Abstract
The pathological aggregation of the tau protein is a common characteristic of many neurodegenerative diseases. There is strong interest in characterizing the potentially toxic nature of tau oligomers. These nonfibrillar, soluble multimers appear to be more toxic than neurofibrillary tangles made up of filamentous tau. However, reliable production, purification, and verification of tau oligomers can provide certain challenges. Here, we provide a series of methods that address these issues. First, recombinant tau is produced using Escherichia coli, purified through affinity, size-exclusion, and anion-exchange chromatography steps and quantified using an SDS Lowry protein quantitation assay. Aggregation of tau is induced using arachidonic acid, and oligomers are purified by centrifugation over a sucrose step gradient. Finally, we describe a sandwich enzyme-linked immunosorbent assay that utilizes the tau oligomerspecific TOC1 antibody to confirm the presence of oligomeric tau. Together, these steps provide a very simple and reliable method for producing tau oligomers that can be used in downstream applications.
1 INTRODUCTION
Aggregation of the tau protein has long been identified as a defining characteristic of Alzheimer’s disease and other tauopathies. Large intracellular inclusions, such as neurofibrillary tangles, are the most prominent pathological markers, but a growing body of evidence suggests that mature tangles are not the most toxic form of tau. For example, a regulatable transgenic mouse expressing a toxic form of tau recovered when the tau expression was turned off despite the continued presence of tangles (Santacruz et al., 2005). Additionally, expression of wild-type and disease-associated forms of tau in mice and Drosophila induced axonal damage and/or neurodegeneration without the formation of tangles (Duff et al., 2000; Spittaels et al., 1999; Wittmann et al., 2001). Importantly, toxic effects can be observed in animal models prior to the formation of tangles, indicating a toxic form of tau precedes them (Andorfer et al., 2005). This toxic species may be oligomeric tau, a nonfibrillar, multimeric, soluble form of the protein that has been identified in a variety of tauopathies, including Alzheimer’s disease, corticobasal degeneration, progressive supranuclear palsy, Pick’s disease, and chronic traumatic encephelopathy (Cox et al., 2016; Gerson et al., 2014; Kanaan et al., 2016; Lasagna-Reeves et al., 2012; Maeda et al., 2006; Patterson et al., 2011). The common presence of tau oligomers suggests that they could act as a toxic molecule across all tauopathies through a similar mechanism.
Recombinant tau protein has proven to be a useful tool to study the biochemistry and effects of tau aggregation. A variety of molecules can be used to induce in vitro aggregation of tau protein, most notably heparin and arachidonic acid (Goedert et al., 1996; Wilson & Binder, 1997). These methods primarily produce tau filaments that are of less relevance for studies aimed to elucidate information about potential tau oligomer toxicity. Typical analyses of tau oligomers include microscopic, immunobased, and biochemical approaches. For example, tau oligomers of various sizes can be imaged through electron or atomic force microscopy (Maeda et al., 2007; Ward et al., 2013; Wille, Drewes, Biernat, Mandelkow, & Mandelkow, 1992). Tau oligomer-specific antibodies, such as TOC1 and T22, have been developed that allow characterization and identification through the application of several immunobased assays like ELISAs, immunohistochemistry, and immunoblotting, among others (Lasagna-Reeves et al., 2012; Patterson et al., 2011; Ward et al., 2013). Biochemical properties of oligomers, such as their solubility or insolubility in buffer or detergents, like sarkosyl, can be used to differentiate them from monomeric and fibrillar tau aggregates or they can be separated based on density using sucrose gradients (Maeda et al., 2006, 2007; also described later). As the focus on multimeric species of tau continues, there is a need for methods to reliably produce and purify tau oligomers to facilitate their biochemical characterization and their effects on cell dysfunction and degeneration. In this chapter, we will discuss methods used in our laboratory to generate samples enriched for recombinant tau oligomers as well as highlight some reagents and assays to characterize and examine them biochemically.
2 PURIFICATION OF RECOMBINANT TAU
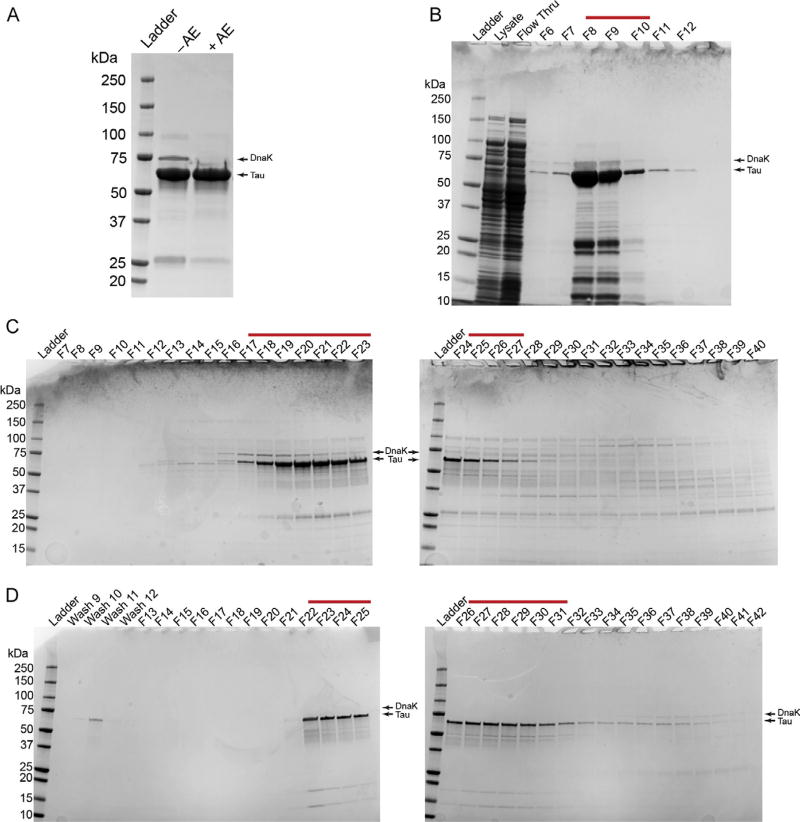
The expression and purification of high quality recombinant tau protein is a critical initial step in the in vitro study of tau and its aggregation. We have adapted a protocol that uses the T7 promoter system in Escherichia coli for IPTG-induced expression of 6× polyhistidine-tagged tau proteins and purification using metal affinity chromatography. This is followed by size-exclusion chromatography purification. We previously established that a bacterial Hsp70 homologue, DnaK, coelutes with recombinant tau from bacteria. Therefore, we added a final anion exchange chromatography step to generate a cleaner tau preparation (Fig. 1A). This protocol uses a GE ÄKTA fast protein liquid chromatography (FPLC) system, but the same principles can be applied using an alternative FPLC system or with a basic pump, column, and fraction collector setup.
