Abstract
Phocoenamicins B and C (1 and 2), together with the known spirotetronate phocoenamicin (3), were isolated from cultures of Micromonospora sp. The acetone extract from a culture of this strain, isolated from marine sediments collected in the Canary Islands, displayed activity against methicillin-resistant Staphylococcus aureus (MRSA), Mycobacterium tuberculosis H37Ra and Mycobacterium bovis. Bioassay-guided fractionation of this extract using SP207ss column chromatography and preparative reversed-phased HPLC led to the isolation of the new compounds 1 and 2 belonging to the spirotetronate class of polyketides. Their structures were determined using a combination of HRMS, 1D and 2D NMR experiments and comparison with the spectra reported for phocoenamicin. Antibacterial activity tests of the pure compounds against these pathogens revealed minimal inhibitory concentration (MIC) values ranging from 4 to 64 µg/mL for MRSA, and 16 to 32 µg/mL for M. tuberculosis H37Ra, with no significant activity found against M. bovis and vancomycin-resistant Enterococcus faecium (VRE) at concentrations below 128 µg/mL, and weak activity detected against Bacillus subtilis grown on agar plates.
Keywords: spirotetronates, antimicrobial activity, Micromonospora sp., structural elucidation
1. Introduction
Antibiotics are compounds mainly produced by microorganisms that specifically kill or inhibit the growth of other microorganisms. Many antibiotics in today’s clinical use are secondary metabolites from actinomycetes or their semisynthetic derivatives [1]. Antibiotic resistance is rising to dangerously high levels all over the world. New resistance mechanisms are emerging and spreading globally, threatening our ability to treat common infectious diseases. Antibiotic resistance is accelerated by the misuse and overuse of antibiotics, as well as poor infection prevention and control. Those microorganisms that develop antimicrobial resistance are sometimes referred to as “superbugs” [2].
According to the World Health Organization (WHO) antimicrobial resistance (AMR) threatens the effective prevention and treatment of an ever-increasing range of infections caused by bacteria, parasites, viruses and fungi. WHO estimated that, in 2014, about 480,000 people developed multidrug-resistant tuberculosis (MDR-TB). Extensively drug-resistant tuberculosis (XDR-TB), a form of tuberculosis that is resistant to at least 4 of the core anti-TB drugs, has been identified in 105 countries [2]. Thus, bacterial infections represent an area of acute unmet medical needs and may become a deadly threat to mankind. Consequently, greater innovation and investment are required in research and development of new antimicrobial drugs, vaccines and diagnostic tools.
Nature is historically the source of new molecules exhibiting antibacterial activities; indeed, a considerable number of drugs have been developed from microorganisms of varied environments [3,4]. For several decades, the marine environment has been drawing attention, because it presents an enormous potential for the discovery of new therapeutic leads in the development of drugs to fight the current antibiotic resistance threats [5,6,7,8,9].
Marine actinobacteria are a great source of natural products with different biological properties including antimicrobial, anticancer, antiviral, insecticidal and enzyme inhibitory activities. They have attracted global interest in the last ten years for their ability to produce pharmacologically active compounds [1,9]. Actinomycetes are the most important biofactories for the production of therapeutic secondary metabolites, and most antibiotic discovery research focuses on the isolation of novel secondary metabolites from actinomycetes. The genus Streptomyces is the most prolific producer, with a 74% share of the natural products isolated from Actinomycetales, while non-Streptomyces species, also called rare actinomycetes, represent 26%. Rare or unusual actinomycetes have the potential to produce diverse, unique, unprecedented and occasionally structurally complicated compounds with excellent antibacterial potency and usually low toxicity [3].
Among the rare actinomycetes group, Micromonospora is one of the most fruitful genera, with more than 740 bioactive microbial metabolites discovered so far [3]. After the discovery of gentamicin [10], the genus Micromonospora became an important source in the drug discovery process. In general, soil and sediments are the main source for the isolation of antibiotic-producing Micromonospora strains. They also have been found to be constituents of endophytic actinobacterial populations recovered from plant tissues. Micromonospora species have also been isolated from water samples, sediments, and aquatic organisms such as ascidians, sponges, soft corals and mollusks [11].
To date, more than 70 spirotetronates have been reported from actinomycetes. These can be classified into three subgroups based on the size of the central ring, small (C11), medium (C13) and large (>C13) [12,13]. Among them, the spirotetronates maklamicin [14], nomimicin [15], together with PA-46101 A and B [16] and the recently isolated phocoenamicin [17] are representatives of the small size spirotetronate class.
During a screening in the search for novel antibiotic-producing microorganisms, we detected antimicrobial activity against the pathogens MRSA, M. tuberculosis H37Ra and M. bovis from the acetone extract of Micromonospora sp. CA-214671, collected in the Canary Islands. The LC–HRMS profile of this extract revealed the presence of potentially new secondary metabolites, which prompted a more detailed investigation of the extract. Extraction of the culture broth followed by a bioassay-guided fractionation of the extract led to the isolation of two new compounds (1 and 2), together with the recently reported phocoenamicin (3) [17], all of which belonged to the spirotetronate class of polyketides.
