system, cells from the HMF spheroid invaded the gel radially, while the surrounding cancer cells migrated towards the spheroid in the same pattern. In this case, instead of defining the path for cancer cells, HMF spheroids attracted cancer cells from the matrix through gradients of signaling molecules. Migration of both cell types followed the same track with a radial orientation, indicating the reorganization of matrix network, and tumor-stromal interactions. This system provides a technology to investigate interactions among gradients of signaling molecules, multiple cell types, and ECM remodeling in cancer cell migration. By further manipulating this system or other similar models, investigators will be able to more precisely identify mechanisms driving tumor progression and test potential therapies to block these steps.
Figure 9.
Collagen-alginate matrix to model breast tumor-stromal interactions. Spheroid of human mammary fibroblasts (HMF, red) that express the chemokine CXCL12 is embedded in a collagen-alginate hydrogel mixed with dissociated triple negative breast cancer cells (green) expressing CXCR4 receptor. After 3 days of culture, cancer cells oriented and migrate toward the spheroid in a sun-burst pattern, indicating the binding of CXCR4 to CXCL12 induces cancer cell invasion. Images were taken using two-photon microscopy and with a 25X objective. (a) Maximum intensity over the z-projection view of the field. (b) 3D reconstruction view of the gel.
Another important consideration for future models is incorporation of tumor vasculature. Angiogenesis, the development of new blood vessels in tumors from surrounding vessles, provides cancer cells access to nutrients and oxygen to support anabolic metabolism and overall tumor growth. Incorporating vasculature in advanced tumor models will help understand regulation of drug responses of cancer cells and develop therapeutics to target angiogenesis. In this progress report, we highlighted that signaling between cancer cells and ECM in 3D tumor models upregulates major pro-angiogenic factors such as VEGF and IL-8. Although there are several 3D co-culture models of cancer and endothelial cells, absence of geometric and physiochemical guidance results in randomly assembled endothelial cells and lack of control of angiogenesis. Recently, a bottom-up approach was used to develop an advanced 3D vascular tumor model that showed increased drug resistance of mammary tumors.[187] Initially, avascular microtumors were formed in alginate collagen microcapsules. These microtumors were then used as a building block for assembling with endothelial cells and stromal cells to create a macroscale 3D vascularized tumor. These macroscale tumors were then cultured in microfluidic channel allowing perfusion of nutrients into the macroscale 3D vascularized tumor and removal of waste products. The vascular macroscale tumor showed 4.7 to 139.5 times greater resistance to doxorubicin than the avascular mammary tumor model. Furthermore, such advanced macroscale 3D vascular models offer a useful tool for discovery of new anti-angiogenic drugs and studies of molecular mechanisms of uncontrolled angiogenesis in cancer. We beleive that further developments of this type of bottom-up-approach and other bioprinting techiniques described in the next section will provide greater opportunities for cancer research and drug discovery.
5.2 Biomaterials and 3D printing
Conventional natural hydrogel materials present a major roadblock in building sophisticated tumor models because of limitations to form a well-defined architecture. Biomaterial-based 3D printing offers a potential solution and allows a more accurate and systematic control to reconstruct the tumor microenvironment.[188, 189] Biomaterials have been incorporated into 3D rapid prototyping to enable a more modular, controlled approach for reproducing intricacies of actual tissues. Bioprinting technology makes it possible to precisely position cells of different types in relation to each other in a 3D environment using hydrogel-based bioinks.[190–192] Consequently, bioprinted organoids can more closely replicate anatomy and functions of target tissues or organs for disease modeling and drug testing. In addition, incorporation of drug release and delivery by hydrogel bioinks will offer a dynamic model to study biomechanical gradients and effects on cell behaviors.[193] We believe that further integration of spheroids and organoids with 3D bioprinting will continue to enhance research applications of these models through spatiotemporal control of specific microenvironmental cues. Using multiple printer heads, cell-laden materials with various bioligands or sequestered soluble signaling molecules can be engineered to elicit targeted cellular assembly, imitating in vivo spatiotemporal dynamics of tissue formation and cancer.
In the last two decades, various bioprinting techniques with a variety of bioink materials have emerged. However, most of the current 3D bioprinting techniques have only shown simultaneous deposition of cell combinations of different types encapsulated within bioinks via a layer-by-layer deposition process.[188] The resultant bioinspired tissue constructs are still in early stages of prototyping and development. Incorporation of organoids into bioinks to substitute dissociated cells provides a promising solution because it offers a secondary hierarchical structure over the self-assembled microstructure of organoids, leading to a more complex tissue structures.[194, 195] Bioprinting of uniluminal vascular spheroids produced elongated structures, due to the fusion of spheroids in the 3D matrix, that resemble segments of intraorgan branched vascular trees.[194] For future engineering of large tissue constructs and tumor environments, bioprinting with more sophisticated control over the fusion of thousands of spheroids will be necessary for automated production and testing. Spheroid- and organoid-based 3D bioprinting not only allows multiscale assembly of tissue units to complex hierarchical organ systems, it also provides a more sophisticated platform for future tumor modeling.
As 3D bioprinting of organoids becomes more sophisticated and follows rational design principles, biomaterials with precisely adjustable properties will play a significant role in directing this process.[196] Therefore, advanced biomaterials are in large demand to accommodate several key features including a large parameter space for properties, stability, and capabilities for drug loading and delivery. In this regard, seminal studies have begun to optimize commonly used bioinks and explore new materials with more specialized, organ-specific properties.[197, 198] Still, more efforts are needed to fabricate novel bioinks that meet both cytocompatibility and mechanical strength requirements for 3D bioprinting.