FIG. 1.
The SDS-PAGE and Coomassie gel staining analysis of a typical purification of recombinant human tau protein. (A) A final preparation of tau without the anion exchange cleanup step (−AE) is compared to a preparation of tau with anion exchange (+AE). Note the clear removal of DnaK (i.e., ~70kDa) when the protein preparation is cleaned using the anion exchange procedure. 10µg purified protein was loaded per lane. (B) A gel showing the bacterial lysate, the column flow through from the sample application (Flow Thru), and elution fractions 6–12 (F6–12) from the Talon column His-tag purification step. Fractions 8–10 were collected for further purification (red line). (C) Two gels showing fractions 7–40 (F7–40) of the S500 column size exclusion chromatography step. Fractions 17–27 were collected for further purification (red line). (D) Two gels showing some of the wash fractions and fractions 13–42 from the anion exchange column cleanup step (F13–42). It is important to avoid fractions containing DnaK at this step; fractions 22–31 were collected for further purification (red line). This is a typical preparation of full-length wild-type human tau (hT40, 2N4R, 441 amino acids+C-terminal 6× His-tag). Note that the tau and DnaK bands are marked by arrows in each panel.
2.1 GROWTH AND INDUCTION OF PROTEINS IN E. coli
2.1.1 Day 1: Transformation of E. coli
- Transform DNA plasmid into T7 Express Competent E. coli cells (New England Biosciences, Ipswich, MA, C2566). In this example, we are using a pT7C ht40 C-His plasmid, but other tau variants can be used as desired.
-
○Mix 50ng of DNA with 25µL of cells and incubate on ice for 10min.
-
○Heat shock at 42°C for 30s and place on ice for 2min.
-
○Add 225µL of Luria Broth media (LB) and incubate for 30–60min at 37°C and 250RPM.
-
○Plate 250µL of cells on LB agar+ampicillin selection plate and residual cells from bacteria spreader onto a second plate.
-
○Incubate overnight at 37°C.
-
○The first plate may be too dense, but the second plate should have separated, individual colonies that can be picked.
-
○
2.1.2 Day 2: Initial Growth of E. coli Culture
Add ampicillin to a final concentration of 100µg/mL to a 250-mL flask containing 80mL LB media.
Pick a single isolated E. coli colony with a sterile glass or metal rod and add to the LB–ampicillin in a 250-mL flask to generate a starter culture.
Shake culture at 250RPM overnight at 25°C.
2.1.3 Day 3: Growth and Induction of Proteins in E. coli
Add 40mL of the starter culture to a 6-L flask containing 2L LB and ampicillin at 100µg/mL.
Grow E. coli at 37°C with shaking at 250RPM until OD600 = 0.8–1.0 is reached.
Induce protein expression by adding 2mL of 1M IPTG to the 6-L flask (1mM final concentration) and shake at 250RPM at 37°C for 2h.
Centrifuge cells (~250mL culture per spin) at 8000 × g at 4°C for 10min in a Sorvall S3000 Rotor.
Discard supernatant and add more of the bacterial culture, repeat centrifugation step until the entire culture is pelleted.
Wash the cells by resuspending them in 40mL ice-cold STE buffer (recipe below).
Transfer cells to a preweighed tube and centrifuge at 8000 × g for 10min at 4°C.
Discard supernatant and weigh to determine the mass of the cell pellet.
Freeze pellet at −80°C for up to 6 months.
2.2 FPLC PURIFICATION OF TAU PROTEINS
2.2.1 Day 1: Purification of Poly-His-Tagged Proteins and Size-Exclusion Chromatography
Degas all chromatography buffers by vacuum before use.
- Prior to beginning purification, equilibrate Hiprep 16/60 Sephacryl S-500 column HR (GE Healthcare, Pittsburgh, PA, 28-9356-06) with two column volumes of 100mM Tris, pH 7.4, at a flow rate of 1 mL/min.
-
○This is typically performed overnight the day before a purification run.
-
○
- Thaw and resuspend the cell pellet in five volumes of ice-cold lysis buffer plus protease inhibitors (e.g., 1 g cells requires 5mL buffer; recipe below).
-
○10µg/mL pepstatin (Fisher Scientific, Pittsburgh, PA, BP267125), 10µg/mL leupeptin (Sigma-Aldrich, St. Louis, MO, L2884), 10µg/mL bestatin (Sigma, 10874515001), 10µg/mL aprotinin (Sigma, A6279), 1mM PMSF (Sigma-Aldrich, 78830)
-
○
Lyse the cells in a French press (alternative lysis methods can be used). Maintain pressure between 500 and 1000psi with a constant flow of material. Pass the cells through the press three times.
Add all protease inhibitors, to same concentrations as earlier, to lysate as well as Brij 35 (Sigma, B4184) to 0.1% (3.3µL/mL of 30% solution).
Centrifuge in a Type 70 Ti rotor at 38,200RPM (76,765 RCF (ave); 107,377 RCF (max)) for 45min at 4°C.
Collect the supernatant and add all protease inhibitors as earlier.
Inject supernatant into 50mL super loop using GE ÄKTA FPLC.
- Purify protein over the metal affinity GE HiTrap Talon Crude column (28-9537-67) as follows:
-
○Equilibrate with 5 column volumes 1X Lysis Buffer, at 5mL/min flow rate.
-
○Inject supernatant from super loop at a flow rate of 3mL/min.
-
○Wash column with 5 CV 1X Lysis Buffer, at 5mL/min flow rate.
-
○Elute protein over 3 column volumes with 100mMimidazole buffer+PMSF at a flow rate of 5mL/min into 2mL fractions, using the upflow setting (to concentrate sample in the early fractions).