2. Results and Discussion
2.1. Isolation and Taxonomy of the Producing Microorganism
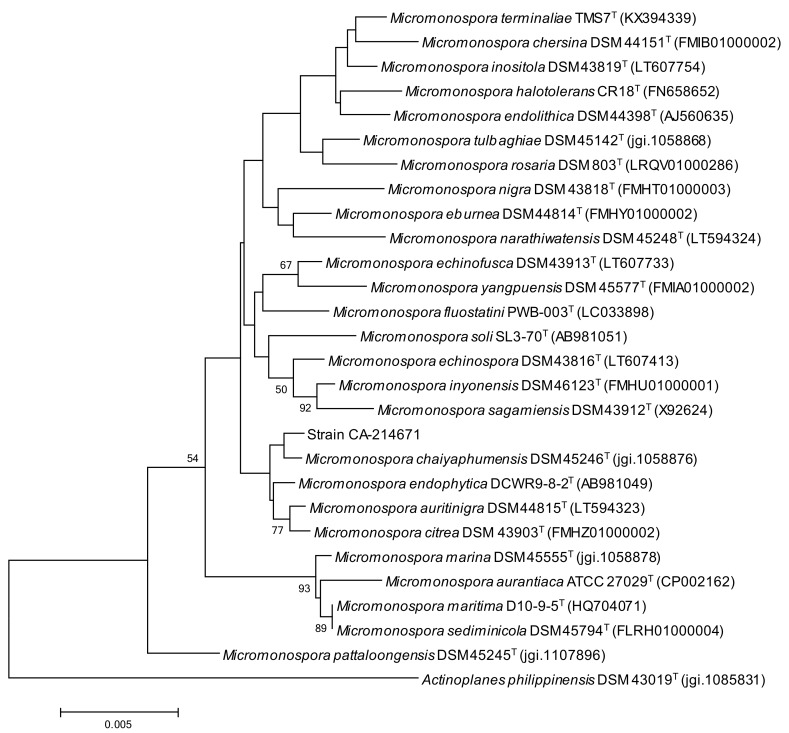
The strain CA-214671 was isolated from a marine cave sediment in Gran Canaria (Spain). A BLASTN search in the Eztaxon database of the PCR-amplified 16S rRNA nucleotide sequence (1290 nt) indicated that the strain is closely related to Micromonospora chaiyaphumensis (99.84 similarity) [18]. A phylogenetic tree was built employing the neighbor-joining (NJ) method corrected with the Jukes–Cantor algorithm [19,20] (Figure 1), which shows the close relationship of strain CA-214671 with M. chaiyaphumensis DSM 45246(T), although this association is not completely supported by the bootstrap value. Further multilocus sequence analysis (MLSA) phylogenetic studies are needed to confirm the species assignment of the Micromonospora strain CA-214671.
Figure 1.
NJ tree built with Mega 6.06 based on 16S rRNA gene sequences of CA-214671 and its closest type strains of the genus Micromonospora. Actinoplanes philippinensis DSM 43019(T) was included as out-group. The scale bar indicates 0.005 substitutions per nucleotide. Numbers at the nodes indicate bootstrap support (%) based on NJ analysis of 1000 replicates (only values ≥ 50 are shown).
2.2. Extraction, Bioassay-Guided Isolation and Structural Elucidation
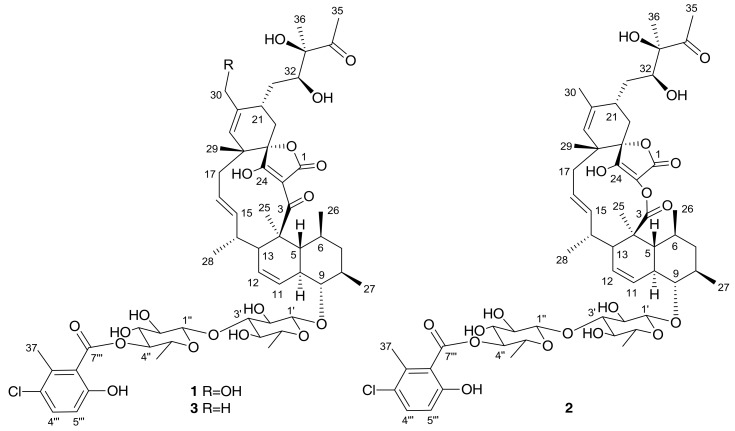
The producing strain was fermented in 3 L of FR23 medium supplemented with sea salts during 14 days at 28 °C. The fermentation broth was extracted with the same volume of acetone, followed by centrifugation, filtration and evaporation of the organic solvent. The remaining aqueous layer was subjected to bioassay-guided fractionation using SP207ss column chromatography and preparative reversed-phase HPLC, which led to the isolation of two new compounds, phocoenamicins B and C (1 and 2) together with the known spirotetronate phocoenamicin (3), recently reported as a potent Clostridium difficile inhibitor also isolated from a Micromonospora strain (Figure 2).
Figure 2.
Structures of phocoenamicins isolated from the Micromonospora sp. (CA-214671) culture.
Compound 1 was obtained as an optically active white amorphous solid ( – 16.7, MeOH). The (+)-ESI-TOF analysis showed an ammonium adduct at m/z 1104.4936 [M + NH4]+, from which a molecular formula of C56H75ClO19 was deduced for the compound. The IR spectrum showed characteristic absorptions of hydroxyl (3402 cm−1), carbonyl (1713 cm−1), and olefinic groups (1619, 1446 cm−1). Compound 1 displayed similar 1H and 13C NMR data to those recently described for phocoenamicin (3) [17]. The major difference between the 1H NMR spectra of both compounds was the presence of two oxygenated methylene protons at δH 3.98 and 4.14 in 1 (Table 1) instead of the methyl group at δH 1.74 ppm (Table S4 in Supplementary Materials) in phocoenamicin (3). The 1H NMR data, together with the HSQC spectrum, revealed that 1 contains a 1,2,3,4-tetrasubstituted benzene ring and two deoxysugar units, in addition to hydroxylated methines, olefinic protons and methyl groups (Table 1).
Table 1.
NMR spectroscopic data (500 MHz, CD3OD) for phocoenamicins B (1) and C (2).
