Abstract
Bangladesh is a cholera endemic country with a population at high risk of cholera. Toxigenic and non-toxigenic Vibrio cholerae (V. cholerae) can cause cholera and cholera-like diarrheal illness and outbreaks. Drinking water is one of the primary routes of cholera transmission in Bangladesh. The aim of this study was to conduct a comparative assessment of the presence of V. cholerae between point-of-drinking water and source water, and to investigate the variability of virulence profile using molecular methods of a densely populated low-income settlement of Dhaka, Bangladesh. Water samples were collected and tested for V. cholerae from “point-of-drinking” and “source” in 477 study households in routine visits at 6 week intervals over a period of 14 months. We studied the virulence profiles of V. cholerae positive water samples using 22 different virulence gene markers present in toxigenic O1/O139 and non-O1/O139 V. cholerae using polymerase chain reaction (PCR). A total of 1,463 water samples were collected, with 1,082 samples from point-of-drinking water in 388 households and 381 samples from 66 water sources. V. cholerae was detected in 10% of point-of-drinking water samples and in 9% of source water samples. Twenty-three percent of households and 38% of the sources were positive for V. cholerae in at least one visit. Samples collected from point-of-drinking and linked sources in a 7 day interval showed significantly higher odds (P < 0.05) of V. cholerae presence in point-of-drinking compared to source [OR = 17.24 (95% CI = 7.14–42.89)] water. Based on the 7 day interval data, 53% (17/32) of source water samples were negative for V. cholerae while linked point-of-drinking water samples were positive. There were significantly higher odds (p < 0.05) of the presence of V. cholerae O1 [OR = 9.13 (95% CI = 2.85–29.26)] and V. cholerae O139 [OR = 4.73 (95% CI = 1.19–18.79)] in source water samples than in point-of-drinking water samples. Contamination of water at the point-of-drinking is less likely to depend on the contamination at the water source. Hygiene education interventions and programs should focus and emphasize on water at the point-of-drinking, including repeated cleaning of drinking vessels, which is of paramount importance in preventing cholera.
Keywords: Vibrio cholerae, drinking water, O1/O139, non-O1/non-O139, household, point-of-drinking, source water
Introduction
Cholera is a life-threatening disease with an estimated 2.9 million cases annually in 69 cholera-endemic countries, including Bangladesh (Ali et al., 2015). A recent review indicated that, in Bangladesh, around 66 million people are at risk for cholera, with an estimated incidence of 1.64 per thousand persons (Ali et al., 2015). In Bangladesh alone, the estimated annual number of cases is 109,000, with a three percent case fatality rate (Ali et al., 2015). Toxigenic and non-toxigenic V. cholerae can cause cholera and cholera-like diarrheal illness and outbreaks. V. cholerae has more than 200 serogroups based on variations in the “O” antigenic lipopolysaccharide (LPS). Cholera toxin-producing serogroups O1 and O139 have been shown to be the etiological agents of epidemic cholera (Kaper et al., 1995). Non-O1/non-O139 and non-toxigenic V. cholerae O1 strains, harboring a range of accessory virulence factors, can cause diarrheal diseases (Morris et al., 1984) and sporadic localized cholera outbreaks (Saha et al., 1996; Faruque et al., 2004; Pang et al., 2007) hence emphasizing the importance of research on both toxigenic and non-toxigenic V. cholerae. Accessory factors that can cause diarrheal diseases are repeats-in-toxin (rtxA) (Lin et al., 1999; Chow et al., 2001), non-O1 (NAG-ST) and O1 (O1-ST) heat-stable enterotoxins encoded by the stn and sto genes, respectively (Ogawa et al., 1990; Dalsgaard et al., 1995; Theophilo et al., 2006), hemolysins encoded by the hlyA gene (Zhang and Austin, 2005; Karlsson et al., 2013), transcriptional activator (toxR) (Waldor and Mekalanos, 1994), hemagglutinin protease encoded by hap (Silva et al., 2006; Mohapatra et al., 2009), ADP ribosylating exotoxin (chxA) (Awasthi et al., 2013), the type VI secretion system (T6SS) (Unterweger et al., 2012), a novel type III secretion system (T3SS) (Dziejman et al., 2005; Shin et al., 2011), and mannose-sensitive hemagglutinin subunit A encoded by mshA (Watnick et al., 1999).
V. cholerae can survive in nutrient limited drinking water for long periods of time in a viable but non-culturable state (VBNC) (Colwell, 2009) and can actively exert its infectious capability when in the human intestine (Colwell et al., 1996). This phenomenon poses serious risks to human health due to its non-detectability of VBNC cells by existing culture methods resulting underestimation of colony forming units (CFU) count of viable cells. V. cholerae can adapt to and persist in unfavorable environments, such as in conditions of nutrient deprivation and fluctuations in salinity and temperature, and can resist predation by heterotrophic protists and bacteriophages by adopting this unique survival strategy of the VBNC state (Ravel et al., 1995; Colwell et al., 1996; Carroll et al., 2001; González-Escalona et al., 2006; Thomas et al., 2006; Jubair et al., 2012; Mishra et al., 2012). Bacteria remain alive, metabolically active and can express virulence factors in this VBNC state; for example, V. cholerae can express tcp encoding a toxin co-regulated pilus (Krebs and Taylor, 2011) and the cholera toxin gene (ctxA) (Mishra et al., 2012). V. cholerae can exert its infectious properties when resuscitation occurs in human and animal digestive tracts (Colwell et al., 1996; Asakura et al., 2007; Senoh et al., 2010). In nutrient limited environments, V. cholerae can enter a starvation state in which cells are non-growing but culturable (Colwell et al., 1996; Thomas et al., 2006) and can survive for prolonged period of time (i.e., >700 days) (Jubair et al., 2012). Furthermore, both pathogenic and non-pathogenic V. cholerae can attach to abiotic surfaces, i.e., borosilicate glass (Watnick et al., 1999) and can survive in fomites in a VBNC state for more than 7 days (Farhana et al., 2016).
