Abstract
Background
Extracellular vesicles (EVs) are membrane bound, secreted by cells, and detected in bodily fluids, including urine, and contain proteins, RNA, and DNA. Our goal was to identify HIV and human proteins (HPs) in urinary EVs from HIV+ patients and compare them to HIV− samples.
Methods
Urine samples were collected from HIV+ (n = 35) and HIV− (n = 12) individuals. EVs were isolated by ultrafiltration and characterized using transmission electron microscopy, tandem mass spectrometry (LC/MS/MS), and nanoparticle tracking analysis (NTA). Western blots confirmed the presence of HIV proteins. Gene ontology (GO) analysis was performed using FunRich and HIV Human Interaction database (HHID).
Results
EVs from urine were 30–400 nm in size. More EVs were in HIV+ patients, P < 0.05, by NTA. HIV+ samples had 14,475 HPs using LC/MS/MS, while only 111 were in HIV−. HPs in the EVs were of exosomal origin. LC/MS/MS showed all HIV+ samples contained at least one HIV protein. GO analysis showed differences in proteins between HIV+ and HIV− samples and more than 50% of the published HPs in the HHID interacted with EV HIV proteins.
Conclusion
Differences in the proteomic profile of EVs from HIV+ versus HIV− samples were found. HIV and HPs in EVs could be used to detect infection and/or diagnose HIV disease syndromes.
1. Introduction
Extracellular vesicles (EVs) are membrane bound vesicles, between 30 nm and 1 μm in size, are secreted into blood, urine, saliva, semen, and other bodily fluids, and have been suggested as a potential source of biomarkers for disease progression [1, 2]. These EVs, microparticles and/or exosomes, are secreted by cells normally or while they are undergoing stress or apoptosis [3] and contain proteins, mRNA, and miRNA [4] that are involved in cell to cell communication, transfer of antigens to cells, and intracellular communication. EVs are described in cancer disease pathogenesis [5] in HIV infection [6], other viral infections [7], and other disease states such as cardiovascular, renal, liver, and metabolic disease [8–11].
EVs from urine are an attractive noninvasive source for biomarkers of diseases [12, 13]. In healthy individuals, protein only accounts for 0.01% of urine components; however, in certain disease states, the protein content and EV numbers can increase in urine [12–16]. The glomerular capsule filters blood that is passed into the renal tubule and accounts for thirty percent of the urinary protein content [14–16]. The remaining seventy percent of proteins in urine is derived from the kidney [17], and thus, urinary EVs are comprised of both renal and efferent components.
HIV proteins are detected in EVs of HIV+ patients and HIV Nef is the most prevalent protein found [18–21]. Other reports of HIV proteins in EVs are from in vitro transfected or HIV infected cultured cells and are not from HIV+ patient samples [6, 18, 19, 22, 23].
Biomarkers in urinary EVs are suggested for use in the diagnosis of many disease states [12, 13, 24–30]. The objectives of this study were to determine the differences in proteins from urinary EVs from HIV+ patients and HIV− individuals using proteomics and mass spectrometry. The analysis of more patient samples could identify specific EV urinary proteins as biomarkers of HIV infection, treatment efficacy, and/or disease progression.
2. Methods
2.1. Sample Collection
Urine was collected from thirty-five (35) HIV+ patients and twelve (12) HIV− individuals in sterile collection cups. The subjects were recruited from clinics in the metropolitan Atlanta area, GA. Patient demographics are described in Table 1. The study was approved by the Institutional Review Board of Morehouse School of Medicine and written informed consent was obtained from all participants.
Table 1.
Patient demographics.
| Characteristics | HIV-positive (N = 35) | HIV-negative (N = 12) |
|---|---|---|
| Age (median ± IQR) | 41.5 ± 14.25 | 59 ± 18 |
| Sex (n, %) | ||
| Male | 25 (71.4%) | 7 (58.3%) |
| Female | 10 (28.6%) | 5 (41.7%) |
| Race (n, %) | ||
| African American/Black | 28 (80%) | 12 (100%) |
| White | 7 (20%) | - |
| Hispanic | - | - |
| Asian | - | - |
| Viral loads (copies/ml) (median ± IQR) | 50 ± 0 | - |
| CD4+ T cell (cells/µl) (median ± IQR) | 66.5 ± 46.5 | - |
| Antiretroviral therapy (n, %) | 34 (97.1%) | - |
2.2. EV Isolation
Urine samples were centrifuged at 1000 ×g to remove cells and sediment then frozen at −80°C. Samples, 4 ml, were thawed and the EVs isolated followed by centrifugal filtration using Amicon Ultra-4 100 kDa centrifugal filter unit (Millipore, Billerica, MA), at 3000 ×g for 15 minutes at 4°C. The retentate, containing EVs, was collected from the top of the filter and resuspended in 200 μl phosphate buffered saline (PBS) for use in the transmission electron microscopy and tandem mass spectrometry (LC/MS/MS) analysis.
2.3. Transmission Electron Microscopy Analysis
Transmission electron microscopy (TEM) was used to identify EVs in two HIV-1 positive and two HIV-1 negative samples. Urinary EVs were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 hours at 4°C followed by 2 washes with 0.1 M cacodylate buffer, 5 minutes each. Samples were stained with 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 hour at 4°C followed by 2 washes with the cacodylate buffer and 3 washes with deionized water, 5 minutes each. Samples were subsequently stained with 0.5% aqueous uranyl acetate for 2 hours at room temperature and subsequently viewed with a JEOL 1200EX transmission electron microscope (JEOL, Peabody, MA).
2.4. Nanoparticle Tracking Analysis (NTA)
Urine samples from HIV-negative (n = 8) and positive individuals (n = 11), 15 ml, were centrifuged at 300 ×g for 10 min at 4°C to remove cell debris. The supernatant was collected and centrifuged at 16,500 ×g for 20 min at 4°C and the supernatant collected and ultracentrifuged at 120,000 ×g at 4°C for 1.5 hr. The pellet was resuspended in 500 μl of PBS. The size and quantification of the EVs were analyzed using the NanoSight NS500 (NanoSight NTA 2.3 Nanoparticle Tracking and Analysis Release version build 0025). Particles were automatically tracked and sized based on Brownian motion and the diffusion coefficient. The NTA measurement conditions were temperature 21.0 +/− 0.5°C, viscosity 0.99 +/− 0.01 cP, frames per second 24.99–25, and measurement time 30 s. The detection threshold was similar in all samples. Two recordings were performed for each sample.
2.5. Mass Spectrometry Analysis
Thirty-five (35) HIV+ and twelve (12) HIV− EV samples were lysed and trypsinized and the sequence of peptides was determined by tandem mass spectrometry (LC/MS/MS), using an LTQ Ion Trap Mass Spectrometer (Thermo Fischer Scientific, Waltham, MA). Peptides were first reduced in DTT 10 mM at 56°C for at least 30 min and alkylated with 15 mM iodoacetic acid for 30 min at room temperature in the dark. Samples were then digested with mass spectrometry grade trypsin 20 ng/μl for 4 hours at 37°C. Just before analysis, the sample was acidified by the addition of formic acid to 0.1%. Peptides were separated by reverse phase HPLC (Agilent) on a 0.5 × 75 mm C-18 column (Michrom) at a flow rate of 500 nl/min using a linear gradient of acetonitrile (5–35%) over 100 min. Ions were directly introduced by nanospray and spectra were collected using Xcalibur 2.0 software using an intensity threshold of 200 counts. The resulting spectra were analyzed using Bioworks 1.1 software to search a hybrid Human-HIV database created from the complete nonredundant peptide database from NCBI. The threshold for inclusion in the search is a minimal S/N ratio of 3. False discovery rates were determined and set based on the control HIV− samples. An initial protein identification list was generated from matches with an Xcorr score versus charge state of 1.0 (+1) 1.5 (+2) and 1.7 (+3) and consensus scores greater than 10.0.
Bioinformatics techniques for analysis of HIV EV proteins were used on the LC/MS/MS detected proteins [31]. Functional enrichment analysis was performed using FunRich (Functional Enrichment analysis tool, http://funrich.org/index.html) [32] against a human database to detect proteins involved in biological processes, cellular components, sites of expression, and biological pathways. Only processes with a P value < 0.05, using the Benjamini-Hochberg False Discovery rate, were reported. The human proteins detected were compared to the top 100 EV proteins in ExoCarta (http://exocarta.org/exosome_markers_new) [33, 34], sixty EV proteins in the EV array [35], and proteins identified in EVs from HIV infected lymphocytic cells [36].
Pathway analysis comparing HIV+ samples with CD4+ T cells greater than 300 (n = 15) to those with less than 500 (n = 15) and HIV high VL, greater than 200 copies (n = 10), compared to HIV low viral loads, less than 200 copies (n = 10), was done using Pathway Studio version 11.4 Mammal Plus (Elsevier, Inc., Atlanta, GA). Gene Set Enrichment Analysis (GSEA) was used to identify the top 10 curated pathways for the proteins in the each of the patient groups. No comparisons were done between patients not on ART or undergoing ART because there was only one patient not on ART.
The HIV proteins, Nef, Vpr, Vpu, and Vif, were searched using the HIV-1 Human Interaction database (https://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/). This database contains all the known, published interactions of HIV-1 gene products with human proteins [37]. Proteins from the search were compared to the human proteins detected in the HIV EVs.
2.6. Western Blot Analysis
To validate the presence of HIV proteins in urinary EVs, western blot analysis (WB) was performed on twenty (20) randomly selected HIV+ and three (3) HIV− control urine samples. Recombinant HIV-1 Nef and HIV-1 p24 were used as positive controls, while HIV-negative urine and HIV-positive filtrate were used as negative controls. Samples were heated at 85°C for two minutes in a tris-glycine SDS sample buffer, were loaded into a 4–20% TGX gradient gel (Bio-Rad, Hercules, CA), and run for 50 mins at 200 V. A semidry transfer unit (Hoefer Scientific, Holliston, MA) was used to transfer the separated proteins onto a PVDF membrane (Bio-Rad) at 15 V for 50 mins. The filter was blocked for nonspecific binding using 5% nonfat dry milk in 1x tris buffered saline (TBS) with Tween 20. The membrane was incubated overnight in pooled plasma from twenty HIV+ patients as the primary antibody at a 1 : 500 dilution and rabbit anti-human IgG conjugated HRP antibody (1 : 1000, Bio-Rad, Hercules, CA) was used as secondary antibody. Super Signal West Femto (Thermo Fischer Scientific, Waltham, MA) was used as a chemiluminescent substrate for detection. The membrane was developed and imaged using the LAS 4000 biomolecular imager (GE Healthcare Life Sciences, Pittsburgh, PA). Recombinant HIV-1 Nef and p24 WB analyses were detected using anti-Nef and p24 monoclonal antibodies (1 : 500, EMD Millipore, Billerica, MA) and anti-mouse IgG conjugated HRP antibodies (1 : 1000, Bio-Rad, Hercules, CA) were used.
2.7. HIV p24 ELISA
Twenty-six (26) HIV+ and eleven (11) HIV− urine samples were tested for the presence of HIV p24 by ELISA (ImmunoDX, Woburn, MA).