-
○
Remove 20µL of each fraction, add it to 4µLof 6 × Laemmli sample buffer (final 1 × composition: 20mM Tris, pH 6.8, 2% SDS, 6% glycerol, 1% β-mercaptoethanol, 0.002% Bromophenol Blue), and boil for 5min.
Run the samples (all 24µL) on an SDS-PAGE gel. Stain with Coomassie Brilliant Blue to identify the fractions that contain protein (Fig. 1B).
Combine the fractions containing protein and load onto equilibrated Hiprep 16/60 Sephacryl S-500 column HR (28-9356-06) using 5-mL capillary loop. Do not load more than 4.8mL sample onto column, concentrate if necessary using an Amicon Ultra-15 Centrifugal Filter Unit (10kDa NMWL, EMD Millipore, Billerica, MA, UFC9010).
After applying the sample to the column, wash loop with 15mL of 100mM Tris buffer at a flow rate of 1mL/min.
Elute the protein over the column using 1.25 column volumes. Delay fractionation collection by 0.15 column volumes and collect the remaining elution volume in 2mL fractions.
Take 20µL of each fraction and analyze using SDS-PAGE and Coomassie Brilliant Blue staining as above (Fig. 1C). Tau typically starts eluting around fraction 20 using this procedure.
Pool all tau-containing fractions, add PMSF to 1mM, and store at 4°C until the following day, or continue with the protocol as outlined later.
2.2.2 Day 2: Anion Exchange Cleanup of Purified Tau
Equilibrate a 5-mL GE HiTrap Q HP column (17-1154-01) with five column volumes of 100mM Tris buffer (pH 7.4) at a flow rate of 5mL/min.
Load tau sample from S-500 column into a 50-mL super loop.
Run sample over the HiTrap column at a flow rate of 5mL/min.
Wash column with five column volumes of 100mM Tris buffer (pH 7.4; 5mL/min flow rate).
Elute protein using a linear gradient of sodium chloride (NaCl) (0–250mM) over 12 column volumes at a flow rate of 5mL/min and collect in 2mL fractions.
Collect 20µL of each fraction and analyze using SDS-PAGE and Coomassie Brilliant Blue as above (Fig. 1D).
Select fractions containing tau protein and without DnaK contaminants (runs at ~70 kDa). The anion exchange step allows cleaner preparations without DnaK as can be seen by comparing a preparation that was obtained without and with this step (Fig. 1A).
Concentrate in Amicon Ultra-15 Centrifugal Filters (10kDaNMWL) by spinning at 3000 × g for 15min intervals until desired volume is reached (usually around 1mL). Our typical yields range between 1.5 and 5mg of purified tau protein. We routinely scale up to double and triple preparations using this procedure to obtain larger protein yields.
Add dithiothreitol (DTT) to a final concentration of 1mMto reduce disulfide bonds.
Aliquot in 10–15µL volumes and store at −80°C
The protein concentration can then be calculated using an SDS-Lowry assay (see below).
2.3 TAU PROTEIN PURIFICATION BUFFERS
Note: All buffers should be 0.22-µm filtered, and degassed 2–3h (ideally overnight) before use.
- Lysis D buffer (10×, 1L)
- 292.2g NaCl (5 M)
- 12.2g Tris (100mM)
- 3.4g Imidazole (50mM)
- pH to 8.0
- 500mM Imidazole buffer (1 L)
- 34.1g Imidazole
- pH to 8.0
- STE buffer (1 L)
- 5.84g NaCl (0.1M)
- 1.21g Tris (10mM)
- 290mg Ethylenediaminetetraacetic acid (EDTA) (1mM)
- pH to 8.0 at room temperature
- 100mM Imidazole (elution) buffer (100mL)
- 81mL 1× Lysis buffer D
- 19mL 500mM Imidazole buffer
- 40µL 1M PMSF
- 100mM Tris (1 L)
- 12.11g Tris base
- 1L sdH2O
- pH to 7.4
- 500mM NaCl(1 L)
- 29.22g NaCl
- 1L sdH2O
3 SDS-LOWRY PROTEIN QUANTITATION ASSAY PROTOCOL
We find the best protein assay for recombinant tau proteins that produces consistent and reproducible results is the SDS-Lowry. Absorbance at 280nm is not suitable for quantifying tau, and other protein assays such as the BCA assay are not as reliable with recombinant tau proteins in our hands.
- Prepare three replicates of protein standard solutions from a 2-mg/mL stock of bovine serum albumin (BSA, ThermoFisher Scientific, 23209).
-
○2µg: 1µL stock in 99µL SDS solution
-
○4µg: 2µL stock in 98µL SDS solution
-
○8µg: 4µL stock in 96µL SDS solution
-
○16µg: 8µL stock in 92µL SDS solution
-
○32µg: 16µL stock in 84µL SDS solution
-
○
Prepare experimental samples by adding an appropriate amount of purified tau to SDS solution for a total volume of 100µL. The experimental samples should fall within the standard curve.
Add 1mL 10% perchloric acid/1% phosphotungstic acid solution to each sample.
Vortex to mix and briefly spin down in microcentrifuge.
Incubate on ice for at least 1h.
Centrifuge at 18,000 × g for 15min at 4°C.
Carefully pour off supernatant and blot end of tubes with a paper towel to completely wick away residual liquid. The tube must be dry before proceeding.
Dissolve protein pellet in 1mL of freshly prepared Lowry solution and incubate at room temperature for 10 min.
Add 100µL of 1× Folin reagent to each sample.
Incubate at room temperature for 30–45 min.
Read absorbance values at 750nm with spectrophotometer and interpolate concentration from standard curve.