Cholera is endemic in Dhaka city (Patel et al., 2012), and low-income urban communities are particularly vulnerable to cholera and diarrheal diseases due to lack of hygiene and access to clean drinking water (Rafique et al., 2016). Drinking water is considered as one of the primary routes of cholera transmission in Bangladesh (Colwell et al., 2003; Huq et al., 2005; Akanda et al., 2009; Jutla et al., 2011). A recent study in Dhaka city established the association of cholera pathogen and its virulence in drinking water from households with confirmed or suspected cholera case patients (Rafique et al., 2016). There is, however, no known comprehensive evaluation of the burden of V. cholerae in source and point-of-drinking water in households in a cholera endemic community. Point of use or household water treatment can be an effective intervention in the prevention of diarrhea (Fewtrell et al., 2005). The World Health Organization has recognized that household water treatment and safe storage can provide rapid and significant health impacts (http://www.who.int/water_sanitation_health/publications/2011/9789241548151_toc.pdf). Therefore, investigating the contamination of drinking water in a population at risk for cholera will be useful to developing specific interventions to protect high risk populations from cholera and cholera-like illnesses. Studies that have investigated water quality at the point of use have focused primarily on water treatment, i.e., filtration, chlorination, flocculation, and solar disinfection of water stored in households (Clasen, 2015; Taylor et al., 2015). Few studies have investigated the microbiological water quality at the point of consumption/drinking (i.e., the quality of water in a drinking vessel immediately before consumption) (Rufener et al., 2010). The aim of this study is to conduct a comparative assessment of the presence of V. cholerae between point-of-drinking water and source water and to investigate the variability of virulence profile using molecular methods of a densely populated low-income settlement of Dhaka, Bangladesh.
Methods and materials
Study design
The study was conducted in Arichpur, located in Tongi Township of Dhaka, Bangladesh. Arichpur is an urban community with an area of 1.2 km2, population density of more than 100,000 residents per km2, and approximately 129,000 residents living in 29,000 households (Azman et al., 2015). Residents of this area use water from two types of communal pumps: “WASA (Water Supply and Sewerage Authority) pump” installed by the municipal government and connected to households through underground networks of pipes, and/or “submersible pump” installed by individuals or groups of residents and connected to households through over ground networks of pipes. The area around the pumps is not usually protected with a wall and floor made of concrete. These pumps extract water at a depth of approximately 75–140 m.
Data collection
A total of 477 households were enrolled in this study. Water samples were collected both at the point-of-drinking and at the source in each study household during routine visits at 6 week intervals from September 2014 to October 2015. Depending on the availability of the caretaker (i.e., the female or male family member who spent the most time in the house), point-of-drinking samples were taken from the drinking vessels (i.e., a mug, glass, bottle, jug, or pitcher) that household members used to drink water. Samples from sources were taken from the communal water source point used by each study household. On average, 20 samples were collected at each weekly visit from point-of-drinking and sources. Caretakers were asked if they treated the water (i.e., boiled, filtered, added alum, etc.) prior to consuming the drinking water. The water samples from sources were taken directly from taps attached to the communal pumps. In the absence of such a tap, samples were collected from taps attached to the nearest closed over-ground reservoir that was connected to the pump. The coordinates of sample collection sites (households and communal sources) were obtained using a global positioning system (GPS). Q-GIS software was used to locate the sites on a Google map.
Sample collection and enrichment
Each sample contained 100 mL of water that was collected in sterile bottles and transported in a cool box to the Environmental Microbiology Laboratory, University of Dhaka, within 2–4 h of collection. Aliquots of water were added to 10 mL of alkaline peptone water (APW), enrichment medium (1 L distilled H2O, 10 gL−1 peptone, 10 gL−1 sodium chloride; pH 8.5) followed by incubation at 37°C for 18–24 h (Alam et al., 2014).
Extraction of total DNA and confirmation of V. cholerae
After overnight incubation, DNA was extracted from 1 mL of each enriched culture using the method described by De Medici et al. (2003). The presence of V. cholerae in water samples was confirmed by detection of the V. cholerae species-specific gene ompW (Nandi et al., 2000) by PCR. Due to the non-detectability of VBNC cells by existing culture methods, PCR was chosen to reliably detect all forms of V. cholerae (both VBNC and culturable).
PCR reaction mix and primer sequences
V. cholerae virulence genes were detected in 143 samples found positive for the V. cholerae species-specific gene ompW using PCR. A total of 22 V. cholerae virulence genes were selected for detection. PCR was performed using an MJ Research PTC-200 Peltier Thermal Cycler (Mexico). The 25-μL reaction mixture contained 2 μL of 10× PCR buffer, 20 mM MgCl2, 0.4 μL of 10 mM deoxynucleoside triphosphates (dNTP) mix (Thermo Scientific, USA), 0.1 μL of 5 U Dream Taq DNA Polymerase (Thermo Scientific, USA) per μL, and 1.25 μL of each 25 μM primer (Tag Copenhagen A/S, Denmark). Sequences of the primers and target genes and their amplicon sizes are presented in S1 Table.
Real-time PCR was performed to detect the V. cholerae ctxA and rtxA genes using an Applied Biosystems StepOne (48-well) Real-Time PCR system. Real-time PCR was used as it provides higher sensitivity and specificity compared to conventional PCR. The fluorogenic probe and primer set (Tag Copenhagen A/S, Denmark) targeting the ctxA and rtxA genes are described in S2 Table. The formula of reaction mixture and cycling conditions for detection of ctxA gene were maintained as per supplier's instruction. The 25-μL reaction mixture containing 12.5 μL 2× TaqMan Universal Master Mix II with UNG (Applied Biosystems USA, with AmpliTaq Gold DNA Polymerase, dNTPs, ROX passive reference, Uracil-N glycosylase), 2.5 μL of each 100 nM of primer, 2.5 μL of 250 nM probe, and 5 μL of template. To detect the rtxA gene, a reaction mixture (25 μL) containing 12.5 μL 2× Power SYBR green PCR master mix (with a propriety version of ROX dye), 2.5 μL of each 100 nM sense and antisense primer, 2.5 μL of DEPC-treated H2O, and 5 μL of template DNA was used. V. cholerae O1 N16961 genomic DNA was used as a positive control, and PCR grade water was used as a no template control for PCR screening.