| Position | Phocoenamicin B (1) | Phocoenamicin C (2) | ||
|---|---|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | |
| 1 | 178.1, C a | 172.7, C a | ||
| 2 | 107.4, C | n.d * | ||
| 3 | 201.3, C a | 176.2, C | ||
| 4 | 51.2, C | 47.1, C | ||
| 5 | 43.9, CH | 1.85, m | 44.1, CH | 1.60, m |
| 6 | 39.6, CH | 1.42, m | 38.6, CH | 1.52, m |
| 7α | 46.0, CH2 | 1.70, m | 46.0, CH2 | 1.75, m |
| 7β | 1.20, m | 1.11, m | ||
| 8 | 40.9, CH | 1.60, m | 40.8, CH | 1.63, m |
| 9 | 88.9, CH | 3.03, t (9.2) | 88.0, CH | 3.03, t (9.8) |
| 10 | 48.4, CH | 1.93, m | 48.3, CH | 1.92, m |
| 11 | 126.4, CH | 6.22, d (10.4) | 126.4, CH | 6.27, dd (10.3, 2.5) |
| 12 | 127.9, CH | 5.55, dd (9.8, 5.9) | 128.2, CH | 5.63, ddd (10.3, 6.2, 2.4) |
| 13 | 42.1, CH | 2.72, m | 50.6, CH | 2.00, m |
| 14 | 39.8, CH | 2.10, m | 40.7, CH | 2.21, m |
| 15 | 141.5, CH | 5.27, m | 136.3, CH | 4.90, m |
| 16 | 123.7, CH | 5.18, dd (15.2, 11.8) | 129.3, CH | 5.27, ddd (15.7, 11.6, 3.1) |
| 17a | 44.1, CH2 | 2.34, m | 43.1, CH2 | 2.32, m |
| 17b | 1.85, m | 1.70, m | ||
| 18 | 41.1, C | 44.3, C | ||
| 19 | 133.6, CH | 5.26, s | 133.5, CH | 4.99, brs |
| 20 | 138.0, C | 134.8, C | ||
| 21 | 30.1, CH | 2.58, m | 34.6, CH | 2.33, m |
| 22α | 30.5, CH2 | 1.68, m | 29.8, CH2 | 1.73, m |
| 22β | 2.26, m | 2.38, m | ||
| 23 | 87.1, C | 87.0, C | ||
| 24 | 204.4, C a | n.d * | ||
| 25 | 16.8, CH3 | 1.53, s | 17.3, CH3 | 1.31, s |
| 26 | 24.0, CH3 | 0.80, brs | 22.1, CH3 | 0.93, d (6.8) |
| 27 | 20.2, CH3 | 1.02, d (6.2) | 20.0, CH3 | 1.02, d (6.4) |
| 28 | 21.5, CH3 | 0.80, brs | 22.5, CH3 | 0.89, d (7.2) |
| 29 | 24.6, CH3 | 1.23, brs | 23.7, CH3 | 1.26, s |
| 30a | 65.4, CH2 | 4.14, d (13.2) | 22.4, CH3 | 1.73, s |
| 30b | 3.98, d (13.2) | |||
| 31a | 33.5, CH2 | 2.07, m | 33.9, CH2 | 1.94, m |
| 31b | 1.69, m | 1.73, m | ||
| 32 | 74.1, CH | 3.86, dd (11.2, 1.4) | 74.1, CH | 3.84, dd (10.9, 1.9) |
| 33 | 83.5, C | 83.5, C | ||
| 34 | 215.3, C | 215.2, C | ||
| 35 | 25.7, CH3 | 2.24, s | 25.6, CH3 | 2.23, s |
| 36 | 22.2, CH3 | 1.24, s | 22.1, CH3 | 1.20, s |
| 1′ | 104.0, CH | 4.36, d (7.3) | 104.0, CH | 4.35, d (7.3) |
| 2′ | 75.4, CH | 3.45, m | 75.4, CH | 3.45, m |
| 3′ | 88.6, CH | 3.46, m | 88.6, CH | 3.48, m |
| 4′ | 75.7, CH | 3.11, t (8.8) | 75.6, CH | 3.11, t (8.8) |
| 5′ | 72.8, CH | 3.22, m | 72.9, CH | 3.24, m |
| 6′ | 18.3, CH3 | 1.27, d (6.0) | 18.3, CH3 | 1.28, d (6.0) |
| 1″ | 105.4, CH | 4.61, d (7.8) | 105.4, CH | 4.61, d (7.9) |
| 2″ | 76.1, CH | 3.42, t (8.5) | 76.1, CH | 3.42, t (8.6) |
| 3″ | 75.3, CH | 3.64, t (9.3) | 75.4, CH | 3.64, t (9.4) |
| 4″ | 77.9, CH | 4.89, ** | 77.9, CH | 4.87, ** |
| 5″ | 71.7, CH | 3.69, dq (9.7, 6.1) | 71.7, CH | 3.69, dq (9.7, 6.2) |
| 6″ | 18.0, CH3 | 1.35, d (6.1) | 18.0, CH3 | 1.35, d (6.2) |
| 1′′′ | 124.3, C | 124.3, C | ||
| 2′′′ | 135.6, C | 135.6, C | ||
| 3′′′ | 126.0, C | 126.0, C | ||
| 4′′′ | 132.4, CH | 7.25, d (8.7) | 132.4, CH | 7.25, d (8.7) |
| 5′′′ | 115.9, CH | 6.70, d (8.7) | 115.8, CH | 6.70, d (8.7) |
| 6′′′ | 155.3, C | 155.3, C | ||
| 7′′′ | 169.3, C | 169.3, C | ||
| 37 | 17.3, CH3 | 2.36, s | 17.9, CH3 | 2.36, s |
a Assigned based on 2D correlations. * n.d. = not detected; ** Obscured by the water peak.
The main difference between the 13C NMR spectra of 1 and 3 was the presence of an oxygenated methylene carbon at δC 65.4 (Table 1) in 1, in contrast to the methyl group at δC 22.3 observed for phocoenamicin (3) (Table S4 in Supplementary Materials). Other signals present in the 13C NMR spectrum of 1 accounted for the presence of two esters at δc 169.3 and 178.1 and two ketones at δC 201.3 and 215.3, two non-protonated carbons (δC 155.3 and 204.4), together with methine and quaternary sp2 carbons, quaternary sp3 carbons, methylene sp3, methine sp3, and methyl groups (Table 1). The distinctive resonances at δC 204.4, 201.3, 178.1 and 107.4 in the 13C NMR spectrum were characteristic for tetronic acid carbons [14].