Three-dimensional bioprinted organ models of kidney, liver, and heart are already used for testing and identifiying novel drugs. Such models also are used testing safety and efficacy of drugs in a pharmaceutical setting. Although some pharmaceutical companies have adapted 3D inkject bioprinters for reserach applications, broad utility of bioprinters for drug discovery requires greater investements to increase availability of commercialized products. Use of 3D bioprinting in pharmaceutical industries is limited by various factors. First, bioinks/biomaterials are limited. Discovery of novel bioinks is essential to help manufacturers standardize and automate bioprinters and bioprinting processes. Most methods for bioprinting organs are limited to use of scaffold-based biomaterials, and very few studies focus on identifying potential scaffold-free biomaterials. A hybrid technology that combines scaffold-free and scaffold-based biomaterials could potentially advance development of bioprinters and the bioprinting process. Second, a major limitation of bioprinting technology is the difficulty of producing large tissues of clinically relevant size and shapes. Recent approaches are limited to small tissues and organ models. Scaling up to larger tissues is more challenging due to complexities of architecture and heterogeneity of native tissues. With existing capabilities, producing large constructs requires prolonged printing. Enhanced technological capabilities to expedite the printing process are highly desirable. Third, commercialized biprinters are costly, ranging from approximately $150k to $200k, and lack versalitiy. Existing bioprinters are rarely customizable by individual users. Most bioprinters are not compatibile with available bioinks, or are not able to dispense various bioinks simultaneously. Bioprinting techniques typically lack full automation and require considerable investment of hands-on time to construct 3D tissue constructs. Low spataial resolution of available bioprints in terms of accurate placement of bioinks detracts from their use for high-throughput screening. Therefore, parallel developments of bioinks/biomaterials and technologies for bioprinters are essential to scale-up bioprinting of 3D tissues, including tumors, and facilitate broader use of bioprinting techniques in pharmaceutical industries.
To conclude, in the next decade, we envision that tumor modeling using engineered biomaterials will be essential to understanding basic mechanisms of cancer and advancing precision medicine to cure more patients of cancer.
Supplementary Material
Acknowledgments
This work was supported by funding from National Institutes of Health R21CA182333, R01CA196018, and U01CA210152. We thank Ms. Ramila Joshi for help to edit the manuscript.
Biographies

Mr. Pradip Shahi Thakuri is a PhD student in the Department of Biomedical Engineering at The University of Akron, Ohio. He received his Bachelor’s degree in Biomedical Engineering in Nepal. His current research interests include anti-cancer drug screening with tumor spheroid model, developing 3D models for cancer drug resistance, and investigating cancer drug treatment regimens with the 3D cancer models.

Dr. Gary Luker is an Associate Professor of Radiology, Biomedical Engineering, and Microbiology and Immunology and the Associate Chair for Clinical Research in Radiology at the University of Michigan. He is a physician-scientist and member of the American Society for Clinical Investigation. His research focuses on tumor-stromal signaling in tumor progression, metastasis, dormancy, and response to therapy.

Dr. Hossein Tavana is an Associate Professor of Biomedical Engineering at The University of Akron. His primary research interests include engineering 3D tumor models that enable cancer drug testing and resistance studies, developing cell migration assays to identify novel compounds against cancer cell motility, and engineering stem cells microenvironment to control differentiation to neural cells.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Pradip Shahi Thakuri, 260 S. Forge St., Akron, OH 44325, USA.
Dr. Chun Liu, 109 Zina Pitcher Place, Ann Arbor, MI 48109, USA
Prof. Gary D. Luker, 109 Zina Pitcher Place, Ann Arbor, MI 48109, USA
Prof. Hossein Tavana, 260 S. Forge St., Akron, OH 44325, USA
References
- 1.Goodspeed A, Heiser LM, Gray JW, Costello JC. Mol. Cancer Res. 2016;14:3. doi: 10.1158/1541-7786.MCR-15-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma SV, Haber DA, Settleman J. Nat. Rev. Cancer. 2010;10:241. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 3.Hughes J, Rees S, Kalindjian S, Philpott K. Br. J. Pharmacol. 2011;162:1239. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav B, Gopalacharyulu P, Pemovska T, Khan SA, Szwajda A, Tang J, Wennerberg K, Aittokallio T. Dis. Model. Mech. 2015;8:1255. doi: 10.1242/dmm.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foley GE, Eagle H. Cancer Res. 1958;18 [PubMed] [Google Scholar]