Data analysis
The proportions of samples positive for V. cholerae in point-of-drinking and source water were calculated. Logistic regression test was employed to examine the association of V. cholerae (and virulence genes) between point-of-drinking and sources, treated and non-treated water, drinking vessel type and all the virulence genes. We also examined the association of V. cholerae by logistic regression analysis of a set of stratified samples of point-of-drinking water and their linked sources that were collected within 7 days (before/after 7 days) of interval from each other.
Ethics statement
The Ethical Review Committee (ERC) of icddr,b, Bangladesh reviewed and approved the study protocol. Informed written consent for collecting samples was obtained from caretaker of each household for “point of use” and from pump operator for “source” water.
Results
A total of 1,463 water samples were collected: 1,082 from the point-of-drinking and 381 from the 66 sources for the 388 enrolled households. Most of the households used mugs (249/388), and/or glasses (195/388), and/or small bottles (75/388) to drink water. Drinking water was treated in 24% (93/388) of the households, and the majority of these households reported boiling (77/93) as the mode of treatment. Twelve households out of 93 reported filtration and three households reported both “boiling and filtration” as the mode of water treatment. Among the 66 water sources for these households, there were three communal “WASA pumps” installed by the government and 63 “submersible pumps” installed by individuals or groups. Of the 66 sources, 31 had direct taps attached to the communal pumps, and 51 had taps attached to the reservoir connected to the pumps.
V. cholerae in “point-of-drinking” and “source” water
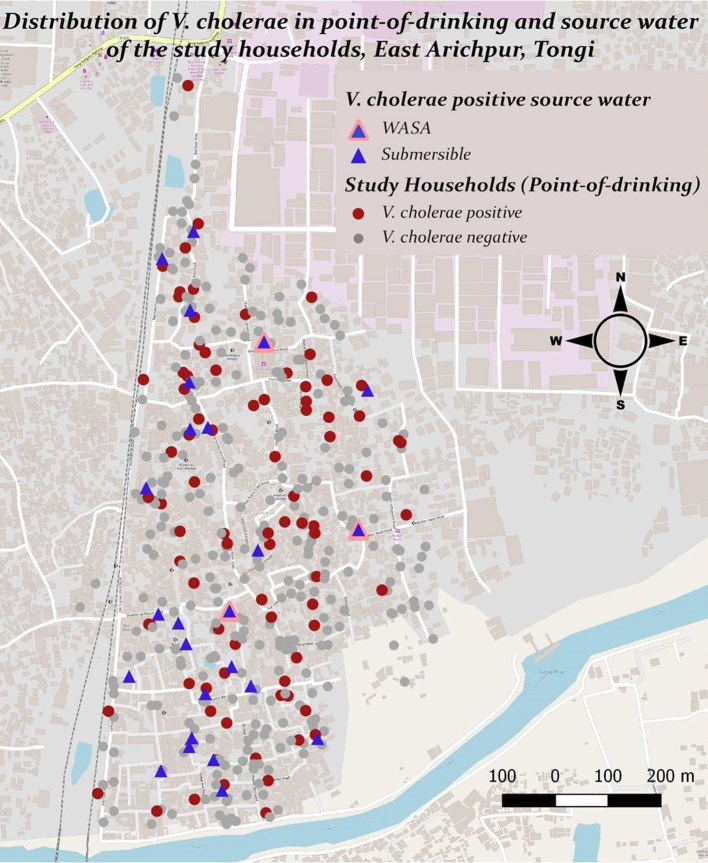
V. cholerae was detected in 10% (110/1082) of point-of-drinking water samples and in 9% (33/381) of source water samples (Table 1). Point-of-drinking water from 23% of households (88/388) and source water for 38% (25/66) of households were positive for V. cholerae at least once in the visits conducted at 6 week intervals. Most (76%, 67/88) households with point-of-drinking water samples positive for V. cholerae were also connected to 19 of 25 V. cholerae positive sources, irrespective of timing of collection. However, from the stratified data in 7 day intervals, 53% (17/32, [(95% CI = 0.360–0.70)]) of the sources were negative for V. cholerae, while point-of-drinking water samples linked to these sources were positive. The percentage of samples positive for V. cholerae was higher in the point-of-drinking water (11% [32/299], P = 0.000) compared to the sources water (9%, 28/299) in the 7 day-interval stratified data. The V. cholerae positive households were distributed throughout the study area, whereas the V. cholerae positive sources were mainly clustered in the southern part of the study area, which is adjacent to a water body (Figure 1).
Table 1.
Presence of V. cholerae in point-of-drinking and source water samples from the study households in Arichpur, Bangladesh, September 2014–October 2015.
| Characteristics of point-of-drinking water | Sample | |
|---|---|---|
| N (%) | V. cholerae positive n (%) | |
| [N = 1082] | [n = 110] | |
| Treated water | 165 (15) | 20 (12) |
| Non-treated water | 917 (85) | 90(10) |
| Types of treatment carried out | [n = 165] | |
| Boiling | 125 (76) | 14 (13) |
| Filtration | 31 (19) | 4 (4) |
| Boiling and filtration | 4 (2) | 2 (2) |
| Types of drinking vessels used at the point-of-drinking | [n = 1069]* | |
| Mug | 575 (54) | 52 (47) |
| Glass | 334 (31) | 41 (37) |
| Bottle | 125 (12) | 8 (7) |
| Jug | 30 (3) | 6 (6) |
| Pitcher | 5 (1) | 1 (1) |
| Characteristics of source water | N (%) [N = 381] | n (%) [n = 33] |
| By types of collection points | ||
| Taps attached to the communal pumps | 146 (38) | 6 (15) |
| Taps attached to the reservoir connected to the pumps | 235 (62) | 27 (82) |
| By types of pumps | ||
| WASA pump | 36 (9) | 4 (12) |
| Submersible pump | 345 (91) | 29 (88) |
For some samples, the types of vessels used were not known.
Figure 1.
Presence and distribution of V. cholerae in point-of-drinking and source water samples from the study households in Arichpur, Bangladesh, September 2014–October 2015.