3.1 REAGENTS
Stock solutions
- 2% Na2CO3 in 0.1N NaOH (Fisher Scientific, S263)
- 8g Na2CO3 in 400mL 0.1N NaOH (4mL 10N NaOH+396mL dH2O)
- Filter (0.22-µm) and store at room temperature
- 1% CuSO4 (Fisher Scientific, AC19772)
- 4g in 400mL dH2O
- Filter (0.22-µm) and store at room temperature
- 2% NaKC4H4O6 (Sodium Potassium Tartrate, Sigma-Aldrich, S2377)
- 8g in 400mL dH2O
- Filter (0.22-µm) and store at room temperature
- 10% perchloric acid/1% phosphotungstic acid
- 57mL 70% perchloric acid (Sigma-Aldrich, 244252)
- 4g phosphotungstic acid (Sigma-Aldrich, P4006)
- 343mL dH2O
SDS solution (2% SDS/5% 2-mercaptoethanol/10% glycerol in 0.0625M Tris, pH 6.8)
50mL 20% SDS (BioRad, Hercules, CA, 161-0418)
25mL 2-mercaptoethanol (Sigma-Aldrich, M3148)
50mL glycerol (Sigma-Aldrich, G5516)
375mL 0.0625M Tris pH 6.8 (56.25mL 0.5M Tris+393.75mL dH2O)
Filter (0.22-µm), aliquot, and store at −20°C
Lowry solution
0.5mL 1% CuSO4 stock solution
0.5mL 2% NaK Tartrate stock solution
50mL 2% Na2CO3 in 0.1N NaOH stock solution
1× Folin reagent
Dilute Folin–Ciocalteu’s Phenol Reagent 1:1 w/dH2O (Sigma-Aldrich, F9252)
4 ARACHIDONIC ACID-INDUCED FORMATION OF TAU OLIGOMERS
Aggregation of tau protein is greatly enhanced by the addition of an inducer molecule, such as arachidonic acid, a polyunsaturated fatty acid normally enriched in the brain (Martinez & Mougan, 1998; Wilson & Binder, 1997). The addition of arachidonic acid to a mixture containing tau at near physiological levels (e.g., 2µM) can induce a robust aggregation response that results in tau aggregates within minutes and is typically completed within 6–18h. Several alterations to various parameters within the standard optimized protocol can result in modifications to filament morphology, including the enhanced formation of oligomeric species. Some of these modifications include changes to the tau protein itself, such as changing the tau isoform content or inserting specific point mutations (Combs & Gamblin, 2012; Cox et al., 2016; King, Gamblin, Kuret, & Binder, 2000). Increasing the arachidonic acid to tau ratio can result in a nearly homogenous population of tau oligomers (Carlson et al., 2007). Performing the reaction in oxidizing or reducing conditions (i.e., with or without DTT) can change aggregate morphology (Gamblin, King, Kuret, Berry, & Binder, 2000). Here, we describe a simple method that uses the traditional arachidonic acid-induced tau aggregation paradigm to generate a mixed population of tau aggregates, followed by separation over a sucrose gradient to obtain a sample enriched in tau oligomers.
Prepare stock solutions of 250mM DTT, 1MNaCl, 250mM Hepes (pH 7.6), and 1mM EDTA in filtered ultrapure water.
A standard aggregation reaction consists of 2µMtau and 75µMarachidonic acid in 200µL buffer containing 5mM DTT, 100mM NaCl, 10mM Hepes (pH 7.6), and 0.1mM EDTA.
Note: These parameters can effectively be adjusted up to 8µMtau or a larger reaction volume as needed. Maintaining the standard 37.5:1Mratio of arachidonic acid to tau is recommended but increasing this ratio can facilitate rapid nucleation and subsequently results in the formation of more oligomers (Carlson et al., 2007).
Prepare reaction samples in 1.5-mL microcentrifuge tubes by adding reagents to water in the following order: DTT, NaCl, Hepes, and EDTA. Mix thoroughly by pipetting up and down with each addition.
Add tau protein to its final concentration and gently mix by pipetting up and down and finger tapping while avoiding the introduction of bubbles. Care should be taken to standardize the mixing procedure to enhance reproducibility.
- Make up the arachidonic acid working stock solution immediately before use.
-
○Clean a Hamilton syringe (model 701) by repeatedly pulling up and expelling 100% ethanol (VWR, Radnor, PA, V1001G).
-
○Use the syringe to add arachidonic acid (peroxide-free, Cayman Chemicals, Ann Arbor, MI, 90010.1) to 100% ethanol for a final concentration of 2mM. Arachidonic acid bottles should be stored at −20°C and can be used for a period of 2–4 weeks before oxidation begins to affect reproducibility.
-
○Gently mix the arachidonic acid solution by inverting tube several times, taking care not to introduce air bubbles. Keep on ice until needed.
-
○Repeat the cleaning of the syringe by repeatedly taking up and expelling 100% ethanol.
-
○
Add arachidonic acid from 2mM stock solution directly to reaction by carefully dispensing the solution while gently swirling pipet tip in the solution—do not pipette up and down—carefully mix solution further by rocking tube/cuvette by hand until solution is thoroughly mixed (only 5–10s should be sufficient).
Allow reaction to proceed for 6–18h at room temperature.
Note: Differences between 6 and 18h incubations are negligible for four-repeat tau isoforms but can lead to greater aggregate formation with three-repeat tau isoforms (Combs, Voss, & Gamblin, 2011).
Freeze the sample at −80°C until used for oligomer enrichment.
5 SUCROSE STEP GRADIENT FRACTIONATION TO ISOLATE OLIGOMERS
The arachidonic acid-induced tau aggregation reactions produce a mixture of long, intermediate, and short filaments as well as globular oligomeric structures. One simple and straightforward method to isolate the oligomeric structures from these mixtures is by separation over a step gradient of sucrose (20%, 30%, 40%, and 50% sucrose). Here, we will describe how we purify oligomeric tau, which is enriched in the 30% sucrose solution using this technique. This technique is described using a TLS 55 swinging bucket rotor for a Beckman tabletop ultracentrifuge; however, alternative systems with similar characteristics can be used. While fixed angle rotors are not ideal for this purpose, they can be used with success as well.
Pipet 200µL of 50% sucrose solution into the bottom of the thick-walled polycarbonate tube (Beckman, 343778)
- Carefully layer 200µL of 40% sucrose solution over the 50% solution, and continue the same process until the 20% solution is on the top layer.
-
○Tip: Tilting the tube to the right and dispensing the solutions using a continuous but slow flow on the left side of the tube with the pipet tip at the level of the solution (avoid adding drop by drop) provide an easy way to layer the steps with little to no disturbance of the other layers.
-
○
- Carefully layer 200µL of the tau aggregate sample over the 20% solution.
-
○By dispensing sucrose solutions and samples carefully and avoiding disturbing the other layers, a clear phase boundary should be noticeable between each step.