- 6.Ham SL, Joshi R, Thakuri PS, Tavana H. Exp. Biol. Med. (Maywood) 2016:1535370216643772. doi: 10.1177/1535370216643772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchinson L, Kirk R. Nat. Rev. Clin. Oncol. 2011;8:189. doi: 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 8.Horvath P, Aulner N, Bickle M, Davies AM, Del Nery E, Ebner D, Montoya MC, Östling P, Pietiäinen V, Price LS, Shorte SL, Turcatti G, von Schantz C, Carragher NO. Nat. Rev. Drug Discov. 2016;15:751. doi: 10.1038/nrd.2016.175. [DOI] [PubMed] [Google Scholar]
- 9.Lindner MD. Pharmacol. Ther. 2007;115:148. doi: 10.1016/j.pharmthera.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Pammolli F, Magazzini L, Riccaboni M. Nat. Rev. Drug Discov. 2011;10:428. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- 11.DiMasi JA, Hansen RW, Grabowski HG. J. Health Econ. 2003;22:151. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 12.Lindsay MA. Nat. Rev. Drug Discov. 2003;2:831. doi: 10.1038/nrd1202. [DOI] [PubMed] [Google Scholar]
- 13.Mullard A. Nat. Rev. Drug Discov. 2015;14:373. doi: 10.1038/nrd4730. [DOI] [PubMed] [Google Scholar]
- 14.Weigelt B, Ghajar CM, Bissell MJ. Adv. Drug Deliv. Rev. 2014;69–70:42. doi: 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu T-Y, Wang Z, Bai J, Sun W, Peng WK, Huang RY-J, Thiery J-P, Kamm RD. Adv. Healthc. Mater. 2014;3:609. doi: 10.1002/adhm.201300151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haisler WL, Timm DM, Gage JA, Tseng H, Killian TC, Souza GR. Nat. Protoc. 2013;8:1940. doi: 10.1038/nprot.2013.125. [DOI] [PubMed] [Google Scholar]
- 17.Pavesi A, Adriani G, Tay A, Warkiani ME, Yeap WH, Wong SC, Kamm RD. Sci. Rep. 2016;6:26584. doi: 10.1038/srep26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiswald L-B, Bellet D, Dangles-Marie V. Neoplasia. 2015;17:1. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao AY, Torisawa Y, Tung Y-C, Sud S, Taichman RS, Pienta KJ, Takayama S. Biomaterials. 2009;30:3020. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patra B, Chen Y-H, Peng C-C, Lin S-C, Lee C-H, Tung Y-C. Biomicrofluidics. 2013;7:54114. doi: 10.1063/1.4824480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naipal KAT, Verkaik NS, Sánchez H, van Deurzen CHM, den Bakker MA, Hoeijmakers JHJ, Kanaar R, Vreeswijk MPG, Jager A, van Gent DC. BMC Cancer. 2016;16:78. doi: 10.1186/s12885-016-2119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutmacher DW. Nat. Mater. 2010;9:90. doi: 10.1038/nmat2619. [DOI] [PubMed] [Google Scholar]
- 23.Pradhan S, Hassani I, Clary JM, Lipke EA. Tissue Eng. Part B Rev. 2016;22:470. doi: 10.1089/ten.TEB.2015.0567. [DOI] [PubMed] [Google Scholar]
- 24.Asti A, Gioglio L. Int. J. Artif. Organs. 2014;37:187. doi: 10.530/ijao.5000307. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Farach-Carson MC, Jia X. Biotechnol. Adv. 2014;32:1256. doi: 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paschos KA, Canovas D, Bird NC. Cell. Signal. 2009;21:665. doi: 10.1016/j.cellsig.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Friedl P, Wolf K. Nat. Rev. Cancer. 2003;3:362. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 28.Fa R, Legate KR, Wickstro SA, Wickstrom SA, Fassler R. Genes Dev. 2009;23:397. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 29.Vivanco I, Sawyers CL. Nat. Rev. Cancer. 2002;2:489. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 30.Van Dijk M, Göransson SA, Strömblad S. Exp. Cell Res. 2013;319:1663. doi: 10.1016/j.yexcr.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Borah A, Raveendran S, Rochani A, Maekawa T, Kumar DS. Oncogenesis. 2015;4:e177. doi: 10.1038/oncsis.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibue T, Weinberg RA. Nat. Rev. Clin. Oncol. 2017 doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swietach P, Hulikova A, Patiar S, Vaughan-Jones RD, Harris AL. PLoS One. 2012;7:1. doi: 10.1371/journal.pone.0035949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenzel C, Riefke B, Gründemann S, Krebs A, Christian S, Prinz F, Osterland M, Golfier S, Räse S, Ansari N, Esner M, Bickle M, Pampaloni F, Mattheyer C, Stelzer EH, Parczyk K, Prechtl S, Steigemann P. Exp. Cell Res. 2014;323:131. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Thakuri P, Ham S, Luker G, Hossein T. Mol. Pharm. 2016 [Google Scholar]
- 36.Shahi Thakuri P, Tavana H. SLAS Discov. Adv. Life Sci. R&D. 2017;22:507. doi: 10.1177/2472555217698817. [DOI] [PubMed] [Google Scholar]
- 37.Hagemann JAN, Jacobi C, Hahn M, Schmid V, Welz C, Schwenk-Zieger S, Stauber R, Baumeister P, Becker S. Anticancer Res. 2017;37:2201. doi: 10.21873/anticanres.11555. [DOI] [PubMed] [Google Scholar]