In point-of-drinking water, V. cholerae was detected twice in 15% (13/88) of households, three times in 2% (2/88) of households, and six times in 1% (1/88) of households. The probability of the presence of V. cholerae was higher in glasses than mugs and bottles (Table 2). There was a higher probability of the presence of V. cholerae in non-treated water compared to treated water (P = 0.22; Table 2). V. cholerae was detected twice in 8% (2/25) of sources, three times in 4% (1/25) of sources, and five times in 4% (1/25) of sources. Samples from all three WASA pumps' were positive for V. cholerae at least once (Figure 1). Most (82%, 29/33) of the V. cholerae detected in source water came from taps attached to the reservoir connected to the pumps (P = 0.008), rather the taps directly attached to the communal pumps (Table 2).
Table 2.
Logistic regression of factors associated with the presence of V. cholerae in water samples from Arichpur, Bangladesh, September 2014–October 2015.
| Factors | OR (95% CI) | P |
|---|---|---|
| PRESENCE OF V. cholerae | ||
| Point-of-drinking vs. source (irrespective of the timing of sample collection) | 1.19 (0.79–1.79) | 0.230 |
| Point-of-drinking vs. source (samples collected within 7 day interval) | 17.24 (7.14–42.89) | 0.000* |
| Taps attached to the reservoir connected to the pumps vs. taps attached to the communal pumps | 3.03 (1.22–7.53) | 0.008* |
| Non-treated vs. treated point-of-drinking water | 1.27 (0.76–2.12) | 0.220 |
| Point-of-drinking water in glass vs. mug | 1.41 (0.91–2.17) | 0.076 |
| Glass vs. bottle | 2.05 (0.93–4.50) | 0.046 |
| Mug vs. bottle | 1.45 (0.67–3.14) | 0.221 |
| PRESENCE OF TOXIGENIC V. cholerae O1/O139 | ||
| Source vs. point-of-drinking | 6.22 (2.54–15.25) | 0.000* |
| Taps attached to the reservoir connected to the pumps vs. taps attached to the communal pumps | 1.74 (0.55–5.58) | 0.254 |
OR, odds ratio; CI, confidence interval;
significance at a level of P ≤ 0.05.
Distribution of virulence genes
A total of 143 V. cholerae positive samples were identified, and virulence genes other than ompW were detected in these positive samples. In total, 11% (15/143) of V. cholerae samples were positive for the rfb O1 gene and 6% (9/143) of V. cholerae samples were positive for the rfb O139 gene (Table 3). The percentages of serogroups O1 and O139 were higher in source water compared to point-of-drinking water. There was a higher probability of having V. cholerae O1 [OR = 9.13 (95% CI = 2.85–29.26)] and V. cholerae O139 [OR = 4.73 (95% CI = 1.19–18.79)] in source water compared to point-of-drinking water (Table 3). Of the samples with non-O1/non-O139 serogroups, the ctxA gene was found in three of the point-of-drinking water samples and two of the source samples. The percentage of samples in which the hlyA gene was detected was higher in point-of-drinking water compared to source water, and this difference was statistically significant (Table 3).
Table 3.
Presence and logistic regression of V. cholerae virulence genes in source and point-of-drinking water samples from Arichpur, September 2014–October 2015.
| Genes | Total samples positive for V. cholerae n = 143, n (%) | Point-of-drinking water samples positive for V. cholerae n = 110, n (%) | Source water samples positive for V. cholerae n = 33, n (%) | Odds ratio of source vs. point-of-drinking water samples (95% CI) | P-value |
|---|---|---|---|---|---|
| ctxA | 8 (6) | 7 (5) | 4 (9) | 2.10 (0.47–9.30) | 0.271 |
| rfbO1 | 15 (11) | 5 (5) | 10 (31) | 9.13 (2.85–29.26) | 0.000* |
| rfbO139 | 9 (6) | 4 (4) | 5 (16) | 4.73 (1.19–18.79) | 0.031* |
| cep | 62 (44) | 50 (46) | 12 (38) | 0.67 (0.31–1.53) | 0.235 |
| ace | 7 (5) | 5 (5) | 2 (6) | 1.36 (0.25–7.32) | 0.510 |
| msh1 | 45 (32) | 34 (31) | 11 (34) | 1.12 (0.49–2.56) | 0.475 |
| stn/sto | 9 (6) | 3 (3) | 6 (19) | 7.93 (1.86–33.75) | 0.005* |
| rtxA | 36 (25) | 24 (22) | 12 (38) | 2.05 (0.88–4.75) | 0.075 |
| toxR | 97 (68) | 77 (70) | 20 (63) | 0.66 (0.29–1.48) | 0.210 |
| tcpI | 3 (2) | 2 (2) | 1 (3) | 1.69 (0.15–19.22) | 0.548 |
| hlyA | 121 (85) | 99 (90) | 22 (69) | 0.22 (0.09–0.58) | 0.002* |
| ompU | 10 (7) | 7 (6) | 3 (9) | 1.47 (0.36–6.04) | 0.417 |
| nag-st | 3 (2) | 3 (3) | 0 (0) | – | – |
| rtxC | 61 (43) | 47 (43) | 14 (44) | 0.98 (0.45–2.17) | 0.569 |
| hap | 88 (62) | 70 (64) | 18 (56) | 0.69 (0.31–1.51) | 0.229 |
| chxA | 56 (39) | 39 (36) | 17 (53) | 1.93 (0.88–4.24) | 0.074 |
| vcsC2 | 8 (6) | 6 (5) | 2 (6) | 1.11 (0.22–5.82) | 0.589 |
| vcsN2 | 8 (6) | 6 (5) | 2 (6) | 1.11 (0.22–5.82) | 0.589 |
| vopF | 12 (8) | 9 (8) | 3 (9) | 1.12 (0.29–4.41) | 0.554 |
| vasK | 141 (99) | 109 (99) | 32 (100) | – | – |
| vasA | 140 (99) | 109 (99) | 31 (97) | 0.14 (0.01–1.62) | 0.133 |
| vasH | 140 (99) | 109 (99) | 31 (97) | 0.14 (0.01–1.62) | 0.133 |
P ≤ 0.05.