3. Results
3.1. HIV Proteins Are Present in Urinary EVs of HIV-Positive Patients
LC/MS/MS mass spectrometry HIV EV protein results are presented in Table 2. Urinary EV proteins meeting the false discovery rate and Xcorr score criteria as HIV-1 proteins included Nef, Gag, Pol, Protease, gp120, gp160, gp41, Rev, reverse transcriptase, Tat, Vif, Vpr, and Vpu. All HIV+ urine samples (n = 35) contained at least one HIV-1 protein in EVs, while no HIV proteins were found in the HIV− samples (n = 12) (Table 3). HIV-1 Nef was detected in twenty-six of thirty-five (26 of 35) (74.3%) HIV+ urine samples. Three (3) patients' urine samples, #173, #174, and #196, were tested 203, 311, and 35 days, respectively, after their first EV sample was analyzed. No difference in the HIV proteins detected in sample #196, 35 days after his previous sample, was found. #173's sample, tested 203 days after the first analysis, had a similar profile, except that Rev and Tat were not detected. In addition, #174's EVs examined 311 days after the first sampling found Rev and RT missing from the profile.
Table 2.
LC/MS/MS analysis of EV HIV proteins.
| Accession | # AAs | MW [kDa] | Calc. pI | Description | Σ Coverage | Σ# peptides | Score A0 | Coverage A0 | # peptides A0 |
|---|---|---|---|---|---|---|---|---|---|
| gi38491705 | 192 | 22.7 | 10.1 | Vif protein [human immunodeficiency virus 1] | 13.54 | 12 | 9.22 | 13.54 | 4 |
| gi73913089 | 104 | 11.7 | 10.1 | Gag protein [human immunodeficiency virus 1] | 14.42 | 15 | 6.27 | 14.42 | 4 |
| gi58374258 | 869 | 98.1 | 8.8 | Envelope glycoprotein [human immunodeficiency virus 1] | 1.5 | 5 | 5.63 | 1.5 | 3 |
| gi183197180 | 404 | 45.8 | 8.4 | Pol protein [human immunodeficiency virus 1] | 3.47 | 3 | 5.27 | 3.47 | 3 |
| gi255984636 | 160 | 18.1 | 5.3 | Reverse transcriptase [human immunodeficiency virus 1] | 7.5 | 4 | 4.80 | 7.5 | 2 |
| gi256012108 | 114 | 13.5 | 5.7 | Nef protein [human immunodeficiency virus 1] | 14.04 | 3 | 4.53 | 14.04 | 2 |
| gi9756252 | 524 | 60.4 | 8.7 | Pol precursor [human immunodeficiency virus 1] | 4.01 | 2 | 4.46 | 4.01 | 2 |
| gi67082579 | 191 | 22.3 | 9.4 | Reverse transcriptase [human immunodeficiency virus 1] | 10.47 | 2 | 4.43 | 10.47 | 2 |
| gi2290009 | 852 | 96.7 | 8.5 | Envelope glycoprotein [human immunodeficiency virus 1] | 7.16 | 11 | 4.33 | 5.87 | 3 |
| gi167886806 | 25 | 2.7 | 8.7 | Rev protein [human immunodeficiency virus 1] | 56 | 4 | 4.29 | 56 | 2 |
| gi23344577 | 99 | 10.6 | 9.4 | Protease [human immunodeficiency virus 1] | 12.12 | 5 | 4.36 | 12.12 | 2 |
| gi4324808 | 1437 | 161.9 | 8.3 | Gag-pol polyprotein [human immunodeficiency virus 1] | 2.51 | 7 | 4.05 | 1.6 | 2 |
| gi222533599 | 73 | 8.0 | 9.1 | Env C2V3 protein [human immunodeficiency virus 1] | 23.29 | 4 | 3.85 | 23.29 | 2 |
| gi71060450 | 206 | 23.7 | 6.3 | Negative factor [human immunodeficiency virus 1] | 4.85 | 4 | 3.84 | 4.85 | 2 |
| gi37935985 | 85 | 10.3 | 4.8 | Vpu protein [human immunodeficiency virus 1] | 11.76 | 4 | 3.84 | 11.76 | 2 |
| gi108860432 | 870 | 98.8 | 8.5 | gp160 [human immunodeficiency virus 1] | 3.33 | 2 | 3.74 | 3.33 | 2 |
| gi114801226 | 209 | 24.4 | 10.1 | Tat protein [human immunodeficiency virus 1] | 5.26 | 2 | 3.70 | 5.26 | 2 |
| gi22596451 | 341 | 38.6 | 8.0 | Truncated envelope glycoprotein [human immunodeficiency virus 1] | 3.81 | 4 | 3.68 | 3.81 | 2 |
| gi183200570 | 342 | 38.7 | 9.2 | Truncated pol protein [human immunodeficiency virus 1] | 3.51 | 2 | 3.68 | 3.51 | 2 |
| gi34786230 | 176 | 19.8 | 9.6 | gp120 protein [human immunodeficiency virus 1] | 10.8 | 3 | 3.60 | 10.8 | 3 |
| gi1002239 | 104 | 11.5 | 8.9 | Envelope glycoprotein, v3 region [human immunodeficiency virus 1] | 18.27 | 2 | 3.54 | 18.27 | 2 |
| gi77168129 | 95 | 11.3 | 7.6 | Vpr protein [human immunodeficiency virus 1] | 13.68 | 3 | 3.40 | 13.68 | 2 |
| gi222532593 | 129 | 14.7 | 8.4 | Gag p17 protein [human immunodeficiency virus 1] | 8.53 | 3 | 2.61 | 8.53 | 1 |
| gi219688191 | 132 | 14.9 | 10.2 | Matrix protein [human immunodeficiency virus 1] | 9.09 | 6 | 2.56 | 9.09 | 1 |
| gi255687141 | 288 | 32.1 | 7.7 | Integrase [human immunodeficiency virus 1] | 9.03 | 1 | 2.52 | 9.03 | 1 |
| gi222532161 | 132 | 15.0 | 9.5 | Gag p17 protein [human immunodeficiency virus 1] | 9.09 | 6 | 2.45 | 9.09 | 1 |
| gi37934078 | 573 | 65.0 | 9.0 | Gag-pol fusion polyprotein [human immunodeficiency virus 1] | 2.79 | 2 | 2.42 | 2.79 | 2 |
| gi405003 | 207 | 23.1 | 7.7 | gp120 [human immunodeficiency virus 1] | 12.08 | 1 | 2.41 | 12.08 | 1 |
| gi54792352 | 213 | 23.7 | 5.6 | Gag polyprotein [human immunodeficiency virus 1] | 9.39 | 2 | 2.34 | 9.39 | 2 |
| gi3885826 | 132 | 14.9 | 9.6 | p17 matrix [human immunodeficiency virus 1] | 11.36 | 2 | 2.31 | 11.36 | 1 |
Accession # = NCBI NR database, #AAs = total number of amino acids in the protein entry, MW = molecular weight of the protein in kDa, Description: description from the NCBI database, and Peptides = total number of unique peptides found.
Table 3.
Presence of HIV-1 proteins in HIV+ patient urinary EVs.
| ID | ART | AIDS | Viral load | CD4 cells/ul | Nef | Gag | Pol | Protease | Rev | RT | Tat | Vif | p1 | p24 | p17 | Poly | Vpu | Env | Vpr | Vif |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| copies/ml | ||||||||||||||||||||
| 22 | No | 224 | X | X | X | X | X | X | X | X | X | X | ||||||||
| 27 | Yes | AIDS | <50 | 134 | X | |||||||||||||||
| 28 | Yes | AIDS | 280100 | 22 | X | X | X | X | X | |||||||||||
| 30 | Yes | AIDS | >10000 | <20 | X | X | X | X | X | X | X | X | X | X | ||||||
| 41 | Yes | 29187 | 440 | X | X | X | ||||||||||||||
| 46 | Yes | <50 | 689 | X | X | X | X | |||||||||||||
| 45 | Yes | 400 | 345 | X | X | X | X | X | ||||||||||||
| 48 | Yes | 4974 | 454 | X | X | X | X | |||||||||||||
| 51 | NA | NA | X | X | X | X | X | |||||||||||||
| 52 | Yes | 51 | 574 | X | X | X | X | X | X | X | X | |||||||||
| 61 | <50 | 655 | X | X | ||||||||||||||||
| 62 | Yes | AIDS | <50 | 232 | X | |||||||||||||||
| 63 | No | 2023 | 83 | X | ||||||||||||||||
| 65 | No | NA | NA | X | ||||||||||||||||
| 66 | No | NA | NA | X | ||||||||||||||||
| 67 | Yes | 75 | 509 | X | ||||||||||||||||
| 68 | No | NA | NA | X | ||||||||||||||||
| 69 | Yes | <50 | 187 | X | ||||||||||||||||
| 70 | Yes | <50 | 399 | X | ||||||||||||||||
| 71 | Yes | <50 | 456 | X | ||||||||||||||||
| 74 | NA | NA | X | |||||||||||||||||
| 86 | Yes | <75 | 1642 | X | X | X | X | |||||||||||||
| 103 | Yes | AIDS | 150 | 560 | X | X | X | |||||||||||||
| 104 | Yes | AIDS | 77 | 313 | X | X | ||||||||||||||
| 108 | Yes | AIDS | <50 | 653 | X | X | X | |||||||||||||
| 110 | Yes | <50 | 379 | X | X | |||||||||||||||
| 111 | Yes | AIDS | <50 | 182 | X | X | ||||||||||||||
| 112 | Yes | AIDS | >200 | 581 | X | X | ||||||||||||||
| 142 | Yes | <50 | 487 | X | X | X | X | X | X | X | X | |||||||||
| 173-1 | Yes | <50 | 398 | X | X | X | X | X | X | X | X | X | X | |||||||
| 173-2 | Yes | <50 | 398 | X | X | X | X | X | X | |||||||||||
| 174-1 | Yes | 48 | 315 | X | X | X | X | X | X | |||||||||||
| 174-2 | Yes | 48 | 315 | X | X | X | X | |||||||||||||
| 196-1 | Yes | AIDS | <50 | 113 | X | X | X | X | X | X | X | X | X | X | X | X | X | |||
| 196-2 | Yes | AIDS | <50 | 113 | X | X | X | X | X | X | X | X | X | X | X | X | X |
An initial protein identification list was generated from matches with an Xcorr score versus charge state of 1.0 (+1), 1.5 (+2), and 1.7 (+3) and consensus scores greater than 10.0; NA = not available.
HIV p24 antigen was only detected in five of thirty-five (5 of the 35 patient) (14%) samples by LC/MS/MS, but of the twenty-six (26) HIV+ and eleven (11) HIV-negative samples tested by ELISA, no p24 was detected. There was no statistical correlation of the number of HIV proteins detected with CD4+ T cell counts, viral loads, or ART therapy.
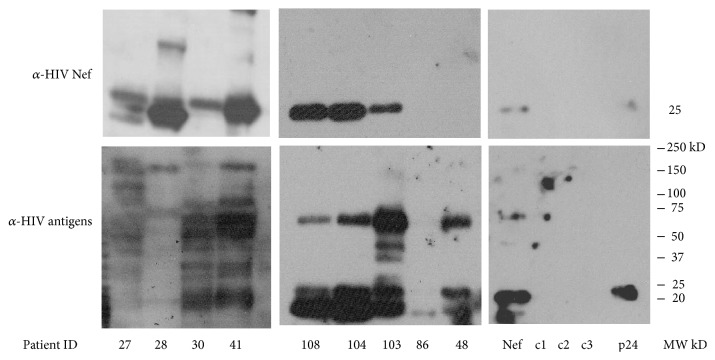
Validation by WB analysis using polyclonal pooled patient serum and monoclonal antibodies against HIV Nef and HIV p24 indicated the presence of HIV proteins. Figure 1 is a WB using polyclonal pooled HIV+ serum used as the detection antibody. All the HIV+ patient samples contained HIV-1 proteins and the top panel shows patient samples reacting to anti-HIV Nef. HIV+ urine samples, 7 of 9 (77.7%), showed HIV-1 Nef bands at 27 kD.