- 38.Lemmo S, Atefi E, Luker GD, Tavana H. Cell. Mol. Bioeng. 2014;7:344. doi: 10.1007/s12195-014-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szot CS, Buchanan CF, Freeman JW, Rylander MN. Biomaterials. 2011;32:7905. doi: 10.1016/j.biomaterials.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jokinen J, Dadu E, Nykvist P, Käpylä J, White DJ, Ivaska J, Vehviläinen P, Reunanen H, Larjava H, Häkkinen L, Heino J. J. Biol. Chem. 2004;279:31956. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- 42.Pickup MW, Mouw JK, Weaver VM. EMBO Rep. 2014;15:1243. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzesiak JJ, Bouvet M. Br. J. Cancer. 2006;94:1311. doi: 10.1038/sj.bjc.6603088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong HH, De Sonneville J, Ghotra VPS, Xiong J, Price L, Hogendoorn PCW, Spaink HH, Van De Water B, Danen EHJ. Biomaterials. 2012;33:181. doi: 10.1016/j.biomaterials.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Xiao Z, Meng Y, Zhao Y, Han J, Su G, Chen B, Dai J. Biomaterials. 2012;33:1437. doi: 10.1016/j.biomaterials.2011.10.056. [DOI] [PubMed] [Google Scholar]
- 46.Lee S-C, Xu X, Lim Y-W, Iau P, Sukri N, Lim S-E, Yap HL, Yeo W-L, Tan P, Tan S-H, McLeod H, Goh B-C. Pharmacogenet. Genomics. 2009;19:181. doi: 10.1097/FPC.0b013e32831ebb5d. [DOI] [PubMed] [Google Scholar]
- 47.Lam CRI, Wong HK, Nai S, Chua CK, Tan NS, Tan LP. Mol. Pharm. 2014;11:2016. doi: 10.1021/mp500059q. [DOI] [PubMed] [Google Scholar]
- 48.Fenner J, Stacer AC, Winterroth F, Johnson TD, Luker KE, Luker GD. Sci. Rep. 2014;4:5512. doi: 10.1038/srep05512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krause S, Maffini MV, Soto AM, Sonnenschein C. BMC Cancer. 2010;10:263. doi: 10.1186/1471-2407-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolznig H, Rupp C, Puri C, Haslinger C, Schweifer N, Wieser E, Kerjaschki D, Garin-chesa P. AJPA. 2011;179:487. doi: 10.1016/j.ajpath.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nyga A, Loizidou M, Emberton M, Cheema U. Acta Biomater. 2013;9:7917. doi: 10.1016/j.actbio.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eastwood M, Porter R, Khan U, McGrouther G, Brown R. J. Cell. Physiol. 1996;166:33. doi: 10.1002/(SICI)1097-4652(199601)166:1<33::AID-JCP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 53.Yip D, Cho CH. Biochem. Biophys. Res. Commun. 2013;433:327. doi: 10.1016/j.bbrc.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Rodenhizer D, Gaude E, Cojocari D, Mahadevan R, Frezza C, Wouters BG, McGuigan AP. Nat. Mater. 2015;15:227. doi: 10.1038/nmat4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bissell MJ, Kenny PA, Radisky DC. Cold Spring Harb. Symp. Quant. Biol. 2005;70:343. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9064. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. Mol. Oncol. 2007;1:84. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Härmä V, Virtanen J, Mäkelä R, Happonen A, Mpindi JP, Knuuttila M, Kohonen P, Lötjönen J, Kallioniemi O, Nees M. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, Krieg A, Stoecklein NH. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stankevicius V, Vasauskas G, Bulotiene D, Butkyte S, Jarmalaite S. BMC Cancer. 2016:1. doi: 10.1186/s12885-016-2825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. PLoS One. 2008;3:1. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maglott A, Bartik P, Cosgun S, Klotz P, Rondé P, Fuhrmann G, Takeda K, Martin S, Dontenwill M. Cancer Res. 2006;66:6002. doi: 10.1158/0008-5472.CAN-05-4105. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Z, Vuori K, Reed JC, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6161. doi: 10.1073/pnas.92.13.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, McCaffrey MW, Ozanne BW, Norman JC. Dev. Cell. 2007;13:496. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 65.Nam JM, Onodera Y, Bissell MJ, Park CC. Cancer Res. 2010;70:5238. doi: 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. Cancer Res. 2006;66 doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Fournier MV, Ware JL, Bissell MJ, Jacoub A, Zehner ZE. Mol. Cancer Ther. 2009;8:499. doi: 10.1158/1535-7163.MCT-08-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. J. Cell Biol. 1997;137:231. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. Breast Cancer Res. Treat. 2010;122:35. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Windus LCE, Glover TT, Avery VM. Mol. Cancer. 2013;12:112. doi: 10.1186/1476-4598-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takai A, Fako V, Dang H, Forgues M, Yu Z, Budhu A, Wang XW. Sci. Rep. 2016;6:21174. doi: 10.1038/srep21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verbridge SS, Choi NW, Zheng Y, Brooks DJ, Stroock AD, Fischbach C. Tissue Eng. Part A. 2010;16:2133. doi: 10.1089/ten.tea.2009.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q, Li S, Xie Y, Yu W, Xiong Y, Ma X, Yuan Q. Hepatol. Res. 2006;35:96. doi: 10.1016/j.hepres.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Rao W, Zhao S, Yu J, Lu X, Zynger DL, He X. Biomaterials. 2014;35:7762. doi: 10.1016/j.biomaterials.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agarwal P, Zhao S, Bielecki P, Rao W, Choi JK, Zhao Y, Yu J, Zhang W, He X. Lab Chip. 2013;13:4525. doi: 10.1039/c3lc50678a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao S, Agarwal P, Rao W, Huang H, Zhang R, Liu Z, Yu J, Weisleder N, Zhang W, He X. Integr. Biol. (Camb) 2014;6:874. doi: 10.1039/c4ib00100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao S, Xu Z, Wang H, Reese BE, Gushchina LV, Jiang M, Agarwal P, Xu J, Zhang M, Shen R, Liu Z, Weisleder N, He X. Nat. Commun. 2016;7:13306. doi: 10.1038/ncomms13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Croisier F, Jérôme C. Eur. Polym. J. 2013;49:780. [Google Scholar]