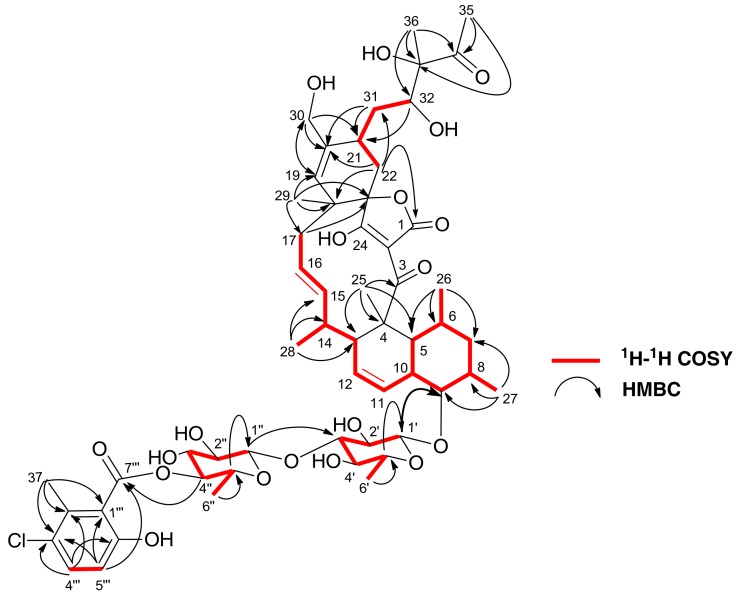
The full planar structure of 1 was assigned based on its 1D and 2D NMR spectroscopic data. The 1H-1H COSY spectrum of 1 displayed a series of correlations establishing the presence of four main fragments (Figure 3), similar to those found in the structure of 3. Correlations in the HMBC spectrum allowed the connection of the 1H-1H COSY-defined fragments as shown in Figure 3 and confirmed a carbon skeleton identical to that of phocoenamicin (3). The new oxygenated methylene group with respect to the structure of phocoenamicin was located at C-30 based on HMBC correlations from both H2-30 to C-19, C-20 and C-21, and from H-19 to C-30. Fragments related to the glycosyl moieties and their linkage positions were deduced based on the HMBC cross-peaks between the anomeric proton H-1′ of the first unit of 6-d-deoxyglucose and the oxygenated carbon C-9 of the decalin ring system. The long-range correlation from the anomeric proton H-1″ to C-3′ revealed that C-1″ of the second 6-deoxyglucose unit is attached to C-3′ of the first unit through an oxygen bridge linkage. Finally, a 3-chloro-6-hydroxy-2-methylbenzoate moiety, identical to that found in phocoenamicin [17] and similar to that of 2′′′-hydroxychlorothricin [21] was also present in the structure. HMBC correlation from H-4″ to C-7′′′ indicated that this aromatic ring was connected through an ester linkage to 4″ of the second 6-deoxyglucose unit.
Figure 3.
1H-1H COSY and key HMBC correlations for 1.
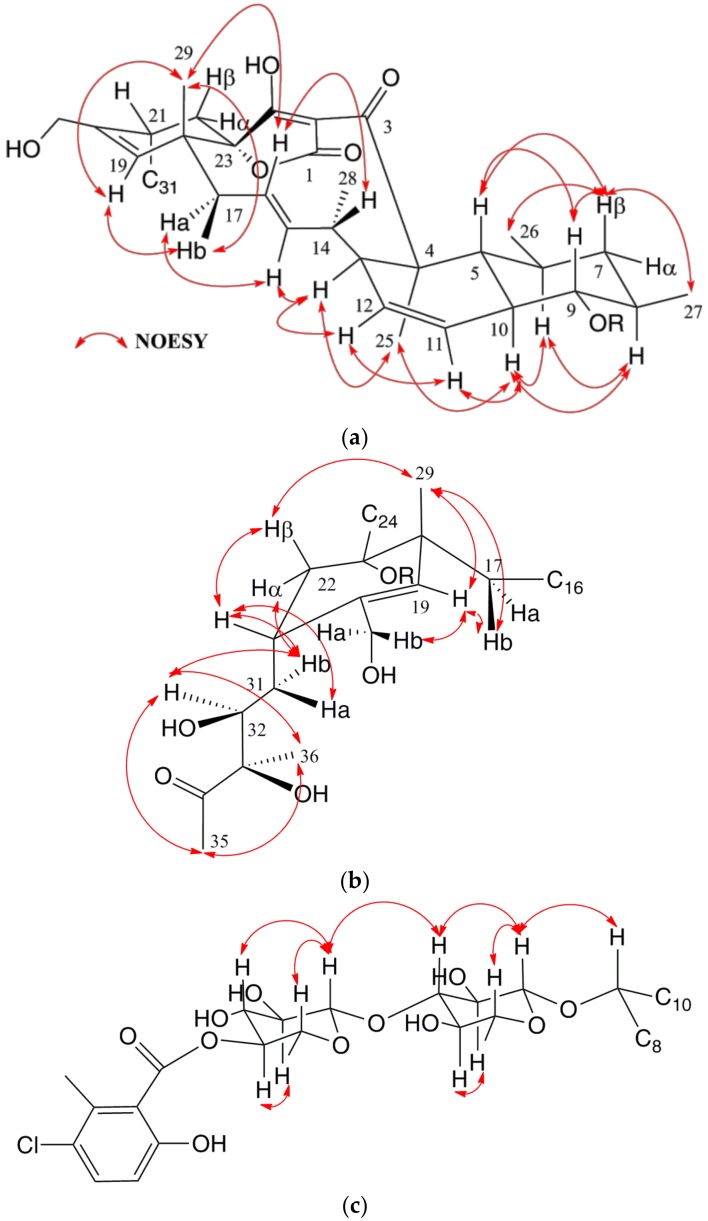
The relative configuration of 1 was established based on NOESY experiments (Figure 4a–c). Correlations observed around the decalin moiety indicated 1,3-diaxial relationships for H-5, H-7β and H-9, as well as for H-6, H-8 and H-10, a trans ring fusion of this decalin unit, and the equatorial orientation of the methyl groups at C-6 and C-8 (Figure 4a). A zig-zag conformation was suggested for the C-13 to C-18 chain based on the NOEs, along with the large 3JHH coupling constants of 15.2 and 11.8 Hz between H-15/H-16 and H-16/H-17a, respectively, which in turn defined H-13, H-15 and H3-25 to be in the bottom face of the molecule (Figure 4a). Additional NOESY correlations (Figure 4b) placed H3-29, H-22β and H-21 on the top face of the cyclohexene ring present in the molecule and confirmed its chair conformation. Conformation of the side chain at C-21 was deduced from the NOESY correlations for H-22α/H-31b, H-31b/H-32 and H-32/H3-36 setting an anti-relationship between H-31a and H-32, and between H-32 and the hydroxy group at C-33, which in turn indicated a configuration S*/R* at C-32 and C-33 carbons, respectively (Figure 4b), the same configuration recently described for phocoenamicin [17]. The configuration at the spirocarbon C-23 could not be determined experimentally due to the overlapped signals, so it was hypothesized that the tetronic acid stereogenic center should be assigned a S* configuration in alignment with other spirotetronates [14,17].
Figure 4.
Conformation and configuration of the decalin (a), cyclohexene (b) and sugars moieties (c) of 1 determined by NOESY data and J-based analysis.