-
○
Counter balance the centrifuge appropriately.
Centrifuge in TLS 55 rotor at 51,000RPM (172,894 RCF (avg); 222,854 RCF (max)) for 2h at 24°C.
- After the spin, very carefully remove fractions 1–5 in 200µL aliquots
-
○Tip: Tilting the tube to the right and drawing up the solutions on the left side of the tube with the pipet tip just under the surface of the solution provides an easy way to collect the fractions.
-
○
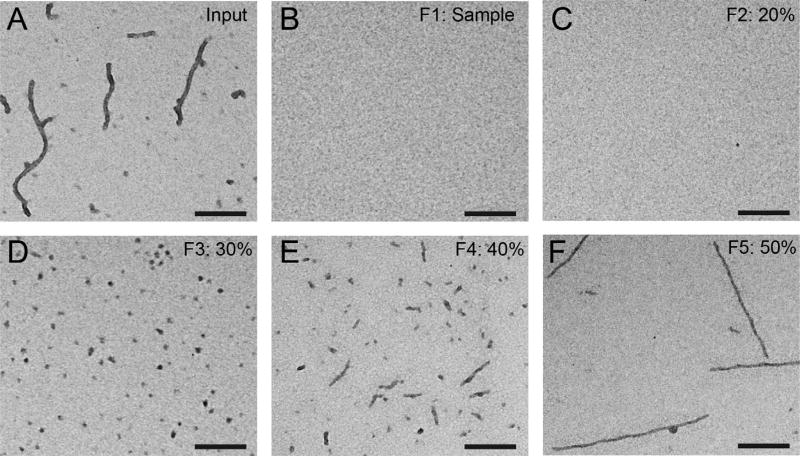
There should now be little to no tau in the “input” sample (compare the prespin “input” in Fig. 2A to the same sample postspin, F1, Fig. 2B), the 20% sucrose fraction will contain relatively low tau levels (F2, Fig. 2C), the 30% sucrose fraction (F3, Fig. 2D) will contain a highly-enriched amount of oligomeric tau structures, the 40% sucrose fraction (F4, Fig. 2E) will contain a mixture of oligomeric tau and short tau filaments, the 50% sucrose fraction (F5, Fig. 2E) will contain mostly short to long tau filaments. With these centrifugation settings, little to no tau is pelleted.
The samples are now ready for a number of downstream analyses and assays.
- Further Purification of oligomeric tau:
-
○If desired, the 30% sucrose fraction can be pelleted and oligomeric tau species can be resuspended in a buffer of choice.
-
○Dilute 200µL fraction F3 with 400µL buffer (e.g., TBS), mix thoroughly with a pipet.
-
○Centrifuge in TLS 55 rotor at 55,000RPM (201,078 RCF (avg); 259,182 RCF (max)) for 3 h at 24°C. A fixed angle rotor can be used as an alternative, and higher speeds may increase recovery of oligomeric aggregates.
-
○Carefully remove supernatant.
-
○Resuspend the pellet in an appropriate volume of desired buffer.
-
○The pellet fraction will contain oligomeric tau structures.
-
○
The presence of oligomers can be confirmed by examination via transmission electron microscopy (TEM) or a sandwich ELISA as described later.
FIG. 2.
Sucrose step gradient enrichment of tau oligomers from an arachidonic acid-induced tau aggregation reaction sample. (A) The typical arachidonic acid-induced preparation produces a mixed population of long, intermediate, and short filaments as well as globular oligomeric structures. (B–F) The mixed population of aggregates can be separated over a sucrose step gradient consisting of the (B) sample (0% sucrose, F1), (C) 20% sucrose (F2), (D) 30% sucrose (F3), (E) 40% sucrose (F4), and (F) 50% sucrose (F5). Note that fraction 3 containing 30% sucrose contains a highly enriched population of oligomeric tau structures with very little to no contamination of filamentous structures. The short filaments and longer filaments are isolated in the 40% and 50% sucrose fractions, respectively. Scale bars are 200nm.
6 ELECTRON MICROSCOPY IMAGING OF TAU OLIGOMERS
Tau aggregates are routinely analyzed using standard TEM techniques. Here, we provide a quick and simple method for imaging tau aggregates generated in vitro with recombinant tau protein. The protocol below describes TEM using unfixed samples, but glutaraldehyde fixation can be used as an alternative approach as described before (Cox et al., 2016). Using these standard methods tau oligomers are easily visualized with TEM and typically range from6 to 16nm in diameter (mean = ~12nm; Fig. 2D).
6.1 SAMPLE PREPARATION FOR TEM
Tape a piece of parafilm to the bench and have a Whatman #1 filter ready for wicking solutions off the grids.
Place a 10-µL drop of the sample on the parafilm
Above the sample place 10µL drops of the following: (1) deionized water (dH2O), (2) 2% uranyl acetate (UA), and (3) 2% UA
Using ultra fine tweezers, remove the grid from the holder—only hold grids on the edges.