Two of the V. cholerae positive point-of-drinking water samples carried virulence genes- ctxA, as well as, rtxA, rtxC, toxR, hlyA, hap, msh1, chxA, T6SS but lacked tcpI, ompU, ace, nag-st. One point-of-drinking water sample was found positive for hlyA, rtxA, toxR, hap, ompU, cep, chxA, and T6SS, but negative for ctxA, rtxC, tcp, and ace. One of the source water samples exhibited hlyA, tcp, hap, cep, mshA, chxA, T3SS, and T6SS, but not ctxA, rtxA, or ompU. However, most of the 121 non-O1/O139 V. cholerae positive samples carried hlyA, rtxA, hap, and toxR, as well as genes encoding T6SS.
Discussion
Toxigenic and non-toxigenic V. cholerae were widely distributed in point-of-drinking and source waters throughout the low-income urban community of Arichpur. The estimated probability of the presence of V. cholerae in point-of-drinking water when absent in linked sources was 0.53 (95% CI = 0.36–0.70) within 7 day intervals, which suggests that post-contamination of point-of-drinking water might have occurred. The probability of the presence of V. cholerae O1 [OR = 9.13 (95% CI = 2.85–29.26)] and O139 [OR = 4.73 (95% CI = 1.19–18.79)] in source water was significantly higher than that in the point-of-drinking water, suggesting that the quality of point-of-drinking water might not be affected by the quality of sources.
Similar to other studies (Wright et al., 2004; Rufener et al., 2010), our study showed that the contamination of water was higher at the point-of-drinking compared to the source. In an observational study in Pakistan, Jensen et al. (2002) showed that water stored inside the household was more often contaminated than the source water when the source water contained < 100 E. coli per 100 mL (Jensen et al., 2002). In this same study, the researchers performed a 5 week intervention using narrow-necked water pitcher (that prevent utensils or hands from retrieving water) to prevent water contamination and found a significant improvement in in-house water quality (Jensen et al., 2002). A systematic review indicated that water quality improvement at sources were ineffective, because water from a good quality source was often contaminated at the point of use through poor hygiene practices in households (Taylor et al., 2015). Another study conducted in Bolivia showed that pathogen-free water at the source is not a guarantee for safe and pathogen-free drinking water at the point-of-consumption (Rufener et al., 2010), supporting our findings that the quality of water at the point-of-drinking did not depend on the presence or absence of V. cholerae in the source water.
Although there are reports that treatment type (boiling, chlorination) (Momba and Notshe, 2003; Levy et al., 2008), have significant impact on drinking water quality, our study did not evidence any significant association of V. cholerae with specific treatment type (boiling or filtration). A study conducted at the household level in rural areas of Peru reported that 69% of jars in which drinking water was stored had fecal coliforms though the water was treated by boiling (Gil et al., 2014). The absence of a holding shaft on a glass might play role in reducing direct hand contamination of drinking water to some extent and might explain the higher probabilities of V. cholerae contamination of water in drinking glasses compared to mugs. Compared to glasses and mugs, bottles were less frequently contaminated with V. cholerae, suggesting that narrow-necked vessels can prevent contamination, as shown by Jensen et al. (2002).
The higher prevalence of toxigenic V. cholerae O1/O139 [OR = 6.22 (2.54–15.25)] in sources compared to point-of-drinking water in this study matched findings of other studies conducted in Dhaka (Rafique et al., 2016) and in northern coastal Ecuador (Levy et al., 2008). In the source water, the number of V. cholerae was significantly higher in the water samples collected from the taps attached to the reservoir connected to the pumps compared to taps attached to the communal pumps which was also in agreement with a study conducted in Ecuador (Chalchisa et al., 2017). Larger storage tanks allowing longer storage times without regular cleaning (Schafer and Mihelcic, 2012) may potentially increase the risk of contamination and allow the persistence of bacteria by inducing the VBNC state (Colwell et al., 1996; Thomas et al., 2006).
V. cholerae lacking the tcpI gene was found in 5% ctxA positive samples. This is consistent with results of a study in Bangladesh (Hasan et al., 2013), where environmental O1 toxigenic strains were found to lack the tcpA and tcpI genes. Furthermore, we obtained O1 positive samples that did not carry ctxA, tcpI but carried hlyA, hap, rtxA genes. A research showed that variant virulence profile can be observed, since environmental strains are more heterogeneous than clinical strains (Hasan et al., 2013).
We found that non-O1/non-O139 V. cholerae was widely distributed throughout both source and point-of-drinking water samples. These strains are recognized to be of public health relevance, because they have been associated with sporadic cases or outbreaks of cholera-like disease (Crump et al., 2003; Dutta, 2013) and many extra-intestinal infections (Akoachere and Mbuntcha, 2014). While it is true that most epidemic cholera cases are caused by toxigenic V. cholerae O1/O139, a large proportion of diarrheal cases do not have a defined etiology where surveys take place (Islam et al., 2013).
After analyzing the genetic profiles of V. cholerae in samples, 85% of the V. cholerae in positive samples possessed hlyA, a gene whose product is an exotoxin related to CT, and rtxA, a heat-stable enterotoxin, both of which can be found in non-O1 strains isolated from patients with cholera (Saka et al., 2008) and from environmental strains from endemic areas (Faruque et al., 2004; Kumar et al., 2008; Mohapatra et al., 2009). These samples also possessed toxR, a 32-kDa transmembrane protein that acts as a master regulator of the ctxAB gene (DiRita et al., 1991). Finally, 10% of V. cholerae positive samples possessed ompU, whose product has been implicated in colonization and can also be found in some environmental isolates from endemic regions (Karunasagar et al., 2003). A gene mshA, also implicated in colonization, encoding a type IV pilus and biofilm formation on abiotic (borosilicate glass) and biotic surfaces (cellulose) (Watnick et al., 1999), was present in approximately half of the V. cholerae positive samples. This might explain the higher frequency of V. cholerae detection in the reservoir tanks.
Recently, fatal diarrheal disease caused by non-O1/O139 strains of V. cholerae has been shown to be associated with T3SS (Tam et al., 2010; Shin et al., 2011), a system absent in common pandemic O1 strains. Six percent of V. cholerae drinking water samples were positive for the presence of T3SS, implying the potential to cause fatal diarrhea via drinking water. Two other genes, chxA (encoding cholix toxin) was present in 39% and hap was present in 62% of the samples, which are also known to be associated with virulence in non-pandemic strains (Islam et al., 2013).