Figure 1.
Detection of HIV-1 proteins by western blot. Extracellular vesicles were isolated from four ml of urine from HIV-1+ patients and HIV-1 negative individuals by Amicon ultrafiltration (MW cutoff = 100,000 kD). The western blot is representative of 9 HIV+ and 3 HIV-negative samples (c1, c2, and c3). Recombinant HIV Nef and p24 were added as positive controls (last panels on the right). Samples were isolated in a 4–20% gradient SDS gel and transferred to a PVDF membrane. The filter was incubated with the primary antibody, pooled HIV-1 positive plasma (bottom panels), or a monoclonal anti-HIV Nef (top panels). The secondary antibody, goat anti-mouse IgG for the anti-Nef blots or rabbit anti-human IgG for the anti-HIV antibodies, conjugated to horseradish peroxidase. Super Signal West Femto was used as chemiluminescent substrate for detection.
3.2. TEM and NTA Analysis of EVs
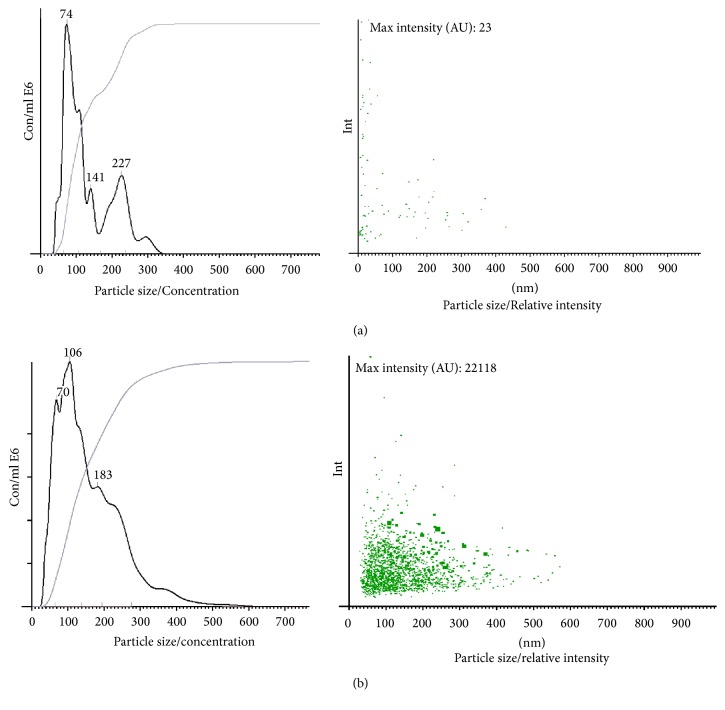
TEM analysis of urine from HIV+ patients showed multiple EVs, ranging in size from 50 nm to 300 nm (Figure 2(a)), while two HIV-negative controls had fewer EVs present (Figure 2(b)). NTA analysis showed that there were significantly more EVs from HIV+ patients than healthy controls, 4.96 ± 0.0733 and 3.69 ± 0.075, respectively (P < 0.05). No significant differences were found in the size of the EVs, 110–227 nm for HIV-negative donors and 54–448 nm HIV+ samples. Representative Nanosight analyses for HIV-negative and HIV+ urine samples are shown in Figure 3.
Figure 2.
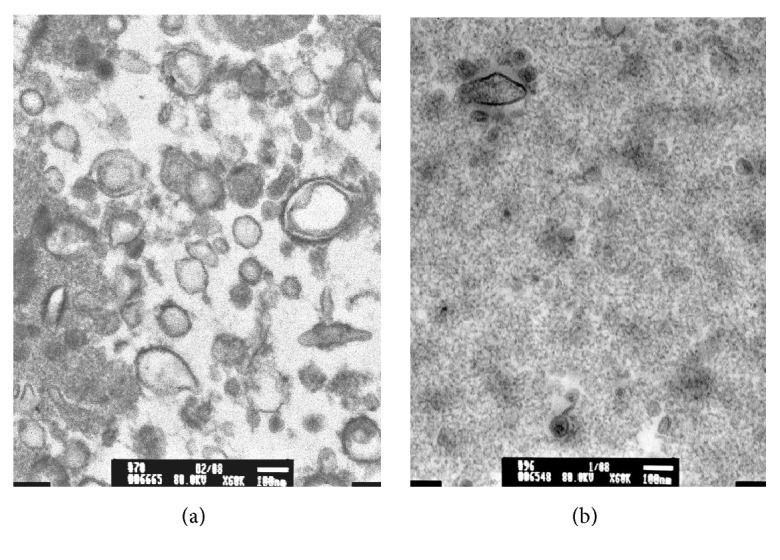
Transmission electron microscopy of urinary extracellular vesicles. Four mls of urine was used to isolate EVs by Amicon ultrafiltration (MW cutoff = 100,000 kD). EVs were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer. Samples were stained with 1% osmium tetroxide in 0.1 M cacodylate buffer and subsequently stained with 0.5% aqueous uranyl acetate. A JEOL 1200EX transmission electron microscope (JEOL, Peabody, MA) was used for observation and photography. 1A. EVs from HIV-1 posi.
Figure 3.
Nanosight analysis (representative analysis). (a) NTA analysis of an HIV-negative urine sample had 0.4 × 108 particles per ml (left panel) while (b) depicts an urine sample from a HIV+ patient that had 8.7 × 108 particles per ml and has a greater relative intensity profile (right panel (a) and (b)) when compared to the HIV-negative sample. The Rank Sum T test showed that HIV+ patient urine samples had more particles per ml than the negative control urine (P < 0.05).
3.3. Human Proteins in HIV+ and Negative EV Urine Samples
EV proteins from the HIV+ patients, 14,475, which entered into FunRich, functional enrichment analysis software, showed 29.44% or 1,932 proteins were associated with exosomes (Table 4). These EV identified proteins were compared to top 100 EV proteins in the ExoCarta database with 83% matching (http://exocarta.org/exosome_markers_new) [33], 22 EV proteins in the EV array [35] were similar, and 7 of 14 EV proteins identified in exosomes from HIV infected lymphocytes [36] were found and are highlighted in Table 4. Exosomal proteins found in the control samples are listed in Table 5.
Table 4.
Exosomal proteins found in urinary EVs from HIV+ patients.
| Genes in our analysis | Genes in the FunRich database | Percentage of genes | Fold enrichment | |
|---|---|---|---|---|
| Exosomal proteins | 1932 | 2001 | 29.44 | 2.11 |
|
| ||||
| A1BG, A2M, AARS, ABCA7, ABCB1, ABCB11, ABCB6, ABCC1, ABCC11, ABCC9, ABCG2, ABHD8, ACAA2, ACAT1, ACAT2, ACE, ACE2, ACLY, ACO1, ACOT11, ACP2, ACSL3, ACSL4, ACSM1, ACTA1, ACTA2, ACTB, ACTBL2, ACTC1, ACTG1, ACTL6A, ACTN1, ACTN2, ACTN4, ACTR1A, ACTR1B, ACTR2, ACTR3, ACY1, ACY3, ADAM10, ADAMTS3, ADCY1, ADH5, ADH6, ADK, ADSL, AEBP1, AGAP2, AGR2, AGR3, AGRN, AGT, AHCTF1, AHCYL1, AHNAK, AHSA1, AHSG, AK1, AK2, AKAP9, AKR1A1, AKR1B10, ALAD, ALB, ALCAM, ALDH16A1, ALDH1A1, ALDH1L1, ALDH2, ALDH3B1, ALDH8A1, ALDOA, ALDOB, ALDOC, ALK, ALOX12, ALPL, ALPP, ALYREF, AMBP, AMN, ANGPT1, ANGPTL1, ANGPTL4, ANKFY1, ANKRD11, ANO1, ANO6, ANPEP, ANXA1, ANXA11, ANXA13, ANXA3, ANXA4, ANXA6, ANXA7, AOX1, AP1M1, AP2A1, AP2A2, AP2M1, AP4M1, APAF1, APLP2, APOA1, APOA2, APOB, APOD, APOE, APOL1, APP, APPL1, APPL2, APRT, AQP2, ARF5, ARFIP1, ARHGAP1, ARHGAP23, ARHGDIA, ARHGDIB, ARHGEF12, ARHGEF18, ARL15, ARL3, ARL8B, ARMC3, ARMC9, ARPC1A, ARPC1B, ARPC2, ARPC3, ARPC5, ARRDC1, ARSE, ARSF, ARVCF, ASAH1, ASB6, ASL, ASNA1, ASNS, ATAD2, ATIC, ATP1A1, ATP1A2, ATP1A3, ATP2B1, ATP2B2, ATP2B4, ATP4A, ATP5A1, ATP5B, ATP5L, ATP6AP1, ATP6AP2, ATP6V0A1, ATP6V0A4, ATP6V0C, ATP6V0D1, ATP6V0D2, ATP6V1A, ATP6V1B1, ATP6V1C1, ATP6V1C2, ATP6V1D, ATP6V1E1, ATP6V1H, ATRN, AUP1, AZGP1, AZU1, B2M, B3GAT3, B4GALT1, B4GALT3, BAIAP2, BAIAP2L1, BASP1, BAZ1B, BCAM, BCR, BDH2, BGN, BHLHB9, BHMT, BHMT2, BLMH, BLOC1S5, BLVRA, BLVRB, BMP3, BPI, BPIFB1, BPTF, BRI3BP, BROX, BSG, BTG2, BTN1A1, C11orf52, C11orf54, C16orf80, C16orf89, C17orf80, C19orf18, C1GALT1C1, C1orf116, C1QC, C1QTNF1, C1QTNF3, C1R, C2orf16, C3, C4BPA, C5, C9, CAB39L, CACNA2D1, CACYBP, CAD, CALM1, CALML3, CALR, CAMK4, CAMP, CAND1, CANX, CAP1, CAPN1, CAPN2, CAPN5, CAPN7, CAPNS1, CAPS, CAPZA2, CAPZB, CARD11, CASP9, CAV1, CBR3, CC2D1A, CCDC105, CCDC132, CCL28, CCPG1, CCT2, CCT3, CCT4, CCT5, CCT6A, CCT7, CCT8, CD101, CD14, CD163L1, CD19, CD2, CD22, CD274, CD2AP, CD300A, CD36, CD37, CD40, CD44, CD53, CD55, CD58, CD59, CD63, CD70, CD74, CD79B, CD80, CD81, CD9, CD97, CDC42, CDC42BPA, CDC42BPB, CDH1, CDH17, CDHR2, CDHR5, CDK1, CDK5RAP2, CDKL1, CEACAM5, CELSR2, CEMIP, CEP250, CES2, CETP, CFD, CFH, CFI, CFL1, CHGB, CHID1, CHMP1A, CHMP2B, CHMP4B, CHRDL2, CHST1, CHST14, CIB1, CKAP4, CKB, CLASP1, CLCA4, CLDN3, CLDN4, CLDN7, CLIC1, CLIC4, CLIC5, CLIC6, CLIP2, CLSTN1, CLTC, CLTCL1, CLU, CMPK1, CNDP2, CNKSR2, CNTLN, COASY, COBLL1, COL12A1, COL15A1, COL18A1, COL6A1, COL6A2, COL6A3, COLEC10, COLGALT1, COMT, COPA, COPB1, COPB2, COPS8, CORO1A, CORO1B, COX4I1, COX5B, CP, CPD, CPN2, CPNE1, CPNE3, CPNE5, CPNE8, CPVL, CR1, CR2, CRB2, CREB5, CRISPLD1, CRNN, CRTC2, CRYAB, CRYZ, CS, CSE1L