Place the grid shiny side onto the sample (each grid has a dull side and shiny side) and incubate for 2min
Pickup the grid, wick away excess solution on the filter paper
Touch the grid shiny side onto the dH2O drop to “pickup” the water rinse droplet, and wick away the solution
Touch the shiny side of the grid onto the UA #1 drop to “pickup” the UA droplet, and wick away the solution
Place the grid shiny side down onto the UA #2 drop and incubated for 2min, and then wick away the solution
Store grids in grid holder (allow the grid to dry before imaging)
6.2 MATERIALS
Uranyl acetate (EMS, Hatfield, PA, Cat.# 22400)
Formvar/Carbon 300 mesh grids (EMS, Cat.# FCF-300-Cu)
- 2% UA solution
-
○20mg of UA
-
○1mL sterile dH2O
-
○Filter UA solution with precleaned filter (preclean filter with sterile dH2O)
-
○Store at room temperature protected from light
-
○UA can be used for ~1 week
-
○
7 SANDWICH ELISA FOR OLIGOMER DETECTION
Tau oligomers and related conformational changes, such as exposure of a motif in the N-terminus called the phosphatase-activating domain (or PAD), occur early during the evolution of pathognomonic tau inclusions in AD and other tauopathies (Combs, Hamel, & Kanaan, 2016; Combs & Kanaan, 2017; Kanaan et al., 2011; Lasagna-Reeves et al., 2012; Maeda et al., 2006; Patterson et al., 2011). To quantify oligomer formation and other related conformational changes we have developed a sandwich enzyme-linked immunosorbant assay (sELISA). These sELSIAs utilize either a tau oligomer selective antibody called tau oligomeric complex 1 (TOC1, available upon request) or one of the PAD-specific antibodies called tau N-terminal 1 or 2 (TNT1 or TNT2, both available at Millipore) as the capture antibodies to detect tau oligomers or PAD exposed tau species, respectively. TOC1 is a monoclonal mouse IgM raised against tau dimers and binds between amino acids 209–224 (Patterson et al., 2011; Ward et al., 2013). TNT1 and TNT2 are monoclonal mouse IgG1 antibodies directed against amino acids 2–18 (AEPFQEFEVMEDHAGTY), and recently we identified that the epitope for both of these reagents more specifically lies between amino acids 7–12 (Combs et al., 2016; Kanaan et al., 2011). Since all of these reagents appear to recognize linear epitopes that are displayed in a conformation-dependent fashion, they are suitable for nondenaturing assays such as sELI-SAs, dot blots, and immunohistochemistry. Our experience with different nondenaturing assays suggest sELISAs provide the best nondenaturing conditions as a physical interaction between tau proteins and a solid-state substrate (i.e., membranes or plastic plates) is not required. Dot blots are an acceptable alternative. These reagents will detect tau in denaturing assays (e.g., Western blotting or antigen-retrieved tissue sections) due to their linear epitopes; however, the signal in such assays does not provide useful information regarding tau conformational differences. Here, we detail the step-bystep protocol for our sELISA, using TOC1 as the example capture antibody (Fig. 3). We most commonly use the rabbit polyclonal pan-tau antibody R1 as a detection antibody (Berry et al., 2004; Combs et al., 2016; Cox et al., 2016; Kanaan et al., 2015); however, other detection antibodies can be used provided they recognize all forms of tau and are either a different species or different mouse immunoglobulin isotype than the capture antibody. We have successfully used Tau5 and Tau7 (IgG1; Horowitz, Lapointe, Guillozet-Bongaarts, Berry, & Binder, 2006; LoPresti, Szuchet, Papasozomenos, Zinkowski, & Binder, 1995) as a detection antibody with TOC1 capture assays. Other commercially available rabbit polyclonal tau antibodies have not been tested in our hands but should be useful as detection reagents in these assays.
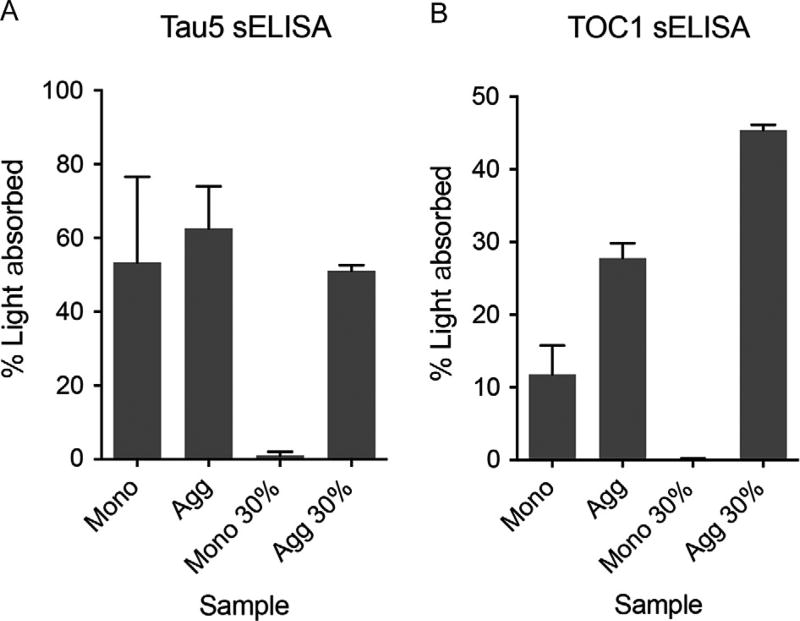
FIG. 3.
Total and oligomeric tau sandwich ELISAs of 30% sucrose fraction from the sucrose step gradient enrichment of tau oligomers. (A) A representative Tau5 sELISA (total tau) with the monomeric and tau aggregates (arachidonic acid-induced) samples and the corresponding 30% sucrose fractions (F3) from the monomer and aggregate samples run over the sucrose step gradient. (B) A representative TOC1 sELISA (oligomeric tau) with the monomeric and tau aggregates (arachidonic acid-induced) samples and the corresponding 30% sucrose fractions (F3) from the monomer and aggregate samples run over the sucrose step gradient. Note the strong TOC1 reactivity in the 30% sucrose fraction from the tau aggregate fractionation procedure, which confirms the presence of oligomeric tau in this fraction. The absorbance values were converted to percent light absorbed as described in the text.
7.1 TOC1 sELISA PROTOCOL
Dilute TOC1 capture antibody in borate saline buffer to a final concentration of 2ng/µL.
Coat wells with 50µL/well TOC1 capture antibody for 1h. All incubations are performed at room temperature on a shaker at low speed.
Wash wells 2× by adding 200µL ELISA wash buffer per well (all washing steps use the same procedure).
Block with 200µL/well 5% nonfat dry milk (NFDM, diluted in ELISA wash buffer) for 1h, followed by two washes.
Dilute recombinant tau protein samples in polymerization buffer (or other suitable buffers, such as TBS). The final concentration of tau protein required for the assay will be dependent on the capture antibody used. In our hands, 20nM recombinant tau protein is sufficient when using TOC1 as a capture antibody. However, we recommend performing an initial optimization experiment with a range of final protein concentrations (e.g., 2–200nM), especially when using a different capture antibody (e.g., TNT1 or TNT2).
Add 50µL/well diluted sample and incubate for 1.5 h, followed by four washes.
Dilute R1 detection antibody 1:20,000 (50ng/mL) in 5% NFDM.
Add 50µL/well R1 detection antibody and incubate for 1.5 h, followed by four washes.