Finally, the relative configurations of the sugars were determined by NOESY correlations (Figure 4c), establishing the axial orientation of all protons and confirming two units of β-6-deoxyglucopyranoside whose absolute configuration is proposed as D based on that previously determined for the related compound phocoenamicin [17]. These data confirmed the structure of compound 1 as a new member of the spirotetronate family comprising a spiro-tetronic acid motif embedded within an eleven-membered macrocyclic core linked to a cyclohexene ring and to a trans-decalin moiety. Compound 1 also contains a side chain at C-21 and a disaccharide connected to a substituted benzoic acid moiety with 23 chiral centers in its structure. NOESY correlations confirmed the same configuration of the chiral centers of our molecule with respect to those proposed for phocoenamicin, and the isolation of the latter compound from our culture broth was also in favor of this stereochemical proposal. Therefore, the structure of compound 1 was confirmed as 30-hydroxyphocoenamicin, for which we propose the name phocoenamicin B.
Compound 2 was obtained as an optically active white amorphous solid ( + 24.3, MeOH). The (+)-HRESIMS data showed an adduct ion at m/z 1104.4929 [M + NH4]+, accounting for a molecular formula C56H75ClO19, isomeric with that of 1. The IR spectrum showed characteristic absorptions of hydroxyl (3425 cm−1), carbonyl (1713 cm−1), and olefinic groups (1590, 1446 cm−1). 1H and 13C chemical shifts of 2 were similar to those of 1 for the trans-decalin, the disaccharide, and the benzoate moiety. The major difference between the 1H NMR data of both compounds was the presence of a methyl group at δH 1.73 instead of the two methylene protons at δH 3.98 and 4.14 (Table 1). Additional minor chemical shift differences were observed for most of the protons present in the molecule (Table 1).
The major differences in the 13C NMR spectra were the presence of a methyl group at δC 22.4 and the lack of the methylene group at δC 65.4 in 2 with respect to 1. Additionally, instead of a ketone carbonyl signal of C-3, a signal of an ester carbonyl carbon was observed at δC 176.2 which was supported by the HMBC correlation from H3-25 and this carbon (Figure S1). HMBC correlations were observed from the methyl protons at δH 1.73 to the carbons at δC 133.5 (C-19) and 134.8 (C-20) indicating that the methyl group was attached to the cyclohexene ring at C-20 (Supplementary Information Figure S1). The relative configurations of 2 were established based on NOESY experiments (Supplementary Information Figure S2). The 1H and 13C chemical shifts observed in 2 suggested the presence of a 12-membered core macrocycle, consisting of a macrolactone ring closely related to that found in chlorothricins and PA-46101 A and B antibiotics [12,16,21]. Accordingly, 2 was identified as a macrolactone core ring, spiro-tetronic acid derivative linked to a cyclohexene ring and to a trans-decalin moiety, which was named phocoenamicin C. Compound 2 most probably derives from the enzymatic Baeyer–Villiger oxidation of phocoenamicin (3). Similar oxidative biotransformations have also been described in the biosynthesis of mithramycin [22].
Compound 3 was obtained as an optically active white amorphous solid ( – 5.9, MeOH). The (+)-HRESIMS showed an adduct ion at m/z 1088.4979 [M + NH4]+, from which the molecular formula C56H75ClO18 was deduced. Its NMR data (Table S4 in Supplementary Materials) were similar to those of 1 and 2 and, although recorded in a different solvent, were in good agreement with those previously published for phocoenamicin [17].
The isolated compounds share a small macrocycle framework, like the aforementioned spirotetronates maklamicin [14] and nomimicin [15], together with PA-46101 A and B (whose aglycone structure shares important features with the tetronate antibiotic chlorothricin) [16]. Unique structural features of the two new compounds herein described are the presence of two 6-deoxyglucose moieties similar to that observed in 2′′′-hydroxychlorothricin [21] together with the diol side chain detected so far only in phocoenamicin [17].
2.3. Antimicrobial Activity
All the isolated compounds were evaluated for their antibacterial activity against several Gram-positive bacteria, methicillin-resistant S. aureus (MRSA) MB5393, M. tuberculosis (H37Ra) ATCC 25177, M. bovis ATCC 35734, vancomycin-resistant E. faecium (VRE) MB5571 and B. subtilis MB964 (Table 2). The results showed that 1 and 3 exhibited a significant activity against MRSA, with minimum inhibitory concentration (MIC) values ranging from 4 to 8 µg/mL to 8 to 16 µg/mL for 3 and 1, respectively. Compound 2 was significantly less active, inhibiting the growth of MRSA (MIC 32–64 µg/mL). In contrast, moderate to poor activity was observed against both Mycobacterium strains. Compound 3 displayed the highest inhibitory effect against M. tuberculosis H37Ra with a MIC value ranging from 16 to 32 µg/mL. On the contrary, negligible activity was observed against M. bovis for all compounds. Only 3 displayed weak activity against vancomycin-resistant E. faecium, with a MIC value of 32–64 µg/mL whereas phocoenamicin B (1) showed the best inhibitory activity against B. subtilis in agar plates with a diameter of the zone of inhibition of 7 mm when 2 μg of the compound was applied to the agar surface.
Table 2.
Antimicrobial bioassay results of phocoenamicins 1–3.
| Compounds | MIC (μg/mL) | ZOI * mm(μg) | |||
|---|---|---|---|---|---|
| S. aureus MB5393 | M. tuberculosis ATCC 25177 | M. bovis ATCC 35734 | E. faecium MB5571 | B. subtilis MB964 | |
| phocoenamicin B (1) | 8–16 | >128 | >128 | >128 | 7 (2) |
| phocoenamicin C (2) | 32–64 | 32 | >128 | >128 | 7 (4) |
| phocoenamicin (3) | 4–8 | 16–32 | >128 | 32–64 | 7 (4) |
| vancomycin | 2–4 | >128 | |||
| streptomycin | 1.6–3.2 | 0.4-0.8 | |||
| gentamicin | 8 (0.25) | ||||
| penicillin G | 19 (0.06) | ||||
* ZOI = zone of inhibition.