, CSK, CSPG4, CSRP1, CST4, CSTB, CTDSPL, CTNNA1, CTNNB1, CTNND1, CTSB, CTSC, CTSG, CTTN, CUBN, CUL3, CUL4B, CUTA, CUX2, CXCR4, CYB5R1, CYBRD1, CYFIP1, CYFIP2, CYP2J2, DAAM2, DAG1, DAK, DARS, DBNL, DCD, DCTN2, DCXR, DDAH1, DDAH2, DDB1, DDC, DDR1, DDX11, DDX19A, DDX19B, DDX21, DDX23, DDX3X, DDX5, DERA, DHCR7, DHX34, DHX36, DHX9, DIAPH2, DIP2A, DIP2B, DIP2C, DLD, DLG1, DMBT1, DNAH7, DNAH8, DNAJA1, DNAJA2, DNAJB1, DNAJB9, DNAJC13, DNAJC3, DNAJC7, DNHD1, DNM2, DNPH1, DOCK10, DOCK2, DOPEY2, DPEP1, DPP3, DPP4, DPYS, DPYSL2, DSC2, DSG2, DSG3, DSP, DSTN, DUOX2, DUSP26, DUT, DYNC1H1, DYNC2H1, DYSF, ECE1, ECH1, ECM1, EDIL3, EEA1, EEF1A1, EEF1A2, EEF1D, EEF1G, EEF2, EFEMP1, EFEMP2, EGF, EGFR, EHD1, EHD2, EHD3, EHD4, EIF2S1, EIF2S3, EIF3A, EIF3B, EIF3E, EIF3L, EIF4A1, EIF4A2, EIF4A3, EIF4E, EIF4G1, EIF4H, ELANE, EML5, ENO1, ENO2, ENO3, ENPP3, ENPP4, ENPP6, ENTPD1, EPB41L2, EPCAM, EPHA2, EPHA5, EPHB1, EPHB2, EPHB3, EPHB4, EPHX2, EPN3, EPPK1, EPRS, EPS8, EPS8L1, EPS8L2, EPS8L3, ERAP1, ERBB2, ERMN, ERO1L, ERP44, ESD, ETFA, EVPL, EXOC4, EXOSC10, EXT2, EYS, EZR, F11, F11R, F5, F7, FABP1, FABP3, FAH, FAM129A, FAM129B, FAM151A, FAM208B, FAM209A, FAM20A, FAM20C, FAM49B, FAM65A, FAS, FASLG, FASN, FAT1, FAT2, FBL, FBP1, FBP2, FCGBP, FCN1, FCN2, FERMT3, FGA, FGB, FGG, FGL2, FGR, FH, FIGNL1, FKBP1A, FKBP4, FKBP5, FLNA, FLNB, FLNC, FLOT1, FLOT2, FMNL1, FN1, FOLH1, FRK, FSCN1, FTCD, FUCA1, FURIN, FUS, FUT2, FUT3, FUT6, FUT8, FUZ, G6PD, GAA, GABRB2, GAL3ST4, GALK1, GALM, GALNT3, GANAB, GARS, GART, GATSL3, GBE1, GBP6, GCN1L1, GCNT2, GCNT3, GDF2, GDI2, GDPD3, GEMIN4, GFPT1, GGCT, GGH, GGT1, GHITM, GIPC1, GK, GK2, GLB1, GLDC, GLG1, GLIPR2, GLO1, GLUD1, GLUL, GNA13, GNAI1, GNAI2, GNAQ, GNAS, GNB1, GNB2, GNB2L1, GNB3, GNB4, GNB5, GNG12, GNPDA1, GNPTG, GOLGA4, GOLGA7, GOT1, GOT2, GPC1, GPC4, GPD1, GPI, GPM6A, GPR155, GPR64, GPR98, GPRASP1, GPRC5A, GPRC5B, GPT, GREB1, GRHPR, GRID1, GRIN1, GRK4, GSN, GSR, GSS, GSTA3, GSTCD, GSTK1, GSTO1, GSTP1, GUSB, H1FOO, H2AFY, H2AFY2, HADHA, HAPLN3, HAUS5, HBB, HBD, HBS1L, HDHD2, HEBP1, HEBP2, HEPH, HGD, HGS, HINT1, HIRA, HIST1H1B, HIST1H2BA, HIST1H2BL, HIST2H2AC, HLA-A, HLA-B, HLA-DPB1, HLA-DQB1, HLA-DRB1, HLA-DRB5, HLAE, HNMT, HNRNPA1, HNRNPA2B1, HNRNPC, HNRNPF, HNRNPK, HNRNPL, HP, HPD, HPGD, HPR, HPRT1, HRG, HRNR, HSD17B10, HSD17B4, HSP90AB1, HSP90B1, HSPA12A, HSPA13, HSPA1L, HSPA2, HSPA4, HSPA5, HSPA6, HSPA8, HSPA9, HSPB1, HSPB8, HSPD1, HSPG2, HSPH1, HTATIP2, HTRA1, HUWE1, HYOU1, IARS, ICAM1, ICAM3, IDH1, IFITM2, IFITM3, IGF2R, IGFALS, IGSF3, IGSF8, IKZF5, IMPDH2, INADL, INSR, IQCB1, IQCG, IQGAP1, IQGAP2, IRF6, IST1, ITFG3, ITGA1, ITGA2, ITGA2B, ITGA3, ITGA4, ITGA6, ITGAL, ITGAV, ITGB1, ITGB2, ITGB3, ITGB4, ITGB7, ITGB8, ITIH2, ITIH4, ITM2C, ITSN1, ITSN2, IVL, JADE2, JUP, KALRN, KCNG2, KHK, KIAA1324, KIF12, KIF15, KIF18B, KIF3A, KIF3B, KIF9, KIFC3, KL, KNG1, KPNB1, KPRP, KRT1, KRT10, KRT12, KRT14, KRT15, KRT16, KRT17, KRT18, KRT19, KRT2, KRT20, KRT24, KRT25, KRT27, KRT28, KRT3, KRT5, KRT6C, KRT7, KRT73, KRT75, KRT76, KRT77, KRT78, KRT79, KRT8, KRT9, L1CAM, LAD1, LAMA3, LAMA4, LAMA5, LAMB2, LAMB3, LAMC1, LAMC2, LAMP1, LAMP2, LAMTOR3, LBP, LCK, LCP1, LDHA, LDHB, LEPRE1, LFNG, LGALS3, LGALS3BP, LGALS4, LIMA1, LIN7A, LIN7C, LMAN1, LMAN2, LOXL4, LPO, LRP1, LRP1B, LRP2, LRP4, LRPPRC, LRRC15, LRRC16A, LRRC57, LRRK2, LRSAM1, LSP1, LSR, LTA4H, LTBP3, LTF, LUZP1, LYPLA2, MAGI3, MAL2, MAN1A1, MAN1A2, MAN2A1, MAP4K4, MAP7, MARCKSL1, MARK3, MARS, MARVELD2, MASP1, MASP2, MBD5, MBLAC2, MCAM, MCPH1, MDH1, MDH2, MEGF8, MEP1A, MEST, METRNL, MFGE8, MFI2, MGAM, MGAT1, MGAT4A, MID2, MIF, MINK1, MLLT3, MLLT4, MME, MMP24, MMP25, MMRN1, MMRN2, MNDA, MOB1A, MOB1B, MOGS, MPO, MPP5, MPP6, MS4A1, MSH6, MSN, MSRA, MTA1, MTAP, MTCH2, MTHFD1, MTMR11, MTMR2, MUC13, MUC16, MUC4, MUM1L1, MVB12A, MVB12B, MVP, MX1, MXRA5, MXRA8, MYADM, MYH10, MYH11, MYH13, MYH14, MYH3, MYH8, MYH9, MYL6B, MYO15A, MYO1B, MYO1C, MYO1D, MYO1E, MYO1G, MYO5B, MYO6, MYOF, N4BP2L2, NAA16, NAA50, NACA, NAGLU, NAMPT, NAP1L4, NAPA, NAPG, NAPRT, NAPSA, NARS, NBR1, NCALD, NCCRP1, NCKAP1, NCKAP1L, NCL, NCOA3, NCSTN, NDRG1, NDRG2, NEB, NEBL, NEDD4, NEDD4L, NEDD8, NEU1, NID1, NIN, NIPBL, NIT2, NKX61, NONO, NOTCH1, NOX3, NPC1, NPEPPS, NPHS1, NPHS2, NPM1, NPNT, NQO2, NT5C, NT5E, NUCB1, NUCB2, NUDT5, NUMA1, NXPE4, OLA1, OPTN, OR2A4, OS9, OSBPL1A, OXSR1, P2RX4, P4HB, PA2G4, PACSIN2, PACSIN3, PADI2, PAFAH1B1, PAFAH1B2, PAGE2, PAICS, PAM, PARD6B, PARP4, PBLD, PCBP1, PCDHGB5, PCK1, PCLO, PCNA, PCSK9, PCYOX1, PDCD2, PDCD5, PDCD6, PDCD6IP, PDDC1, PDE8A, PDIA2, PDIA3, PDIA4, PDIA6, PDLIM7, PDZK1, PEBP1, PECAM1, PEF1, PEPD, PEX1, PFAS, PFKL, PFKP, PGAM1, PGD, PGK1, PGLYRP1, PGM1, PHB2, PHGDH, PI4KA, PIGR, PIK3C2A, PIK3C2B, PILRA, PIP, PIP4K2C, PKD1, PKD1L3, PKD2, PKHD1, PKLR, PKM, PKN2, PKP3, PLAT, PLAU, PLCB1, PLCD1, PLCG2, PLD3, PLEC, PLEKHA1, PLEKHA7, PLEKHB2, PLG, PLIN2, PLOD1, PLOD2, PLOD3, PLS1, PLSCR1, PLTP, PLVAP, PLXNA1, PLXNB2, PM20D1, PMEL, PNP, PODXL, POFUT2, PON1, PON3, POTEE, POTEF, POTEI, POTEM, PPA1, PPARG, PPFIA2, PPIA, PPIB, PPL, PPM1L, PPP1CB, PPP1R7, PPP2CA, PPP2R1A, PPP2R1B, PRCP, PRDX1, PRDX3, PRDX4, PRDX5, PRG4, PRKAR2A, PRKCA, PRKCD, PRKCH, PRKCI, PRKCZ, PRKDC, PRKRIP1, PRNP, PROM1, PROM2, PROS1, PROZ, PRRC2A, PRSS23, PRTN3, PSAP, PSAT1, PSMA2, PSMA3, PSMA5, PSMA7, PSMB1, PSMB3, PSMB4, PSMB5, PSMB6, PSMB8, PSMB9, PSMC2, PSMC4, PSMC6, PSMD11, PSMD12, PSMD13, PSMD2, PSME1, PSME2, PSME3, PTBP1, PTER, PTGFRN, PTGR1, PTGS1, PTPN13, PTPN23, PTPRA, PTPRC, PTPRF, PTPRJ, PTPRO, PTRF, PTX3, PYGB, PYGL, QDPR, QPCT, QPRT, QSOX1, RAB10, RAB11B, RAB17, RAB1A, RAB1B, RAB22A, RAB25, RAB29, RAB2A, RAB34, RAB3B, RAB3GAP1, RAB43, RAB4B, RAB6B, RAB7A, RAB8A, RAB8B, RAB9A, RAC1, RACGAP1, RALA, RALB, RAP1A, RAP1B, RAP1GDS1, RAP2A, RAPGEF3, RARRES1, RARS, RASAL3, RASSF9, RBL2, RCC2, REG4, RELN, RENBP, RFC1, RFTN1, RHEB, RHOB, RHOF, RIMS2, RLF, RNASE7, RNF213, RNH1, RNPEP, ROBO2, ROCK2, RP2, RPL10, RPL10A, RPL14, RPL15, RPL23, RPL3, RPL30, RPL34, RPL35A, RPL4, RPL5, RPL6, RPL8, RPLP2, RPN1, RPS11, RPS14, RPS15A, RPS16, RPS18, RPS2, RPS20, RPS21, RPS27A, RPS3A, RPS4X, RPS4Y1, RPS4Y2