Add 50µL/well of the detection secondary antibody HRP-conjugated goat antirabbit (Vector Labs, Burlingame, CA, PI-1000; diluted 1:5000 in 5% NFDM) and incubate for 1h, followed by 4 washes. The secondary antibody used during this step recognizes the detection antibody. Thus, if an alternative detection antibody is used above, a complementary secondary antibody should be used here. The secondary antibody should always be HRP-conjugated, as the TMB reagent is a peroxidase substrate.
Add 50µL/well TMB solution.
Incubate for 10–20min, until reaction has produced sufficient signal. Ideally, the reaction will be stopped within the linear portion of an absorbance curve (i.e., A450<0.8–1.0), and absorbance data should be handled appropriately (see below).
Stop reaction with 50µL/well H2SO4 stop solution.
Read immediately at 450nm (for TMB) on spectrophotometer microplate reader.
7.2 sELISA QUANTIFICATION
Data from an sELISA in this context should be handled one of two ways. First, the total amount of tau applied in this assay is known and equivalent between groups because these are recombinant protein samples (Fig. 3). Thus, a standard curve is not necessary, but only relative differences in signal between samples can be obtained without a standard curve. Absorbance (A) is not a linear measurement (i.e., A = Log10 (1/transmittance)) and not useful for comparing directly across samples (e.g., A = 0.3 is 50% light absorbed, A = 1 is 90% light absorbed, and A = 2 is 99% light absorbed). Thus, absorbance data should be converted to percent light absorbed (a linear scale) using the following equation:
where x is absorbance. The percent light absorbed can be used for statistical comparisons between groups.
Second, a recombinant tau protein standard can be used to more quantitatively evaluate the amount of tau across samples. Since the measured signal in the sELISA is based upon the detection antibody (in this case R1 tau antibody), performing a standard indirect ELISA where a range of recombinant taumonomer protein is bound to the plate is the best option to provide a standard curve that will be indicative of maximal R1 signal per a known amount of tau. In this approach, the amount of tau in the standard samples is known, the species of tau is uniform (e.g., monomeric), and it is unmodified by posttranslational modifications. It is worth noting that there are no perfect standards for these assays as all variations contain some caveats; thus, these assays can only be semiquantitative. To perform this standard, wells are coated with recombinant tau monomers (0–200 nM), when the other experimental sample wells are coated with the capture antibody as outlined above in the sELISA protocol. The standard curve wells are kept in blocking buffer until application of the detection primary antibody (i.e., R1), at which point they are processed following all of the steps as described earlier along with the experimental sample wells.
7.3 MATERIALS
96-well EIA/RIA High Binding 96-well plate (Corning, Tewksbury, MA, 3590).
- Borate saline buffer (1 L)
- 6.2g Boric acid (Sigma-Aldrich, B6768)
- 9.5g Sodium tetraborate decahydrate (Na2B4O7·10H2O; VWR, MK745706)
- 4.4g NaCl (Sigma-Aldrich, S9888)
- 0.1g Thimerosal (Sigma-Aldrich, T5125)
- 1000mL H2O
- Filter (0.22-µm) and store at 4°C
- ELISA washing solution (2L)
- 12.4g Boric acid
- 19g Sodium borate (Na2B4O7·10H2O)
- 8.8g NaCl
- 0.2g Thimerosal
- 8g Bovine serum albumin (VWR, 0332)
- 10mL 10% Tween-20 (Fisher Scientific, BP337)
- 1990mL H2O
- Filter (0.22-µm) and store at 4°C
- 5% NFDM
- 5g NFDM
- 100mL ELISA wash buffer
- Store at 4°C
- Polymerization buffer (50mL)
- 5mL 1M NaCl
- 2mL 250mM HEPES
- 5mL 1mM EDTA
- 38mL H2O
TMB (3,3,5′,5-tetramethylbenzidine; Sigma-Aldrich, T8665)
- Stop solution (500mL)
- 18mL H2SO4 (Sigma-Aldrich, 258105)
- 482mL H2O
- Store at room temperature
8 CONCLUSION
One of the great challenges in elucidating the toxic role of tau aggregation in neurodegenerative diseases will be to understand how its various forms exert toxicity. Based on recent advances, it is evident that we cannot treat all aggregates as if they were the same and, with that, there is a need to develop techniques that allow simple and reproducible generation and purification of oligomeric populations of tau. This also requires methods to validate these processes. We have described methods at each step of this process that we have found useful to study oligomeric tau. The combined chromatographic approach to recombinant tau production and purification provides a relatively straightforward protocol that results in high quality preparations. Incubation of tau with arachidonic acid followed by separation over a sucrose step gradient allows for simple and quick production of oligomeric tau species that can be used in a variety of downstream assays. It is also noteworthy that shortened incubations (e.g., 15min) with arachidonic acid (Ward, Himmelstein, Lancia, & Binder, 2012; Ward et al., 2013), and use of 3R tau isoforms using the protocol above produces oligomeric-enriched samples with very little to no filamentous aggregates. Finally, reagents such as TOC1 and the TNT antibodies can be used to identify and characterize the presence and characteristics of tau oligomers in a variety of settings, including a sensitive sandwich ELISA. However, it is noteworthy that oligomeric or multimeric tau species likely exist in a heterogeneous set of conformational states, which remain to be comprehensively defined and TOC1 and TNT reagents are unlikely to detect all of tau’s potential oligomeric conformational states. The methods described earlier should prove useful in a variety of applications, such as structural and biochemical assessment of tau oligomers, cell toxicity assays, and in vivo applications.
Acknowledgments
This work was supported by NIH/National Institute on Aging R01 AG044372 (N.M.K.), NIH/ National Institute of Neurological Diseases and Stroke R01 NS082730 (N.M.K.), the Jean P. Schultz Biomedical Research Endowment (N.M.K.), and the Secchia Family Foundation Research Fund (N.M.K.).