Based on the structure–activity relationship findings, similarly to other related spirotetronates, phocoenamicins B and C showed antibacterial activity against Gram-positive bacteria. Although phocoenamicin C possess a similar structure to that of phocoenamicin, they showed different biological activities that are likely due to the macrocycle core bearing a carboxylic ester instead of a ketone group, possibly the requirement for the activity. Findings related to the structure–activity relationships in other small spirotetronates revealed that conformational rigidity plays a key role in achieving the proper orientation of the enone moiety for interaction with the target [12].
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were measured using a Jasco P-2000 polarimeter (JASCO Corporation, Tokyo, Japan). UV spectra were obtained with an Agilent 1100 DAD (Agilent Technologies, Santa Clara, CA, USA). IR spectra were recorded on a JASCO FT/IR-4100 spectrometer (JASCO Corporation, Tokyo, Japan) equipped with a PIKE MIRacleTM single reflection ATR accessory (PIKE Thecnologies Inc., Madison, WI, USA). NMR spectra were recorded on a Bruker Avance III spectrometer (500 and 125 MHz for 1H and 13C NMR, respectively) equipped with a 1.7 mm TCI MicroCryoProbeTM (Bruker Biospin, Falländen, Switzerland). Chemical shifts were reported in ppm using the signals of the residual solvent as internal reference (δH 3.31 and δC 49.15 for CD3OD). LC–MS and LC–HRMS analyses were performed as described previously [23].
3.2. Taxonomical Identification of the Producing Microorganism
The 16S rRNA gene was PCR-amplified and sequenced as previously described [24]. The resulting DNA sequence lectures were aligned and visually inspected with Bionumerics 6.6 (Applied Maths, Sint-Martens-Latem, Belgium) to obtain the 16S rRNA sequence (1290 nt) (Supplementary Information Text S3).
3.3. Fermentation of the Producing Microorganism
A 3 L fermentation of the producing microorganism was generated as follows: a fresh seed culture of Micromonospora sp. CA-214671 was obtained inoculating a 25 × 150 mm tube containing 16 mL of ATCC-2-M medium (soluble starch 20 g/L, glucose 10 g/L, NZ Amine Type E 5 g/L, meat extract 3 g/L, peptone 5 g/L, yeast extract 5 g/L, sea salts 30 g/L, calcium carbonate 1 g/L, pH 7) with a freshly thawed inoculum stock of the strain. The tube was incubated in an orbital shaker at 28 °C, 220 rpm, 70% relative humidity for 7 days. The grown culture was then used to inoculate two 250 mL flasks each containing 50 mL of ATCC-2-M medium (5% v/v). The flasks were incubated in an orbital shaker at 28 °C, 220 rpm and 70% humidity for 7 days. The content of both flasks was then mixed, and the mixture was used to inoculate sixty 250 mL flasks each containing 50 mL of FR23 fermentation medium supplemented with sea salts (glucose 5 g/L, soluble starch from potato 30 g/L, cane molasses 20 g/L, cottonseed flour 20 g/L, sea salts 30 g/L, pH 7) (2.5% v/v). The flasks were incubated in an orbital shaker at 28 °C, 220 rpm and 70% humidity during 14 days before harvesting.
3.4. Extraction and Bioassay-Guided Isolation
After 14 days of fermentation, 50 mL of acetone was added to each Erlenmeyer flask (60 Erlenmeyer flasks which were shaken for 1 h in a Kühner at 200 rpm and 25 °C). Afterwards, the mixture was centrifuged at 8500 rpm for 10 min, and the supernatant was separated from the mycelium by decantation. Supernatant was then filtered under vacuum and the extract was concentrated in a rotary evaporator to remove the acetone. The aqueous residue was loaded onto a SP207ss resin column (65 g, 32 × 100 mm) and eluted with an acetone/H2O stepped gradient (10/90 for 7.2 min, 20/80 for 7.2 min, 40/60 for 7.1 min, 60/40 for 7.1 min, 80/20 for 6 min and 100/0 for 14.3 min, 10 mL/min, 20 mL/fraction) to give twenty-five fractions. Fractions containing the compounds of interest from this chromatography (19 to 22) were pooled and subjected to preparative reversed-phase HPLC (Waters™ XBridge® Prep C18 (Waters Corporation, Milford, MA, USA), 19 × 250 mm, 5 µm, 14 mL/min, UV detection at 210 and 280 nm, 8.75 mL/fraction) using H2O as solvent A and CH3CN as solvent B. Elution was carried out using isocratic conditions of 5% B for 5 min and then a linear gradient from 5% to 100% B for 52 min, held at 100% B for 5 min, yielding 84 fractions that were tested against MRSA, M. tuberculosis H37Ra and M. bovis.
Subfractions 59–76 were active against MRSA, M. tuberculosis H37Ra and M. bovis. Subfractions 59 to 61 were further purified by semi-preparative reversed-phase HPLC (Waters™ XBridge® Semiprep C18, 10 × 150 mm, 5 µm, 3.8 mL/min, UV detection at 210 and 230 nm, 1.82 mL/fraction) using H2O as solvent A and CH3CN as solvent B. Elution was carried out using isocratic conditions of 40% B for 5 min, linear gradient from 40% B to 100% B in 35 min to yield 1 (tR 22 min, 0.8 mg).
Subfractions 68 and 69 were further purified by semi-preparative reversed-phase HPLC (Waters™ XBridge® Semiprep C18, 10 × 150 mm, 5 µm, 3.8 mL/min, UV detection at 210 and 230 nm, 1.82 mL/fraction) using the same chromatographic conditions to yield 2 (tR 26 min, 1.2 mg).
Subfractions 74 to 76 were further purified by semi-preparative reversed-phase HPLC (Waters™ XBridge® Semiprep C18, 10 × 150 mm, 5 µm, 3.8 mL/min, UV detection at 210 and 230 nm, 1.82 mL/fraction) using the same chromatographic conditions to yield 3 (tR 31 min, 1.3 mg).
Phocoenamicin B (1): white amorphous solid; – 16.7 (c 0.16, MeOH); UV (DAD) λmax 230, sh 290, 320 nm; IR (ATR) νmax 3402, 2924, 1713, 1619, 1446, 1393, 1068, 1024 cm−1; for 1H and 13C NMR data see Table 1; (+)-HRESIMS m/z 1104.4936 [M + NH4]+ (calcd for C56H79ClNO19+, 1104.4929).