, RPS7, RPS9, RRAS, RREB1, RSU1, RTN4, RUSC2, RUVBL1, RUVBL2, RYR1, S100A11, S100A6, S100P, SAA1, SAFB2, SAMM50, SARS, SBSN, SCAMP2, SCAMP3, SCARB1, SCARB2, SCEL, SCIN, SCN10A, SCN11A, SCPEP1, SCRIB, SCRN2, SDCBP, SDF4, SEC31A, SELENBP1, SELP, SEMA3G, SEPP1, SERBP1, SERINC1, SERINC2, SERINC5, SERPINA1, SERPINA3, SERPINA4, SERPINA5, SERPINA7, SERPINB1, SERPINB13, SERPINB6, SERPINB9, SERPING1, SETD4, SFI1, SFN, SFRP1, SFT2D2, SH3BP4, SHMT1, SHMT2, SHROOM2, SIAE, SIRPA, SIT1, SLAMF1, SLAMF6, SLC12A1, SLC12A2, SLC12A3, SLC12A7, SLC12A9, SLC13A2, SLC13A3, SLC15A2, SLC16A1, SLC1A1, SLC1A4, SLC1A5, SLC20A2, SLC22A11, SLC22A12, SLC22A13, SLC22A2, SLC22A5, SLC22A6, SLC23A1, SLC25A1, SLC25A3, SLC25A4, SLC25A6, SLC26A11, SLC26A4, SLC26A9, SLC27A2, SLC29A1, SLC2A1, SLC2A3, SLC34A2, SLC35D1, SLC36A2, SLC37A2, SLC38A1, SLC39A5, SLC3A1, SLC3A2, SLC44A1, SLC44A2, SLC44A4, SLC46A3, SLC4A1, SLC4A4, SLC5A1, SLC5A10, SLC5A2, SLC5A5, SLC5A6, SLC5A8, SLC5A9, SLC6A13, SLC6A14, SLC6A19, SLC7A5, SLC9A1, SLC9A3, SLC9A3R1, SLC9A3R2, SLCO4C1, SLIT2, SLK, SMC2, SMC3, SMIM22, SMIM24, SMO, SMPDL3B, SMURF1, SNCG, SND1, SNRNP200, SNX12, SNX18, SNX25, SNX33, SNX9, SOD1, SOGA1, SORD, SORL1, SORT1, SPAG9, SPAST, SPEN, SPINK1, SPON2, SPRR3, SPTAN1, SPTBN1, SQSTM1, SRC, SRPR, SRSF7, ST13, ST3GAL1, ST3GAL6, STAMBP, STAU1, STIP1, STK10, STK11, STK24, STOM, STRIP1, STX3, STX4, STX7, STXBP1, STXBP2, STXBP3, STXBP4, SUB1, SUCLA2, SUSD2, SYAP1, SYNE1, SYNE2, TAB3, TACSTD2, TAF6L, TALDO1, TAOK1, TARS, TAX1BP1, TAX1BP3, TBC1D10A, TBC1D21, TC2N, TCP1, TECTA, TEKT3, TEX14, TF, TFRC, TGFB1, TGFBI, TGFBR3, TGM1, TGM2, TGM3, TGM4, THBS1, THBS2, THRAP3, THSD4, THY1, TIAM2, TINAGL1, TJP2, TKT, TLN1, TLR2, TM7SF3, TM9SF2, TMBIM1, TMC6, TMC8, TMED2, TMED9, TMEM109, TMEM192, TMEM2, TMEM256, TMEM27, TMEM63A, TMPRSS11B, TMPRSS11D, TMPRSS2, TNFAIP3, TNFRSF8, TNFSF10, TNFSF13, TNIK, TNKS1BP1, TNPO3, TOLLIP, TOM1, TOM1L2, TOMM70A, TOR1A, TOR1B, TOR3A, TPI1, TPM3, TPP1, TPRG1L, TRAP1, TREH, TRIP10, TSNAXIP1, TSPAN1, TSPAN15, TSPAN3, TSSK3, TSTA3, TTC17, TTC18, TTLL3, TTN, TTR, TUBA1B, TUBA4A, TUBB3, TUBB4A, TUBB8, TUFM, TWF2, TXNDC16, TXNDC8, TXNRD1, TYK2, TYRP1, UACA, UBA1, UBAC1, UBASH3A, UBE2N, UBE2V2, UBL3, UBXN6, UEVLD, UGDH, UGGT1, UGP2, ULK3, UMOD, UPB1, UPK1A, UPK3A, UQCRC2, UTRN, UXS1, VAMP1, VAMP3, VAMP7, VAPA, VASN, VASP, VAT1, VCL, VCP, VDAC3, VIL1, VIM, VMO1, VPS13C, VPS13D, VPS28, VPS35, VPS36, VPS37B, VPS37C, VPS37D, VPS4A, VPS4B, VTA1, VWA2, VWF, WARS, WAS, WASF2, WASL, WDR1, WIZ, WNT5B, XDH, XPNPEP2, XPO1, XRCC5, XRCC6, YBX1, YES1, YWHAE, YWHAG, YWHAH, YWHAZ, ZCCHC11, ZDHHC1, ZFYVE20, ZG16B, ZMPSTE24, ZNF114, ZNF486, ZNF571, and ZNHIT6. | ||||
GENE: ExoCarta (http://exocarta.org/exosome_markers_new) [33]; GENE: EV antibody array [35]; GENE: HIV exosomal proteins [36].
Table 5.
Exosomal proteins found in urinary EVs from uninfected controls.
| Genes in our analysis | Genes in the FunRich database | Percentage of genes | Fold enrichment | |
|---|---|---|---|---|
| Exosomal proteins | 37 | 2001 | 72.54 | 5.26 |
A1BG, ACTA1, ACTA2, ACTB, ACTBL2, ACTC1, ACTG1, ACTG2, ALB, AMBP, APOA1, APOD, AZGP1, B2M, CDH1, CLU, CP, CRNN, DCTN2, EGF, HP, HPR, HSPB1, ITIH4, KNG1, LAMA3, LMAN2, POTEE, POTEF, POTEI, S100A8, SERPINA1, SERPING1, TF, TTR, UMOD, and VASN.
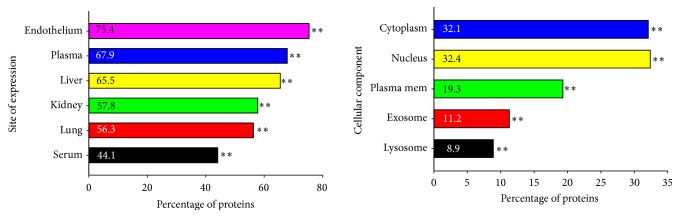
The GO results of the FunRich analysis of the EVs from the HIV+ samples are summarized in Table 6 and Figure 4. The top five (P < 0.01) EV sites of expression were endothelial cells, plasma, liver, serum, and kidney and the most significant cellular components were lysosomes, exosomes, membranes, plasma membranes, the nucleus, and the cytoplasm (P < 0.01) (Figure 4). The top five ontologies (Table 6) were protein serine/threonine kinase activity, catalytic activity, GTPase activator activity, guanyl-nucleoside exchange factor activity, and cell adhesion molecule activity (P < 0.0001), the top biological process was regulation of nucleobase, nucleoside, and nucleic acid (P < 0,0001), and the most prominent biological pathway was integrin cell surface interactions (P < 0.03).
Table 6.
Functional enrichment analysis of HIV+ EV proteins.
| Genes in the dataset | Genes in the Bkg. database | Percentage of genes | Fold enrichment | Corrected P value (BH FDR) | |
|---|---|---|---|---|---|
| Molecular function | |||||
| Protein serine/threonine kinase activity | 272 | 5,602 | 30 | 1.18 | 1.04−08 |
| Catalytic activity | 456 | 827 | 4.9 | 1.1 | 1.12−05 |
| GTPase activator activity | 131 | 836 | 4.7 | 1.2 | 8.14−05 |
| Guanyl-nucleoside exchange factor activity | 105 | 614 | 3.6 | 1.2 | 8.54−05 |
| Cell adhesion molecule activity | 307 | 531 | 3.3 | 1.1 | 0.0001 |
| Biological process | |||||
| Regulation of nucleobase, nucleoside, and nucleic acid | 2,236 | 4,658 | 24.8 | 1.05 | 3.24−05 |
| Biological pathway | |||||
| Integrin cell surface interactions | 69 | 1,366 | 23.3 | 1.2 | 0.03 |
Figure 4.
Percentage of proteins found in HIV+ urinary EVs. FunRich analysis of the LC/MS/MS proteins from HIV+ EVs determined the most likely tissue expressing the proteins, site of expression, and the cellular component from which the protein is derived. Data is graphed as the percentage of proteins found. ∗∗ denotes significance, P < 0.01.
LC/MS/MS identified 15,571 proteins in EVs from HIV+ patients with CD4+ T cells greater than 300, 2,115 from CD4+ T cells less than 300, 15,028 proteins from patients with low VL, and 2486 from patients with high VLs. Pathway analysis was similar between EV proteins from patients with greater than 300 CD4+ T cells and low VLs and different between the low CD4+ T cells and high VLs (summarized in Table 7). The pathways found are detailed in Supplementary Material 1. Interleukin proteins detected were IL10, IL10RA, IL16, IL17RC, IL18, IL18BP, IL1RAP, IL1RL2, IL1RN, IL33, IL4I1, IL6, and IL6ST. Immunomodulatory molecules, HOXB4, CD81, CD9, TGF-β1, IDO, Notch1, ADAM17, Rab4, and HGF, were also found by LC/MS/MS in addition to MHC Class I and II antigens.
Table 7.