Our study had some limitations. Data presented here did not consider the inclusion of isolates. However, PCR performed directly on DNA samples allowed us to detect toxigenic genes of V. cholerae in both the culturable and non-culturable state, the latter of which can explain cholera or cholera-like diarrheal illness resulting from drinking water. In addition, PCR provides rapid detection with reduced cost compared to the culture method, and so might be useful for identifying the pathogen in outbreak settings. Although we found significantly higher presence of toxigenic V. cholerae O1/O139 in source water compared to point-of-drinking water, this assumption should be interpreted carefully, since the number of samples is low in this study.
Conclusion
Our study findings showed that contamination of water at point-of-drinking was less likely to depend on the contamination at sources and presence of V. cholerae in point-of-drinking water possibly did not depend on home-based water treatment suggesting that different routes (by hand, drinking vessel, flies) might have facilitated the contamination of drinking water at point-of-drinking. Hygiene education intervention and program should focus and emphasize on point-of-drinking including repeated cleaning of drinking vessels (such as mug, glass, bottle), which is of paramount importance in the prevention of cholera and cholera-like diarrheal illness. Data obtained in our study will serve as the baseline for the future investigations of V. cholerae in the environment, particularly in water.
Author contributions
JF designed the study concept, conducted the study in the laboratory, performed statistical analysis, and wrote the manuscript. RS contributed to framing the manuscript, data analysis, and writing and critical revision of the manuscript. RR contributed to the study concept, laboratory work, and data acquisition. MT contributed to the laboratory work and data acquisition. AN performed part of the statistical analysis. AB was the principal investigator of the project, contributed reagents, and approved the final version of the manuscript to be submitted. PJ was the functional principal investigator of the project and contributed to manuscript development, critical revision, and approval of the final version to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We specially thank Nadia Ali Rimi for language editing and helping to improve the clarity of the manuscript by highlighting specific results. We thank the research assistants Md. Akhtarujjaman, Sheikh Shariful Islam of icddr,b who conducted the field work for this study and helped in field data clarification. We thank the laboratory assistants Md. Mashud Parvez and Md. Rajib Hossain of Environmental Microbiology Laboratory, University of Dhaka, for their continuous support. RS from icddr, b is a co-author and icddr,b acknowledges the governments of Bangladesh, Canada, Sweden, and the UK for providing core/unrestricted support to icddr,b.
Footnotes
Funding. This study was supported by the project entitled Combating Cholera Caused by Climate Change in Bangladesh, C5 (Grant no. 12-040KU) of Danish International Development Agency (DANIDA).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00489/full#supplementary-material
References
- Akanda A. S., Jutla A. S., Islam S. (2009). Dual peak cholera transmission in Bengal Delta: a hydroclimatological explanation. Geophys. Res. Lett. 36:L19401 10.1029/2009GL039312 [DOI] [Google Scholar]
- Akoachere J. F., Mbuntcha C. K. (2014). Water sources as reservoirs of Vibrio cholerae O1 and non-O1 strains in Bepanda, Douala (Cameroon): relationship between isolation and physico-chemical factors. BMC Infect. Dis. 14:421. 10.1186/1471-2334-14-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. T., Weppelmann T. A., Weber C. D., Johnson J. A., Rashid M. H., Birch C. S., et al. (2014). Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerging Infect. Dis. 20, 356–363. 10.3201/eid2003.131293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M., Nelson A. R., Lopez A. L., Sack D. A. (2015). Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9:e0003832. 10.1371/journal.pntd.0003832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura H., Ishiwa A., Arakawa E., Makino S., Okada Y., Yamamoto S., et al. (2007). Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbiol. 9, 869–879. 10.1111/j.1462-2920.2006.01206.x [DOI] [PubMed] [Google Scholar]
- Awasthi S. P., Asakura M., Chowdhury N., Neogi S. B., Hinenoya A., Golbar H. M., et al. (2013). Novel cholix toxin variants, ADP-ribosylating toxins in Vibrio cholerae non-O1/non-O139 strains, and their pathogenicity. Infect. Immun. 81, 531–541. 10.1128/IAI.00982-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azman A. S., Lessler J., Satter S. M., Mckay M. V., Khan A., Ahmed D., et al. (2015). Tracking cholera through surveillance of oral rehydration solution sales at pharmacies: insights from urban Bangladesh. PLoS Negl. Trop. Dis. 9:e0004230. 10.1371/journal.pntd.0004230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. W., Mateescu M. C., Chava K., Colwell R. R., Bej A. K. (2001). Response and tolerance of toxigenic Vibro cholerae O1 to cold temperatures. Antonie Van Leeuwenhoek 79, 377–384. 10.1023/A:1012004725373 [DOI] [PubMed] [Google Scholar]
- Chalchisa D., Megersa M., Beyene A. (2017). Assessment of the quality of drinking water in storage tanks and its implication on the safety of urban water supply in developing countries. Environ. Syst. Res. 6:12 10.1186/s40068-017-0089-2 [DOI] [Google Scholar]
- Chow K., Ng T. K., Yuen K., Yam W. (2001). Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39, 2594–2597. 10.1128/JCM.39.7.2594-2597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen T. (2015). Household water treatment and safe storage to prevent diarrheal disease in developing countries. Curr. Environ. Health Rep. 2, 69–74. 10.1007/s40572-014-0033-9 [DOI] [PubMed] [Google Scholar]
- Colwell R. R., Brayton P., Herrington D., Tall B., Huq A., Levine M. M. (1996). Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12, 28–31. 10.1007/BF00327795 [DOI] [PubMed] [Google Scholar]
- Colwell R. R., Huq A., Islam M. S., Aziz K., Yunus M., Khan N. H., et al. (2003). Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. U.S.A. 100, 1051–1055. 10.1073/pnas.0237386100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell R. R. (2009). Viable but not cultivable bacteria, in Uncultivated Microorganisms. Microbiology Monographs, Vol. 10, ed Epstein S. (Heidelberg: Springer; ), 121–129. [Google Scholar]