- 79.Chang P, Sekine K, Chao H, Hsu S, Chern E. Nat. Publ. Gr. 2017:1. [Google Scholar]
- 80.Dhiman HK, Ray AR, Panda AK. Biomaterials. 2005;26:979. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 81.Rao W, Wang H, Han J, Zhao S, Dumbleton J, Agarwal P, Zhang W, Zhao G, Yu J, Zynger DL, Lu X, He X. ACS Nano. 2015;9:5725. doi: 10.1021/nn506928p. [DOI] [PubMed] [Google Scholar]
- 82.Li Z, Ramay HR, Hauch KD, Xiao D, Zhang M. Biomaterials. 2005;26:3919. doi: 10.1016/j.biomaterials.2004.09.062. [DOI] [PubMed] [Google Scholar]
- 83.Leung M, Kievit FM, Florczyk SJ, Veiseh O, Wu J, Park JO, Zhang M. Pharm. Res. 2010;27:1939. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang HL, Anatelli F, Zhai QJ, Adley B, Chuang S-TT, Yang XJ. Arch. Pathol. Lab. Med. 2008;132:1723. doi: 10.5858/132.11.1723. [DOI] [PubMed] [Google Scholar]
- 85.Kievit FM, Florczyk SJ, Leung MC, Veiseh O, Park JO, Disis ML, Zhang M. Biomaterials. 2010;31:5903. doi: 10.1016/j.biomaterials.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kievit FM, Florczyk SJ, Leung MC, Wang K, Wu JD, Silber JR, Ellenbogen RG, Lee JSH, Zhang M. Biomaterials. 2014;35:9137. doi: 10.1016/j.biomaterials.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao S, Zhang L, Han J, Chu J, Wang H, Chen X, Wang Y, Tun N, Lu L, Bai X-F, Yearsley M, Devine S, He X, Yu J. ACS Nano. 2016;10:6189. doi: 10.1021/acsnano.6b02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anttila MA, Tammi RH, Tammi MI, Syrja KJ, Saarikoski SV. Cancer Res. 2000:150. [PubMed] [Google Scholar]
- 89.Liu N, Lapcevich RK, Underhill CB, Han Z, Gao F, Swartz G, Plum SM, Zhang L, Green SJ. Cancer Res. 2001;61:1022. [PubMed] [Google Scholar]
- 90.Xu X, Jha AK, Harrington DA, Farach-Carson MC, Jia X. Soft Matter. 2012;8:3280. doi: 10.1039/C2SM06463D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang H, Agarwal P, Zhao S, Xu RX, Yu J, Lu X, He X. Biomaterials. 2015;72:74. doi: 10.1016/j.biomaterials.2015.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H, Agarwal P, Zhao S, Yu J, Lu X, He X. Biomaterials. 2016;97:62. doi: 10.1016/j.biomaterials.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H, Agarwal P, Zhao S, Yu J, Lu X, He X. Adv. Mater. 2016;28:347. doi: 10.1002/adma.201504263. [DOI] [PubMed] [Google Scholar]
- 94.Fong ELS, Martinez M, Yang J, Mikos AG, Navone NM, Harrington DA, Farach-Carson MC. Mol. Pharm. 2014;11:2040. doi: 10.1021/mp500085p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hao Y, Zerdoum AB, Stuffer AJ, Rajasekaran AK, Jia X. Biomacromolecules. 2016;17:3750. doi: 10.1021/acs.biomac.6b01266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fong ELS, Wan X, Yang J, Morgado M, Mikos AG, Harrington DA, Navone NM, Farach-Carson MC. Biomaterials. 2016;77:164. doi: 10.1016/j.biomaterials.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wan X, Corn PG, Yang J, Palanisamy N, Starbuck MW, Efstathiou E, Li Ning Tapia EM, Zurita AJ, Aparicio A, Ravoori MK, Vazquez ES, Robinson DR, Wu Y-M, Cao X, Iyer MK, McKeehan W, Kundra V, Wang F, Troncoso P, Chinnaiyan AM, Logothetis CJ, Navone NM, Navone NM. Sci. Transl. Med. 2014;6:252ra122. doi: 10.1126/scitranslmed.3009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu X, Gurski LA, Zhang C, Harrington DA, Farach-Carson MC, Jia X. Biomaterials. 2012;33:9049. doi: 10.1016/j.biomaterials.2012.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang Y-J, Hsu S. Biomaterials. 2014;35:10070. doi: 10.1016/j.biomaterials.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 100.Wang K, Kievit FM, Erickson AE, Silber JR, Ellenbogen RG, Zhang M. Adv. Healthc. Mater. 2016;5:3173. doi: 10.1002/adhm.201600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jacobs DJ, Rader AJ, Kuhn LA, Thorpe MF. Proteins. 2001;44:150. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 102.Ki CS, Park SY, Kim HJ, Jung H-M, Woo KM, Lee JW, Park YH. Biotechnol. Lett. 2008;30:405. doi: 10.1007/s10529-007-9581-5. [DOI] [PubMed] [Google Scholar]
- 103.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. 2003;24:401. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 104.Tan PHS, Aung KZ, Toh SL, Goh JCH, Nathan SS. Biomaterials. 2011;32:6131. doi: 10.1016/j.biomaterials.2011.04.084. [DOI] [PubMed] [Google Scholar]
- 105.Talukdar S, Kundu SC. Adv. Funct. Mater. 2012;22:4778. [Google Scholar]