Comparison of pathways between HIV+ groups from Pathway Studio 11.4.
| HIV group | Pathway |
|---|---|
| CD4+ T cells greater than 300 n = 15 |
Natural killer cell inhibitor receptor signaling Intermediate filament polymerization Ca2+ flux regulation G1/S phase transition G2/M phase transition S/G2 phase transition Protein folding Golgi to endosome transport Endosomal recycling Kinetochore assembly |
|
| |
| CD4+ T cells less than 300 n = 15 |
Neutrophil chemotaxis Vascular motility Platelet activation via GPCR signaling Insulin influence on protein synthesis mTOR signaling overview EDNRA/B → vascular motility Proplatelet maturation Natural killer cell activation through ITAM-containing receptors Taste sensor receptors activates mTOR signaling Natural killer cell activation |
|
| |
| Low VLs n = 14 |
Intermediate filament polymerization Natural killer cell inhibitory receptor signaling golgi to endosome transport Ca+ flux regulation HRH1/3 → synaptic transmission Vascular motility Endosomal recycling G1/S phase transition Golgi transport G2/M phase transition |
|
| |
| High VLs n = 10 |
Metaphase/anaphase phase transition S/G2 phase transition Spindle assembly Natural killer cell activation Histone ubiquitylation Eosinophil survival by cytokine signaling Protein folding G2/M phase transition |
The HIV-1 Human Interaction database search found that HIV Nef interacted with 559 EV proteins of 770 total human proteins (72.6%); HIV Vpr interacted with 437 EV of 598 human (73.1%); HIV Vif interacted with 162 EV of 310 human (52.2%); and HIV Vpu interacted with 165 EV of 244 human proteins which were found in the HIV+ EVs (67.6%) (see Supplementary Material 2, including PMIDs for references).
Functional analysis of the control EVs are listed in Table 8. The major sites of expression were cervicovaginal fluid, neutrophils, and gastric juice (P < 0.0001). The most significant ontologies were molecular function of the proteins and defense/immunity protein activity and principal biological processes were immune response, signal transduction, cell communication, and antigen presentation (P < 0.0073).
Table 8.
Functional enrichment analysis of control EV proteins.
| Genes in the database | Genes in the Bkg. database | Percentage of genes | Fold enrichment | Corrected P value (BH FDR ) | |
|---|---|---|---|---|---|
| Site of expression | |||||
| Cervicovaginal fluid | 16 | 544 | 12.0 | 4.2 | 2.59E − 06 |
| Neutrophils | 13 | 392 | 9.7 | 4.8 | 6.68E − 06 |
| Gastric juice | 9 | 222 | 6.7 | 6.1 | 4E − 05 |
| Molecular function | |||||
| Defense/immunity protein activity | 5 | 52 | 3.7 | 15.7 | 3.96E − 05 |
| Biological process | |||||
| Immune response | 13 | 561 | 9.8 | 3.4 | 0.00026 |
| Signal transduction | 43 | 3907 | 32.5 | 1.5 | 0.0026 |
| Cell communication | 41 | 3687 | 31.1 | 1.5 | 0.0028 |
| Antigen presentation | 1 | 1 | 0.7 | 134.4 | 0.0073 |
Only sixty-four (64) proteins overlapped between the HIV+ and control EV samples and are listed in Table 8. The top fourteen (14) GO ontologies for cellular components include extracellular exosome, extracellular region, extracellular space, hemoglobin complex, and blood microparticle (P < 0.001, Table 9), GO ontologies for molecular function were heparin binding, ion gated activity, and oxygen transporter activity, and the most significant biological processes found were response to yeast, defense response to fungus, macrophage chemotaxis, negative regulation of growth of symbiont in host, oxygen transport, and hydrogen peroxide catabolic process.
Table 9.
Overlapping EV proteins from HIV+ and HIV− samples, LC/MS/MS analysis.
| Gene | |
|---|---|
| ABCB1 | ATP-binding cassette, subfamily B (MDR/TAP), member 1 |
| ANXA8 | Annexin A8 |
| ASIC1 | Acid-sensing (proton-gated) ion channel 1 |
| ASIC2 | Acid-sensing (proton-gated) ion channel 2 |
| AUTS2 | Autism susceptibility candidate 2 |
| AZU1 | Azurocidin 1 |
| BCAT1 | Branched chain amino acid transaminase 1, cytosolic |
| BRD4 | Bromodomain containing 4 |
| CCL5 | Chemokine (C-C motif) ligand 5 |
| CEACAM8 | Carcinoembryonic antigen-related cell adhesion molecule 8 |
| CFH | Complement factor H |
| CHIT1 | Chitinase 1 (chitotriosidase) |
| CLDN7 | Claudin 7 |
| COL16A1 | Collagen, type XVI, alpha 1 |
| CPB2 | Carboxypeptidase B2 (plasma) |
| CRADD | CASP2 and RIPK1 domain containing adaptor with death domain |
| CTSG | Cathepsin G |
| CYP4A11 | Cytochrome P450, family 4, subfamily A, polypeptide 11 |
| DEFA1 | Defensin, alpha 1 |
| DNAH17 | Dynein, axonemal, heavy chain 17 |
| DUSP9 | Dual specificity phosphatase 9 |
| EIF4A1 | Eukaryotic translation initiation factor 4A1 |
| ELANE | Elastase, neutrophil expressed |
| v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | |
| FARP1 | FERM, RhoGEF (ARHGEF), and pleckstrin domain protein 1 (chondrocyte-derived) |
| GDF15 | Growth differentiation factor 15 |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 |
| GNL1 | Guanine nucleotide binding protein-like 1 |
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| HAAO | 3-Hydroxyanthranilate 3,4-dioxygenase |
| HAL | Histidine ammonia-lyase |
| HBA1 | Hemoglobin, alpha 1 |
| HBB | Hemoglobin, beta |
| HBD | Hemoglobin, delta |
| IGKC | Immunoglobulin kappa constant |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 |
| MEF2C | Myocyte enhancer factor 2C |
| MLLT4 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
| MPO | Myeloperoxidase |
| MRC2 | Mannose receptor, C type 2 |
| MYBPC3 | Myosin binding protein C, cardiac |
| NCAM1 | Neural cell adhesion molecule 1 |
| NKTR | Natural killer-tumor recognition sequence |
| NUP93 | Nucleoporin 93 kDa |
| PDE1C | Phosphodiesterase 1C, calmodulin-dependent 70 kDa |
| PDLIM5 | PDZ and LIM domain 5 |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| RAB31 | RAB31, member RAS oncogene family |
| RAP1GAP | RAP1 GTPase activating protein |
| REG1A | Regenerating islet-derived 1 alpha |
| RNASE2 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| RNASE3 | Ribonuclease, RNase A family, 3 |
| RPS14 | Ribosomal protein S14 |
| RUNX2 | Runt-related transcription factor 2 |
| SHBG | Sex hormone-binding globulin |
| SLC22A5 | Solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| SLC6A6 | Solute carrier family 6 (neurotransmitter transporter), member 6 |
| TACC2 | Transforming, acidic coiled-coil containing protein 2 |
| TAF6L | TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65 kDa |
| TNIK | TRAF2 and NCK interacting kinase |
| TRAPPC12 | Trafficking protein particle complex 12 |
| TRIM58 | Tripartite motif containing 58 |
| WNT2B | Wingless-type MMTV integration site family, member 2B |
| WNT6 | Wingless-type MMTV integration site family, member 6 |
4. Discussion
This is the first report of the detection of urinary EVs containing HIV and human proteins from HIV+ patients by mass spectrometry and western blot. EVs provide intercellular communication to cells through the delivery of their cargo, nucleic acids, miRNAs, and proteins, to recipient cells reviewed in [3]. Previous studies have found EVs in plasma of HIV+ patients but did not describe HIV or human proteins within them. Others have described EVs containing HIV proteins but these results were from in vitro HIV infected cell cultures and not from HIV+ patients [18, 20, 22, 23, 36, 38–47]. This study details both the HIV and human proteins found in urinary EVs from HIV+ patients.
According to the International Society for Extracellular Vesicles (ISEV), the minimal requirements for EVs or their presence in samples includes the simultaneous detection of transmembrane proteins and cytosolic proteins with membrane/receptor binding abilities, while major cell organelles are absent [48]. LC/MS/MS analysis identified these proteins and functional enrichment analysis determined a significant number which were of exosomal origin in both the EVs in HIV+, 1,932, and HIV−, only 37. TEM analysis of HIV+ and HIV− urine showed pleiotropic membrane bound vesicles in both groups' urine samples and NTA analysis showed particles ranging in size from 50 nm to 300 nm in both groups, although the HIV+ samples had significantly more particles than uninfected samples. Other studies have found increased numbers of EVs in the plasma of HIV+ patients [43, 49]. Proteins from both the HIV+ and HIV− individuals were significantly associated with exosomal proteins, further substantiating our hypothesis that urine from HIV+ patients contains EVs (Table 10). The FunRich analysis of the sites of expression showed that a significant number of proteins were associated with the endothelium, plasma, serum, kidney, liver, and lung. These findings suggest that EVs from HIV+ patients may be filtered from these sites and concentrated in urine.
Table 10.
Functional analysis of overlapping HIV+ and HIV− EV proteins.
| Genes in the data set | Genes in the Bkg. database | Percentage of genes | Fold enrichment | Corrected P value (Bonferroni method) | |
|---|---|---|---|---|---|
| Site of expression | |||||
| Urine | 31 | 3202 | 51.7 | 3.0 | 6.85E − 07 |
| Cervicovaginal fluid | 12 | 544 | 20.0 | 7.2 | 5.33E − 05 |
| Neutrophils | 9 | 392 | 15.0 | 7.7 | 1.56E − 03 |
| 032403_BALF4_glypep | 4 | 43 | 6.7 | 35.0 | 3.53E − 03 |
| Neutrophil | 19 | 1979 | 31.7 | 3.0 | 3.67E − 03 |
| Monocyte | 23 | 2786 | 38.3 | 2.6 | 3.90E − 03 |
| Cellular component | |||||
| Extracellular | 22 | 1808 | 37.9 | 3.1 | 4.61E − 05 |
| Stored secretory granule | 3 | 19 | 5.2 | 51.0 | 3.27E − 03 |
| Lysosome | 17 | 1609 | 29.3 | 2.8 | 7.73E − 03 |
| Extracellular space | 8 | 399 | 13.8 | 5.6 | 1.05E − 02 |
| Exosomes | 19 | 2001 | 32.8 | 2.5 | 1.14E − 02 |
| Azurophil granule | 2 | 6 | 3.4 | 108.9 | 1.35E − 02 |
HIV has previously been detected in the urine of HIV+ patients; however, it was shown that HIV virions are associated with cell pellets and not in centrifuged urine [50, 51]. p24 is found in replicative HIV infectious virions but was not found in twenty-six of our HIV+ samples by ELISA and only five of thirty-five HIV+ EV urine samples had detectable p24 by LC/MS/MS analysis. p24 in urine pellets is derived from mononuclear cells but was found in only 3 of 80 analyzed samples [51]. This represents a low sensitivity, primarily because the HIV-1 p24 protein is not always present during advanced stages of HIV infection. To further confirm that these HIV proteins were from EVs, we tested the filtrate from ultracentrifugation (MW cutoff 100,000 kD) of HIV-positive urine, and no HIV proteins were present. We did not, however, perform an HIV infectivity assay, MAGI, on the isolated urinary EVs, and thus cannot be totally confident that HIV virions were not present in the EVs. HIV proteins in urinary EVs may be the result of a nonproductive HIV infection in the kidney [52–56] and/or EVs filtered from blood [21, 49, 57]. The type of HIV protein in the EVs remained relatively constant as demonstrated by the resampling of two patients, 203 and 311 days, after the first sample that had similar results. The identification of HIV proteins in urinary EVs may be a potential noninvasive diagnostic tool to monitor HIV disease states as well as treatment efficacy.