References
- Andorfer C, Acker CM, Kress Y, Hof PR, Duff K, Davies P. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. The Journal of Neuroscience. 2005;25(22):5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry RW, Sweet AP, Clark FA, Lagalwar S, Lapin BR, Wang T, et al. Tau epitope display in progressive supranuclear palsy and corticobasal degeneration. Journal of Neurocytology. 2004;33(3):287–295. doi: 10.1023/B:NEUR.0000044190.96426.b9. [DOI] [PubMed] [Google Scholar]
- Carlson SW, Branden M, Voss K, Sun Q, Rankin CA, Gamblin TC. A complex mechanism for inducer mediated tau polymerization. Biochemistry. 2007;46(30):8838–8849. doi: 10.1021/bi700403a. [DOI] [PubMed] [Google Scholar]
- Combs B, Gamblin TC. FTDP-17 tau mutations induce distinct effects on aggregation and microtubule interactions. Biochemistry. 2012;51(43):8597–8607. doi: 10.1021/bi3010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs B, Hamel C, Kanaan NM. Pathological conformations involving the amino terminus of tau occur early in Alzheimer’s disease and are differentially detected by monoclonal antibodies. Neurobiology of Disease. 2016;94:18–31. doi: 10.1016/j.nbd.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs B, Kanaan NM. Exposure of the amino terminus of tau is a pathological event in multiple tauopathies. The American Journal of Pathology. 2017;187(6):1222–1229. doi: 10.1016/j.ajpath.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs B, Voss K, Gamblin TC. Pseudohyperphosphorylation has differential effects on polymerization and function of tau isoforms. Biochemistry. 2011;50(44):9446–9456. doi: 10.1021/bi2010569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K, Combs B, Abdelmesih B, Morfini G, Brady ST, Kanaan NM. Analysis of isoform-specific tau aggregates suggests a common toxic mechanism involving similar pathological conformations and axonal transport inhibition. Neurobiology of Aging. 2016;47:113–126. doi: 10.1016/j.neurobiolaging.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff K, Knight H, Refolo LM, Sanders S, Yu X, Picciano M, et al. Characterization of pathology in transgenic mice over-expressing human genomic and cDNA tau transgenes. Neurobiology of Disease. 2000;7(2):87–98. doi: 10.1006/nbdi.1999.0279. [DOI] [PubMed] [Google Scholar]
- Gamblin TC, King ME, Kuret J, Berry RW, Binder LI. Oxidative regulation of fatty acid-induced tau polymerization. Biochemistry. 2000;39(46):14203–14210. doi: 10.1021/bi001876l. [DOI] [PubMed] [Google Scholar]
- Gerson JE, Sengupta U, Lasagna-Reeves CA, Guerrero-Munoz MJ, Troncoso J, Kayed R. Characterization of tau oligomeric seeds in progressive supranuclear palsy. Acta Neuropathologica Communications. 2014;2:73. doi: 10.1186/2051-5960-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996;383(6600):550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- Horowitz PM, Lapointe N, Guillozet-Bongaarts AL, Berry RW, Binder LI. N-terminal fragments of tau inhibit full-length tau polymerization in vitro. Biochemistry. 2006;45(42):12859–12866. doi: 10.1021/bi061325g. [DOI] [PubMed] [Google Scholar]
- Kanaan NM, Cox K, Alvarez VE, Stein TD, Poncil S, McKee AC. Characterization of early pathological tau conformations and phosphorylation in chronic traumatic encephalopathy. Journal of Neuropathology and Experimental Neurology. 2015;75:19–34. doi: 10.1093/jnen/nlv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan NM, Morfini GA, LaPointe NE, Pigino GF, Patterson KR, Song Y, et al. Pathogenic forms of tau inhibit kinesin-dependent axonal transport through a mechanism involving activation of axonal phosphotransferases. The Journal of Neuroscience. 2011;31(27):9858–9868. doi: 10.1523/JNEUROSCI.0560-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ME, Gamblin TC, Kuret J, Binder LI. Differential assembly of human tau isoforms in the presence of arachidonic acid. Journal of Neurochemistry. 2000;74(4):1749–1757. doi: 10.1046/j.1471-4159.2000.0741749.x. [DOI] [PubMed] [Google Scholar]
- Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Sarmiento J, Troncoso J, Jackson GR, et al. Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. The FASEB Journal. 2012;26(5):1946–1959. doi: 10.1096/fj.11-199851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, Binder LI. Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(22):10369–10373. doi: 10.1073/pnas.92.22.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Sahara N, Saito Y, Murayama S, Ikai A, Takashima A. Increased levels of granular tau oligomers: An early sign of brain aging and Alzheimer’s disease. Neuroscience Research. 2006;54(3):197–201. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Maeda S, Sahara N, Saito Y, Murayama M, Yoshiike Y, Kim H, et al. Granular tau oligomers as intermediates of tau filaments. Biochemistry. 2007;46(12):3856–3861. doi: 10.1021/bi061359o. [DOI] [PubMed] [Google Scholar]
- Martinez M, Mougan I. Fatty acid composition of human brain phospholipids during normal development. Journal of Neurochemistry. 1998;71(6):2528–2533. doi: 10.1046/j.1471-4159.1998.71062528.x. [DOI] [PubMed] [Google Scholar]
- Patterson KR, Remmers C, Fu Y, Brooker S, Kanaan NM, Vana L, et al. Characterization of prefibrillar tau oligomers in vitro and in Alzheimer disease. The Journal of Biological Chemistry. 2011;286(26):23063–23076. doi: 10.1074/jbc.M111.237974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309(5733):476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittaels K, Van Den Haute C, Van Dorpe J, Bruynseels K, Vandezande K, Laenen I, et al. Prominent axonopathy in the brain and spinal cord of transgenic mice over-expressing four-repeat human tau protein. The American Journal of Pathology. 1999;155(6):2153–2165. doi: 10.1016/S0002-9440(10)65533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Himmelstein DS, Lancia JK, Binder LI. Tau oligomers and tau toxicity in neurodegenerative disease. Biochemical Society Transactions. 2012;40(4):667–671. doi: 10.1042/BST20120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Himmelstein DS, Lancia JK, Fu Y, Patterson KR, Binder LI. TOC1: Characterization of a selective oligomeric tau antibody. Journal of Alzheimer’s Disease. 2013;37(3):593–602. doi: 10.3233/JAD-131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Drewes G, Biernat J, Mandelkow EM, Mandelkow E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. The Journal of Cell Biology. 1992;118(3):573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, Binder LI. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer’s disease. The American Journal of Pathology. 1997;150(6):2181–2195. [PMC free article] [PubMed] [Google Scholar]
- Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, et al. Tauopathy in Drosophila: Neurodegeneration without neurofibrillary tangles. Science. 2001;293(5530):711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]