- 106.Talukdar S, Mandal M, Hutmacher DW, Russell PJ, Soekmadji C, Kundu SC. Biomaterials. 2011;32:2149. doi: 10.1016/j.biomaterials.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 107.Subia B, Dey T, Sharma S, Kundu SC. ACS Appl. Mater. Interfaces. 2015;7:2269. doi: 10.1021/am506094c. [DOI] [PubMed] [Google Scholar]
- 108.Cheung A, Bax HJ, Josephs DH, Ilieva KM, Pellizzari G, Opzoomer J, Bloomfield J, Fittall M, Grigoriadis A, Figini M, Canevari S, Spicer JF, Tutt AN, Karagiannis SN. Oncotarget. 2016;7:52553. doi: 10.18632/oncotarget.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Biomaterials. 2010;31:8494. doi: 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 110.Almany L, Seliktar D. Biomaterials. 2005;26:2467. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 111.Pradhan S, Hassani I, Seeto WJ, Lipke EA. J. Biomed. Mater. Res. - Part A. 2017;105:236. doi: 10.1002/jbm.a.35899. [DOI] [PubMed] [Google Scholar]
- 112.Pradhan S, Clary JM, Seliktar D, Lipke EA. Biomaterials. 2017;115:141. doi: 10.1016/j.biomaterials.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 113.Moriyama K, Naito S, Wakabayashi R, Goto M, Kamiya N. Biotechnol. J. 2016;11:1452. doi: 10.1002/biot.201600087. [DOI] [PubMed] [Google Scholar]
- 114.Engel BJ, Constantinou PE, Sablatura LK, Doty NJ, Carson DD, Farach-Carson MC, Harrington DA, Zarembinski TI. Adv. Healthc. Mater. 2015;4:1664. doi: 10.1002/adhm.201500258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Albertsson P. Wiley. p. 1986. [Google Scholar]
- 116.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat. Mater. 2009;8:736. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tavana H, Mosadegh B, Takayama S. Adv. Mater. 2010;22:2628. doi: 10.1002/adma.200904271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tavana H, Mosadegh B, Zamankhan P, Grotberg JB, Takayama S. Microprinted feeder cells guide embryonic stem cell fate. Biotechnol. Bioeng. 2011;108:2509–2516. doi: 10.1002/bit.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tavana H, Kaylan K, Bersano-Begey T, Luker KE, Luker GD, Takayama S. Adv. Funct. Mater. 2011;21:2920. doi: 10.1002/adfm.201002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lemmo S, Nasrollahi S, Tavana H. Biotechnol. J. 2014;9:426. doi: 10.1002/biot.201300227. [DOI] [PubMed] [Google Scholar]
- 121.Ham SL, Nasrollahi S, Shah KN, Soltisz A, Paruchuri S, Yun YH, Luker GD, Bishayee A, Tavana H. Integr. Biol. (Camb) 2015;7:792. doi: 10.1039/c5ib00121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Atefi E, Lemmo S, Fyffe D, Luker GD, Tavana H. Adv. Funct. Mater. 2014;24:6509. doi: 10.1002/adfm.201401302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Atefi E, Joshi R, Mann JA, Tavana H. ACS Appl. Mater. Interfaces. 2015;7:21305. doi: 10.1021/acsami.5b05757. [DOI] [PubMed] [Google Scholar]
- 124.Ham SL, Joshi R, Luker GD, Tavana H. Adv. Healthc. Mater. 2016 doi: 10.1002/adhm.201600644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ham SL, Atefi E, Fyffe D, Tavana H. J. Vis. Exp. 2015:e52754. doi: 10.3791/52754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Labet M, Thielemans W. Chem. Soc. Rev. 2009;38:3484. doi: 10.1039/b820162p. [DOI] [PubMed] [Google Scholar]
- 127.Fong ELS, Lamhamedi-Cherradi S-E, Burdett E, Ramamoorthy V, Lazar AJ, Kasper FK, Farach-Carson MC, Vishwamitra D, Demicco EG, Menegaz BA, Amin HM, Mikos AG, Ludwig JA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6500. doi: 10.1073/pnas.1221403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Garofalo C, Manara MC, Nicoletti G, Marino MT, Lollini P, Astolfi A, Pandini G, López-Guerrero Ja, Schaefer K-L, Belfiore A, Picci P, Scotlandi K. Oncogene. 2011;30:2730. doi: 10.1038/onc.2010.640. [DOI] [PubMed] [Google Scholar]
- 129.Palomeras S, Rabionet M, Ferrer I, Sarrats A, Garcia-Romeu ML, Puig T, Ciurana J. Molecules. 2016;21:1. doi: 10.3390/molecules21040537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sims-Mourtada J, Niamat RA, Samuel S, Eskridge C, Kmiec EB. Int. J. Nanomedicine. 2014;9:995. doi: 10.2147/IJN.S55720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou Y, Chen F, Ho ST, Woodruff MA, Lim TM, Hutmacher DW. Biomaterials. 2007;28:814. doi: 10.1016/j.biomaterials.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 132.Sieh S, Lubik AA, Clements JA, Nelson CC, Hutmacher DW. Organogenesis. 2010;6:181. doi: 10.4161/org.6.3.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dong Z, Bonfil RD, Chinni S, Deng X, Carlos J, Filho T, Bernardo M, Vaishampayan U, Che M, Sloane BF, Sheng S, Fridman R, Cher ML. Am. J. Pathol. 2005;166:1173. doi: 10.1016/S0002-9440(10)62337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sieh S, Taubenberger AV, Lehman ML, Clements JA, Nelson CC, Hutmacher DW. Bone. 2014;63:121. doi: 10.1016/j.bone.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 135.Gustavsson H, Jennbacken K, Welén K, Damber J-E. Prostate. 2008;68:161. doi: 10.1002/pros.20672. [DOI] [PubMed] [Google Scholar]