Different proteins and pathways were found in EVs from (1) CD4+ T cell > 300 versus <300 and (2) VLs < 200 versus >200 copies. It is interesting that EVs from HIV+ patients with low VLs and high CD4+ T cells, usually indicative of better health, had more proteins detected than EVs from high VLs and low CD4+ T cells (high VLs = 2486 vrs low = 15028; low CD4+ T cells = 2115 versus high CD4+ = 15761). These groups also had overlapping pathway results; however, proteins from high VLs and low CD4+ T cells did not have similar pathway results. Further comparison and analysis of the EV protein profile between the low VL/high CD4+ T cells and high VL/low CD4+ T cells may reveal more mechanisms involved in the evolving pathology of HIV infection.
Proteins contained in EVs can both enhance and inhibit host responses from innate, inflammatory, and adaptive reactions. Proteins from HIV+ patients showed a predominantly immunosuppressive profile. IL10 is a Th2 cytokine that downregulates macrophage function and inhibits T cell proliferation while IL6 can stimulate IL10 production and inhibit the effects of TNF-α and IL1. Both these cytokines were present in the EVs from HIV+ patients while TNF-α and IL1 were not detected suggesting an immunomodulatory effect may be elicited by the EVs. Other immune downregulating factors, IDO, HOXB4, HGF, and TGFβ1, were found. IDO [58], HLA-G [59], and HGF [60] can inhibit natural killer cell activation which was one of the top biological processes found in the pathway analysis of the EV proteins in patients with high CD4+ T cells and low VLs. TGFβ-1, an inhibitor of immune function, is induced by HIV Tat [61] and is a mediator of immune suppression in HIV infection [62–64]. These proteins were found in EVs from HIV+ patients while proinflammatory cytokines were not. New studies show that HIV+ nonprogressors have lower plasma TGFβ-1 and IL10 than patients with progressive disease [65] and it is possible that EVs may sequester TGFβ-1 and IL10 and remove them from circulation. The presence of over 16 different MHC Class I and II antigens in the EVs from HIV+ patients may support the hypothesis that this mechanism is used by intracellular pathogens to evade the immune response by decreasing cytotoxic T cell activity [66]. Herpes Simplex Virus-1 binds to HLA-DR inhibiting antigen presentation that leads to immune evasion [67]. Future studies should focus on the correlation of the concentration of these factors to HIV+ patients' clinical status.
In this study, we showed that structural, regulatory, and accessory HIV proteins could be detected in urinary EVs of HIV+ patients. Our WB analysis using polyclonal and monoclonal antibodies confirmed the presence of HIV proteins in the EVs from HIV+ patients. The most prevalent protein was HIV Nef. EVs from both in vitro and patient samples have been previously reviewed in [6]. HIV Nef induces an alternative pathway for TNF induction utilizing Notch-1, ADAM17, and Rab4+, all found in EVs from HIV+ patients, which leads to high plasma TNF levels [68]. Whether the isolation of these factors in EVs represents a diminishing or enhancement of TNF production remains to be examined.
The HIV Human Interaction database found significant interactions between HIV Nef, Vpr, Vif, and Vpu and human proteins. Serine/threonine protein kinases are important in T cell receptor signaling [69]. These kinases as well as CD4 and MHC antigens were found in EVs from the HIV+ samples; however, further studies are needed to determine the mechanisms involved with EV function in HIV infections. Cell adhesion molecules, ICAM, VCAM, and PECAM, were also found in the EVs from patients. Others have reported these molecules are present in HIV+ blood samples and may represent biomarkers from inflamed endothelium due to HIV infection [70].
One of the limitations of this study was a small sample size of specific HIV syndromes such as comorbidities, AIDS, HIV-associated nephropathy, and HIV-associated dementia as well as patients on or naïve to antiretroviral therapy. Increasing the numbers of HIV+ patients in these categories may allow us to determine whether specific HIV proteins as well as human proteins in urinary EVs could be associated with these conditions. Future studies will also quantify the amount of HIV proteins as well as human proteins to determine if a correlation exists between different HIV conditions and the amount of proteins detected.
HIV infection is usually detected by antibodies to HIV and can take up to three months to develop or by measuring VLs in blood whereas we can detect HIV-1 proteins in urinary EVs. In summary, urinary proteins in EVs from HIV+ patients may allow a noninvasive method to (1) rapidly screen for infection and identification of patients eligible for antiretroviral treatment (ART); (2) monitor ART treatment efficacy; and (3) diagnose HIV comorbidities.
Acknowledgments
The authors acknowledge Jane Chu and Mahfuz Khan of Morehouse School of Medicine for technical assistance and Dr. Douglas Paulsen for his support and editorial suggestions. This study was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award no. UL1TR000454. Other funds were received from the Minority Biomedical Research Support (MBRS) of the Research Initiative for Scientific Advancement (RISE) Program 5R25GM058268 funded by NIGMS and NIH Research Endowment S21MD000101 funded by the National Institute on Minority Health and Health Disparities (NIH/NIMHD). The authors also acknowledge the Research Centers in Minority Institutions (RCMI) G12 funded by the NIH/NIMHD, #8G12MD0076202. MEB Core facility was constructed with support from the Research Facilities Improvement Grant C06 RR18386 from NIH/NCRR. The newly renovated Core Resources space was funded by G20 RR031196 from NIH/NCRR. The R-CENTER was funded by Grant no. U54MD007588 from NIH/NIMHD and NIH/NCRR 5P20R R0111044 pilot for study support.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Top 10 biological function pathways using Pathway Studio 11 Mammal Plus, Elsevier, Inc., for HIV+ EV proteins from HIV+ patients with (1) CD4+ T cells greater than 300, (2) CD4+ T cells less than 300, (3) viral loads less than 200 copies, and (4) viral loads greater than 200 copies.
EV HIV protein interactions (Nef, Vif, Vpr, and Vpu) with human proteins identified using HIV-1 Human Interaction database (https://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/). The file includes the gene symbol, human protein name, interaction keywords, protein accession ID, and PMID of references citing the interaction.
References
- 1.Schorey J. S., Cheng Y., Singh P. P., Smith V. L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Reports. 2015;16:24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schorey J. S., Harding C. V. Extracellular vesicles and infectious diseases: New complexity to an old story. The Journal of Clinical Investigation. 2016;126(4):1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreyer F., Baur A. Biogenesis and functions of exosomes and extracellular vesicles. Methods in Molecular Biology. 2016;1448:201–216. doi: 10.1007/978-1-4939-3753-0_15. [DOI] [PubMed] [Google Scholar]
- 4.Février B., Raposo G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Current Opinion in Cell Biology. 2004;16(4):415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Ciardiello C., Cavallini L., Spinelli C., et al. Focus on extracellular vesicles: New frontiers of cell-to-cell communication in cancer. International Journal of Molecular Sciences. 2016;17(2) doi: 10.3390/ijms17020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellwanger J. H., Veit T. D., Chies J. A. B. Exosomes in HIV infection: A review and critical look. Infection, Genetics and Evolution. 2017;53:146–154. doi: 10.1016/j.meegid.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Chahar H. S., Bao X., Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses. 2015;7(6):3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen F., Nickenig G., Werner N. Extracellular vesicles in cardiovascular disease. Circulation Research. 2017;120(10):1649–1657. doi: 10.1161/CIRCRESAHA.117.310752. [DOI] [PubMed] [Google Scholar]
- 9.Karpman D., Ståhl A.-L., Arvidsson I. Extracellular vesicles in renal disease. Nature Reviews Nephrology. 2017;13(9):545–562. doi: 10.1038/nrneph.2017.98. [DOI] [PubMed] [Google Scholar]
- 10.Martínez M. C., Andriantsitohaina R. Extracellular vesicles in metabolic syndrome. Circulation Research. 2017;120(10):1674–1686. doi: 10.1161/CIRCRESAHA.117.309419. [DOI] [PubMed] [Google Scholar]
- 11.Szabo G., Momen-Heravi F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nature Reviews Gastroenterology & Hepatology. 2017;14(8):455–466. doi: 10.1038/nrgastro.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salih M., Zietse R., Hoorn E. J. Urinary extracellular vesicles and the kidney: biomarkers and beyond. American Journal of Physiology-Renal Physiology. 2014;306(11):F1251–F1259. doi: 10.1152/ajprenal.00128.2014. [DOI] [PubMed] [Google Scholar]
- 13.Barreiro K., Holthofer H. Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell and Tissue Research. 2017;369(1):217–227. doi: 10.1007/s00441-017-2621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thongboonkerd V., Malasit P. Renal and urinary proteomics: current applications and challenges. Proteomics. 2005;5(4):1033–1042. doi: 10.1002/pmic.200401012. [DOI] [PubMed] [Google Scholar]
- 15.Thongboonkerd V., McLeish K. R., Arthur J. M., Klein J. B. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney International. 2002;62(4):1461–1469. doi: 10.1046/j.1523-1755.2002.00565.x. doi: 10.1046/j.1523-1755.2002.00565.x.4493245 [DOI] [PubMed] [Google Scholar]
- 16.Pieper R., Gatlin C. L., McGrath A. M., et al. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4(4):1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 17.Christensen E. I., Birn H. Megalin and cubilin: Multifunctional endocytic receptors. Nature Reviews Molecular Cell Biology. 2002;3(4):258–268. doi: 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- 18.Aqil M., Mallik S., Bandyopadhyay S., Maulik U., Jameel S. Transcriptomic analysis of mRNAs in human monocytic cells expressing the HIV-1 nef protein and their exosomes. BioMed Research International. 2015;2015:10. doi: 10.1155/2015/492395.492395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aqil M., Naqvi A. R., Mallik S., Bandyopadhyay S., Maulik U., Jameel S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. Journal of Extracellular Vesicles (JEV) 2014;3 doi: 10.3402/jev.v3.23129.23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenassi M., Cagney G., Liao M., et al. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010;11(1):110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan M. B., Lang M. J., Huang M.-B., et al. Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Aβ1–42 secretion in SH-SY5Y neural cells. Journal of NeuroVirology. 2016;22(2):179–190. doi: 10.1007/s13365-015-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arenaccio C., Anticoli S., Manfredi F., Chiozzini C., Olivetta E., Federico M. Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology. 2015;12, article 87 doi: 10.1186/s12977-015-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arenaccio C., Chiozzini C., Columba-Cabezas S., et al. Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. Journal of Virology. 2014;88(19):11529–11539. doi: 10.1128/jvi.01712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruh-Garcia N. A., Wolfe L. M., Dobos K. M. Deciphering the role of exosomes in tuberculosis. Tuberculosis. 2015;95(1):26–30. doi: 10.1016/j.tube.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Marcilla A., Martin-Jaular L., Trelis M., et al. Extracellular vesicles in parasitic diseases. Journal of Extracellular Vesicles (JEV) 2014;3 doi: 10.3402/jev.v3.25040.25040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang D. Y. P., King H. W., Li J. Y. Z., Gleadle J. M. Exosomes and the kidney: blaming the messenger. Nephrology. 2013;18(1):1–10. doi: 10.1111/nep.12005. [DOI] [PubMed] [Google Scholar]