Phocoenamicin C (2): white amorphous solid; + 24.3 (c 0.17, MeOH); UV (DAD) λmax sh 230, sh 250, 290 nm; IR (ATR) νmax 3425, 2926, 1713, 1590, 1446, 1378, 1070, 1026 cm−1; for 1H and 13C NMR data see Table 1; (+)-HRESIMS m/z 1104.4929 [M + NH4]+ (calcd for C56H79ClNO19+, 1104.4929).
Phocoenamicin (3): white amorphous solid; – 5.9 (c 0.35, MeOH); UV (DAD) λmax 230, sh 290, 320 nm; IR (ATR) νmax 3404, 2925, 1714, 1618, 1446, 1391, 1295, 1069, 1023 cm−1; for 1H and 13C NMR data see Table S4 Supplementary information; (+)-HRESIMS m/z 1088.4979 [M + NH4]+ (calcd for C56H79ClNO18+, 1088.4980).
3.5. Antibacterial Activity Assay
The antibacterial activity of the isolated compounds was evaluated using sequential 2-fold serial dilutions of each compound in DMSO to provide 10 concentrations starting at 128 µg/mL for all the assays. Three Gram-positive bacteria including methicillin-resistant S. aureus (MRSA) MB5393, M. tuberculosis (H37Ra) ATCC 25177, M. bovis ATCC 35734, vancomycin-resistant E. faecium MB5571 and B. subtilis MB964 were used in this study, following previously described methodologies [25,26,27].
4. Conclusions
Two new spirotetronates have been isolated from the marine actinomycete Micromonospora sp. The new structures comprise a spiro-tetronic acid motif embedded within an eleven-membered macrocyclic core linked to a cyclohexene ring and to a trans-decalin moiety. Unique structural features of the two new compounds are the presence of two 6-d-deoxyglucose moieties together with the diol side chain only detected in phocoenamicin to date [17].
Concerning their biological activity, phocoenamicins B (1) and C (2) showed antibacterial activity against Gram-positive bacteria. Compounds 1 and 3 displayed significant activity against MRSA, one of the most common causes of hospital-acquired infections, while moderate to negligible activity was observed against M. tuberculosis H37Ra, M. bovis, B. subtilis and E. faecium VRE. Additionally, moderate anti-tuberculosis activity against M. tuberculosis was displayed by 2 and 3. Apparently, conformational rigidity to achieve the proper orientation of the enone moiety could play a key role in the interaction with the target, revealing the possible structure–activity relationship for these compounds.
Our study also confirms that Micromonospora species producing spirotetronates are widely spread within the marine environment, and current studies by LC–MS of our strain collection point out that other Micromonospora strains producers of phocoenamicins have also been isolated from terrestrial environments. Furthermore, the present study highlights that the marine environment offers an enormous potential for the discovery of compounds with new chemical scaffolds, thus representing a great opportunity to find new antibiotics to confront multi-resistant bacteria. The challenge in the future will be the discovery of more chemically and hopefully mechanistically new agents to satisfy these needs.
Acknowledgments
The authors acknowledge Ignacio Pérez-Victoria for assistance with NMR experiments and Pilar Sánchez for assistance with antimicrobial assays. This work has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement No. 312184 (PharmaSea), and the H2020 program under grant agreement No. 721421 (MarPipe). The polarimeter, HPLC, IR, and NMR equipment, pipetting station and plate reader used in this work were purchased via grants for scientific and technological infrastructures from the Ministerio de Ciencia e Innovación [Grants No. PCT-010000-2010-4 (NMR), INP-2011-0016-PCT-010000 ACT6 (polarimeter, HPLC and IR), and PCT-01000-ACT7, 2011-13 (pipetting station and plate reader)].
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/16/3/95/s1, Figure S1: 1H-1H COSY and key HMBC correlations for 2; Figure S2: Conformation and configuration of the decalin, cyclohexene and sugars moieties of 2 determined by NOESY data and J-based analysis; Text S3: 16S rRNA gene sequence from strain CA-214671; Table S4: NMR spectroscopic data (500 Mz, CD3OD) for phocoenamicin (3); Figure S5: 1H-NMR (500 MHz, methanol-d4) spectrum of phocoenamicin B (1); Figure S6: 13C-NMR (125 MHz, methanol-d4) spectrum of phocoenamicin B (1); Figure S7: HSQC (methanol-d4) spectrum of phocoenamicin B (1); Figure S8: HMBC (methanol-d4) spectrum of phocoenamicin B (1); Figure S9: COSY (methanol-d4) spectrum of phocoenamicin B (1); Figure S10: NOESY (methanol-d4) spectrum of phocoenamicin B (1); Figure S11: TOCSY (methanol-d4) spectrum of phocoenamicin B (1); Figure S12: 1H-NMR (500 MHz, methanol-d4) spectrum of phocoenamicin C (2); Figure S13: 13C-NMR (125 MHz, methanol-d4) spectrum of phocoenamicin C (2); Figure S14: HSQC (methanol-d4) spectrum of phocoenamicin C (2); Figure S15: HMBC (methanol-d4) spectrum of phocoenamicin C (2); Figure S16: COSY (methanol-d4) spectrum of phocoenamicin C (2); Figure S17: NOESY (methanol-d4) spectrum of phocoenamicin C (2); Figure S18: TOCSY (methanol-d4) spectrum of phocoenamicin C (2).