- 136.Mishra S, Tang Y, Wang L, deGraffenried L, Yeh I-T, Werner S, Troyer D, Copland JA, Sun L-Z. Prostate. 2011;71:1441. doi: 10.1002/pros.21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang T, Zhang Q, Chen J, Fang K, Dou J, Gu N. Colloids Surfaces A Physicochem. Eng. Asp. 2014;452:115. [Google Scholar]
- 138.Ho WJ, Pham EA, Kim JW, Ng CW, Kim JH, Kamei DT, Wu BM. Cancer Sci. 2010;101:2637. doi: 10.1111/j.1349-7006.2010.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim JW, Ho WJ, Wu BM. Anticancer Res. 2011;31:3237. [PubMed] [Google Scholar]
- 140.Girard YK, Wang C, Ravi S, Howell MC, Mallela J, Alibrahim M, Green R, Hellermann G, Mohapatra SS, Mohapatra S. PLoS One. 2013;8:e75345. doi: 10.1371/journal.pone.0075345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat. Methods. 2007;4:855. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 142.Maschler S, Wirl G, Spring H, Bredow DV, Sordat I, Beug H, Reichmann E. Oncogene. 2005;24 VN-r:2032. doi: 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- 143.Desmoulière A, Guyot C, Gabbiani G. Int. J. Dev. Biol. 2004;48:509. doi: 10.1387/ijdb.041802ad. [DOI] [PubMed] [Google Scholar]
- 144.Wang D, Cheng D, Guan Y, Zhang Y. Biomacromolecules. 2011;12:578. doi: 10.1021/bm101187b. [DOI] [PubMed] [Google Scholar]
- 145.Wu Y, Zhao Z, Guan Y, Zhang Y. Acta Biomater. 2014;10:1965. doi: 10.1016/j.actbio.2013.12.044. [DOI] [PubMed] [Google Scholar]
- 146.Dutta D, Heo I, Clevers H. Trends Mol. Med. 2017;23:393. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 147.Girda E, Huang EC, Leiserowitz GS, Smith LH. Int. J. Gynecol. Cancer. 2017:1. doi: 10.1097/IGC.0000000000001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim G-A, Spence JR, Takayama S. Curr. Opin. Biotechnol. 2017;47:51. doi: 10.1016/j.copbio.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shah SB, Singh A. Acta Biomater. 2017;53:29. doi: 10.1016/j.actbio.2017.01.075. [DOI] [PubMed] [Google Scholar]
- 150.Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, van Houdt WJ, Pronk A, van Gorp J, Siersema PD, Clevers H. Gastroenterology. 2011;141:1762. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 151.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B. Proc. Natl. Acad. Sci. U. S. A. 2015;112:15672. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Cancer Discov. 2017;7:462. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jardé T, Lloyd-Lewis B, Thomas M, Kendrick H, Melchor L, Bougaret L, Watson PD, Ewan K, Smalley MJ, Dale TC. Nat. Commun. 2016;7:13207. doi: 10.1038/ncomms13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Y-W, Huang SX, de Carvalho ALRT, Ho S-H, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova J-L, Bhattacharya J, Liang AF, Palermo LM, Porotto M, Moscona A, Snoeck H-W. Nat. Cell Biol. 2017;19:542. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kimura M, Endo H, Inoue T, Nishino K, Uchida J, Kumagai T, Kukita Y, Kato K, Imamura F, Inoue M. J. Thorac. Oncol. 2015;10:527. doi: 10.1097/JTO.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 156.Yan H, Konstorum A, Lowengrub JS. Bull. Math. Biol. 2017 doi: 10.1007/s11538-017-0294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, Cantrell MA, Rack PG, Neal JT, Chan CW-M, Yeung T, Gong X, Yuan J, Wilhelmy J, Robine S, Attardi LD, Plevritis SK, Hung KE, Chen C-Z, Ji HP, Kuo CJ. Nat. Med. 2014;20:769. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, Macdonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Cell. 2014;159:176. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. Cell Stem Cell. 2010;6:25. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 160.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W, Koike N, Aoyama S, Adachi Y, Taniguchi H. Nature. 2013;499:481. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 161.Fan Y, Tajima A, Goh SK, Geng X, Gualtierotti G, Grupillo M, Coppola A, Bertera S, Rudert WA, Banerjee I, Bottino R, Trucco M. Mol. Ther. 2015;23:1262. doi: 10.1038/mt.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boj SF, Il Hwang C, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, Van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Öhlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FHM, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RGJ, Clevers H, Tuveson DA. Cell. 2015;160:324. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, Crawford HC, Arrowsmith C, Kalloger SE, Renouf DJ, Connor AA, Cleary S, Schaeffer DF, Roehrl M, Tsao M-S, Gallinger S, Keller G, Muthuswamy SK. Nat. Med. 2015;21:1364. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Science (80-. ) 2016;352:816. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 165.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Nature. 2017;545:48. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suematsu S, Watanabe T. Nat. Biotechnol. 2004;22:1539. doi: 10.1038/nbt1039. [DOI] [PubMed] [Google Scholar]