- 27.Junker K., Heinzelmann J., Beckham C., Ochiya T., Jenster G. Extracellular Vesicles and Their Role in Urologic Malignancies. European Urology. 2016;70(2):323–331. doi: 10.1016/j.eururo.2016.02.046. [DOI] [PubMed] [Google Scholar]
- 28.Krause M., Samoylenko A., Vainio S. J. Exosomes as renal inductive signals in health and disease, and their application as diagnostic markers and therapeutic agents. Frontiers in Cell and Developmental Biology. 2015;3 doi: 10.3389/fcell.2015.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawaz M., Camussi G., Valadi H., et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nature Reviews Urology. 2014;11(12):688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 30.Pomatto M. A., Gai C., Bussolati B., Camussi G. Extracellular Vesicles in Renal Pathophysiology. Frontiers in Molecular Biosciences. 2017;4 doi: 10.3389/fmolb.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Ramratnam B. Proteomic characterization of exosomes from HIV-1-lnfected cells. Methods in Molecular Biology. 2016;1354:311–326. doi: 10.1007/978-1-4939-3046-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathan M., Keerthikumar S., Ang C.-S., et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 33.Simpson R. J., Kalra H., Mathivanan S. ExoCarta as a resource for exosomal research. Journal of Extracellular Vesicles. 2012;1 doi: 10.3402/jev.v1i0.18374.18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathivanan S., Fahner C. J., Reid G. E., Simpson R. J. Exocarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Research. 2012;40(1):D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jørgensen M. M., Bæk R., Varming K. Potentials and capabilities of the Extracellular Vesicle (EV) Array. Journal of Extracellular Vesicles (JEV) 2015;4 doi: 10.3402/jev.v4.26048.26048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M., Aliotta J. M., Asara J. M., et al. Quantitative proteomic analysis of exosomes from HIV-1-infected lymphocytic cells. Proteomics. 2012;12(13):2203–2211. doi: 10.1002/pmic.201100376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ako-Adjei D., Fu W., Wallin C., et al. HIV-1, Human Interaction database: Current status and new features. Nucleic Acids Research. 2015;43(1):D566–D570. doi: 10.1093/nar/gku1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arenaccio C., Chiozzini C., Columba-Cabezas S., Manfredi F., Federico M. Cell activation and HIV-1 replication in unstimulated CD4+ T lymphocytes ingesting exosomes from cells expressing defective HIV-1. Retrovirology. 2014;11(1, article 46) doi: 10.1186/1742-4690-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Booth A. M., Fang Y., Fallon J. K., et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. The Journal of Cell Biology. 2006;172(6):923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chertova E., Chertov O., Coren L. V., et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. Journal of Virology. 2006;80(18):9039–9052. doi: 10.1128/jvi.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izquierdo-Useros N., Naranjo-Gómez M., Archer J., et al. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood. 2009;113(12):2732–2741. doi: 10.1182/blood-2008-05-158642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadiu I., Narayanasamy P., Dash P. K., Zhang W., Gendelman H. E. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. The Journal of Immunology. 2012;189(2):744–754. doi: 10.4049/jimmunol.1102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.-H., Schierer S., Blume K., et al. HIV-Nef and ADAM17-Containing Plasma Extracellular Vesicles Induce and Correlate with Immune Pathogenesis in Chronic HIV Infection. EBioMedicine. 2016;6:103–113. doi: 10.1016/j.ebiom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park I.-W., He J. J. HIV-1 is budded from CD4+ T lymphocytes independently of exosomes. Virology Journal. 2010;7, article no. 234 doi: 10.1186/1743-422X-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahimian P., He J. J. Exosome-associated release, uptake, and neurotoxicity of HIV-1 Tat protein. Journal of NeuroVirology. 2016;22(6):774–788. doi: 10.1007/s13365-016-0451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth W., Huang M., Addae Konadu K., Powell M., Bond V. Micro RNA in Exosomes from HIV-Infected Macrophages. International Journal of Environmental Research and Public Health. 2016;13(12):p. 32. doi: 10.3390/ijerph13010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampey G. C., Saifuddin M., Schwab A., et al. Exosomes from HIV-1-infected cells stimulate production of pro-inflammatory cytokines through trans-activating response (TAR) RNA. The Journal of Biological Chemistry. 2016;291(3):1251–1266. doi: 10.1074/jbc.M115.662171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lötvall J., Hill A. F., Hochberg F., et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles (JEV) 2014;3 doi: 10.3402/jev.v3.26913.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubert A., Subra C., Jenabian M.-A., et al. Elevated abundance, size, and MicroRNA content of plasma extracellular vesicles in viremic HIV-1+ patients: Correlations with known markers of disease progression. Journal of Acquired Immune Deficiency Syndromes. 2015;70(3):219–227. doi: 10.1097/QAI.0000000000000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skolnik P. R., Kosloff B. R., Bechtel L. J., et al. Concise communications absence of infectious hiv-1 in the urine of seropositive viremic subjects. The Journal of Infectious Diseases. 1989;160(6):1056–1060. doi: 10.1093/infdis/160.6.1056. [DOI] [PubMed] [Google Scholar]
- 51.Li J. J., Huang Y. Q., Poiesz B. J., Zaumetzger-Abbot L., Friedman-Kien A. E. Detection of human immunodeficiency virus type 1 (HIV-1) in urine cell pellets from HIV-1-seropositive individuals. Journal of Clinical Microbiology. 1992;30(5):1051–1055. doi: 10.1128/jcm.30.5.1051-1055.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khatua A. K., Taylor H. E., Hildreth J. E. K., Popik W. Non-productive HIV-1 infection of human glomerular and urinary podocytes. Virology. 2010;408(1):119–127. doi: 10.1016/j.virol.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marras D., Bruggeman L. A., Gao F., et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nature Medicine. 2002;8(5):522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 54.Bruggeman L. A., Ross M. D., Tanji N., et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. Journal of the American Society of Nephrology. 2000;11(11):2079–2087. doi: 10.1681/ASN.V11112079. [DOI] [PubMed] [Google Scholar]
- 55.Tanji N., Ross M. D., Tanji K., et al. Detection and localization of HIV-1 DNA in renal tissues by in situ polymerase chain reaction. Histology and Histopathology. 2006;21(4-6):393–401. doi: 10.14670/HH-21.393. [DOI] [PubMed] [Google Scholar]
- 56.Canaud G., Dejucq-Rainsford N., Avettand-Fenoël V., et al. The kidney as a reservoir for HIV-1 after renal transplantation. Journal of the American Society of Nephrology. 2014;25(2):407–419. doi: 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raymond A. D., Campbell-Sims T. C., Khan M., et al. HIV type 1 Nef is released from infected cells in CD45+ microvesicles and is present in the plasma of HIV-infected individuals. AIDS Research and Human Retroviruses. 2011;27(2):167–178. doi: 10.1089/aid.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kai S., Goto S., Tahara K., Sasaki A., Tone S., Kitano S. Indoleamine 2,3-Dioxygenase is Necessary for Cytolytic Activity of Natural Killer Cells. Scandinavian Journal of Immunology. 2004;59(2):177–182. doi: 10.1111/j.0300-9475.2004.01378.x. [DOI] [PubMed] [Google Scholar]
- 59.Rouas-Freiss N., Moreau P., Menier C., LeMaoult J., Carosella E. D. Expression of tolerogenic HLA-G molecules in cancer prevents antitumor responses. Seminars in Cancer Biology. 2007;17(6):413–421. doi: 10.1016/j.semcancer.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Wang D., Saga Y., Sato N., et al. The hepatocyte growth factor antagonist NK4 inhibits indoleamine-2, 3-dioxygenase expression via the c-Met-phosphatidylinositol 3-kinase-AKT signaling pathway. International Journal of Oncology. 2016;48(6):2303–2309. doi: 10.3892/ijo.2016.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zauli G., Davis B. R., Re M. C., Visani G., Furlini G., La Placa M. tat Protein stimulates production of transforming growth factor-β1 by marrow macrophages: A potential mechanism for human immunodeficiency virus- 1-induced hematopoietic suppression. Blood. 1992;80(12):3036–3043. [PubMed] [Google Scholar]
- 62.Kekow J., Wachsman W., McCutchan J. A., Cronin M., Carson D. A., Lotz M. Transforming growth factor β and noncytopathic mechanisms of immunodeficiency in human immunodeficiency virus infection. Proceedings of the National Acadamy of Sciences of the United States of America. 1990;87(21):8321–8325. doi: 10.1073/pnas.87.21.8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kekow J., Wachsman W., Allen McCutchan J., et al. Transforming growth factor-β and suppression of humoral immune responses in HIV infection. The Journal of Clinical Investigation. 1991;87(3):1010–1016. doi: 10.1172/JCI115059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazdins J. K., Klimkait T., Woods-Cook K., et al. In vitro effect of transforming growth factor-β on progression of HIV-1 infection in primary mononuclear phagocytes. The Journal of Immunology. 1991;147(4):1201–1207. [PubMed] [Google Scholar]
- 65.Maina E. K., Abana C. Z., Bukusi E. A., Sedegah M., Lartey M., Ampofo W. K. Plasma concentrations of transforming growth factor beta 1 in non-progressive HIV-1 infection correlates with markers of disease progression. Cytokine. 2016;81:109–116. doi: 10.1016/j.cyto.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Synowsky S. A., Shirran S. L., Cooke F. G. M., Antoniou A. N., Botting C. H., Powis S. J. The major histocompatibility complex class I immunopeptidome of extracellular vesicles. The Journal of Biological Chemistry. 2017;292(41):17084–17092. doi: 10.1074/jbc.M117.805895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neumann J., Eis-Hübinger A. M., Koch N. Herpes simplex virus type 1 targets the MHC class II processing pathway for immune evasion. The Journal of Immunology. 2003;171(6):3075–3083. doi: 10.4049/jimmunol.171.6.3075. [DOI] [PubMed] [Google Scholar]
- 68.Ostalecki C., Wittki S., Lee J.-H., et al. HIV Nef- and Notch1-dependent Endocytosis of ADAM17 Induces Vesicular TNF Secretion in Chronic HIV Infection. EBioMedicine. 2016;13:294–304. doi: 10.1016/j.ebiom.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Navarro M. N., Cantrell D. A. Serine-threonine kinases in TCR signaling. Nature Immunology. 2014;15(9):808–814. doi: 10.1038/ni.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Gaetano Donati K., Rabagliati R., Iacoviello L., Cauda R. HIV infection, HAART, and endothelial adhesion molecules: Current perspectives. The Lancet Infectious Diseases. 2004;4(4):213–222. doi: 10.1016/S1473-3099(04)00971-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top 10 biological function pathways using Pathway Studio 11 Mammal Plus, Elsevier, Inc., for HIV+ EV proteins from HIV+ patients with (1) CD4+ T cells greater than 300, (2) CD4+ T cells less than 300, (3) viral loads less than 200 copies, and (4) viral loads greater than 200 copies.
EV HIV protein interactions (Nef, Vif, Vpr, and Vpu) with human proteins identified using HIV-1 Human Interaction database (https://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/). The file includes the gene symbol, human protein name, interaction keywords, protein accession ID, and PMID of references citing the interaction.