- Crump J. A., Bopp C. A., Greene K. D., Kubota K. A., Middendorf R. L., Wells J. G., et al. (2003). Toxigenic Vibrio cholerae serogroup O141–associated cholera-like diarrhea and bloodstream infection in the United States. J. Infect. Dis. 187, 866–868. 10.1086/368330 [DOI] [PubMed] [Google Scholar]
- Dalsgaard A., Albert M. J., Taylor D. N., Shimada T., Meza R., Serichantalergs O., et al. (1995). Characterization of Vibrio cgolerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 33, 2715–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Medici D., Croci L., Delibato E., Di Pasquale S., Filetici E., Toti L. (2003). Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl. Environ. Microbiol. 69, 3456–3461. 10.1128/AEM.69.6.3456-3461.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRita V. J., Parsot C., Jander G., Mekalanos J. J. (1991). Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 88, 5403–5407. 10.1073/pnas.88.12.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D. (2013). Vibrio cholerae non-O1, Non-O139 serogroups and cholera-like Diarrhea, Kolkata, India. Emerg. Infect. Dis. 9, 464–467. 10.3201/eid1903.121156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M., Serruto D., Tam V. C., Sturtevant D., Diraphat P., Faruque S. M., et al. (2005). Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 102, 3465–3470. 10.1073/pnas.0409918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana I., Hossain Z. Z., Tulsiani S. M., Jensen P. K., Begum A. (2016). Survival of Vibrio cholerae O1 on fomites. World J. Microbiol. Biotechnol. 32:146. 10.1007/s11274-016-2100-x [DOI] [PubMed] [Google Scholar]
- Faruque S. M., Chowdhury N., Kamruzzaman M., Dziejman M., Rahman M. H., Sack D. A., et al. (2004). Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. U.S.A. 101, 2123–2128. 10.1073/pnas.0308485100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewtrell L., Kaufmann R. B., Kay D., Enanoria W., Haller L., Colford J. M. (2005). Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect. Dis. 5, 42–52. 10.1016/S1473-3099(04)01253-8 [DOI] [PubMed] [Google Scholar]
- Gil A. I., Lanata C. F., Hartinger S. M., Mäusezahl D., Padilla B., Ochoa T. J. (2014). Fecal contamination of food, water, hands, and kitchen utensils at the household level in rural areas of Peru. J. Environ. Health 76:102. [PubMed] [Google Scholar]
- González-Escalona N., Fey A., Höfle M. G., Espejo R. T., A Guzmán C. (2006). Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ. Microbiol. 8, 658–666. 10.1111/j.1462-2920.2005.00943.x [DOI] [PubMed] [Google Scholar]
- Hasan N. A., Ceccarelli D., Grim C. J., Taviani E., Choi J., Sadique A., et al. (2013). Distribution of virulence genes in clinical and environmental Vibrio cholerae strains in Bangladesh. Appl. Environ. Microbiol. 79, 5782–5785. 10.1128/AEM.01113-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq A., Sack R. B., Nizam A., Longini I. M., Nair G. B., Ali A., et al. (2005). Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71, 4645–4654. 10.1128/AEM.71.8.4645-4654.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Labbate M., Djordjevic S. P., Alam M., Darling A., Melvold J., et al. (2013). Indigenous Vibrio cholerae strains from a non-endemic region are pathogenic. Open Biol. 3:120181. 10.1098/rsob.120181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. K., Ensink J. H., Jayasinghe G., van der Hoek W., Cairncross S., Dalsgaard A. (2002). Domestic transmission routes of pathogens: the problem of in-house contamination of drinking water during storage in developing countries. Trop. Med. Int. Health 7, 604–609. 10.1046/j.1365-3156.2002.00901.x [DOI] [PubMed] [Google Scholar]
- Jubair M., Morris J. G., Jr., Ali A. (2012). Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS ONE 7:e45187. 10.1371/journal.pone.0045187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutla A. S., Akanda A. S., Griffiths J. K., Colwell R., Islam S. (2011). Warming oceans, phytoplankton, and river discharge: implications for cholera outbreaks. Am. J. Trop. Med. Hyg. 85, 303–308. 10.4269/ajtmh.2011.11-0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Morris J. G., Jr., Levine M. M. (1995). Cholera. Clin. Microbiol. Rev. 8, 48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson E. K., Harris J. B., Tabrizi S., Rahman A., Shlyakhter I., Patterson N., et al. (2013). Natural selection in a bangladeshi population from the cholera-endemic ganges river delta. Sci. Transl. Med. 5, 192ra186–192ra186. 10.1126/scitranslmed.3006338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunasagar I., Rivera I., Joseph B., Kennedy B., Shetty V. R., Huq A., et al. (2003). ompU genes in non-toxigenic Vibrio cholerae associated with aquaculture. J. Appl. Microbiol. 95, 338–343. 10.1046/j.1365-2672.2003.01984.x [DOI] [PubMed] [Google Scholar]
- Krebs S. J., Taylor R. K. (2011). Nutrient-dependent, rapid transition of Vibrio cholerae to coccoid morphology and expression of the toxin co-regulated pilus in this form. Microbiology 157, 2942–2953. 10.1099/mic.0.048561-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Peter W. A., Thomas S. (2008). Detection of virulence genes in Vibrio cholerae isolated from aquatic environment in Kerala, Southern India. Appl. Biochem. Biotechnol. 151, 256–262. 10.1007/s12010-008-8184-5 [DOI] [PubMed] [Google Scholar]
- Levy K., Nelson K. L., Hubbard A., Eisenberg J. N. (2008). Following the water: a controlled study of drinking water storage in northern coastal Ecuador. Environ. Health Perspect. 116:1533. 10.1289/ehp.11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Fullner K. J., Clayton R., Sexton J. A., Rogers M. B., Calia K. E., et al. (1999). Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. U.S.A. 96, 1071–1076. 10.1073/pnas.96.3.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Taneja N., Sharma M. (2012). Viability kinetics, induction, resuscitation and quantitative real-time polymerase chain reaction analyses of viable but nonculturable Vibrio cholerae O1 in freshwater microcosm. J. Appl. Microbiol. 112, 945–953. 10.1111/j.1365-2672.2012.05255.x [DOI] [PubMed] [Google Scholar]