- 167.Alves-Lopes JP, Söder O, Stukenborg J-B. Biomaterials. 2017;130:76. doi: 10.1016/j.biomaterials.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 168.Sailer V, Pauli C, Merzier EC, Mosquera JM, Beltran H, Rubin MA, Rao RA. Anticancer Res. 2017;37:1569. doi: 10.21873/anticanres.11486. [DOI] [PubMed] [Google Scholar]
- 169.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Nature. 2009;459:262. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 170.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Nat. Med. 2009;15:701. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Usui T, Sakurai M, Enjoji S, Kawasaki H, Umata K, Ohama T, Fujiwara N, Yabe R, Tsuji S, Yamawaki H, Hazama S, Takenouchi H, Nakajima M, Tsunedomi R, Suzuki N, Nagano H, Sato K. Stem Cells Int. 2016;2016:1. doi: 10.1155/2016/7053872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Nat. Med. 2015;21:256. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 173.Fujii M, Matano M, Nanki K, Sato T. Nat. Protoc. 2015;10:1474. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- 174.Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, Weiss R. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.O’Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, Simon J, Romesser PB, Leach B, Han T, Pauli C, Beltran H, Rubin MA, Dow LE, Lowe SW. Nat. Biotechnol. 2017;35:577. doi: 10.1038/nbt.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, Peng W, Sandhu SK, Olmos D, Riisnaes R, McCormack R, Burzykowski T, Kheoh T, Fleisher M, Buyse M, de Bono JS. J. Clin. Oncol. 2015;33:1348. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Toyohara T, Mae S-I, Sueta S-I, Inoue T, Yamagishi Y, Kawamoto T, Kasahara T, Hoshina A, Toyoda T, Tanaka H, Araoka T, Sato-Otsubo A, Takahashi K, Sato Y, Yamaji N, Ogawa S, Yamanaka S, Osafune K. Stem Cells Transl. Med. 2015;4:980. doi: 10.5966/sctm.2014-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kleinman HK, Martin GR. Semin. Cancer Biol. 2005;15:378. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 179.Hughes CS, Postovit LM, Lajoie GA. Proteomics. 2010;10:1886. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 180.Vukicevic S, Kleinman HK, Luyten FP, Roberts AB, Roche NS, Reddi AH. Exp. Cell Res. 1992;202:1. doi: 10.1016/0014-4827(92)90397-q. [DOI] [PubMed] [Google Scholar]
- 181.Cruz-Acuña R, García AJ. Matrix Biol. 2017;57–58:324. doi: 10.1016/j.matbio.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wells JM, Spence JR. Development. 2014;141:752. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.DiMarco RL, Su J, Yan KS, Dewi R, Kuo CJ, Heilshorn SC. Integr. Biol. (Camb) 2014;6:127. doi: 10.1039/c3ib40188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ. Biomaterials. 2013;34:9486. doi: 10.1016/j.biomaterials.2013.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.DiMarco RL, Dewi RE, Bernal G, Kuo C, Heilshorn SC. Biomater. Sci. 2015;3:1376. doi: 10.1039/c5bm00108k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chun LLDL, Mejia Benjamin, Chiang Kathy, Luker Gary. Adv. Funct. Mater. 2017 [Google Scholar]
- 187.Agarwal P, Wang H, Sun M, Xu J, Zhao S, Liu Z, Gooch KJ, Zhao Y, Lu X, He X. ACS Nano. 2017;11:6691. doi: 10.1021/acsnano.7b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Atala A, Yoo JJ. Essentials of 3D Biofabrication and Translation. Elsevier Inc.; 2015. [Google Scholar]
- 189.Pereira BA, Lister NL, De-Juan-Pardo EM, Risbridger GP, Ellem SJ. Comprehensive Biomaterials II. Elsevier; 2017. pp. 303–328. [Google Scholar]
- 190.Klebe RJ. Exp. Cell Res. 1988;179:362. doi: 10.1016/0014-4827(88)90275-3. [DOI] [PubMed] [Google Scholar]
- 191.Melchels FPW, Domingos MAN, Klein TJ, Malda J, Bartolo PJ, Hutmacher DW. Prog. Polym. Sci. 2012;37:1079. [Google Scholar]
- 192.Nakamura M, Iwanaga S, Henmi C, Arai K, Nishiyama Y. Biofabrication. 2010;2:14110. doi: 10.1088/1758-5082/2/1/014110. [DOI] [PubMed] [Google Scholar]
- 193.Goole J, Amighi K. Int. J. Pharm. 2016;499:376. doi: 10.1016/j.ijpharm.2015.12.071. [DOI] [PubMed] [Google Scholar]
- 194.Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, Markwald RR. Biomaterials. 2009;30:2164. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Okudaira T, Amimoto N, Mizumoto H, Kajiwara T. J. Biosci. Bioeng. 2016;122:213. doi: 10.1016/j.jbiosc.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 196.Hospodiuk M, Dey M, Sosnoski D, Ozbolat IT. Biotechnol. Adv. 2017;35:217. doi: 10.1016/j.biotechadv.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 197.Jang J, Kim TG, Kim BS, Kim S-W, Kwon S-M, Cho D-W. Acta Biomater. 2016;33:88. doi: 10.1016/j.actbio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 198.Pourchet LJ, Thepot A, Albouy M, Courtial EJ, Boher A, Blum LJ, Marquette CA. Adv. Healthc. Mater. 2017;6:1601101. doi: 10.1002/adhm.201601101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.