Author Contributions
M.P.-B. performed the extraction of the culture broth, the purification of the compounds and LC–MS analysis. D.O.-C. carried out the fermentation and the taxonomic identification of the microbial strain. M.d.l.C. and F.V. performed the antibacterial assays. M.K. contributed to the isolation of the compounds. M.P.-B. and F.R. analyzed the spectroscopic data and elucidated the structure of the new molecules. J.M. performed the LC–MS analysis and interpretation of the ESI-TOF spectra. F.R., O.G. and F.V. conceived and supervised the work. M.P.-B., D.O.-C. and F.R. wrote the paper, which was revised and approved by all the authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Manivasagan P., Kanga K.-H., Sivakumarb K., Li-Chanc E.C.Y., Oha H.-M., Kim S.-K. Marine actinobacteria: An important source of bioactive natural products. Environ. Toxicol. Pharmacol. 2014;38:172–188. doi: 10.1016/j.etap.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) WHO Fact Sheet 194, Updated November 2017. [(accessed on 17 January 2018)]; Available online: http://who.int/mediacentre/factsheets/fs194/en/
- 3.Bérdy J. Bioactive Microbial Metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 4.Newman D.J., Cragg G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 5.Rahman H., Austin B., Mitchell W.J., Morris P.C., Jamieson D.J., Adams D.R., Spragg A.M., Schweizer M. Novel Anti-Infective Compounds from Marine Bacteria. Mar. Drugs. 2010;8:498–518. doi: 10.3390/md8030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pramanik A., Saha M., Sana B. Antimicrobial Agents from Marine Cyanobacteria and Actinomycetes. Mar. Microbiol. 2013:191–205. doi: 10.1002/9783527665259.ch11. [DOI] [Google Scholar]
- 7.Crowley S.P., O’Gara F., O’Sullivan O., Cotter P.D., Dobson A.D.W. Marine Pseudovibrio sp. as a Novel Source of Antimicrobials. Mar. Drugs. 2014;12:5916–5929. doi: 10.3390/md12125916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R.P., Kumari P., Reddy C.R.K. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol. 2015;99:1571–1586. doi: 10.1007/s00253-014-6334-y. [DOI] [PubMed] [Google Scholar]
- 9.Schinke C., Martins T., Queiroz S.C.N., Melo I.S., Reyes F.G.R. Antibacterial Compounds from Marine Bacteria, 2010–2015. J. Nat. Prod. 2017;80:1215–1228. doi: 10.1021/acs.jnatprod.6b00235. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein M.J., Luedemann G.M., Oden E.M., Wagman G.H., Rosselet J.P., Marquez J.A., Coniglio C.T., Charney W., Herzog H.L., Black J. Gentamicin, a new antibiotic complex from Micromonospora. J. Med. Chem. 1963;6:463–464. doi: 10.1021/jm00340a034. [DOI] [PubMed] [Google Scholar]
- 11.Trujillo M.E., Hong K., Genilloud O. The Family Micromonosporaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes. 4th ed. Springer; New York, NY, USA: 2014. pp. 499–569. [Google Scholar]
- 12.Vieweg L., Reichau S., Schobert R., Leadlayc P.F., Sussmuth R.D. Recent advances in the field of bioactive tetronates. Nat. Prod. Rep. 2014;31:1554–1584. doi: 10.1039/C4NP00015C. [DOI] [PubMed] [Google Scholar]
- 13.Lacoske M.H., Theodorakis E.A. Spirotetronate Polyketides as Leads in Drug Discovery. J. Nat. Prod. 2015;78:562–575. doi: 10.1021/np500757w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igarashi Y., Ogura H., Furihata K., Oku N., Indananda C., Thamchaipenet A. Maklamicin, an Antibacterial Polyketide from an Endophytic Micromonospora sp. J. Nat. Prod. 2011;74:670–674. doi: 10.1021/np100727h. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi Y., Iida T., Odu N., Watanabe H., Furihata K., Miyanouchi K. Nomimicin, a new spirotetronate-class polyketide from an actinomycete of the genus Actinomadura. J. Antibiot. 2012;65:355–359. doi: 10.1038/ja.2012.30. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto M., Kawamura Y., Yoshimura Y., Terui Y., Nakai H., Yoshida T., Shoji J. Isolation, characterization and structures of PA-46101 A and B. J. Antibiot. 1990;43:739–747. doi: 10.7164/antibiotics.43.739. [DOI] [PubMed] [Google Scholar]
- 17.Ochoa J.L., Sanchez L.M., Koo B.-M., Doherty J.S., Rajendram M., Huang K.C., Gross C.A., Linington R.G. Marine Mammal Microbiota Yields Novel Antibiotic with Potent Activity Against Clostridium difficile. ACS Infect. Dis. 2018;4:59–67. doi: 10.1021/acsinfecdis.7b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim O.S., Cho Y.J., Lee K., Yoon S.H., Kim M., Na H., Park S.C., Jeon Y.S., Lee J.H., Yi H., et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 19.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Jukes T.H., Cantor C. Evolution of proteins molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. Academic Press; New York, NY, USA: 1969. pp. 21–132. [Google Scholar]
- 21.Yamamoto I., Nakagawa M., Hayakawa Y., Adachi K., Kobayashi E. Hydroxychlorothricin, a New Antitumor Antibiotic. J. Antibiot. 1987;40:1452–1454. doi: 10.7164/antibiotics.40.1452. [DOI] [PubMed] [Google Scholar]
- 22.Gibson M., Nur-e-alam M., Lipata F., Oliveira M.A., Rohr J. Characterization of Kinetics and Products of the Baeyer-Villiger Oxygenase MtmOIV, The Key Enzyme of the Biosynthetic Pathway toward the Natural Product Anticancer Drug Mithramycin from Streptomyces argillaceus. J. Am. Chem. Soc. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 23.Martín J., Crespo G., González-Menéndez V., Pérez-Moreno G., Sánchez-Carrasco P., Pérez-Victoria I., Ruiz-Pérez L.M., González-Pacanowska D., Vicente F., Genilloud O., et al. MDN-0104, an Antiplasmodial Betaine Lipid from Heterospora chenopodii. J. Nat. Prod. 2014;77:2118–2123. doi: 10.1021/np500577v. [DOI] [PubMed] [Google Scholar]
- 24.Lacret R., Perez-Victoria I., Oves-Costales D., de la Cruz M., Domingo E., Martin J., Diaz C., Vicente F., Genilloud O., Reyes F. MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078. Mar. Drugs. 2016;14:188–199. doi: 10.3390/md14100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín J., Sousa T.S., Crespo G., Palomo S., González I., Tormo J.R., de la Cruz M., Anderson M., Hill R.T., Vicente F., et al. Kokurin, the True Structure of PM181104, an Anti-Methicillin-Resistant Staphylococcus aureus (MRSA) Thiazolyl Peptide from the Marine-Derived Bacterium Kokuria pallustris. Mar. Drugs. 2013;11:387–398. doi: 10.3390/md11020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palomino J.C., Martin A., Camacho M., Guerra H., Swings J., Portaels F. Resazurin microtiter assay plate: Simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suay I., Arenal F., Asensio F.J., Basilio A., Angeles Cabello M., Teresa Díez M., García J.B., González del Val A., Gorrochategui J., Hernández P., et al. Screening of basidiomycetes for antimicrobial activities. Antonie Leeuwenhoek. 2000;78:129–140. doi: 10.1023/A:1026552024021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.