- Mohapatra S. S., Ramachandran D., Mantri C. K., Colwell R. R., Singh D. V. (2009). Determination of relationships among non-toxigenic Vibrio cholerae O1 biotype El Tor strains from housekeeping gene sequences and ribotype patterns. Res. Microbiol. 160, 57–62. 10.1016/j.resmic.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Momba M. N., Notshe T. (2003). The microbiological quality of groundwater-derived drinking water after long storage in household containers in a rural community of South Africa. J. Water Supply 52, 67–77. [Google Scholar]
- Morris J. G., Jr., Picardi J. L., Lieb S., Lee J. V., Roberts A., Hood M., et al. (1984). Isolation of nontoxigenic Vibrio cholerae O group 1 from a patient with severe gastrointestinal disease. J. Clin. Microbiol. 19, 296–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi B., Nandy R. K., Mukhopadhyay S., Nair G. B., Shimada T., Ghose A. C. (2000). Rapid method for species-specific identification ofVibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38, 4145–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A., Kato J., Watanabe H., Nair B., Takeda T. (1990). Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler's diarrhea. Infect. Immun. 58, 3325–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Yan M., Cui Z., Ye X., Diao B., Ren Y., et al. (2007). Genetic diversity of toxigenic and nontoxigenic Vibrio cholerae serogroups O1 and O139 revealed by array-based comparative genomic hybridization. J. Bacteriol. 189, 4837–4849. 10.1128/JB.01959-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. M., Rahman M. A., Mohasin M., Riyadh M. A., Leung D. T., Alam M. M., et al. (2012). Memory B cell responses to Vibrio cholerae O1 lipopolysaccharide are associated with protection against infection from household contacts of patients with cholera in Bangladesh. Clin. Vacc. Immunol. 19, 842–848. 10.1128/CVI.00037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique R., Rashid M. U., Monira S., Rahman Z., Mahmud M. T., Mustafiz M., et al. (2016). Transmission of infectious Vibrio cholerae through drinking water among the household contacts of cholera patients (CHoBI7 Trial). Front. Microbiol. 7:635. 10.3389/fmicb.2016.01635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J., Knight I. T., Monahan C. E., Hill R. T., Colwell R. R. (1995). Temperature-induced recovery of Vibrio cholerae from the viable but nonculturable state: growth or resuscitation? Microbiology 141, 377–383. 10.1099/13500872-141-2-377 [DOI] [PubMed] [Google Scholar]
- Rufener S., Mäusezahl D., Mosler H. J., Weingartner R. (2010). Quality of drinking-water at source and point-of-consumption—drinking cup as a high potential recontamination risk: a field study in Bolivia. J. Health Popul. Nutr. 28, 34–41. 10.3329/jhpn.v28i1.4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P. K., Koley H., Mukhopadhyay A. K., Bhattacharya S. K., Nair G. B., Ramakrishnan B., et al. (1996). Nontoxigenic Vibrio cholerae 01 serotype Inaba biotype El Tor associated with a cluster of cases of cholera in southern India. J. Clin. Microbiol. 34, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka H. A., Bidinost C., Sola C., Carranza P., Collino C., Ortiz S., et al. (2008). Vibrio cholerae cytolysin is essential for high enterotoxicity and apoptosis induction produced by a cholera toxin gene-negative V. cholerae non-O1, non-O139 strain. Microbial Pathogen. 44, 118–128. 10.1016/j.micpath.2007.08.013 [DOI] [PubMed] [Google Scholar]
- Schafer C. A., Mihelcic J. R. (2012). Effect of storage tank material and maintenance on household water quality. J. Am. Water Works Assoc. 104, E521–E529. 10.5942/jawwa.2012.104.0125 [DOI] [Google Scholar]
- Senoh M., Ghosh-Banerjee J., Ramamurthy T., Hamabata T., Kurakawa T., Takeda M., et al. (2010). Conversion of viable but nonculturable Vibrio cholerae to the culturable state by co-culture with eukaryotic cells. Microbiol. Immunol. 54, 502–507. 10.1111/j.1348-0421.2010.00245.x [DOI] [PubMed] [Google Scholar]
- Shin O. S., Tam V. C., Suzuki M., Ritchie J. M., Bronson R. T., Waldor M. K., et al. (2011). Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio 2:e00106-11. 10.1128/mBio.00106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., Leitch G. J., Camilli A., Benitez J. A. (2006). Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 74, 2072–2079. 10.1128/IAI.74.4.2072-2079.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V. C., Suzuki M., Coughlin M., Saslowsky D., Biswas K., Lencer W. I., et al. (2010). Functional analysis of VopF activity required for colonization in Vibrio cholerae. MBio 1:e00289-10. 10.1128/mBio.00289-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. L., Kahawita T. M., Cairncross S., Ensink J. H. (2015). The impact of water, sanitation and hygiene interventions to control cholera: a systematic review. PLoS ONE 10:e0135676. 10.1371/journal.pone.0135676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theophilo G. N., Rodrigues Ddos P., Leal N. C., Hofer E. (2006). Distribution of virulence markers in clinical and environmental Vibrio cholerae non-O1/non-O139 strains isolated in Brazil from 1991 to 2000. Rev. Inst. Med. Trop. Sao Paulo 48, 65–70. 10.1590/S0036-46652006000200002 [DOI] [PubMed] [Google Scholar]
- Thomas K. U., Joseph N., Raveendran O., Nair S. (2006). Salinity-induced survival strategy of Vibrio cholerae associated with copepods in Cochin backwaters. Mar. Pollut. Bull. 52, 1425–1430. 10.1016/j.marpolbul.2006.04.011 [DOI] [PubMed] [Google Scholar]
- Unterweger D., Kitaoka M., Miyata S. T., Bachmann V., Brooks T. M., Moloney J., et al. (2012). Constitutive type VI secretion system expression gives Vibrio cholerae intra-and interspecific competitive advantages. PLoS ONE 7:e48320. 10.1371/journal.pone.0048320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. (1994). ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick P. I., Fullner K. J., Kolter R. (1999). A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181, 3606–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J., Gundry S., Conroy R. (2004). Household drinking water in developing countries: a systematic review of microbiological contamination between source and point-of-use. Trop. Med. Int. Health 9, 106–117. 10.1046/j.1365-3156.2003.01160.x [DOI] [PubMed] [Google Scholar]
- Zhang X. H., Austin B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019. 10.1111/j.1365-2672.2005.02583.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.